Abstract
Recent decades have seen an explosion in the amount of data available on all aspects of biodiversity, which has led to data-driven approaches to understand how and why diversity varies in time and space. Global repositories facilitate access to various classes of species-level data including biogeography, genetics and conservation status, which are in turn required to study different dimensions of diversity. Ensuring that these different data sources are interoperable is a challenge as we aim to create synthetic data products to monitor the state of the world's biodiversity. One way to approach this is to link data of different classes, and to inventory the availability of data across multiple sources. Here, we use a comprehensive list of more than 200 000 marine animal species, and quantify the availability of data on geographical occurrences, genetic sequences, conservation assessments and DNA barcodes across all phyla and broad functional groups. This reveals a very uneven picture: 44% of species are represented by no record other than their taxonomy, but some species are rich in data. Although these data-rich species are concentrated into a few taxonomic and functional groups, especially vertebrates, data are spread widely across marine animals, with members of all 32 phyla represented in at least one database. By highlighting gaps in current knowledge, our census of marine diversity data helps to prioritize future data collection activities, as well as emphasizing the importance of ongoing sustained observations and archiving of existing data into global repositories.
This article is part of the theme issue ‘Integrative research perspectives on marine conservation’.
Keywords: marine biodiversity, ecoinformatics, global occurrences, conservation status, linked data
1. Introduction
The explosion in the availability of data describing the natural world has, in recent decades, transformed the kinds of questions that we can now ask as ecologists. Efforts to reconstruct the evolutionary relationships between all living species (e.g. Open Tree of Life; [1,2]) can draw upon over 200 M sequences (https://www.ncbi.nlm.nih.gov/genbank/statistics/) from over 170 000 metazoan species stored in GenBank [3,4]. In 2018, the Global Biodiversity Information Facility (GBIF; [5]) passed a billion species occurrence records (https://www.gbif.org/news/5BesWzmwqQ4U84suqWyOQy/big-data-for-biodiversity-gbiforg-surpasses-1-billion-species-occurrences), providing an unparalleled resource for students of biogeography. The conservation status of more than 116 000 species has now been formally assessed [6]. Significant efforts are underway to collate data on biological, physiological, metabolic and thermal traits [7–11] across multiple species, as well as information on animal movement [12,13] and ecological interactions [14].
Against this background of increased data availability, the oceans are still often characterized as the data-poor relative of the data-rich land. Various autonomous platforms operating throughout the world's oceans do now enable vast quantities of physical and biogeochemical data to be transmitted [15] but marine biodiversity data remain more challenging to collect. In part, the vastness of the oceans precludes routine and casual observation by the citizen scientists who have contributed so much to the collection of terrestrial biodiversity data [16,17], except in some more accessible coastal areas [18–20]. However, coordinated global initiatives have made enormous progress in collating existing data and promoting systematic new data collection. The Census of Marine Life [21] drove this effort from 2000 to 2010, and its legacies include the Ocean Biodiversity Information System (OBIS; [22]), which currently holds nearly 60 M occurrence records from over 120 000 marine species. Initiatives like this have built on sustained observations of marine ecosystems [23] and continue to be developed to deliver the Essential Biodiversity Variables that we need to monitor progress towards Sustainable Development Goals (e.g. [24]). Application of technologies from satellites and drones to biologgers and molecular methods such as eDNA continue to expand the range of data available to marine biodiversity scientists [25]. Crucially, the accumulation of data has proceeded in parallel with massive improvements in data infrastructure, and much better tools (taking advantage of the improved computing power available even to casual users) with which to access and analyse it [26,27]. This is important because the challenge now is to extract meaning from the sea of data, to deliver effective outcomes for marine conservation and monitoring of the state of the global ocean [19,24].
Although access to biodiversity data of different types is now much improved, to extract full value from existing data requires linking together different datasets that were often collected for different purposes, by different organizations and at different times. This kind of interoperability of diversity data is central to the vision of a ‘macroscope’ to sample and monitor the entire biosphere [25], and is a fundamental principle of the Bari Manifesto of best practice in biodiversity informatics [28]. Progress towards such interoperability requires comparable coverage across multiple classes of data and dimensions of diversity, as well as parallel measures of the abiotic environment and of human pressures. An exemplar of successful data integration for terrestrial plant communities is the Botanical Information and Ecology Network [29], which combines standardized information on plant distributions, traits and evolutionary relationships with the computational tools needed to work with them. An important step towards this kind of model is to fully understand the gaps and biases in available data. In the marine environment, key gaps in the overall knowledge of marine biodiversity have been documented [30–32], including estimates of the extent of unknown biodiversity [33] and undocumented extinction risk [34]. Efforts to quantify these gaps across different key variables and data sources have been limited to the regional scale, but have shown for instance that the species and taxonomic groups that we know most in one dimension (e.g. global occurrences) tend to be those that we also know most about in another (e.g. biological traits, extinction risk; [34,35]). To date, we lack a global overview of how data (and gaps) are co-distributed across axes of marine diversity, to compare for example with previous global analyses of terrestrial plants [36].
Such a task is feasible, however, given the availability of a standardized global taxonomy of marine species, the World Register of Marine Species (WoRMS; [37]), which includes links out to other key biodiversity datasets (table 1). In this paper, we focus on key data sources that, when linked to robust taxonomy, individually or in combination can be used to construct different dimensions of marine diversity. We consider geographical occurrences and nucleotide sequences to be the fundamental building blocks of the spatial and phylogenetic dimensions of diversity, which interact to structure the distribution of key ecological traits across species [39]. A first step to adding the functional dimension of diversity is to classify species into broad ecological guilds, similar to the way in which species can be classified in global theories and models of biodiversity [40,41]. Supplementing these with information on conservation status and molecular taxonomy provides insights into how marine diversity is changing, and how we might efficiently monitor this. Throughout we use open-source computational tools to link data across these components of marine diversity to take stock of the current state of data availability, identifying gaps and priorities for future work. In this way, we summarize data availability across multiple axes for more than 200 000 marine animal species from 32 phyla and across broad ecological guilds (e.g. benthos, zooplankton and seabirds) and we assess the extent to which this availability is correlated across different classes of diversity data. Above all, our aim is to highlight the wealth of marine biodiversity data that we have amassed as a community over centuries, and the opportunities that we now have to link different classes of data in order to better understand the dimensions of marine diversity.
Table 1.
Data sources used to link different dimensions of diversity across all marine animals.
| dimension of diversity | data source | data type | reference |
|---|---|---|---|
| taxonomy | WoRMS | authoritative classification and catalogue of marine taxonomic names | [37] |
| functional groups | WoRMS | classification of marine species into broad ecological groups | [37] |
| biogeography | OBIS | global database of marine species occurrence records | [22] |
| genetics | GenBank | the NIH genetic sequence database, an annotated collection of all publicly available DNA sequences | [3] |
| molecular taxonomy | BOLD | barcode of life data system for DNA barcodes | [38] |
| conservation status | IUCN Red List | the IUCN red list of threatened species | [6] |
2. Methods
To provide an overview of the state of knowledge of marine animal biodiversity, we mine the World Register of Marine Species (WoRMS; [37]), the most comprehensive source of taxonomic information on marine species, consisting of over half a million distinct names checked by expert taxonomic editors. We focus our investigation on marine animals, and so filtered the WoRMS database to Kingdom Animalia, retaining only those species considered to be marine by WoRMS (flag isMarine is TRUE), and excluding any species only known from fossils. We consider only taxa identified at the species rank, with a current accepted name and valid WoRMS identifier (Aphia ID).
In addition to taxonomy, WoRMS has aggregated data on species attributes including broad ‘functional groups’. In reality, these are closer to ecological guilds, defining habitat affinity (e.g. benthos, zooplankton) rather than ecological function, but for transparency, we retain the terminology employed by WoRMS. We use these attributes to assign each species to a functional group, using a dedicated R function (https://github.com/tomjwebb/WoRMS-functional-groups) that accesses the WoRMS API using the worrms R package [42]. We supplement these functional groups with taxonomic groups to identify fish (using the WoRMS paraphyletic Superclass Pisces; [43]), marine mammals, seabirds and reptiles. We consolidate functional groups into broad categories for maximum coverage; for example, our ‘benthos’ group includes all species categorized in WoRMS as endobenthos, epibenthos, hyperbenthos, macrobenthos, meiobenthos and microbenthos, as well as those originally classified simply as benthos. When separate functional groups are recorded for different life stages, we always use the group for the adult stage. We group together categories with very few species (including meso, macro and neuston) and species with no functional group classification into the single category ‘other/unknown’. For fish, we include an additional grouping variable based on the broad habitat categories recorded in FishBase [10] accessed using the rfishbase package [44], classifying 17 568 of 18 261 species as bathydemersal, bathypelagic, benthopelagic, demersal, pelagic-oceanic, pelagic-nertitic or reef-associated.
The WoRMS database includes links to other major biodiversity databases (table 1), and we exploit these to compare the state of biodiversity information availability across axes of biogeography, genetics, conservation and molecular taxonomy. Specifically, we record for each species its total number of occurrences in the ocean biogeographic information system (OBIS; [22]) and its total number of nucleotide sequences in GenBank. The taxonomy in OBIS is standardized to WoRMS, making these links straightforward, and GenBank's taxonomic information is generally reliable for marine animals [4] meaning that links between WoRMS and GenBank are likely to robustly associate relevant sequences with the correct taxonomic identifier. We also record for each species its IUCN conservation assessment category (if available) and whether or not it has DNA barcodes listed in the barcode of life data system (BOLD).
Using our tidy database linking the diversity data sources shown in table 1, we then summarize the availability of biodiversity data across all marine animals as follows. First, we consider the two major quantitative databases, OBIS and GenBank. We calculate the proportion of species within each phylum with records in each of these databases, and the distribution of records between species within each phylum. To derive an indication of relative data availability across functional groups, highlighting groups that are particularly highly likely (or unlikely) to occur in the dataset, and those that tend to have more records when they are present, we model data availability across functional groups. We apply a two-step hurdle process because of the high degree of zero-inflation in our data [45]. To assess whether certain functional groups were better represented in the databases than others, we model the presence of species in OBIS or GenBank using a binomial GLM of the form species presence∼functional group, and we model the distribution of counts (OBIS records or GenBank nucleotides) between functional groups, for those species present in the data source, using a zero-truncated negative binomial GLM. These hurdle models are implemented using the hurdle function in the pscl package [45,46]. For visualization, we plot the exponentiated binomial coefficients from the zero component of the model, which shows the ratio of the probability of getting a non-zero to a zero observation within a functional group. We also plot the predicted counts for the subset of species in each functional group with non-zero counts.
To assess whether data availability is correlated across data sources, we use categorical scales of numbers of records per species in both OBIS and GenBank, using categories bounded by upper limits of 0, 1, 10, 100, 1000, 10 000, 100 000 and a final category of greater than 100 000 records. We use mosaic plots [47], created using the ggmosaic R package [48], to illustrate the distribution of GenBank count categories for each OBIS count category. We also consider how IUCN conservation assessments are distributed across species in different functional groups, and between species present and absent in OBIS, and we compare the number of OBIS occurrence records between species in different IUCN categories. To simplify this analysis, we aggregate to the following IUCN assessment categories: not assessed, data deficient (i.e. formally assessed but insufficient data to assign the species to a threat category), threatened (formally assessed as vulnerable, endangered, critically endangered, conservation dependent, extinct in the wild or extinct) and non-threatened (formally assessed as near threatened or least concern). We perform a similar analysis comparing species presence or absence in the Barcode of Life database with presence in OBIS and number of OBIS records.
All data and links were extracted from WoRMS on 11 January 2020 and the statistics we report are correct as of that date. Manipulation, visualization and analysis is performed in R 3.6.2 [49] using RStudio 1.2.5033 [50] and the tidyverse suite of packages [51] as well as worrms [42] to access the WoRMS API and rfishbase [44] to access FishBase, and the plotting packages ggmosaic [48], ggbeeswarm [52] and patchwork [53].
3. Results
Our final dataset consisted of 206 849 valid marine animal species, from 32 phyla and 89 classes. Of these, 106 213 (51%) have at least one occurrence record listed in OBIS (table 2). 18 869 (18% of species in OBIS, 9% of all species) are represented by just a single occurrence record (table 2), while one species (Atlantic Cod, Gadus morhua) has over a million occurrence records (1 108 463). Overall, there are 45 974 726 OBIS occurrence records across all species. 36 094 (17%) of all species have at least one nucleotide recorded in GenBank, while eight species (five fish, the Antarctic minke whale Balaenoptera bonaerensis, the tunicate Ciona intestinalis and the California sea hare Aplysia californica) have more than a million. Overall, the species in our database total 56 846 294 GenBank nucleotides. Furthermore, 13 179 species have had their conservation status assessed by the IUCN, and 25 272 have at least one DNA barcode in the Barcode of Life database.
Table 2.
Breakdown of 206 849 marine animal species by number of global occurrence records in OBIS, and numbers of nucleotide sequences in GenBank.
| number of OBIS records | number of GenBank nucleotides |
totals | in OBIS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2–10 | 11–100 | 101–1000 | 1001–10 000 | 10 001–100 000 | 100 001–1 000 000 | >1 000 000 | |||
| 0 | 93 519 | 1312 | 4484 | 1164 | 116 | 13 | 15 | 13 | 0 | 100 636 | |
| 1 | 16 905 | 356 | 1253 | 314 | 33 | 3 | 3 | 2 | 0 | 18 869 | 106 213 |
| 2–10 | 35 613 | 1086 | 3714 | 990 | 122 | 17 | 11 | 8 | 0 | 41 561 | |
| 11–100 | 19 998 | 1392 | 5931 | 2733 | 351 | 32 | 30 | 26 | 2 | 30 495 | |
| 101–1000 | 4274 | 594 | 3334 | 2917 | 512 | 51 | 35 | 37 | 1 | 11 755 | |
| 1001–10 000 | 402 | 86 | 630 | 1113 | 315 | 33 | 53 | 33 | 4 | 2669 | |
| 10 001–100 000 | 42 | 4 | 107 | 406 | 167 | 31 | 22 | 31 | 1 | 811 | |
| 100 001–1 000 000 | 2 | 0 | 0 | 14 | 20 | 5 | 3 | 8 | 0 | 52 | |
| >1 000 000 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Totals | 170 755 | 4830 | 19 453 | 9651 | 1636 | 185 | 173 | 158 | 8 | 206 849 | |
| In GenBank | 36 094 | ||||||||||
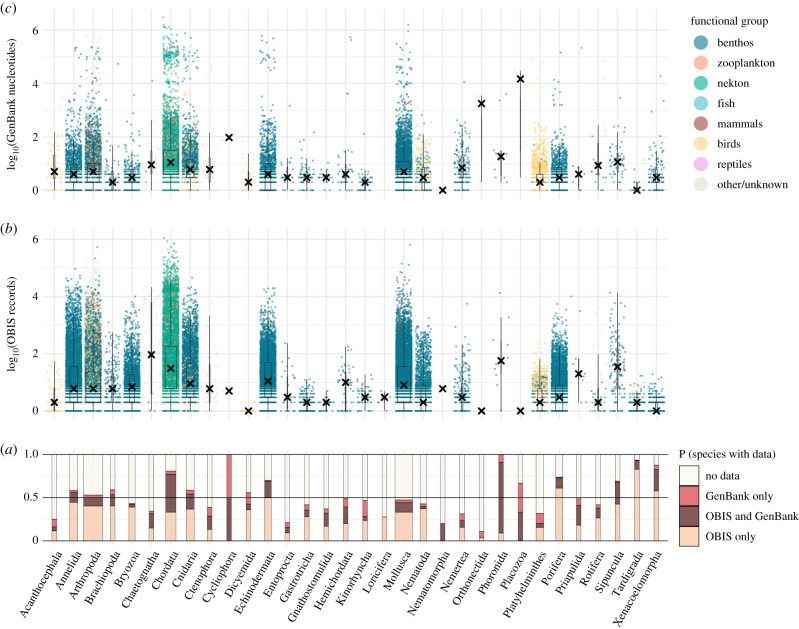
The distribution of OBIS and GenBank records across animal phyla and functional groups is shown in figure 1. At least one species from every phylum has records in either OBIS or GenBank, with all phyla except Loricifera (which has just 29 species) represented in both databases (figure 1a). Across all phyla, just over half (55%) of all species are represented in one or other database. Most species that are present in OBIS have only a few occurrence records, with median values of records ranging from 1 to 92 across phyla (figure 1b). A similar pattern is observed for GenBank nucleotides (figure 1c), with median values between 1 and 94 except in phyla Orthonectida and Placozoa, both of which have only two species represented in GenBank, one of which has several thousand nucleotides (in Orthonectida, Intoshia linei has 3522, in Placozoa, Trichoplax adhaerens has 29 176).
Figure 1.
Availability of biogeographic (over 45 M OBIS occurrence records) and genetic (over 56 M GenBank nucleotides) data across 206 849 marine animal species, summarized by phylum and by broad functional group. (a) Proportion of species in each phylum with data in either database, both databases, or neither. Bar width is proportional to the number of species in each phylum. The number of (b) OBIS occurrence records and (c) GenBank nucleotide sequences are shown for species that occur in the respective database. Each point represents a species, coloured by functional group. Box plots are superimposed with X marking the median number of records within each phylum. Phylum size varies from two species (Cycliophora) to 57 336 species (Arthropoda).
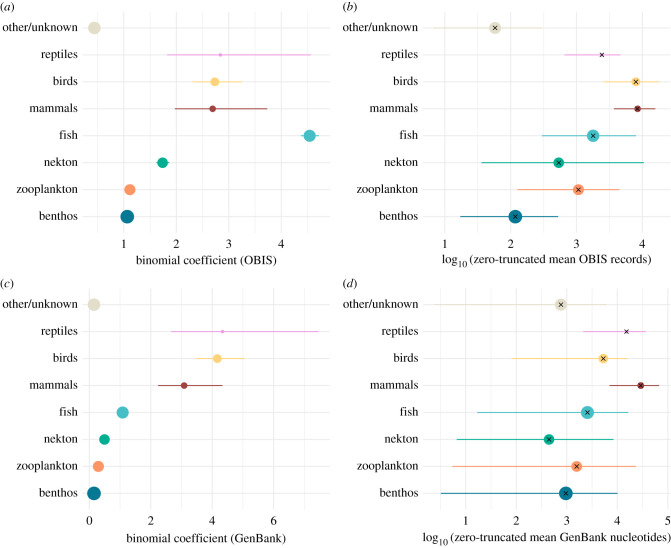
Data availability is variable across functional groups (figures 1b,c and 2). Modelling the presence or absence of species in OBIS in a binomial GLM shows that species of fish, mammal, bird and reptile are much more likely to have occurrences in OBIS than are benthic or zooplankton species, with nekton falling in between, and species with unknown or other functional group classification the least likely to have occurrence records (figure 2a). A broadly similar pattern holds when modelling the number of occurrence records for those species with at least 1 (figure 2b), with the vertebrate taxa again tending to have most records, although distinctions between vertebrates and other groups are less stark. Benthic invertebrates typically have few OBIS records, but zooplankton that do occur in OBIS tend to have more records than nekton. In GenBank, birds, reptiles and mammals are most likely to be present in the database, followed by fish, nekton and zooplankton, with benthos and other/unknown functional groups least likely to be represented (figure 2c). The rank order changes somewhat when considering number of nucleotides across species present in GenBank (figure 2d), with most records from mammals and reptiles, followed by birds and fish. Nekton tends to have fewest records, but there is considerable variability within all major groups. Data availability in both major databases is broadly similar across fish habitat groupings (electronic supplementary material, figure S1).
Figure 2.
Coefficients from the hurdle models of data availability across functional groups, first modelling presence in a database with a binomial model, and then non-zero counts of records in a database with a negative binomial model. Species presence in OBIS (a) or GenBank nucleotide database (c) across functional groups is indicated with binomial coefficients (with 95% confidence intervals) on the response scale, representing the ratio of the probabilities of species within a group having records in the database versus not having records in the database. For the subset of species present in (b) OBIS or (d) GenBank, the empirical mean number of records per species is plotted together with bootstrapped 95% confidence intervals. For each group, the predicted non-zero count from the hurdle model is indicated with an X. Point size is scaled to the total number of species in each functional group (a,c, ranging from 96 reptiles to 146 551 benthos) and to the number of species in each group with records in OBIS (b, 71 reptiles to 75 604 benthos) or GenBank (d, 78 reptiles to 19 235 benthos).
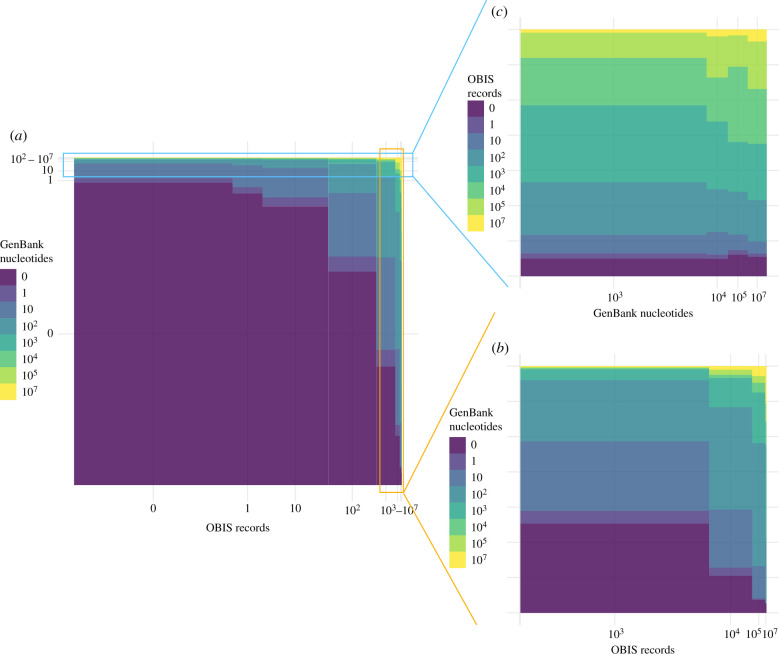
Considering the joint distribution of species across OBIS and GenBank categorical scales, 93 519 (45%) species have no records in either database (table 2 and figure 3a). In general, species with more records in OBIS also tend to have more nucleotides in GenBank (table 2 and figure 3), indicating that these different biodiversity data aggregators have similar biases in terms of the known marine biodiversity that they encompass. There are exceptions though: in particular, several species have many (more than 100 000) GenBank nucleotides but very few (if any) OBIS records (table 3).
Figure 3.
Mosaic plot showing the joint distribution of species between categories of OBIS records and GenBank nucleotides. Panel (a) shows all species, and is dominated by species with no records in either database, (b) zooms in on species with high numbers (greater than 100) of OBIS records and (c) reverses the axes and zooms in on species with high numbers (greater than 100) of GenBank nucleotides. Axis labels indicate the number of records at the right-hand bound of each category.
Table 3.
Species with high numbers of GenBank nucleotide records but few OBIS occurrences.
| species | phylum | class | functional group | GenBank nucleotides | OBIS records |
|---|---|---|---|---|---|
| Olavius algarvensis | Annelida | Clitellata | benthos | 173 609 | 0 |
| Capitella teleta | Annelida | Polychaeta | benthos | 208 794 | 1 |
| Platynothrus peltifer | Arthropoda | Arachnida | other/unknown | 106 099 | 0 |
| Caligus rogercresseyi | Arthropoda | Hexanauplia | other/unknown | 628 843 | 0 |
| Proasellus racovitzai | Arthropoda | Malacostraca | benthos | 127 716 | 0 |
| Proasellus ibericus | Arthropoda | Malacostraca | benthos | 150 798 | 0 |
| Bragasellus molinai | Arthropoda | Malacostraca | benthos | 209 419 | 0 |
| Proasellus beticus | Arthropoda | Malacostraca | benthos | 228 033 | 0 |
| Seriola quinqueradiata | Chordata | Actinopterygii | fish | 105 911 | 6 |
| Theragra finnmarchica | Chordata | Actinopterygii | fish | 130 916 | 0 |
| Takifugu flavidus | Chordata | Actinopterygii | fish | 138 301 | 0 |
| Takifugu rubripes | Chordata | Actinopterygii | fish | 466 790 | 5 |
| Molgula tectiformis | Chordata | Ascidiacea | benthos | 106 904 | 0 |
| Halocynthia roretzi | Chordata | Ascidiacea | benthos | 116 123 | 4 |
| Pelecanus crispus | Chordata | Aves | birds | 231 775 | 0 |
| Balaenoptera acutorostrata | Chordata | Mammalia | mammals | 238 976 | 0 |
| Emydocephalus ijimae | Chordata | Reptilia | reptiles | 157 876 | 0 |
| Hemicentrotus pulcherrimus | Echinodermata | Echinoidea | benthos | 153 541 | 3 |
| Apostichopus parvimensis | Echinodermata | Holothuroidea | benthos | 166 764 | 1 |
| Apostichopus japonicus | Echinodermata | Holothuroidea | benthos | 401 310 | 4 |
| Cumia reticulata | Mollusca | Gastropoda | benthos | 144 517 | 2 |
| Amphimedon queenslandica | Porifera | Demospongiae | benthos | 142 554 | 9 |
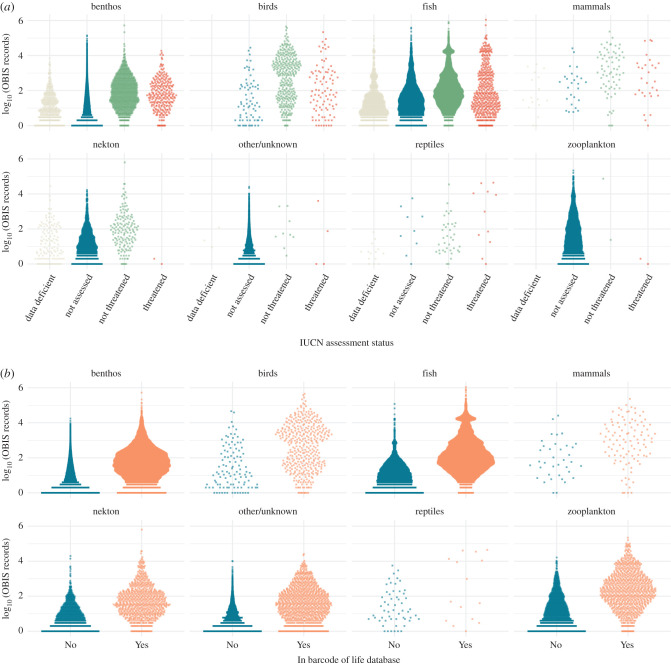
A similar pattern is evident when examining the distribution of OBIS records across different IUCN assessment categories. In general, and across functional groups, the proportion of species with records in OBIS is higher in assessed species (threatened and non-threatened) than it is in unassessed or data-deficient species: overall, 84% of threatened and 94% of non-threatened species have occurrence records in OBIS, compared to 75% of data-deficient and 49% of unassessed species (table 4). Considering only those species with records in OBIS, there is considerable variation within and between IUCN categories in the number of occurrence records per species, but a general tendency is apparent in all functional groups for species in threatened and non-threatened categories to have more occurrence records than those in data-deficient and unassessed categories (figure 4a).
Table 4.
Breakdown of marine animal species by functional group and IUCN assessment status. Listed for each IUCN assessment status are the total number of species per functional group, the number of these species with occurrences in OBIS and the associated percentage.
| functional group | IUCN assessment status |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| not assessed |
data deficient |
threatened |
non-threatened |
|||||||||
| N (species) | N (species in OBIS) | % species in OBIS | N (species) | N (species in OBIS) | % species in OBIS | N (species) | N (species in OBIS) | % species in OBIS | N (species) | N (species in OBIS) | % species in OBIS | |
| benthos | 144 097 | 73 610 | 51 | 749 | 530 | 71 | 305 | 258 | 85 | 1400 | 1 206 | 86 |
| zooplankton | 5742 | 3027 | 53 | 0 | 0 | — | 4 | 2 | 50 | 2 | 2 | 100 |
| nekton | 3076 | 1878 | 61 | 160 | 127 | 79 | 7 | 2 | 29 | 156 | 151 | 97 |
| fish | 8599 | 6161 | 72 | 1780 | 1350 | 76 | 523 | 457 | 87 | 7359 | 6997 | 95 |
| mammals | 66 | 26 | 39 | 20 | 17 | 85 | 36 | 29 | 81 | 70 | 68 | 97 |
| birds | 179 | 71 | 40 | 1 | 0 | 0 | 125 | 92 | 74 | 382 | 340 | 89 |
| reptiles | 20 | 9 | 45 | 21 | 14 | 67 | 11 | 11 | 100 | 44 | 37 | 84 |
| other/unknown | 31 891 | 9725 | 31 | 3 | 3 | 100 | 10 | 4 | 40 | 11 | 9 | 82 |
| totals | 193 670 | 94 507 | 49 | 2734 | 2041 | 75 | 1021 | 855 | 84 | 9424 | 8810 | 94 |
Figure 4.
Distribution of occurrence records across 106 213 marine animal species present in OBIS by functional group and by (a) IUCN assessment status and (b) presence in the Barcode of Life Data System. Each point represents a species.
Species with DNA barcodes are disproportionately likely to also have occurrence records in OBIS: 45% of species with no record in the Barcode of Life database have at least one occurrence record in OBIS, compared to 89% of species with a barcode (table 5). In addition, in all functional groups, species with barcodes tend to have more OBIS records than those that do not (figure 4b).
Table 5.
Breakdown of marine animal species by functional group and presence in the BOLD DNA Barcode database. Listed for species absent from or present in BOLD are the total number of species per functional group, the number of these species with occurrences in OBIS, and the associated percentages.
| functional group | in barcode of life database? |
|||||
|---|---|---|---|---|---|---|
| no |
yes |
|||||
| N (species) | N (species in OBIS) | % species in OBIS | N (species) | N (species in OBIS) | % species in OBIS | |
| benthos | 131 390 | 62 316 | 47 | 15 161 | 13 288 | 88 |
| zooplankton | 4768 | 2117 | 44 | 980 | 914 | 93 |
| nekton | 2506 | 1355 | 54 | 893 | 803 | 90 |
| fish | 8683 | 5842 | 67 | 9578 | 9123 | 95 |
| mammals | 85 | 37 | 44 | 107 | 103 | 96 |
| birds | 238 | 108 | 45 | 449 | 395 | 88 |
| reptiles | 80 | 59 | 70 | 16 | 15 | 94 |
| other/unknown | 30 292 | 8692 | 29 | 1623 | 1049 | 65 |
| totals | 178 042 | 80 526 | 45 | 28 807 | 25 690 | 89 |
4. Discussion
Using the taxonomic backbone of the World Register of Marine Species [37] we have summarized data availability across axes of biogeography, genetics, molecular taxonomy and conservation status for 206 849 marine animal species. This presents a mixed picture. One the one hand, 91 828 (44%) species have no records in any of these databases, and are represented only by their name. This is considerably higher than the 27% of plant species with no information other than their name [36], although of course, the marine environment represents far larger habitable volume [54] and marine animals are a much more diverse taxonomic group. Only 6688 marine animal species (3%) have records in all four of the datasets that we consider – again, rather lower than the 18% of broadly covered plant species [36]. At the same time, it is important to remember that presence in a dataset does not imply extensive knowledge: among the 106 203 species with records in OBIS, for example, the median number of recorded occurrences is just 7, and 18% of these species (18 869 species) are known from only a single occurrence. Nonetheless, the distribution of biogeographic and genetic information across the animal tree of life is extensive, with all animal phyla represented in at least one database (figure 1). Data availability tends to be biased towards well-known taxa and functional groups (especially vertebrates; figures 1, 2, 4), in agreement with previous assessments (e.g. [32]), but the subset of 225 species with more than 1000 occurrences in OBIS and more than 1000 nucleotides in GenBank is drawn from 10 phyla and 27 classes, representing all major functional groups, and most of them have a barcode in BOLD (214 species) and have been assessed by the IUCN as something other than data deficient (102 non-threatened, 23 threatened species). For these diverse marine animal species, then, it is reasonable to propose that the information available across multiple sources can be translated into knowledge about their distribution, evolutionary relationships and conservation status.
The broad positive correlation between data availability across different sources (tables 2 and 4 and figure 3) reinforces previous findings that species with good information on one facet of their biology and ecology tend to be well represented in other databases too, both in plants [36] and in marine species [35]. These information-rich species are likely to be those most easily and frequently observed, or those of high economic or cultural value, and so will not be a random subset of all species. However, the consequences of biases towards data availability from these common species will vary depending on the specific question of interest. For instance, ecosystem function may be driven largely by just those common species that tend to be so well known [55]; but rare species will clearly be of great interest to conservationists, and may indeed sometimes contribute unique trait combinations to marine communities [56].
In terrestrial conservation, considerable concern has been expressed over the likely conservation status of species too poorly known to formally assess, as they tend to have characteristics (rarity, small ranges, occurring in poorly studied regions) that will predispose them to be at risk [57]. For some marine taxa, this appears to be the case too, with high rates of extinction risk predicted for European sharks and rays formally assessed as Data Deficient [58], and low levels of conservation assessment in poorly known marine groups may contribute to low overall documented levels of extinction risk [59]. On the other hand, the fact that the biggest data gaps in marine biodiversity tend to be in remote habitats largely inaccessible to humans (e.g. the deep pelagic ocean; [60]), and the highest rates of discoveries of new species and habitats are also in the deep sea [61,62], provides some contrast with the terrestrial situation, and may insulate these poorly known species somewhat from human pressures. However, some patterns still hold in the deep sea, such as the tendency for widespread species to be encountered and described first [63], meaning that many of the species not yet present in major databases may be genuinely rare. Given the acceleration of human activities into previously unexploited regions of the oceans [64], with new threats including deep sea mining [65] and exploitation of the mesopelagic [66], it seems unwise to assume that the large fraction of marine biodiversity that remains poorly known is not at risk. Given the fact that Data Deficient conservation assessments are twice as frequent in marine versus non-marine taxa [34], data-driven predictive conservation assessments [58,67,68], which rely on some of the kinds of data we consider here (spatial distribution, evolutionary relationships and ecological guilds) combined with biological traits, may prove to be especially valuable tools.
The aim of this study was to flag priorities for future work. One important point is that the major publicly available databases on which we draw do not constitute the sum total of data ever collected on marine species. This is particularly the case for occurrence data, as globally researchers have yet to adopt the routine deposition of species occurrences in OBIS as a cultural norm, in the way that genetic sequence data are deposited in GenBank. To this end, improving incentives for researchers to add their data to global repositories in an important goal [25], while data archaeology and rescue initiatives can help to ensure that historical data are captured [69]. Equally, it remains vital that ongoing survey schemes are properly valued [23], even as novel exploration is planned. At the same time, our quantification exercise can help to identify groups of species where a little additional research effort in one area would quickly result in a more valuable dataset. One candidate set of species might be those that are frequently observed but poorly represented in other databases. For instance, 1216 species have more than 1000 OBIS records but fewer than 10 GenBank nucleotides; and over half of the 3533 species with more than 1000 OBIS occurrences are either not assessed by the IUCN (1876 species) or data-deficient (82 species). The fact that almost 90% (3163) of these species have DNA barcodes in BOLD is encouraging, however, suggesting considerable potential for an increasing role for molecular studies to address a wide range of questions in marine ecology [70].
Mining the spatial information already present in other databases also has potential for supplementing existing occurrence datasets. In this study, we relied on existing links between WoRMS and GenBank and BOLD, which simply summarize the number of nucleotides or barcodes present for each species. The spatial meta-data stored in the sequence databases provide an additional source of information, although in GenBank these data are relatively unstructured. Searching the GenBank nucleotide database, we found just 1437 records for animals that contained a lat-lon field; matching this to our list of marine animals reduced this further to 183 records from 42 species. Nonetheless, even from this small set of species, 21 do not have occurrence records in OBIS, suggesting that mining GenBank for spatial data would likely add valuable information for a small number of species. Various methods have been developed to attempt this, based around mining spatial information from the full text of associated publications [71,72], with initiatives such as the Genomic Observatories MetaDatabase (GEOME, https://geome-db.org) also seeking to simplify access to meta-data from sequence datasets.
BOLD typically does store spatial data for individual specimens in a well-structured manner, only some of which have been harvested by OBIS. In our dataset, 3117 species have BOLD barcodes but no OBIS records. Several of these are parasites, which we know are not well recorded in OBIS (e.g. Schistocephalus solidus, 718 barcodes; Anguillicoloides crassus, 508 barcodes) but there are free-living marine species too, such as the Gastropod mollusc Littoraria sinensis (257 barcodes) and the Copepod Calanoides natalis (183 barcodes). Accessing the specimen data from BOLD using the bold R package [73] for these two species reveals that none of the L. sinensis specimens have information in the latitude and longitude fields, but full geographical information is available for 227 specimens for Calanoides natalis. Although none of these locations are currently recorded in OBIS, some are in GBIF, highlighting the often complex pipelines from data providers to global data aggregators. Improving pipelines from genetic databases to occurrence databases is currently a priority for OBIS (W. Appeltans, OBIS Project Manager 2020, personal communication).
Finally, the dimensions of diversity that we summarize in this study are somewhat limited. We did not consider the traits of species, for instance, beyond functional groups that indicate habitat affiliation in very broad terms (e.g. benthic versus planktonic). These groupings are already useful as global patterns of diversity are known to differ between them [40], and they can also be used to refine methods of matching species occurrences to global sea temperature datasets [74], helping to predict species responses to climate change [75]. Beyond these coarse functional groups, however, traits data remain scarce even in reasonably common marine species in well-studied regions [35], and despite many efforts at collating traits—including within WoRMS [76]—there is still no widely adopted central standard [77]. Certain groups are well covered by existing initiatives (e.g. FishBase [10], the Coral Trait Database [11]), and whether a single overarching portal to cover the immense diversity of marine lifeforms is possible—or even desirable—remains open for discussion. However, it is certainly the case that multiple smaller-scale projects collect valuable traits data for a subset of species that is typically made available (if at all) via supplementary material or bespoke web portals, at risk of being lost to the community. A wider adoption of principles embedded in initiatives like the Open Traits Network [7] would ensure interoperability of these small, project-specific traits datasets, maximizing the availability of information on key traits for the largest possible fraction of marine diversity. Readily available information on even just a few traits (e.g. body size, longevity, fecundity, planktonic larval duration) would help to test predictions from biodiversity models, embed life-history theory into marine conservation and predict the consequences of human activities for marine diversity [40,78–80].
The stocktake of marine biodiversity data availability that we have undertaken here adds to previous efforts focused on occurrence data [19,32,81]. While we reveal a similar story of gaps and biases across other data sources, there is considerable overlap in coverage too, and overall the potential to link dimensions of marine animal diversity is now high. The priority now should be to build on the substantial community-built foundations and to improve the pipeline from raw data to interoperable data products, both as a resource for fundamental macroecological research and to facilitate effective stewardship of our blue planet.
Supplementary Material
Acknowledgements
The ideas for this study were conceived while T.J.W. was a Royal Society University Research Fellow, and developed over the course of the Natural Environment Research Council and Department for Environment, Food and Rural Affairs Marine Ecosystems Research Programme (grant no. NE/L003279/1) and EMODnet Biology. The data underpinning this work rely on support from the Research Foundation Flanders (FWO - the Flemish contribution to LIfewatch). Thanks to Helmut Hillebrand for the invitation to present this work at the 2019 HIFMB Symposium on Functional Marine Biodiversity and for the opportunity to contribute to this theme issue. Constructive and insightful comments from two anonymous reviewers have helped to greatly improve this work.
Data accessibility
All data used in this article are publicly available via WoRMS. The processed summary data we use for our analysis is openly available under a Creative Commons Attribution 4.0 International License as Webb, T.J.; Vanhoorne, B. (2020): WoRMS Marine Animals with links to external databases. Marine Data Archive. https://doi.org/10.14284/417. R code to replicate our analyses and figures is available via https://github.com/tomjwebb/linking_marine_diversity_data and is archived on Figshare via the University of Sheffield's Online Research Data repository here: https://doi.org/10.15131/shef.data.12833891.
Authors' contributions
T.J.W. conceived the research, conducted analyses and drafted the manuscript. B.V. contributed technical expertise in accessing, formatting and interpreting the database.
Competing interests
We declare we have no competing interests.
References
- 1.Redelings BD, Holder MT. 2017. A supertree pipeline for summarizing phylogenetic and taxonomic information for millions of species. PeerJ 5, e3058 ( 10.7717/peerj.3058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinchliff CE, et al. 2015. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl Acad. Sci. USA 112, 12 764–12 769. ( 10.1073/pnas.1423041112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res. 44, D67–D72. ( 10.1093/nar/gkv1276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leray M, Knowlton N, Ho S-L, Nguyen BN, Machida RJ. 2019. GenBank is a reliable resource for 21st century biodiversity research. Proc. Natl Acad. Sci. USA 116, 22 651–22 656. ( 10.1073/pnas.1911714116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GBIF. 2020. GBIF Home Page. See https://www.gbif.org.
- 6.IUCN. 2020. The IUCN Red List of Threatened Species. Version 2020-1. See httpswww.iucnredlist.org.
- 7.Gallagher RV, et al. 2020. Open science principles for accelerating trait-based science across the tree of life. Nat. Ecol. Evol. 4, 294–303. ( 10.1038/s41559-020-1109-6) [DOI] [PubMed] [Google Scholar]
- 8.Bennett JM, et al. 2018. GlobTherm, a global database on thermal tolerances for aquatic and terrestrial organisms. Sci. Data 5, 180022 ( 10.1038/sdata.2018.22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarieva A, Gorshkov V, Li B. 2005. Biochemical universality of living matter and its metabolic implications. Funct Ecol 19, 547–557. ( 10.1111/j.1365-2435.2005.01005.x) [DOI] [Google Scholar]
- 10.Froese R, Pauly D. 2019. FishBase. World Wide Web electronic publication. version (12/2019). www.fishbase.org.
- 11.Madin JS, et al. 2016. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci. Data 3, 178–122 ( 10.1038/sdata.2016.17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kranstauber B, Cameron A, Weinzerl R, Fountain T, Tilak S, Wikelski M, Kays R. 2011. The Movebank data model for animal tracking. Environ. Model. Softw. 26, 834–835. ( 10.1016/j.envsoft.2010.12.005) [DOI] [Google Scholar]
- 13.Wikelski M, Davidson SC, Kays R.. 2020. Movebank: archive, analysis and sharing of animal movement data. www.movebank.org.
- 14.Poelen JH, Simons JD, Mungall CJ. 2014. Global biotic interactions: an open infrastructure to share and analyze species-interaction datasets. Ecol. Inf. 24, 148–159. ( 10.1016/j.ecoinf.2014.08.005) [DOI] [Google Scholar]
- 15.Tanhua T, et al. 2019. Ocean FAIR data services. Front. Mar. Sci. 6, 92 ( 10.3389/fmars.2019.00440) [DOI] [Google Scholar]
- 16.Silvertown J. 2009. A new dawn for citizen science. Trends Ecol. Evol. 24, 467–471. ( 10.1016/j.tree.2009.03.017) [DOI] [PubMed] [Google Scholar]
- 17.Chandler M, et al. 2017. Contribution of citizen science towards international biodiversity monitoring. Biol. Conserv. 213, 280–294. ( 10.1016/j.biocon.2016.09.004) [DOI] [Google Scholar]
- 18.Hyder K, Townhill B, Anderson LG, Delany J, Pinnegar JK. 2015. Can citizen science contribute to the evidence-base that underpins marine policy? Mar. Policy 59, 112–120. ( 10.1016/j.marpol.2015.04.022) [DOI] [Google Scholar]
- 19.Edgar GJ, Bates AE, Bird TJ, Jones AH, Kininmonth S, Stuart-Smith RD, Webb TJ. 2015. New approaches to marine conservation through scaling up of ecological data. Annu. Rev. Mar. Sci. 8, 150807173619006 ( 10.1146/annurev-marine-122414-033921) [DOI] [PubMed] [Google Scholar]
- 20.Edgar GJ, Stuart-Smith RD. 2014. Systematic global assessment of reef fish communities by the reef life survey program. Sci. Data 1, 1–8. ( 10.1038/sdata.2014.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snelgrove PVR. 2010. Discoveries of the census of marine life: making ocean life count, 1st edn Cambridge, UK; New York, NY: Cambridge University Press. [Google Scholar]
- 22.OBIS. 2020. Ocean Biodiversity Information System. www.iobis.org.
- 23.Mieszkowska N, Sugden H, Firth LB, Hawkins SJ. 2014. The role of sustained observations in tracking impacts of environmental change on marine biodiversity and ecosystems. Phil. Trans. R. Soc. A 372, 20130339 ( 10.1098/rsta.2013.0339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miloslavich P, et al. 2018. Essential ocean variables for global sustained observations of biodiversity and ecosystem changes. Global Change Biol. 105, 10456 ( 10.1111/gcb.14108) [DOI] [PubMed] [Google Scholar]
- 25.Dornelas M, et al. 2019. Towards a macroscope: leveraging technology to transform the breadth, scale and resolution of macroecological data. Global Ecol. Biogeogr. 28, 1937–1948. ( 10.1111/geb.13025) [DOI] [Google Scholar]
- 26.Basset A, Los W. 2012. Biodiversity e-Science: LifeWatch, the European infrastructure on biodiversity and ecosystem research. Plant Biosyst. Int. J. Dealing Aspects Plant Biol. 146, 780–782. ( 10.1080/11263504.2012.740091) [DOI] [Google Scholar]
- 27.La Salle J, Williams KJ, Moritz C. 2016. Biodiversity analysis in the digital era. Phil. Trans. R. Soc. B 371, 20150337 ( 10.1098/rstb.2015.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardisty AR, et al. 2019. The Bari Manifesto: an interoperability framework for essential biodiversity variables. Ecol. Inf. 49, 22–31. ( 10.1016/j.ecoinf.2018.11.003) [DOI] [Google Scholar]
- 29.Maitner BS, et al. 2018. The bien r package: a tool to access the botanical information and ecology network (BIEN) database. Methods Ecol. Evol. 9, 373–379. ( 10.1111/2041-210X.12861) [DOI] [Google Scholar]
- 30.Costello M, Coll M, Danovaro R, Halpin P, Ojaveer H.. 2010. A census of marine biodiversity knowledge, resources, and future challenges. PLoS ONE 5, e12110 ( 10.1371/journal.pone.0012110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snelgrove P, et al. 2016. Global patterns in marine biodiversity. In The first global integrated marine assessment: World Ocean Assessment I (United Nations), pp. 501–524. Cambridge, UK: Cambridge University Press; ( 10.1017/9781108186148.037) [DOI] [Google Scholar]
- 32.Miloslavich P, et al. 2016. Chapter 15: Extent of assessment of marine biological diversity. In The first global integrated marine assessment: World Ocean Assessment I (United Nations), pp. 525–554. Cambridge, UK: Cambridge University Press; ( 10.1017/9781108186148.038) [DOI] [Google Scholar]
- 33.Appeltans W, et al. 2012. The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. ( 10.1016/j.cub.2012.09.036) [DOI] [PubMed] [Google Scholar]
- 34.Webb TJ, Mindel BL. 2015. Global patterns of extinction risk in marine and non-marine systems. Curr. Biol. 25, 506–511. ( 10.1016/j.cub.2014.12.023) [DOI] [PubMed] [Google Scholar]
- 35.Tyler EHM, Somerfield PJ, Berghe EV, Bremner J, Jackson E, Langmead O, Palomares MLD, Webb TJ. 2012. Extensive gaps and biases in our knowledge of a well-known fauna: implications for integrating biological traits into macroecology. Global Ecol. Biogeogr. 21, 922–934. ( 10.1111/j.1466-8238.2011.00726.x) [DOI] [Google Scholar]
- 36.Cornwell WK, Pearse WD, Dalrymple RL, Zanne AE. 2019. What we (don't) know about global plant diversity. Ecography 42, 1819–1831. ( 10.1111/ecog.04481) [DOI] [Google Scholar]
- 37.WoRMS Editorial Board. 2020World Register of Marine Species. See http://www.marinespecies.org ( 10.14284/170) [DOI]
- 38.Ratnasingham S, Hebert P. 2007. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. ( 10.1111/j.1471-8286.2006.01678.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freckleton RP, Jetz W. 2009. Space versus phylogeny: disentangling phylogenetic and spatial signals in comparative data. Proc. R. Soc. B 276, 21–30. ( 10.1098/rspb.2008.0905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worm B, Tittensor DP. 2018. A theory of global biodiversity. Princeton, NJ and Oxford, UK: Princeton University Press. [Google Scholar]
- 41.Harfoot MBJ, Newbold T, Tittensor DP, Emmott S, Hutton J, Lyutsarev V, Smith MJ, Scharlemann JPW, Purves DW. 2014. Emergent global patterns of ecosystem structure and function from a mechanistic general ecosystem model. PLoS Biol. 12, e1001841 ( 10.1371/journal.pbio.1001841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain S. 2019. World Register of Marine Species (WoRMS) Client. R package worrms version 0.4.0. See https://CRAN.R-project.org/package=worrms.
- 43.WoRMS. 2020. Pisces. See http://www.marinespecies.org/aphia.php?p=taxdetails&id=11676.
- 44.Boettiger C, Lang DT, Wainwright PC. 2012. rfishbase: exploring, manipulating and visualizing FishBase data from R. J. Fish Biol. 81, 2030–2039. ( 10.1111/j.1095-8649.2012.03464.x) [DOI] [PubMed] [Google Scholar]
- 45.Zeileis A, Kleiber C, Jackman S. 2008. Regression models for count data in R. J. Stat. Softw. 27, 1–25. ( 10.18637/jss.v027.i08) [DOI] [Google Scholar]
- 46.Jackman S. 2020. Pscl: classes and methods for R developed in the political science computational laboratory. Sydney, New South Wales, Australia: United States Studies Centre, University of Sydney; R package version 1.5.5. See https://github.com/atahk/pscl/. [Google Scholar]
- 47.Hofmann H. 2008. Mosaic plots and their variants. In Handbook of data visualisation (eds Chen C-H, Härdle W, Unwin A), pp. 617–642. Berlin, Germany: Springer Handbooks of Computational Statistics; ( 10.1007/978-3-540-33037-0_24) [DOI] [Google Scholar]
- 48.Jeppson H, Hofmann H, Cook D. 2018. ggmosaic: Mosaic Plots in the ‘ggplot2’ Framework. R package version 0.2.0. See https://CRAN.R-project.org/package=ggmosaic.
- 49.R Core Team. 2019. R: A language and environment for statistical computing. See https://www.R-project.org.
- 50.RStudio Team. 2019. RStudio: Integrated Development for R. See https://rstudio.com.
- 51.Wickham H, et al. 2019. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 ( 10.21105/joss.01686) [DOI] [Google Scholar]
- 52.Clarke E, Sherrill-Mix S. 2017. ggbeeswarm: Categorical Scatter (Violin Point) Plots. R package version 0.6.0. See https://CRAN.R-project.org/package=ggbeeswarm.
- 53.Pedersen TL. 2019. patchwork: The Composer of Plots. R package version 1.0.0. See https://CRAN.R-project.org/package=patchwork.
- 54.Dawson MN. 2012. Species richness, habitable volume, and species densities in freshwater, the sea, and on land. Front. Biogeogr. 4, fb_12675 ( 10.21425/F54312675) [DOI] [Google Scholar]
- 55.Gaston K, Fuller R. 2008. Commonness, population depletion and conservation biology. Trends Ecol. Evol. 23, 14–19. ( 10.1016/j.tree.2007.11.001) [DOI] [PubMed] [Google Scholar]
- 56.Mouillot D, et al. 2013. Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol. 11, e1001569 ( 10.1371/journal.pbio.1001569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don't know about Earth's missing biodiversity. Trends Ecol. Evol. 27, 501–510. ( 10.1016/j.tree.2012.05.008) [DOI] [PubMed] [Google Scholar]
- 58.Walls RHL, Dulvy NK. 2019. Predicting the conservation status of Europe's Data Deficient sharks and rays. bioRxiv276, 614776 ( 10.1101/614776) [DOI]
- 59.Mindel BL, Webb TJ, Neat FC, Blanchard JL. 2016. A trait-based metric sheds new light on the nature of the body size–depth relationship in the deep sea. J. Anim. Ecol. 85, 427–436. ( 10.1111/1365-2656.12471) [DOI] [PubMed] [Google Scholar]
- 60.Webb TJ, Vanden Berghe E, O'Dor R. 2010. Biodiversity's big wet secret: the global distribution of marine biological records reveals chronic under-exploration of the deep pelagic ocean. PLoS ONE 5, e10223 ( 10.1371/journal.pone.0010223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez-Llodra E, et al. 2010. Deep, diverse and definitely different: unique attributes of the world's largest ecosystem. Biogeosciences 7, 2851–2899. ( 10.5194/bg-7-2851-2010) [DOI] [Google Scholar]
- 62.Danovaro R, Snelgrove PVR, Tyler P. 2014. Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475. ( 10.1016/j.tree.2014.06.002) [DOI] [PubMed] [Google Scholar]
- 63.Higgs ND, Attrill M. 2015. Biases in biodiversity: wide-ranging species are discovered first in the deep sea. Front. Mar. Sci. 2, 717 ( 10.3389/fmars.2015.00061) [DOI] [Google Scholar]
- 64.Jouffray JB, Blasiak R, Norström AV, Österblom H, Nyström M. 2020. The blue acceleration: the trajectory of human expansion into the ocean. One Earth 2, 43–54. ( 10.1016/j.oneear.2019.12.016) [DOI] [Google Scholar]
- 65.Jones DOB, Amon DJ, Chapman ASA. 2018. Mining deep-ocean mineral deposits: what are the ecological risks? Elements 14, 325–330. ( 10.2138/gselements.14.5.325) [DOI] [Google Scholar]
- 66.Hidalgo M, Browman HI. 2019. Developing the knowledge base needed to sustainably manage mesopelagic resources. ICES J. Mar. Sci. 76, 609–615. ( 10.1093/icesjms/fsz067) [DOI] [Google Scholar]
- 67.Jetz W, Freckleton RP. 2015. Towards a general framework for predicting threat status of data-deficient species from phylogenetic, spatial and environmental information. Phil. Trans. R. Soc. B 370, 20140016 ( 10.1098/rstb.2014.0016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.González-del-Pliego P, Freckleton RP, Edwards DP, Koo MS, Scheffers BR, Pyron RA, Jetz W. 2019. Phylogenetic and trait-based prediction of extinction risk for data-deficient amphibians. Curr. Biol. 29, 1557–1563.e3. ( 10.1016/j.cub.2019.04.005) [DOI] [PubMed] [Google Scholar]
- 69.Faulwetter S, et al. 2016. EMODnet Workshop on mechanisms and guidelines to mobilise historical data into biogeographic databases. RIO 2, e9774–e9728 ( 10.3897/rio.2.e9774) [DOI] [Google Scholar]
- 70.Goodwin KD, Thompson LR, Duarte B, Kahlke T, Thompson AR, Marques JC, Caçador I. 2017. DNA sequencing as a tool to monitor marine ecological status. Front. Mar. Sci. 4, e1002358 ( 10.3389/fmars.2017.00107) [DOI] [Google Scholar]
- 71.Tahsin T, Weissenbacher D, Rivera R, Beard R, Firago M, Wallstrom G, Scotch M, Gonzalez G. 2016. A high-precision rule-based extraction system for expanding geospatial metadata in GenBank records. J. Am. Med. Inform. Assoc. 23, 934–941. ( 10.1093/jamia/ocv172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tahsin T, Weissenbacher D, O'Connor K, Magge A, Scotch M, Gonzalez-Hernandez G. 2017. GeoBoost: accelerating research involving the geospatial metadata of virus GenBank records. Bioinformatics 34, 1606–1608. ( 10.1093/bioinformatics/btx799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chamberlain S. 2020 bold: Interface to Bold Systems API. R package version 1.1.0. See https://CRAN.R-project.org/package=bold .
- 74.Webb TJ, Lines A, Howarth LM. 2020. Occupancy-derived thermal affinities reflect known physiological thermal limits of marine species. Ecol. Evol. 75, 209 ( 10.1002/ece3.6407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pinsky ML, Selden RL, Kitchel ZJ. 2020. Climate-driven shifts in marine species ranges: scaling from organisms to communities. Annu. Rev. Mar. Sci. 12, 153–179. ( 10.1146/annurev-marine-010419-010916) [DOI] [PubMed] [Google Scholar]
- 76.Costello MJ, Claus S, Dekeyzer S, Vandepitte L, Tuama ÉÓ, Lear D, Tyler-Walters H. 2015. Biological and ecological traits of marine species. PeerJ 3, e1201 ( 10.7717/peerj.1201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beauchard O, Veríssimo H, Queirós AM, Herman PMJ. 2017. The use of multiple biological traits in marine community ecology and its potential in ecological indicator development. Ecol. Indic. 76, 81–96. ( 10.1016/j.ecolind.2017.01.011) [DOI] [Google Scholar]
- 78.Kindsvater HK, Mangel M, Reynolds JD, Dulvy NK. 2016. Ten principles from evolutionary ecology essential for effective marine conservation. Ecol. Evol. 6, 2125–2138. ( 10.1002/ece3.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hiddink JG, et al. 2019. Assessing bottom trawling impacts based on the longevity of benthic invertebrates. J. Appl. Ecol. 56, 1075–1084. ( 10.1111/1365-2664.13278) [DOI] [Google Scholar]
- 80.Álvarez-Noriega M, Burgess SC, Byers JE, Pringle JM, Wares JP, Marshall DJ. 2020. Global biogeography of marine dispersal potential. Nat. Ecol. Evol. 4, 1196–1203. ( 10.1038/s41559-020-1238-y) [DOI] [PubMed] [Google Scholar]
- 81.Menegotto A, Rangel TF. 2018. Mapping knowledge gaps in marine diversity reveals a latitudinal gradient of missing species richness. Nat. Commun. 9, 4713 ( 10.1038/s41467-018-07217-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this article are publicly available via WoRMS. The processed summary data we use for our analysis is openly available under a Creative Commons Attribution 4.0 International License as Webb, T.J.; Vanhoorne, B. (2020): WoRMS Marine Animals with links to external databases. Marine Data Archive. https://doi.org/10.14284/417. R code to replicate our analyses and figures is available via https://github.com/tomjwebb/linking_marine_diversity_data and is archived on Figshare via the University of Sheffield's Online Research Data repository here: https://doi.org/10.15131/shef.data.12833891.