Abstract
In patients with gliomas, isocitrate dehydrogenase 1 (IDH1) mutation status has been studied as a prognostic indicator. Recent advances in machine learning (ML) have demonstrated promise in utilizing radiomic features to study disease processes in the brain. We investigate whether ML analysis of multiparametric radiomic features from preoperative Magnetic Resonance Imaging (MRI) can predict IDH1 mutation status in patients with glioma. This retrospective study included patients with glioma with known IDH1 status and preoperative MRI. Radiomic features were extracted from Fluid-Attenuated Inversion Recovery (FLAIR) and Diffusion-Weighted-Imaging (DWI). The dataset was split into training, validation, and testing sets by stratified sampling. Synthetic Minority Oversampling Technique (SMOTE) was applied to the training sets. eXtreme Gradient Boosting (XGBoost) classifiers were trained, and the hyperparameters were tuned. Receiver operating characteristic curve (ROC), accuracy, and f1-scores were collected. A total of 100 patients (age: 55 ± 15, M/F 60/40); with IDH1 mutant (n = 22) and IDH1 wildtype (n = 78) were included. The best performance was seen with a DWI-trained XGBoost model, which achieved ROC with Area Under the Curve (AUC) of 0.97, accuracy of 0.90, and f1-score of 0.75 on the test set. The FLAIR-trained XGBoost model achieved ROC with AUC of 0.95, accuracy of 0.90, f1-score of 0.75 on the test set. A model that was trained on combined FLAIR-DWI radiomic features did not provide incremental accuracy. The results show that a XGBoost classifier using multiparametric radiomic features derived from preoperative MRI can predict IDH1 mutation status with > 90% accuracy.
Keywords: glioma, radiomics, machine learning, IDH1, DWI
1. Introduction
Gliomas are primary brain tumors that account for nearly 30% of all primary brain tumors and 80% of all malignant brain tumors, and they are accountable for majority of deaths from primary brain tumors, despite advancements in treatment [1]. In patients with gliomas, those with IDH1 mutations, specifically IDH1 R132H, are associated with better prognosis when compared to those with IDH1 wildtype [2,3,4,5]. Currently, IDH1 mutation status is identified by DNA sequencing or immunohistochemistry techniques. When considering how the World Health Organization (WHO) Classification of 2016 encourages routine testing for IDH1 mutational status [6], noninvasive methods of glioma assessment would be highly desirable for patients.
Radiomics may have the potential of providing noninvasive assessment of IDH1 mutational status. The study of radiomics involves the computation of an extensive number of quantitative features, referred to as “radiomic features”, which describe the imaging characteristics, such as intensity and geometry attributed to radiological images. Previous studies have utilized radiomic features to predict diagnosis, prognosis and treatment responses for patients with gliomas [7,8]. The association between radiomic features and IDH1 genotype has also been explored [9,10].
In recent years, utilizing machine learning (ML) methods for characterizing gliomas from medical imaging have attracted attention [11]. With regards to predicting glioma characteristics from MRI radiomic features, studies have primarily explored support vector machines (SVM) and random forest (RF) classifiers [11,12]. Recently, a new open source highly scalable gradient tree boosting model named eXtreme Gradient Boosting (XGBoost) has been introduced with some promising results [13]. Whereas, RF relies on simple averaging to achieve the final ensemble, gradient boosting (GB) involves a more constructive strategy, sequentially adding models to the ensemble [14]. XGBoost is an optimized form of GB. To the best of our knowledge, no study has investigated the utility of XGBoost in identification of IDH1 mutations in grade II, III, and IV gliomas using FLAIR and DWI radiomic features.
We hypothesized that a supervised ML approach using a XGBoost classifier would be able to predict IDH1 mutation status purely from MRI radiomic features. Therefore, the purpose of this study was to train and optimize a XGBoost classifier with preoperative Fluid-Attenuated Inversion Recovery (FLAIR) and Diffusion-Weighted-Imaging (DWI) radiomic features and predict IDH1 mutation status.
2. Results
2.1. Patient Characteristics
A total of 100 patients met the criteria, 60 males and 40 females. The mean ± standard deviation of age (years) was 55 ± 15. There were 19 patients with lower-grade glioma, including grade II glioma (n = 11) and grade III glioma (n = 8) and 81 patients with grade IV glioma. There were 78 IDH1 wildtype and 22 IDH1 mutants. Table 1 summarizes patient characteristics. Table 2 summarizes IDH1 status and characteristics of patients within each of the training, validation, and test sets.
Table 1.
Patient Characteristics.
| Patient Characteristics | n (%) |
|---|---|
| Total Patients | 100 |
| Female | 40 (40) |
| Male | 60 (60) |
| Age (in years) | |
| Mean | 55 ± 15 |
| Median | 57 |
| Range | 28–88 |
| Presence of enhancement on MRI | 82 (82) |
| IDH1 status by immunohistochemistry | |
| Wildtype | 78 (78) |
| Mutant | 22 (22) |
| WHO Grade | |
| Grade II | 11 (11) |
| Grade III | 8 (8) |
| Grade IV | 81 (81) |
Table 2.
IDH1 Status and Characteristics of Patients in Training/Validation/Test Sets.
| Subset | IDH1 Status | Total | Male | Female | Grade II | Grade III | Grade IV |
|---|---|---|---|---|---|---|---|
| Training (n = 60) |
Wildtype | 46 | 25 | 21 | 1 | 0 | 45 |
| Mutant | 14 | 10 | 4 | 5 | 5 | 4 | |
| Validation (n = 20) |
Wildtype | 16 | 10 | 6 | 0 | 0 | 16 |
| Mutant | 4 | 3 | 1 | 2 | 1 | 1 | |
| Test (n = 20) |
Wildtype | 16 | 10 | 6 | 1 | 0 | 15 |
| Mutant | 4 | 2 | 2 | 2 | 2 | 0 |
2.2. Prediction of IDH1 Mutation Status Using XGBoost Models
Our FLAR-trained XGBoost model utilized 33 final radiomic features and achieved a Receiver operating characteristic Area Under the Curve (ROC AUC) of 95%, Accuracy of 90%, Precision/Recall/f1-score of 94%/94%/94% for IDH1 wildtype, and 75%/75%/75% for IDH1 mutants. Of the 20 cases in the test set, it correctly classified 15 wildtype cases, incorrectly classified one wildtype as mutant, correctly classified three mutants, and incorrectly classified one mutant as wildtype.
Our DWI-trained XGBoost model utilized 71 final radiomic features and achieved ROC AUC of 97%, Accuracy of 90%, Precision/Recall/f1-score of 94%/94%/94% for IDH1 wildtype, and 75%/75%/75% for IDH1 mutants. Of the 20 cases in the test set, it correctly classified 15 wildtype cases, incorrectly classified one wildtype as mutant, correctly classified three mutants, and incorrectly classified 1 mutant as wildtype.
Our DWI-FLAIR trained XGBoost model utilized 88 final radiomic features and achieved ROC AUC of 91%, Accuracy of 90%, Precision/Recall/f1-score of 94%/94%/94% for IDH1 wildtype, and 75%/75%/75% for IDH1 mutants. Of the 20 cases in the test set, it correctly classified 15 wildtype cases, incorrectly classified one wildtype as mutant, correctly classified three mutants, and incorrectly classified one mutant as wildtype.
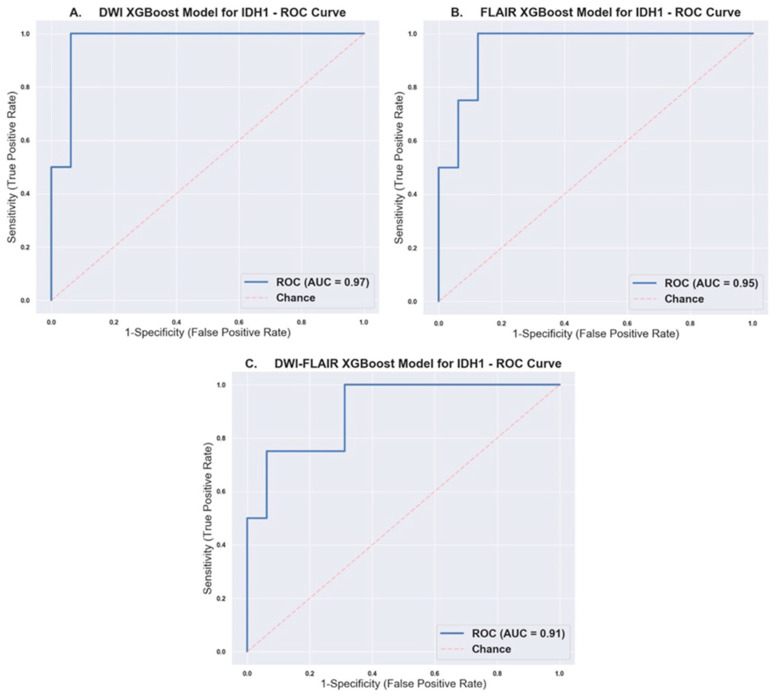
The AUC with 95% confidence interval, Accuracy, Precision, Recall, and f1-score for each model is aggregated in Table 3. Figure 1 shows the ROC curve with the AUC score for each model.
Table 3.
Receiver operating characteristic Area under the Curve (ROC AUC), Accuracy, Precision, Recall, and F1-Score of Trained XGBoost Models.
| Model | ROC AUC [95% CI] | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| DWI | 0.97 [0.898, 1.000] |
0.90 | Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
| FLAIR | 0.95 [0.864, 1.000] |
0.90 | Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
| DWI-FLAIR | [0.741, 1.000] | 0.90 | Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
Wildtype: 0.94 Mutant: 0.75 |
Figure 1.
Receiver Operating Characteristic Curves with calculated AUC for (A) Diffusion-Weighted-Imaging (DWI), (B) Fluid-Attenuated Inversion Recovery (FLAIR), and (C) DWI-FLAIR XGBoost models.
The 10 most important radiomic features ordered by gain for each XGBoost model is aggregated in Table 4. Out of 184 total radiomic features, 153 were non-normally distributed (p < 0.05 on Shapiro–Wilks test). Total of 46 DWI and FLAIR radiomic features with significant difference between IDH1 Wildtype and Mutant are listed in Appendix A Table A1. A total of 98 DWI and FLAIR radiomic features with significant difference between glioblastomas and non-glioblastomas are listed in Appendix A Table A2. The list of final features used ranked by gain are shown in Appendix A Table A3, Table A4 and Table A5 for the DWI-FLAIR, DWI, and FLAIR XGBoost model, respectively. The list of all radiomic features by feature class are shown in Appendix A Table A6. Spearman correlation matrix of DWI and FLAIR radiomic features are shown in Appendix A Figure A1 and Figure A2.
Table 4.
Top 10 Most Important Radiomic Features Ranked By Gain for Each Model.
| DWI-FLAIR | FLAIR | DWI |
|---|---|---|
| DWI_Original First Order Total Energy | Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | Original Gray Level Co-occurrence Matrix Autocorrelation |
| DWI_Original First Order Mean Absolute Deviation | Original First Order Mean Absolute Deviation | Original Gray Level Run Length Matrix Run Entropy |
| FLAIR_Original First Order 90th Percentile | Original Gray Level Co-occurrence Matrix Correlation | Original Gray Level Dependence Matrix Dependence Non Uniformity Normalized |
| FLAIR_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | Original Gray Level Size Zone Matrix Gray Level Variance | Original Gray Level Dependence Matrix Gray Level Variance |
| FLAIR_Original Gray Level Run Length Matrix High Gray Level Run Emphasis | Original Gray Level Size Zone Matrix Low Gray Level Zone Emphasis | Original Gray Level Co-occurrence Matrix Maximum Probability |
| FLAIR_Original Gray Level Size Zone Matrix Gray Level Non Uniformity | Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 | Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis |
| DWI_Original First Order Maximum | Original Gray Level Co-occurrence Matrix Cluster Prominence | Original Gray Level Run Length Matrix Gray Level Non Uniformity |
| DWI_Original Gray Level Run Length Matrix Run Entropy | Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis |
| DWI_Original First Order Skewness | Original Neighboring Gray Tone Difference Matrix Coarseness | Original First Order Total Energy |
| DWI_Original First Order 10th Percentile | Original First Order Range | Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 |
3. Discussion
In patients with glioma, IDH1 mutation has been shown to be an independent positive prognostic biomarker with improved progression-free survival and treatment outcome in comparison to the IDH1 wildtype [15]. Although the genetic biomarkers are determined by histopathological testing, the ability to predict biomarker status noninvasively is of clinical interest, as large tissue specimens are often needed for accurate histopathological diagnosis and potential inaccuracies that are related to tumoral tissue heterogeneity. Furthermore, pre-surgical identification of these biomarkers can help in surgical planning and the determination of the extent of surgical resection.
Qualitative MRI features have been shown to correlate with IDH1 genotypes in high grade gliomas [16,17,18]. More recently, the T2-FLAIR mismatch sign, which is defined as the presence of hyperintense signal on a T2-weighted image and a relatively hypointense signal on FLAIR (except for a hyperintense peripheral rim), has been described as a helpful imaging marker of IDH-mutant gliomas [19,20].
Recent improvements in ML algorithms and computational power provide an attractive venue for exploring MR radiomic features, an excellent fit for ML-approach analysis that considers the large data size and multimodal nature. Therefore, ML methods have been recently explored to predict glioma genetic biomarkers from MRI radiomic features [10,11,12,21,22,23,24,25,26,27,28,29,30,31,32,33].
Recent investigations on the use of ML and MRI-radiomics to predict IDH1 genotype have primarily explored the SVM [21,25,30] and RF [10,22,24,27,28,29] models. Some studies only used conventional MRI sequences and achieved AUC ranging 0.84–0.96 [10,22,23,24,25,30]. Others explored the added value of advanced MRI imaging, such as MR diffusion or perfusion, with mixed results [21,26,27,28,29,31]. The highest performance of predicting IDH1 genotype with AUC of 0.96 was observed with a RF model that was trained with conventional MRI, but this study focused only on patients with glioblastomas [10].
Expanding on above studies, we selected XGBoost as our classifier and trained models using FLAIR and DWI radiomic features, in a diverse cohort of patients, including grade II, III, and IV gliomas. Our DWI model achieved AUC of 0.97 (I: 0.898, 1.000) and 90% accuracy (Figure 1A), and our FLAIR model achieved an AUC of 0.95 (CI: 0.864, 1.000) and 90% accuracy (Figure 1B). XGBoost is a non-linear gradient boosted tree model with superior performance in comparison to conventional ML models [13]. RF and GB are both sets of decision trees. Whereas, RF builds each tree independently and combines the results at the end, GB builds each tree sequentially, and works to correct the error of the previous tree [14]. In GB, there are more hyperparameters than RF to optimize. Therefore, it may be more difficult to optimize a GB algorithm, but a better tuned GB algorithm may outperform a RF. In the training process, XGBoost calculates the importance score of each feature in each iteration, which provides a basis for establishing a new tree with gradient direction in the next iteration [13,34]. When two features are correlated, then, when deciding a split, the tree will only choose the one feature with greater importance, and this process is repeated. This automated feature selection structure is of particular use with high-dimensional data with potential multicollinearity, such as in radiomics. Another advantage of XGBoost is that it provides both L1 and L2 regularization, thus handling sparsity and reduces overfitting [13]. In this study, 92 radiomic features that were calculated by our postprocessing program were used as input to train our DWI and FLAIR classifiers, and a total of 184 radiomic features as input to train our DWI-FLAIR classifier. The DWI-FLAIR model utilized 88 out of the initial 184 features (Appendix A Table A3). The DWI model utilized 71 out of the 92 features (Appendix A Table A4). The FLAIR model utilized 33 out of the initial 92 features (Appendix A Table A5). A comparison of features that were selected by XGBoost and analysis of features with statistically significant differences between IDH1 wildtype and mutants (Appendix A Table A1) and between glioblastomas and non-glioblastomas (Appendix A Table A2) shows the effectiveness of the automated feature selection. In review, all of the features with statistical significance between IDH1 wildtype and mutant were included in at least one of the XGBoost models, except for two features, DWI_Original Gray Level Dependence Matrix High Gray Level Emphasis and FLAIR_Original First Order Maximum. In comparison, there were seven features that had statistical significance between glioblastomas and non-glioblastomas, but they were not included in any of the XGBoost models. This is consistent with the fact that we trained the models based on IDH1 status as opposed to glioma grade. In review of features within the top 10 that were included in our XGBoost model by automatic feature selection, but did not have statistical significant difference between IDH1 wildtype and mutant, the mean p-values were 0.07 in the DWI model, 0.08 in the FLAIR model, and 0.23 in the DWI-FLAIR model. This may partially explain why we did not observe an incremental value in combining DWI and FLAIR in our XGBoost models (Figure 1C).
Our results compared favorably with the results of a recent study conducted by Shboul et al., where XGBoost models were used to predict several glioma biomarkers, including IDH mutation [23]. In this study, a XGBoost model was trained while using conventional MRI sequences in patients with grade II and III gliomas and reported an AUC of 0.83 in predicting IDH status.
In our experience, the diagnostic performance of the combined trained model using combination of DWI and FLAIR (AUC: 91%, accuracy: 90%) was comparable to the isolated model (DWI only or FLAIR only) and with no significant incremental diagnostic value (Table 3). This is likely attributed to the fact that, in the combined model, the number of features was doubled without increasing the number of observations, which can result in overfitting during the training process.
Interestingly, we observed from feature importance assessment that six out of the top 10 important radiomic features for the combined model (DWI-FLAIR) were from the DWI dataset (Table 4). DWI has shown to corelate with physiological characterization of tumors, such as cellularity and proliferation index, as a function of water diffusivity [35]. Prior histopathological studies showed that IDH mutations can decrease glioma proliferation through the upregulation of miR-128a [36]. It is plausible that DWI may hold invaluable information regarding the IDH status, as shown in our results.
Our study has several limitations. Our XGBoost classifiers were trained with single-center data and, thus, the generalizability of our results may be impacted by differences in imaging acquisition protocols and the image postprocessing programs that were used for radiomic feature extraction.
The small total sample size of in combination with the skewed distribution of IDH1 wildtype and mutant is a notable limitation. We utilized stratified sampling when splitting our dataset into train, validation, and test sets in order to account for the small proportion of IDH1 mutants and the sampling error that could be introduced during randomization. Subsequently, as suggested by prior reports [10,32], we applied SMOTE on our train set in order to prevent biased training that favors the majority class. During our hyperparameter tuning step, the parameters were optimized for the highest ROC AUC score, as ROC curves are mathematically insensitive to class distribution unlike accuracy. Nonetheless, the effect of the skewed distribution is observed in our study (Table 2). The test set had 16 wildtype and four mutants. Our XGBoost models correctly classified 15/16 as wildtype, and correctly classified 3/4 as mutants. As the denominator is four for the mutants, one error results in a numerically steep decrease (Table 3). The skewed distribution of IDH1 genotypes have been acknowledged in literature; studies with low grade gliomas involved predominantly IDH1 mutants [33,37] and studies with glioblastomas involved predominantly IDH1 wildtypes [10,28]. This is reflected in our dataset, as 17 out of 19 (89%) low grade gliomas were IDH1 mutants and 76 out of 81 (94%) grade IV gliomas were wildtype (Table 2).
Another limitation is that, within our IDH1 wildtype population, there were two cases with minor IDH2 (p.R172M, p.R172K) mutations. IDH2 mutations are much less common than IDH1 and they are mutually exclusive with IDH1 mutations [3]. To our knowledge, there is no study that has specifically compared the radiomic features between IDH1 and IDH2 mutants in gliomas.
Another limitation is that tumor segmentation to generate VOI was performed by one observer and under supervision of another board-certified neuroradiologist, who made necessary adjustments before the extraction of radiomic features. Therefore, inter-observer variability assessment was not performed.
Another limitation of our ML-approach is that it does not fully explain the physiological significance of radiomic features. The prolonged survival of patients with IDH1 mutations has previously been proposed to be associated with less aggressive biological behavior from the perspective of MRI tumor heterogeneity [16]. In this study, no additional categorical variables, such as list of comorbidities, were included or analyzed in the machine learning process. Because our models were trained purely with radiomic features, future work may focus on studying the relationship between these quantitative radiomic features and tumor heterogeneity across IDH1 genotypes.
In conclusion, training a XGBoost classifier while using multiparametric radiomic features derived from DWI and FLAIR images discriminated IDH1 mutation status with accuracy > 90% and AUC > 0.95, which may provide an approach for noninvasive assessment of IDH1 status in patients with gliomas. Further studies with larger and more diverse MRI datasets are required to validate and improve upon our findings.
4. Materials and Methods
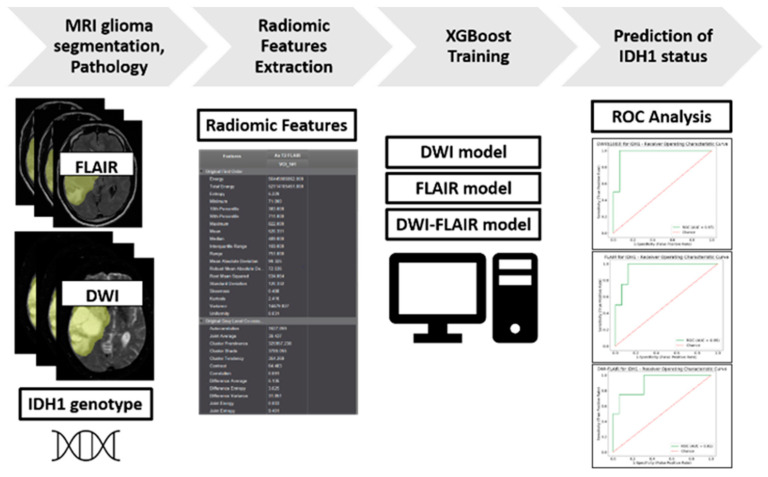
The overall study design is summarized as a diagram in Figure 2.
Figure 2.
Study Design Diagram.
4.1. Patient Population
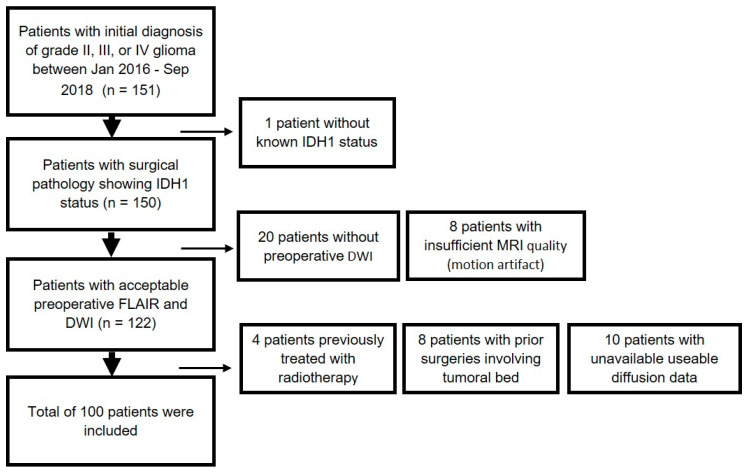
An institutional review board approved this retrospective study and informed consent was waived. Patients with initial diagnosis of grade II, III, or IV glioma between January 2016 to September 2018 were reviewed (n = 151). Patients were included if they 1) had grade II, III, or IV glioma with known IDH1 status from surgical pathology and 2) had preoperative MRI, including FLAIR, T1c+, and diffusion within 30 days of biopsy or surgical resection. One patient was removed due to lack of IDH1 status. 20 patients were excluded due to lack of preoperative DWI. The patients were excluded if they had insufficient MR image quality (motion artifact, n = 8). Patients were excluded if they had prior surgeries involving the tumoral bed (n = 8). Patients were excluded if they had prior radiotherapy treatment (n = 4). In addition, 10 patients were excluded due to unavailable useable diffusion data. This yielded, in total, a cohort of 100 patients (Figure 3). Assuming incidence of >15,000 new cases of malignant gliomas per year in the United States [38], 5% margin of error, 90% confidence interval, and estimated IDH mutation rate of 12% in literature [39], the recommended sample size is 114 [40]. With our sample size of 100, the margin of error is 5.33%.
Figure 3.
Flowchart of Patient Inclusion and Exclusion.
4.2. Histopathological Data
Tissue samples were obtained from patients undergoing MRI-guided tissue biopsy or tumor resection, as part of routine clinical care and diagnostic neuropathology and molecular evaluation. Hematoxylin and eosin (H&E) sections and immunohistochemistry (IHC) slides were re-reviewed by pathologists (N.T) and (F.K). IHC was performed on 5 micron thick sections from paraffin embedded tumor sections of all the evaluated patients. Cut sections were backed in 60 °C for one hour to deparaffinize and enhance tissue adhesion; following deparaffinization, the sections were stained with pre-diluted monoclonal anti-mIDH1 antibody, purified from culture supernatant in PBS (2% BSA, 0.05% NaN3, pH 7.4, DIA-H09L; Dianova, Hamburg, Germany), which has specificity for human IDH1 R132H point mutation. The high frequency and distribution of the IDH1 R132H mutation allow the highly sensitive and specific discrimination of higher-grade gliomas by immunohistochemistry. The staining was performed using Ventana Benchmark XT stainer, (pretreated with CC1 mild, detection DAB ultraview kit; Ventana Medical Systems, Tucson, Arizona). The majority of IDH1 mutations in diffuse gliomas occur at a specific sites and they are characterized by a base exchange of guanine to adenine within codon 132, resulting in an amino acid change from arginine to histidine (R132H). Therefore, a monoclonal antibody has been developed in order to detect the consistent mutant iteration site of IDH mutant protein, allowing for its use in paraffin-embedded specimens (mIDH1R132H). The ability of the antibody to detect a small number of cells as mutant makes IHC more sensitive than sequencing for identifying R132H mutant gliomas. However, mutations in IDH2 and other mutations in IDH1 will not be detected using IHC. Next generation sequencing (NGS) was performed to confirm an IDH1 R132H negative IHC results, and/or if the patient is less than 55years old, as IDH mutations in general are extremely rare in patients over 55 years, as per College of American Pathologists (CAP) recommendations.
4.3. NGS Analysis
Clinical samples after the histological diagnoses of primary CNS glioma were tested for prognostic molecular biomarkers, as outlined in the NCCN guidelines, depending on the clinical and pathological context, NGS analysis was performed according to a licensed protocol in the molecular pathology lab where histological evidence of tumor cellularity of >20% was considered to be acceptable. Extracted DNA was submitted to amplicon-based library preparation and sequencing according to manufacturer’s procedures for the 50-gene Hotspot panel on a (Ion Personal Genome Machine™ (PGM™) System, Thermo Fisher Scientific Inc. Waltham, MA). In each run, a low level VAF (variant allelic frequency) control was included for both SNVs and indels. For each specimen, an average minimum coverage of 500× was considered to be acceptable to proceed. Based on the variants identified by the Torrent Variant Caller, a pipeline was applied to permit annotation, filtering of variants with a VAF of >5% (unless clinically relevant and with a VAF of >10%), and the exclusion of those in Genome Aggregation Database (gnomAD https://gnomad.broadinstitute.org/) with a population frequency >0.05%. Read depths of IDH1, and IDH2 targets were confirmed in all cases to be adequate to rule out false negatives. Molecular pathology workup of Gliomas also included NGS based testing (IHC) of Isocitrate Dehydrogenase. If, through immunohistochemistry, the IDH result was negative then, for those patients with >55 years, NGS was not performed, unless clinically indicated. For patients <55 years, NGS was performed in order to identify other IDH1 or IDH2 mutations.
4.4. Image Acquisition
MR imaging was obtained using seven MRI scanners (2 Skyra 3T and 2 Aera 1.5T from Siemens Healthineers, Erlangen Germany; 2 Signa 1.5T and one Discovery 3T from GE Healthcare, Waukesha, WI) within our Radiology Department. Image acquisition was performed using a standardized preoperative brain tumor MRI protocol within our radiology department, including: FLAIR (TR/TE/TI, 8000–12,000/98–130/2400–2700 ms, voxel size: 0.5 × 0.5 × 1 mm3), DWI (TR/TE: 4025-4600/65-82 ms, with b values of 0 and 1000 s/mm2, voxel size: 0.9 × 0.9 × 5.0 mm3), and post-contrast T1W imaging (TR/TE, 600–1800/9–19 ms, voxel size: 0.5 × 0.5 × 1 mm3). A total volume of 0.1 mmol/kg of gadobenate dimeglumine was intravenously injected for post-contrast T1W imaging.
4.5. Volume Acquisition and Texture Analysis
The tumors were manually segmented with volume-of-interest (VOI) analysis on commercially available FDA-approved software (Olea Sphere software, Olea Medical SAS, La Ciotat, France). T1c+, FLAIR and diffusion images (ADC/b1000) were coregistered on each examination using a 6-df transformation and a mutual information cost function. Subsequently, a VOI was generated while using a voxel-based signal intensity threshold method subsuming the entire region of FLAIR hyperintensity. This VOI was then overlaid onto coregistered T1c+ and diffusion datasets. The segmentation was conducted by 1 radiology resident (S.K) under supervision of an experienced board certified neuroradiologist (K.N.), who made necessary adjustments before radiomic features were extracted. One VOI was segmented from each patient.
Radiomic features were calculated from the VOIs using Olea Sphere software. A total of 92 texture features were collected: 19 first-order metrics, including the mean, standard deviation, skewness, and kurtosis, and 73 second-order metrics consisting of 23 gray level run length matrix [41], 16 gray level run length matrix [42], 15 gray level size zone matrix [43], five neighboring gray tone difference matrix [44], and 14 gray level dependence matrix [45]. Definitions and calculations of these features are explained elsewhere [46].
4.6. Statistical Analysis and ML
All of the statistical analysis was performed using Python. (Python 3.7; Packages: scipy v. 1.3.0; numpy 1.16.4; matplotlib 3.1.1; pandas 0.24.2; sklearn 0.21.2; imblearn 0.5.0; xgboost 0.90.)
4.6.1. Radiomic Features Analysis
MRI radiomic features were tested for normality by the Shapiro–Wilks test. Wilcoxon rank sum test analysis was conducted in order to assess radiomic features with statistically significant (p < 0.05) difference between IDH1 wildtype and mutants. Similarly, Wilcoxon rank sum test analysis was conducted to assess radiomic features with a statistically significant (p < 0.05) difference between glioblastomas and non-glioblastomas.
4.6.2. ML Classifier Procedure
Input: three datasets were created as inputs for our ML methods. (1) A table consisting of FLAIR radiomic features and IDH1 genotype for each patient. (2) A table consisting of DWI radiomic features and IDH1 genotype for each patient. Finally, (3) a third dataset combining the DWI and FLAIR radiomic features was created. We refer to these as the (1) FLAIR dataset, (2) DWI dataset, and (3) DWI-FLAIR dataset.
Sampling: the patients were divided into train, validation, and test sets with a 60:20:20 ratio, thus resulting in 60 cases for the train set, 20 cases for the validation set, and 20 cases for the test set. Stratified random sampling was employed in this process, thus approximately maintaining the ratio of IDH1 wildtype to mutant across the subsets to be equal to the ratio in the original dataset.
Oversampling: the Synthetic Minority Oversampling Technique (SMOTE) was applied to the train sets [47].
Training: three separate XGBoost classifiers [13] were trained: DWI, FLAIR, and DWI-FLAIR.
Hyperparameter tuning: XGBoost hyperparameters were tuned on the validation set, using exhaustive grid search (scikit-learn GridSearchCV) with five-fold cross validation. The hyperparameters were optimized for the highest receiver operating characteristic curve’s area under the curve (ROC AUC) score. The following hyperparameters and ranges for exhaustive grid search were studied: eta, 0–100 with interval of 1; max_depth, 0–100 with interval of 1; min_child_weight, 0–1 with interval of 1/500; gamma 0–1 with interval of 1/500, subsample 0–1 with interval of 1/500; colsample_by_tree, 0–1 with interval of 1/500; colsample_bylevel, 0–1 with interval of 1/500; scale_pos_weight, 0–1 with interval of 1/500; leaning_rate, 0–1 with interval of 1/500; n_estimators, 0–500 with interval of 1; and, reg_alpha, 0–100 with interval of 1. The details of these parameters are accessible elsewhere [48]. Three final models were collected: DWI, FLAIR, and DWI-FLAIR.
Feature importance by XGBoost: the importance of each radiomic feature was assessed and collected, ordered by the average Gain across all splits, the feature was used in. The Gain was calculated by taking each feature’s contribution for each tree in the XGBoost models and, thus, represents the relative contribution of the feature to the model.
Testing: each of the final models were tested using the respective test sets: DWI test set, FLAIR test set, and the DWI-FLAIR test set. For each model, a classification report containing the Accuracy, Precision, Recall, and f1-score was collected. For each model, the confusion matrix depicting the number of true positives, false positives, true negatives, and false negatives was collected. For each model, a ROC curve was drawn and the AUC was calculated. For each AUC, the 95% confidence interval was also calculated.
Abbreviations
| AUC | Area Under the Curve |
| ADC | Apparent Diffusion Coefficient |
| DWI | Diffusion-Weighted-Imaging |
| FLAIR | Fluid-Attenuated Inversion Recovery |
| GB | Gradient Boosting |
| H&E | Hematoxylin and eosin |
| IDH1 | Isocitrate Dehydrogenase 1 |
| IHC | Immunohistochemistry |
| MRI | Magnetic Resonance Imaging |
| NGS | Next Generation Sequencing |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic curve |
| SMOTE | Synthetic Minority Oversampling Technique |
| SVM | Support Vector Machine |
| T1c+ | Post-contrast T1 |
| XGBoost | eXtreme Gradient Boosting |
Appendix A
Table A1.
DWI and FLAIR radiomic features with significant difference by Wilcoxon rank sum test between IDH1 Wildtype and Mutant.
| Radiomic Feature | p-Value | IDH1 Wildtype Mean ± SD | IDH1 Mutant Mean ± SD |
|---|---|---|---|
| FLAIR_Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | <0.001 | 1093.41 ± 372.49 | 1456.95 ± 376.78 |
| FLAIR_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis | <0.001 | 635.48 ± 187.18 | 824.98 ± 1 91.8 |
| FLAIR_Original Gray Level Run Length Matrix High Gray Level Run Emphasis | <0.001 | 1201.22 ± 405.95 | 1602.17 ± 426.86 |
| FLAIR_Original Gray Level Dependence Matrix High Gray Level Emphasis | <0.001 | 1209.73 ± 412.96 | 1613.11 ± 433.94 |
| FLAIR_Original Gray Level Co-occurrence Matrix Cluster Shade | 0.001 | −167.45 ± 1997.12 | −2819.29 ± 5053.17 |
| FLAIR_Original Gray Level Co-occurrence Matrix Autocorrelation | 0.001 | 1228.14 ± 426.63 | 1619.82 ± 455.56 |
| FLAIR_Original Gray Level Co-occurrence Matrix Joint Average | 0.002 | 33.81 ± 6.42 | 39.06 ± 5.76 |
| FLAIR_Original Gray Level Co-occurrence Matrix Sum Average | 0.002 | 67.62 ± 12.85 | 78.12 ± 11.52 |
| FLAIR_Original First Order Skewness | 0.003 | −0.02 ± 0.57 | −56 ± 0.67 |
| DWI_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis | 0.005 | 546.7 ± 250.71 | 743.57 ± 287.76 |
| FLAIR_Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis | 0.006 | 1997.25 ± 716.52 | 2621.33 ± 982.83 |
| DWI_Original Gray Level Run Length Matrix High Gray Level Run Emphasis | 0.007 | 831.51 ± 457.59 | 1205.94 ± 547.74 |
| DWI_Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | 0.007 | 769.52 ± 434.1 | 1119.01 ± 509.77 |
| DWI_Original Gray Level Dependence Matrix High Gray Level Emphasis | 0.007 | 825.3 ± 462.6 | 1201.58 ± 553.71 |
| FLAIR_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | 0.007 | 241.62 ± 118.86 | 316.36 ± 117.92 |
| DWI_Original First Order Skewness | 0.008 | 0.46 ± 1.06 | −0.32 ± 0.9 |
| FLAIR_Original First Order Range | 0.010 | 1214.54 ± 671.87 | 989.59 ± 882 |
| DWI_Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis | 0.010 | 1211.61 ± 574.88 | 1710.77 ± 768.55 |
| DWI_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | 0.012 | 213.92 ± 161.61 | 312.42 ± 172.48 |
| DWI_Original Gray Level Co-occurrence Matrix Autocorrelation | 0.012 | 821.63 ± 471.38 | 1184.7 ± 570.82 |
| DWI_Original Neighboring Gray Tone Difference Matrix Busyness | 0.013 | 1.23 ± 1.02 | 0.68 ± 0.53 |
| DWI_Original First Order Maximum | 0.014 | 1174.11 ± 596.34 | 835.35 ± 453.04 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Busyness | 0.014 | 4.04 ± 3.16 | 2.17 ± 1.59 |
| DWI_Original Gray Level Co-occurrence Matrix Joint Average | 0.015 | 26.59 ± 8.64 | 32.68 ± 8.43 |
| DWI_Original Gray Level Co-occurrence Matrix Sum Average | 0.015 | 53.18 ± 17.29 | 65.35 ± 16.86 |
| DWI_Original First Order Uniformity | 0.022 | 0.05 ± 0.02 | 0.04 ± 0.01 |
| DWI_Original First Order Range | 0.023 | 1101.2 ± 596.03 | 775.94 ± 454.76 |
| DWI_Original Gray Level Dependence Matrix Gray Level Variance | 0.024 | 62.83 ± 36.88 | 78.73 ± 30.73 |
| DWI_Original Gray Level Run Length Matrix Gray Level Non Uniformity Normalized | 0.024 | 0.05 ± 0.02 | 0.04 ± 0.01 |
| DWI_Original Gray Level Run Length Matrix Gray Level Variance | 0.024 | 64.79 ± 36.43 | 80.37 ± 30.18 |
| DWI_Original Gray Level Run Length Matrix Gray Level Non Uniformity | 0.027 | 1106.8 ± 928.02 | 619.32 ± 481.82 |
| DWI_Original Gray Level Dependence Matrix Gray Level Non Uniformity | 0.031 | 1403.34 ± 1300.29 | 728.69 ± 584.29 |
| DWI_Original Neighboring Gray Tone Difference Matrix Contrast | 0.032 | 0.1 ± 0.1 | 0.14 ± 0.11 |
| DWI_Original Neighboring Gray Tone Difference Matrix Coarseness | 0.034 | 0.00 ± 0.0015 | 0.0013 ± 0.0016 |
| DWI_Original Gray Level Dependence Matrix Large Dependence High Gray Level Emphasis | 0.034 | 15,902.19 ± 7204.39 | 21,277.99 ± 11,907.81 |
| FLAIR_Original First Order Maximum | 0.036 | 1461.66 ± 783.13 | 1223.18 ± 953.74 |
| DWI_Original Gray Level Size Zone Matrix Small Area Emphasis | 0.037 | 0.59 ± 0.05 | 0.62 ± 0.05 |
| FLAIR_Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 | 0.037 | 0.77 ± 0.09 | 0.82 ± 0.09 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Strength | 0.038 | 0.53 ± 1.08 | 0.51 ± 0.42 |
| DWI_Original Gray Level Size Zone Matrix Size Zone Non Uniformity Normalized | 0.039 | 0.33 ± 0.05 | 0.36 ± 0.05 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Coarseness | 0.039 | 0.0003 ± 0.0005 | 0.0003 ± 0.0002 |
| FLAIR_Original First Order Standard Deviation | 0.045 | 151.45 ± 77.39 | 132.06 ± 109.62 |
| FLAIR_Original First Order Variance | 0.045 | 28,850.28 ± 32,282.11 | 28,910.57 ± 43,752.27 |
| FLAIR_Original First Order Total Energy | 0.046 | 73,306,599,504.28 ± 90,074,867,196.59 | 78,905,231,446.5 ± 128,228,405,390.59 |
| DWI_Original First Order Entropy | 0.047 | 4.7 ± 0.49 | 4.95 ± 0.29 |
| DWI_Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized | 0.047 | 0.03 ± 0.01 | 0.03 ± 0.005 |
Table A2.
DWI and FLAIR Radiomic Features with Significant Difference by Wilcoxon Rank Sum Test between Glioblastoma and Non-glioblastoma.
| Radiomic Feature | p-Value | Glioblastoma Mean ± SD | Non-glioblastoma Mean ± SD |
|---|---|---|---|
| FLAIR_Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | <0.001 | 1079.74 ± 349.33 | 1572.62 ± 367.53 |
| FLAIR_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis | <0.001 | 628.91 ± 172.61 | 882.89 ± 198.65 |
| FLAIR_Original Gray Level Run Length Matrix High Gray Level Run Emphasis | <0.001 | 1187.83 ± 380.14 | 1722.54 ± 429.06 |
| FLAIR_Original Gray Level Dependence Matrix High Gray Level Emphasis | <0.001 | 1195.65 ± 386.56 | 1736.83 ± 435.68 |
| FLAIR_Original Gray Level Co-occurrence Matrix Autocorrelation | <0.001 | 1212.08 ± 398.97 | 1750.11 ± 460.47 |
| FLAIR_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | <0.001 | 233.31 ± 109.06 | 363.58 ± 120.84 |
| FLAIR_Original Gray Level Co-occurrence Matrix Joint Average | <0.001 | 33.62 ± 6.08 | 40.72 ± 5.82 |
| FLAIR_Original Gray Level Co-occurrence Matrix Sum Average | <0.001 | 67.23 ± 12.17 | 81.43 ± 11.65 |
| FLAIR_Original First Order Skewness | <0.001 | −0.01 ± 0.57 | −0.67 ± 0.63 |
| FLAIR_Original First Order Range | <0.001 | 1265.91 ± 713.65 | 735.05 ± 618.31 |
| FLAIR_Original Gray Level Co-occurrence Matrix Cluster Shade | <0.001 | −219.28 ± 2123.23 | −3017.05 ± 5179.56 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Busyness | <0.001 | 4.1 ± 3.09 | 1.62 ± 1.13 |
| FLAIR_Original First Order Standard Deviation | 0.001 | 158.12 ± 82.19 | 100.57 ± 84.5 |
| FLAIR_Original First Order Variance | 0.001 | 31,674.63 ± 34,622.28 | 16,879.46 ± 34,301.7 |
| FLAIR_Original First Order Mean Absolute Deviation | 0.001 | 126.23 ± 67.02 | 78.74 ± 63.44 |
| DWI_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis | 0.001 | 544.01 ± 249.55 | 786.15 ± 274.83 |
| FLAIR_Original First Order Interquartile Range | 0.001 | 216.27 ± 124.42 | 128.84 ± 97.67 |
| FLAIR_Original First Order Robust Mean Absolute Deviation | 0.001 | 89.61 ± 50 | 54.64 ± 41.84 |
| FLAIR_Original Gray Level Run Length Matrix Long Run Low Gray Level Emphasis | 0.001 | 0.0031 ± 0.0049 | 0.0022 ± 0.0031 |
| DWI_Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | 0.001 | 764 ± 428.7 | 1197.72 ± 498.08 |
| DWI_Original Gray Level Dependence Matrix High Gray Level Emphasis | 0.001 | 819.66 ± 457.07 | 1285.02 ± 542.64 |
| DWI_Original Neighboring Gray Tone Difference Matrix Busyness | 0.001 | 1.24 ± 1.01 | 0.55 ± 0.42 |
| DWI_Original Gray Level Run Length Matrix High Gray Level Run Emphasis | 0.001 | 826.14 ± 452.35 | 1287.92 ± 536.33 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Strength | 0.001 | 0.5 ± 1.05 | 0.64 ± 0.48 |
| DWI_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | 0.001 | 210.01 ± 158.21 | 344.64 ± 170.11 |
| DWI_Original First Order Skewness | 0.002 | 0.46 ± 1.04 | −0.46 ± 0.88 |
| DWI_Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis | 0.002 | 1206.58 ± 570.58 | 1811.05 ± 762.28 |
| FLAIR_Original First Order Maximum | 0.002 | 1510.64 ± 821.7 | 976.74 ± 703.15 |
| DWI_Original Gray Level Co-occurrence Matrix Autocorrelation | 0.002 | 814.72 ± 465.16 | 1271.48 ± 562.39 |
| FLAIR_Original First Order Total Energy | 0.002 | 78,837,906,831.01 ± 92,307,518,260.86 | 56,208,389,465.53 ± 124,951,899,957.93 |
| FLAIR_Original First Order Energy | 0.002 | 121,143,113,125.22 ± 174,604,200,352.47 | 93,788,946,398.58 ± 241,864,772,843.25 |
| DWI_Original Gray Level Co-occurrence Matrix Joint Average | 0.002 | 26.49 ± 8.51 | 34.04 ± 8.2 |
| DWI_Original Gray Level Co-occurrence Matrix Sum Average | 0.002 | 52.99 ± 17.02 | 68.07 ± 16.4 |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Coarseness | 0.003 | 0.0002 ± 0.0004 | 0.0004 ± 0.0003 |
| DWI_Original Gray Level Run Length Matrix Gray Level Non Uniformity | 0.003 | 1119.88 ± 914.66 | 486.56 ± 346.9 |
| DWI_Original Neighboring Gray Tone Difference Matrix Coarseness | 0.003 | 0.001 ± 0.0015 | 0.0015 ± 0.0017 |
| FLAIR_Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis | 0.003 | 0.04 ± 0.04 | 0.02 ± 0.02 |
| DWI_Original Gray Level Dependence Matrix Gray Level Non Uniformity | 0.003 | 1415.7 ± 1279.42 | 569.48 ± 421.22 |
| DWI_Original Neighboring Gray Tone Difference Matrix Contrast | 0.004 | 0.1 ± 0.1 | 0.16 ± 0.11 |
| FLAIR_Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis | 0.006 | 1989.38 ± 660.24 | 2753.4 ± 1121.6 |
| DWI_Original Gray Level Size Zone Matrix Large Area Low Gray Level Emphasis | 0.006 | 48.74 ± 148.12 | 1.35 ± 3.01 |
| FLAIR_Original Gray Level Run Length Matrix Run Length Non Uniformity | 0.006 | 78,567.04 ± 57,195.07 | 41,756.66 ± 37,247.69 |
| FLAIR_Original Gray Level Size Zone Matrix Gray Level Non Uniformity | 0.006 | 719.94 ± 511.92 | 400.45 ± 299.7 |
| DWI_Original Gray Level Run Length Matrix Gray Level Variance | 0.007 | 64.37 ± 36.15 | 84.64 ± 28.49 |
| FLAIR_Original Gray Level Dependence Matrix Low Gray Level Emphasis | 0.007 | 0.002 ± 0.0037 | 0.0016 ± 0.0026 |
| DWI_Original Gray Level Dependence Matrix Gray Level Variance | 0.007 | 62.45 ± 36.62 | 82.85 ± 29.07 |
| FLAIR_Original Gray Level Run Length Matrix Low Gray Level Run Emphasis | 0.007 | 0.002 ± 0.0039 | 0.0016 ± 0.0026 |
| FLAIR_Original Gray Level Dependence Matrix Dependence Non Uniformity | 0.007 | 13,941.11 ± 9449.91 | 7721.06 ± 5691.44 |
| DWI_Original First Order Uniformity | 0.009 | 0.05 ± 0.02 | 0.04 ± 0.01 |
| DWI_Original Gray Level Co-occurrence Matrix Contrast | 0.009 | 41.76 ± 29.66 | 56.34 ± 27.96 |
| DWI_Original Gray Level Size Zone Matrix Small Area Emphasis | 0.009 | 0.59 ± 0.05 | 0.62 ± 0.05 |
| FLAIR_Original Gray Level Size Zone Matrix Low Gray Level Zone Emphasis | 0.009 | 0.0025 ± 0.0059 | 0.0019 ± 0.0025 |
| DWI_Original Gray Level Run Length Matrix Gray Level Non Uniformity Normalized | 0.009 | 0.05 ± 0.02 | 0.04 ± 0.01 |
| DWI_Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized | 0.010 | 0.03 ± 0.01 | 0.03 ± 0.004 |
| DWI_Original Gray Level Size Zone Matrix Size Zone Non Uniformity Normalized | 0.010 | 0.33 ± 0.05 | 0.36 ± 0.05 |
| FLAIR_Original Gray Level Run Length Matrix Short Run Low Gray Level Emphasis | 0.011 | 0.0019 ± 0.0038 | 0.0015 ± 0.0025 |
| FLAIR_Original Gray Level Run Length Matrix Gray Level Non Uniformity | 0.011 | 3783.52 ± 3124.39 | 2072.1 ± 2363.18 |
| DWI_Original Gray Level Co-occurrence Matrix Difference Variance | 0.012 | 20.13 ± 12.63 | 26.75 ± 11.87 |
| DWI_Original Gray Level Co-occurrence Matrix Inverse Difference Moment Normalized | 0.012 | 0.99 ± 0.01 | 0.99 ± 0.01 |
| FLAIR_Original Gray Level Run Length Matrix Run Variance | 0.012 | 0.29 ± 0.15 | 0.22 ± 0.14 |
| FLAIR_Original Gray Level Size Zone Matrix Size Zone Non Uniformity | 0.012 | 6324.41 ± 4176.75 | 4061.53 ± 2756.39 |
| DWI_Original Gray Level Size Zone Matrix Gray Level Variance | 0.013 | 91.84 ± 27.92 | 111.61 ± 32.95 |
| DWI_Original Gray Level Run Length Matrix Run Length Non Uniformity | 0.013 | 16,416.98 ± 10,512.48 | 10,095.05 ± 6915.17 |
| FLAIR_Original Gray Level Dependence Matrix Gray Level Non Uniformity | 0.014 | 4533.69 ± 3896.27 | 2487.02 ± 3005.24 |
| DWI_Original Gray Level Size Zone Matrix Large Area Emphasis | 0.015 | 5713.84 ± 17,261.49 | 559.63 ± 1038.73 |
| DWI_Original Gray Level Size Zone Matrix Zone Variance | 0.015 | 5669.15 ± 17,179.37 | 543.05 ± 1027.67 |
| DWI_Original Gray Level Co-occurrence Matrix Cluster Shade | 0.017 | 657.55 ± 2804.84 | −1369.51 ± 3597.59 |
| DWI_Original First Order Maximum | 0.018 | 1161.89 ± 588.37 | 833.97 ± 489.5 |
| DWI_Original First Order Entropy | 0.019 | 4.7 ± 0.48 | 5 ± 0.27 |
| FLAIR_Original Gray Level Run Length Matrix Long Run Emphasis | 0.020 | 1.67 ± 0.3 | 1.52 ± 0.3 |
| DWI_Original Gray Level Dependence Matrix Dependence Non Uniformity | 0.022 | 2986.94 ± 1884.99 | 1937.84 ± 1175.1 |
| DWI_Original Gray Level Dependence Matrix Small Dependence Emphasis | 0.022 | 0.21 ± 0.09 | 0.26 ± 0.09 |
| DWI_Original Gray Level Size Zone Matrix Gray Level Non Uniformity | 0.024 | 158.2 ± 101.68 | 101.38 ± 57.73 |
| DWI_Original Gray Level Co-occurrence Matrix Cluster Prominence | 0.027 | 203,134.01 ± 165,799.16 | 285,344.44 ± 193,348.11 |
| DWI_Original Gray Level Co-occurrence Matrix Maximum Probability | 0.028 | 0.02 ± 0.02 | 0.01 ± 0.01 |
| DWI_Original First Order Range | 0.029 | 1089.95 ± 588.32 | 772.8 ± 487.35 |
| DWI_Original Gray Level Co-occurrence Matrix Joint Energy | 0.030 | 0.01 ± 0.01 | 0.0038 ± 0.0017 |
| DWI_Original Gray Level Run Length Matrix Run Variance | 0.030 | 0.28 ± 0.22 | 0.18 ± 0.1 |
| DWI_Original Gray Level Co-occurrence Matrix Difference Average | 0.031 | 4.08 ± 1.49 | 4.86 ± 1.48 |
| DWI_Original Gray Level Size Zone Matrix Zone Percentage | 0.031 | 0.24 ± 0.11 | 0.3 ± 0.11 |
| FLAIR_Original Gray Level Size Zone Matrix Small Area Low Gray Level Emphasis | 0.031 | 0.0016 ± 0.0041 | 0.0012 ± 0.0017 |
| DWI_Original Gray Level Run Length Matrix Long Run Emphasis | 0.032 | 1.68 ± 0.47 | 1.45 ± 0.21 |
| FLAIR_Original Gray Level Size Zone Matrix Zone Entropy | 0.032 | 7.64 ± 0.27 | 7.5 ± 0.28 |
| DWI_Original Gray Level Co-occurrence Matrix Sum of Squares | 0.033 | 62.05 ± 36.14 | 77.43 ± 30.25 |
| FLAIR_Original First Order 90th Percentile | 0.034 | 1067.34 ± 552.14 | 837.61 ± 620.08 |
| DWI_Original Gray Level Run Length Matrix Short Run Emphasis | 0.035 | 0.91 ± 0.04 | 0.93 ± 0.02 |
| DWI_Original Gray Level Co-occurrence Matrix Inverse Difference Normalized | 0.036 | 0.95 ± 0.02 | 0.94 ± 0.02 |
| DWI_Original Gray Level Run Length Matrix Run Percentage | 0.036 | 0.87 ± 0.05 | 0.9 ± 0.03 |
| DWI_Original Gray Level Run Length Matrix Run Entropy | 0.036 | 5.34 ± 0.31 | 5.51 ± 0.19 |
| DWI_Original First Order Energy | 0.038 | 52,082,170,32.98 ± 39,247,896,40.04 | 34,436,543,32.49 ± 34,586,473,38.81 |
| DWI_Original Gray Level Dependence Matrix Large Dependence Emphasis | 0.038 | 28.71 ± 19.21 | 19.22 ± 9.25 |
| DWI_Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis | 0.038 | 0.36 ± 0.98 | 0.07 ± 0.1 |
| DWI_Original Gray Level Run Length Matrix Run Length Non Uniformity Normalized | 0.039 | 0.79 ± 0.07 | 0.83 ± 0.05 |
| DWI_Original Gray Level Dependence Matrix Large Dependence High Gray Level Emphasis | 0.041 | 15,954.41 ± 7160.01 | 21,904.17 ± 12,525.77 |
| DWI_Original Neighboring Gray Tone Difference Matrix Complexity | 0.042 | 4919.68 ± 1476.01 | 5651.97 ± 1458.46 |
| DWI_Original Gray Level Dependence Matrix Dependence Non Uniformity Normalized | 0.043 | 0.14 ± 0.04 | 0.16 ± 0.05 |
| DWI_Original Gray Level Co-occurrence Matrix Sum Entropy | 0.046 | 5.55 ± 0.48 | 5.8 ± 0.3 |
| DWI_Original Gray Level Co-occurrence Matrix Difference Entropy | 0.049 | 3.36 ± 0.45 | 3.58 ± 0.35 |
Table A3.
Final Features Used by DWI-FLAIR XGBoost Model Ranked By Gain.
| Radiomic Features (n = 88) |
|---|
| DWI_Original First Order Total Energy |
| DWI_Original First Order Mean Absolute Deviation |
| FLAIR_Original First Order 90th Percentile |
| FLAIR_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis |
| FLAIR_Original Gray Level Run Length Matrix High Gray Level Run Emphasis |
| FLAIR_Original Gray Level Size Zone Matrix Gray Level Non Uniformity |
| DWI_Original First Order Maximum |
| DWI_Original Gray Level Run Length Matrix Run Entropy |
| DWI_Original First Order Skewness |
| DWI_Original First Order 10th Percentile |
| DWI_Original First Order 90th Percentile |
| FLAIR_Original First Order Total Energy |
| DWI_Original Gray Level Size Zone Matrix Large Area Emphasis |
| DWI_Original Gray Level Size Zone Matrix Zone Variance |
| DWI_Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 |
| FLAIR_Original Gray Level Co-occurrence Matrix Joint Entropy |
| DWI_Original Gray Level Co-occurrence Matrix Correlation |
| DWI_Original Gray Level Co-occurrence Matrix Contrast |
| FLAIR_Original Gray Level Run Length Matrix Long Run Low Gray Level Emphasis |
| DWI_Original Gray Level Co-occurrence Matrix Cluster Shade |
| FLAIR_Original Gray Level Co-occurrence Matrix Cluster Shade |
| DWI_Original Gray Level Co-occurrence Matrix Joint Entropy |
| DWI_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis |
| DWI_Original Neighboring Gray Tone Difference Matrix Complexity |
| FLAIR_Original Gray Level Co-occurrence Matrix Joint Average |
| DWI_Original Gray Level Dependence Matrix Small Dependence Emphasis |
| FLAIR_Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 1 |
| DWI_Original Gray Level Size Zone Matrix Zone Entropy |
| FLAIR_Original First Order Energy |
| DWI_Original First Order Kurtosis |
| FLAIR_Original Gray Level Co-occurrence Matrix Sum Average |
| DWI_Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis |
| FLAIR_Original First Order 10th Percentile |
| DWI_Original First Order Variance |
| FLAIR_Original Gray Level Size Zone Matrix Size Zone Non Uniformity |
| FLAIR_Original Gray Level Size Zone Matrix Gray Level Variance |
| FLAIR_Original First Order Skewness |
| FLAIR_Original Gray Level Dependence Matrix Dependence Non Uniformity |
| FLAIR_Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis |
| FLAIR_Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 |
| FLAIR_Original First Order Standard Deviation |
| FLAIR_Original Gray Level Size Zone Matrix Large Area Low Gray Level Emphasis |
| FLAIR_Original Gray Level Co-occurrence Matrix Sum of Squares |
| DWI_Original First Order Energy |
| FLAIR_Original Gray Level Co-occurrence Matrix Maximum Probability |
| FLAIR_Original Gray Level Co-occurrence Matrix Correlation |
| DWI_Original Gray Level Dependence Matrix Gray Level Non Uniformity |
| FLAIR_Original First Order Range |
| FLAIR_Original Gray Level Co-occurrence Matrix Cluster Tendency |
| FLAIR_Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis |
| DWI_Original First Order Entropy |
| DWI_Original First Order Uniformity |
| FLAIR_Original First Order Interquartile Range |
| DWI_Original Gray Level Dependence Matrix Dependence Entropy |
| FLAIR_Original Gray Level Dependence Matrix Large Dependence High Gray Level Emphasis |
| DWI_Original Gray Level Dependence Matrix Gray Level Variance |
| FLAIR_Original Gray Level Dependence Matrix Dependence Entropy |
| DWI_Original Gray Level Run Length Matrix Gray Level Variance |
| DWI_Original First Order Robust Mean Absolute Deviation |
| DWI_Original First Order Median |
| FLAIR_Original Gray Level Co-occurrence Matrix Difference Variance |
| FLAIR_Original Gray Level Run Length Matrix Gray Level Variance |
| FLAIR_Original First Order Minimum |
| FLAIR_Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis |
| DWI_Original First Order Standard Deviation |
| DWI_Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized |
| DWI_Original Gray Level Run Length Matrix Low Gray Level Run Emphasis |
| FLAIR_Original Gray Level Dependence Matrix Dependence Variance |
| DWI_Original Neighboring Gray Tone Difference Matrix Contrast |
| DWI_Original Gray Level Co-occurrence Matrix Inverse Difference |
| DWI_Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis |
| FLAIR_Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized |
| DWI_Original Gray Level Run Length Matrix Run Variance |
| DWI_Original Gray Level Dependence Matrix Low Gray Level Emphasis |
| FLAIR_Original Gray Level Size Zone Matrix Zone Percentage |
| FLAIR_Original Gray Level Size Zone Matrix Small Area Low Gray Level Emphasis |
| DWI_Original Gray Level Co-occurrence Matrix Inverse Variance |
| DWI_Original First Order Minimum |
| DWI_Original Gray Level Co-occurrence Matrix Maximum Probability |
| DWI_Original Gray Level Size Zone Matrix Large Area Low Gray Level Emphasis |
| DWI_Original Gray Level Co-occurrence Matrix Joint Energy |
| FLAIR_Original Neighboring Gray Tone Difference Matrix Complexity |
| FLAIR_Original Gray Level Co-occurrence Matrix Contrast |
| FLAIR_Original Gray Level Run Length Matrix Run Entropy |
| FLAIR_Original Gray Level Co-occurrence Matrix Joint Energy |
| DWI_Original Gray Level Run Length Matrix Run Percentage |
| DWI_Original Gray Level Co-occurrence Matrix Joint Average |
| FLAIR_Original First Order Mean Absolute Deviation |
Table A4.
Final Features Used by DWI XGBoost Model Ranked By Gain.
| Radiomic Features (n = 71) |
|---|
| Original Gray Level Co-occurrence Matrix Autocorrelation |
| Original Gray Level Run Length Matrix Run Entropy |
| Original Gray Level Dependence Matrix Dependence Non Uniformity Normalized |
| Original Gray Level Dependence Matrix Gray Level Variance |
| Original Gray Level Co-occurrence Matrix Maximum Probability |
| Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis |
| Original Gray Level Run Length Matrix Gray Level Non Uniformity |
| Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis |
| Original First Order Total Energy |
| Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 |
| Original Gray Level Size Zone Matrix Large Area High Gray Level Emphasis |
| Original Gray Level Size Zone Matrix Size Zone Non Uniformity |
| Original First Order 10th Percentile |
| Original First Order Root Mean Squared |
| Original Gray Level Co-occurrence Matrix Cluster Shade |
| Original First Order Kurtosis |
| Original Gray Level Size Zone Matrix Low Gray Level Zone Emphasis |
| Original Gray Level Run Length Matrix Run Length Non Uniformity |
| Original Neighboring Gray Tone Difference Matrix Coarseness |
| Original First Order Mean Absolute Deviation |
| Original First Order Skewness |
| Original Gray Level Run Length Matrix Short Run Low Gray Level Emphasis |
| Original Gray Level Co-occurrence Matrix Sum Average |
| Original First Order Robust Mean Absolute Deviation |
| Original Gray Level Dependence Matrix Dependence Non Uniformity |
| Original First Order Range |
| Original Neighboring Gray Tone Difference Matrix Busyness |
| Original First Order Maximum |
| Original Gray Level Co-occurrence Matrix Sum of Squares |
| Original Gray Level Co-occurrence Matrix Inverse Difference Normalized |
| Original First Order Standard Deviation |
| Original Gray Level Dependence Matrix Gray Level Non Uniformity |
| Original Neighboring Gray Tone Difference Matrix Strength |
| Original Gray Level Size Zone Matrix Size Zone Non Uniformity Normalized |
| Original Gray Level Dependence Matrix Dependence Entropy |
| Original Gray Level Size Zone Matrix Zone Entropy |
| Original Gray Level Dependence Matrix Low Gray Level Emphasis |
| Original Gray Level Co-occurrence Matrix Sum Entropy |
| Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized |
| Original Gray Level Co-occurrence Matrix Difference Entropy |
| Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 1 |
| Original Gray Level Run Length Matrix Long Run Emphasis |
| Original Gray Level Run Length Matrix Long Run Low Gray Level Emphasis |
| Original First Order Minimum |
| Original Gray Level Run Length Matrix Low Gray Level Run Emphasis |
| Original Gray Level Size Zone Matrix Zone Percentage |
| Original Gray Level Co-occurrence Matrix Cluster Prominence |
| Original First Order Energy |
| Original Neighboring Gray Tone Difference Matrix Complexity |
| Original Gray Level Co-occurrence Matrix Correlation |
| Original Gray Level Co-occurrence Matrix Joint Energy |
| Original Gray Level Size Zone Matrix Gray Level Non Uniformity |
| Original Gray Level Size Zone Matrix Zone Variance |
| Original Gray Level Size Zone Matrix Small Area Emphasis |
| Original Gray Level Run Length Matrix High Gray Level Run Emphasis |
| Original Gray Level Run Length Matrix Gray Level Non Uniformity Normalized |
| Original Gray Level Co-occurrence Matrix Difference Average |
| Original First Order Median |
| Original First Order Uniformity |
| Original Gray Level Co-occurrence Matrix Joint Entropy |
| Original Gray Level Co-occurrence Matrix Joint Average |
| Original First Order Mean |
| Original First Order Interquartile Range |
| Original Gray Level Size Zone Matrix Large Area Low Gray Level Emphasis |
| Original Gray Level Size Zone Matrix Gray Level Variance |
| Original Neighboring Gray Tone Difference Matrix Contrast |
| Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis |
| Original Gray Level Run Length Matrix Gray Level Variance |
| Original Gray Level Run Length Matrix Run Percentage |
| Original Gray Level Dependence Matrix Large Dependence High Gray Level Emphasis |
| Original Gray Level Co-occurrence Matrix Contrast |
Table A5.
Final Features Used by FLAIR XGBoost Model Ranked By Gain.
| Radiomic Features (n = 33) |
|---|
| Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis |
| Original First Order Mean Absolute Deviation |
| Original Gray Level Co-occurrence Matrix Correlation |
| Original Gray Level Size Zone Matrix Gray Level Variance |
| Original Gray Level Size Zone Matrix Low Gray Level Zone Emphasis |
| Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 |
| Original Gray Level Co-occurrence Matrix Cluster Prominence |
| Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis |
| Original Neighboring Gray Tone Difference Matrix Coarseness |
| Original First Order Range |
| Original Gray Level Size Zone Matrix Size Zone Non Uniformity |
| Original First Order Kurtosis |
| Original First Order Skewness |
| Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis |
| Original First Order Interquartile Range |
| Original Gray Level Size Zone Matrix Gray Level Non Uniformity |
| Original Gray Level Run Length Matrix Gray Level Non Uniformity Normalized |
| Original Gray Level Co-occurrence Matrix Inverse Difference Moment Normalized |
| Original Gray Level Dependence Matrix Small Dependence Low Gray Level Emphasis |
| Original Gray Level Co-occurrence Matrix Cluster Shade |
| Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis |
| Original First Order Energy |
| Original Gray Level Co-occurrence Matrix Difference Entropy |
| Original Gray Level Run Length Matrix Run Entropy |
| Original Gray Level Run Length Matrix Low Gray Level Run Emphasis |
| Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis |
| Original Gray Level Run Length Matrix Run Percentage |
| Original Gray Level Co-occurrence Matrix Joint Average |
| Original Gray Level Run Length Matrix Long Run Emphasis |
| Original Gray Level Run Length Matrix Short Run Emphasis |
| Original First Order Robust Mean Absolute Deviation |
| Original Gray Level Run Length Matrix High Gray Level Run Emphasis |
| Original First Order Total Energy |
Table A6.
All Radiomic Features Listed By Feature Class.
| Feature Class | Radiomic Features |
|---|---|
| First Order Statistics | Original First Order Energy |
| Original First Order Total Energy | |
| Original First Order Entropy | |
| Original First Order Minimum | |
| Original First Order 10th Percentile | |
| Original First Order 90th Percentile | |
| Original First Order Maximum | |
| Original First Order Mean | |
| Original First Order Median | |
| Original First Order Interquartile Range | |
| Original First Order Range | |
| Original First Order Mean Absolute Deviation | |
| Original First Order Robust Mean Absolute Deviation | |
| Original First Order Root Mean Squared | |
| Original First Order Standard Deviation | |
| Original First Order Skewness | |
| Original First Order Kurtosis | |
| Original First Order Variance | |
| Original First Order Uniformity | |
| Gray Level Co-occurrence Matrix | Original Gray Level Co-occurrence Matrix Autocorrelation |
| Original Gray Level Co-occurrence Matrix Joint Average | |
| Original Gray Level Co-occurrence Matrix Cluster Prominence | |
| Original Gray Level Co-occurrence Matrix Cluster Shade | |
| Original Gray Level Co-occurrence Matrix Cluster Tendency | |
| Original Gray Level Co-occurrence Matrix Contrast | |
| Original Gray Level Co-occurrence Matrix Correlation | |
| Original Gray Level Co-occurrence Matrix Difference Average | |
| Original Gray Level Co-occurrence Matrix Difference Entropy | |
| Original Gray Level Co-occurrence Matrix Difference Variance | |
| Original Gray Level Co-occurrence Matrix Joint Energy | |
| Original Gray Level Co-occurrence Matrix Joint Entropy | |
| Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 1 | |
| Original Gray Level Co-occurrence Matrix Informal Measure of Correlation 2 | |
| Original Gray Level Co-occurrence Matrix Inverse Difference Moment | |
| Original Gray Level Co-occurrence Matrix Inverse Difference Moment Normalized | |
| Original Gray Level Co-occurrence Matrix Inverse Difference | |
| Original Gray Level Co-occurrence Matrix Inverse Difference Normalized | |
| Original Gray Level Co-occurrence Matrix Inverse Variance | |
| Original Gray Level Co-occurrence Matrix Maximum Probability | |
| Original Gray Level Co-occurrence Matrix Sum Average | |
| Original Gray Level Co-occurrence Matrix Sum Entropy | |
| Original Gray Level Co-occurrence Matrix Sum of Squares | |
| Gray Level Run Length Matrix | Original Gray Level Run Length Matrix Short Run Emphasis |
| Original Gray Level Run Length Matrix Long Run Emphasis | |
| Original Gray Level Run Length Matrix Gray Level Non Uniformity | |
| Original Gray Level Run Length Matrix Gray Level Non Uniformity Normalized | |
| Original Gray Level Run Length Matrix Run Length Non Uniformity | |
| Original Gray Level Run Length Matrix Run Length Non Uniformity Normalized | |
| Original Gray Level Run Length Matrix Run Percentage | |
| Original Gray Level Run Length Matrix Gray Level Variance | |
| Original Gray Level Run Length Matrix Run Variance | |
| Original Gray Level Run Length Matrix Run Entropy | |
| Original Gray Level Run Length Matrix Low Gray Level Run Emphasis | |
| Original Gray Level Run Length Matrix High Gray Level Run Emphasis | |
| Original Gray Level Run Length Matrix Short Run Low Gray Level Emphasis | |
| Original Gray Level Run Length Matrix Short Run High Gray Level Emphasis | |
| Original Gray Level Run Length Matrix Long Run Low Gray Level Emphasis | |
| Original Gray Level Run Length Matrix Long Run High Gray Level Emphasis | |
| Gray Level Size Zone Matrix | Original Gray Level Size Zone Matrix Small Area Emphasis |
| Original Gray Level Size Zone Matrix Large Area Emphasis | |
| Original Gray Level Size Zone Matrix Gray Level Non Uniformity | |
| Original Gray Level Size Zone Matrix Gray Level Non Uniformity Normalized | |
| Original Gray Level Size Zone Matrix Size Zone Non Uniformity | |
| Original Gray Level Size Zone Matrix Size Zone Non Uniformity Normalized | |
| Original Gray Level Size Zone Matrix Zone Percentage | |
| Original Gray Level Size Zone Matrix Gray Level Variance | |
| Original Gray Level Size Zone Matrix Zone Variance | |
| Original Gray Level Size Zone Matrix Zone Entropy | |
| Original Gray Level Size Zone Matrix Low Gray Level Zone Emphasis | |
| Original Gray Level Size Zone Matrix Small Area Low Gray Level Emphasis | |
| Original Gray Level Size Zone Matrix Small Area High Gray Level Emphasis | |
| Original Gray Level Size Zone Matrix Large Area Low Gray Level Emphasis | |
| Original Gray Level Size Zone Matrix Large Area High Gray Level Emphasis | |
| Neighboring Gray Tone Difference Matrix | Original Neighboring Gray Tone Difference Matrix Coarseness |
| Original Neighboring Gray Tone Difference Matrix Contrast | |
| Original Neighboring Gray Tone Difference Matrix Busyness | |
| Original Neighboring Gray Tone Difference Matrix Complexity | |
| Original Neighboring Gray Tone Difference Matrix Strength | |
| Gray Level Dependence Matrix | Original Gray Level Dependence Matrix Small Dependence Emphasis |
| Original Gray Level Dependence Matrix Large Dependence Emphasis | |
| Original Gray Level Dependence Matrix Gray Level Non Uniformity | |
| Original Gray Level Dependence Matrix Dependence Non Uniformity | |
| Original Gray Level Dependence Matrix Dependence Non Uniformity Normalized | |
| Original Gray Level Dependence Matrix Gray Level Variance | |
| Original Gray Level Dependence Matrix Dependence Variance | |
| Original Gray Level Dependence Matrix Dependence Entropy | |
| Original Gray Level Dependence Matrix Low Gray Level Emphasis | |
| Original Gray Level Dependence Matrix High Gray Level Emphasis | |
| Original Gray Level Dependence Matrix Small Dependence Low Gray Level Emphasis | |
| Original Gray Level Dependence Matrix Small Dependence High Gray Level Emphasis | |
| Original Gray Level Dependence Matrix Large Dependence Low Gray Level Emphasis | |
| Original Gray Level Dependence Matrix Large Dependence High Gray Level Emphasis |
Figure A1.
Spearman Correlation Matrix of DWI Radiomic Features.
Figure A2.
Spearman Correlation Matrix of FLAIR Radiomic Features.
Author Contributions
Conceptualization, K.N., A.H. and Y.S.; methodology, K.N.; software, Y.S., C.Y., S.K.; validation, K.N., A.H., N.T., A.L., T.C.; formal analysis, Y.S., C.Y., S.K., F.K.; investigation, Y.S., C.Y., S.K., F.K., K.N.; resources, K.N., A.H., N.T.; data curation, K.N., Y.S., S.K., A.H., N.T.; writing-original draft preparation, Y.S.; writing-review and editing, Y.S., C.Y., S.K., N.T., F.K., A.H., A.L., T.C., K.N.; visualization, Y.S., C.Y., S.K., K.N.; supervision, K.N.; project administration, K.N. funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
Kambiz Nael is a consultant to Olea, none for others.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weller M., Wick W., Aldape K., Brada M., Berger M., Pfister S.M., Nishikawa R., Rosenthal M., Wen P.Y., Stupp R., et al. Glioma. Nat. Rev. Dis. Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.Bralten L.B.C., Kloosterhof N.K., Balvers R., Sacchetti A., Lapre L., Lamfers M., Leenstra S., De Jonge H., Kros J.M., Jansen E.E.W., et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann. Neurol. 2011;69:455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 3.Wang H.-Y., Tang K., Liang T.-Y., Zhang W.-Z., Li J.-Y., Wang W., Hu H.-M., Li M.-Y., Wang H.-Q., He X.-Z., et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J. Exp. Clin. Cancer Res. 2016;35:1–9. doi: 10.1186/s13046-016-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartmann C., Meyer J., Balss J., Capper D., Mueller W., Christians A., Felsberg J., Wolter M., Mawrin C., Wick W., et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 5.Bent M.J.V.D., Dubbink H.J., Marie Y., Brandes A.A., Taphoorn M.J., Wesseling P., Frenay M., Tijssen C.C., Lacombe D., Idbaih A., et al. IDH1 and IDH2 Mutations Are Prognostic but not Predictive for Outcome in Anaplastic Oligodendroglial Tumors: A Report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin. Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 6.Louis D.N., Perry A., Reifenberger G., Von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Chaddad A., Kucharczyk M.J., Daniel P., Sabri S., Jean-Claude B.J., Niazi T., Abdulkarim B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019;9:374. doi: 10.3389/fonc.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M., Scott J., Chaudhury B., Hall L., Goldgof D., Yeom K., Iv M., Ou Y., Kalpathy-Cramer J., Napel S., et al. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. Am. J. Neuroradiol. 2017;39:208–216. doi: 10.3174/ajnr.A5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita M., Sakai M., Arita H., Shofuda T., Chiba Y., Kagawa N., Watanabe Y., Hashimoto N., Fujimoto Y., Yoshimine T., et al. Introduction of High Throughput Magnetic Resonance T2-Weighted Image Texture Analysis for WHO Grade 2 and 3 Gliomas. PLoS ONE. 2016;11:e0164268. doi: 10.1371/journal.pone.0164268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z.C., Bai H., Sun Q., Zhao Y., Lv Y., Zhou J., Liang C., Chen Y., Liang D., Zheng H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018;7:5999–6009. doi: 10.1002/cam4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotan E., Jain R., Razavian N., Fatterpekar G.M., Lui Y.W. State of the Art: Machine Learning Applications in Glioma Imaging. Am. J. Roentgenol. 2019;212:26–37. doi: 10.2214/AJR.18.20218. [DOI] [PubMed] [Google Scholar]
- 12.Sotoudeh H., Shafaat O., Bernstock J.D., Brooks M.D., Elsayed G.A., Chen J.A., Szerip P., Chagoya G., Gessler F., Sotoudeh E., et al. Artificial Intelligence in the Management of Glioma: Era of Personalized Medicine. Front. Oncol. 2019;9:768. doi: 10.3389/fonc.2019.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T., Guestrin C. XGBoost: A Scalable Tree Boosting System. [(accessed on 22 October 2020)];2016 Mar 1; arXiv e-Prints [Internet] Available online: https://ui.adsabs.harvard.edu/abs/2016arXiv160302754C.
- 14.Natekin A., Knoll A. Gradient boosting machines, a tutorial. Front. Neurorobotics. 2013;7:21. doi: 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wick W., Roth P., Hartmann C., Hau P., Nakamura M., Stockhammer F., Sabel M.C., Wick A., Koeppen S., Ketter R., et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro-Oncol. 2016;18:1529–1537. doi: 10.1093/neuonc/now133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi S., Yu L., Li H., Ou Y., Qiu X., Ding Y., Han H., Zhang X. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol. Lett. 2014;7:1895–1902. doi: 10.3892/ol.2014.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonoda Y., Shibahara I., Kawaguchi T., Saito R., Kanamori M., Watanabe M., Suzuki H., Kumabe T., Tominaga T. Association between molecular alterations and tumor location and MRI characteristics in anaplastic gliomas. Brain Tumor Pathol. 2014;32:99–104. doi: 10.1007/s10014-014-0211-3. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita K., Hiwatashi A., Togao O., Kikuchi K., Hatae R., Yoshimoto K., Mizoguchi M., Suzuki S., Yoshiura T., Honda H. MR Imaging–Based Analysis of Glioblastoma Multiforme: Estimation ofIDH1Mutation Status. Am. J. Neuroradiol. 2015;37:58–65. doi: 10.3174/ajnr.A4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel S.H., Poisson L.M., Brat D.J., Zhou Y., Cooper L., Snuderl M., Thomas C., Franceschi A.M., Griffith B., Flanders A.E., et al. T2–FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin. Cancer Res. 2017;23:6078–6085. doi: 10.1158/1078-0432.CCR-17-0560. [DOI] [PubMed] [Google Scholar]
- 20.Broen M.P., Smits M., Wijnenga M.M., Dubbink H.J., Anten M.H., Schijns O.E., Beckervordersandforth J., Postma A.A., van den Bent M.J. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: A validation study. Neuro-oncology. 2018;20:1393–1399. doi: 10.1093/neuonc/noy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bisdas S., Shen H., Thust S., Katsaros V., Stranjalis G., Boskos C., Brandner S., Zhang J. Texture analysis- and support vector machine-assisted diffusional kurtosis imaging may allow in vivo gliomas grading and IDH-mutation status prediction: A preliminary study. Sci. Rep. 2018;8:6108. doi: 10.1038/s41598-018-24438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S., Meng J., Yu Q., Li P., Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019;145:543–550. doi: 10.1007/s00432-018-2787-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shboul Z.A., Chen J., Iftekharuddin K.M. Prediction of Molecular Mutations in Diffuse Low-Grade Gliomas using MR Imaging Features. Sci. Rep. 2020;10:3711. doi: 10.1038/s41598-020-60550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H., Chang K., Bai H.X., Xiao B., Su C., Bi W.L., Zhang P.J., Senders J.T., Vallières M., Kavouridis V.K., et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neuro-Oncol. 2019;142:299–307. doi: 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macyszyn L., Akbari H., Pisapia J.M., Da X., Attiah M., Pigrish V., Bi Y., Pal S., Davuluri R.V., Roccograndi L., et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology. 2015;18:417–425. doi: 10.1093/neuonc/nov127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M.H., Kim J., Kim S.-T., Shin H.-M., You H.-J., Choi J.W., Seol H.J., Nam D.-H., Lee J.-I., Kong D.-S. Prediction of IDH1 Mutation Status in Glioblastoma Using Machine Learning Technique Based on Quantitative Radiomic Data. World Neurosurg. 2019;125:e688–e696. doi: 10.1016/j.wneu.2019.01.157. [DOI] [PubMed] [Google Scholar]
- 27.Alis D., Bagcilar O., Senli Y.D., Yergin M., Isler C., Kocer N., Islak C., Kizilkilic O. Machine learning-based quantitative texture analysis of conventional MRI combined with ADC maps for assessment of IDH1 mutation in high-grade gliomas. Jpn. J. Radiol. 2019;38:135–143. doi: 10.1007/s11604-019-00902-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B., Chang K., Ramkissoon S., Tanguturi S., Bi W.L., Reardon D.A., Ligon K.L., Alexander B.M., Wen P.Y., Huang R.Y. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-oncology. 2016;19:109–117. doi: 10.1093/neuonc/now121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calabrese E., Villanueva-Meyer J.E., Cha S. A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci. Rep. 2020;10:11852. doi: 10.1038/s41598-020-68857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Tian Q., Wang L., Liu Y., Li B., Liang Z., Gao P., Zheng K., Zhao B., Lu H. Radiomics Strategy for Molecular Subtype Stratification of Lower-Grade Glioma: Detecting IDH andTP53Mutations Based on Multimodal MRI. J. Magn. Reson. Imaging. 2018;48:916–926. doi: 10.1002/jmri.25960. [DOI] [PubMed] [Google Scholar]
- 31.Eichinger P., Alberts E., Delbridge C., Trebeschi S., Valentinitsch A., Bette S., Huber T., Gempt J., Meyer B., Schlegel J., et al. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci. Rep. 2017;7:13396. doi: 10.1038/s41598-017-13679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kickingereder P., Bonekamp D., Nowosielski M., Kratz A., Sill M., Burth S., Wick A., Eidel O., Schlemmer H.-P., Radbruch A., et al. Radiogenomics of Glioblastoma: Machine Learning–based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology. 2016;281:907–918. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 33.Yu J., Shi Z., Lian Y., Li Z., Liu T., Gao Y., Wang Y., Chen L., Mao Y. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur. Radiol. 2016;27:3509–3522. doi: 10.1007/s00330-016-4653-3. [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Xu J., Zhao C.-A., Peng Y., Wang H. An ensemble feature selection method for high-dimensional data based on sort aggregation. Syst. Sci. Control. Eng. 2019;7:32–39. doi: 10.1080/21642583.2019.1620658. [DOI] [Google Scholar]
- 35.Calvar J.A., Meli F.J., Romero C., Yánez M.L.C.P., Martinez A.R., Lambre H., Taratuto A.L., Sevlever G.E. Characterization of brain tumors by MRS, DWI and Ki-67 labeling index. J. Neuro-Oncol. 2005;72:273–280. doi: 10.1007/s11060-004-3342-2. [DOI] [PubMed] [Google Scholar]
- 36.Nie Q.-M., Lin Y.-Y., Yang X., Shen L., Guo L.-M., Que S.-L., Li X.-X., Ge J.-W., Wang G.-S., Xiong W.-H., et al. IDH1R132H decreases the proliferation of U87 glioma cells through upregulation of microRNA-128a. Mol. Med. Rep. 2015;12:6695–6701. doi: 10.3892/mmr.2015.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H., Vallières M., Bai H.X., Su C., Tang H., Oldridge D., Zhang Z., Xiao B., Liao W., Tao Y., et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-oncology. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostrom Q.T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N.E., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006–2010. Neuro-oncology. 2013;15:ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons D.W., Jones S., Zhang X., Lin J.C.-H., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.-M., Gallia G.L., et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raosoft Sample Size Online Calculator. [(accessed on 22 October 2020)]; Available online: http://www.raosoft.com/samplesize.html.
- 41.Haralick R.M., Shanmugam K., Dinstein I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973;SMC-3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 42.Tang X. Texture information in run-length matrices. IEEE Trans. Image Process. 1998;7:1602–1609. doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 43.Thibault G., Fertil B., Navarro C., Pereira S., Cau P., Lévy N., Sequeira J., Mari J.-L. SHAPE AND TEXTURE INDEXES APPLICATION TO CELL NUCLEI CLASSIFICATION. Int. J. Pattern Recognit. Artif. Intell. 2013;27 doi: 10.1142/S0218001413570024. [DOI] [Google Scholar]
- 44.Chen S., Harmon S., Perk T., Li X., Chen M., Li Y., Jeraj R. Using neighborhood gray tone difference matrix texture features on dual time point PET/CT images to differentiate malignant from benign FDG-avid solitary pulmonary nodules. Cancer Imaging. 2019;19:1–8. doi: 10.1186/s40644-019-0243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao X., Cai B., Tian B., Luo Y., Song W., Li Y. Machine-learning based radiogenomics analysis of MRI features and metagenes in glioblastoma multiforme patients with different survival time. J. Cell. Mol. Med. 2019;23:4375–4385. doi: 10.1111/jcmm.14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parekh V., Jacobs M.A. Radiomics: A new application from established techniques. Expert Rev. Precis. Med. Drug Dev. 2016;1:207–226. doi: 10.1080/23808993.2016.1164013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chawla N., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 48.Ling J., Wang X., Sun Y. Research of Android Malware Detection based on ACO Optimized Xgboost Parameters Approach; Proceedings of the 3rd International Conference on Mechatronics Engineering and Information Technology (ICMEIT 2019); Dalian, China. 29–30 March 2019. [Google Scholar]