Abstract
Regulator of G protein signaling (RGS) proteins are multifunctional proteins expressed in peripheral and neuronal cells, playing critical roles in development, physiologic processes, and pharmacological responses. RGS proteins primarily act as GTPase accelerators for activated Gα subunits of G-protein coupled receptors, but they may also modulate signal transduction by several other mechanisms. Over the last two decades, preclinical work identified members of the RGS family with unique and critical roles in intracellular responses to drugs of abuse. New information has emerged on the mechanisms by which RGS proteins modulate the efficacy of opioid analgesics in a brain region– and agonist-selective fashion. There has also been progress in the understanding of the protein complexes and signal transduction pathways regulated by RGS proteins in addiction and analgesia circuits. In this review, we summarize findings on the mechanisms by which RGS proteins modulate functional responses to opioids in models of analgesia and addiction. We also discuss reports on the regulation and function of RGS proteins in models of psychostimulant addiction. Using information from preclinical studies performed over the last 20 years, we highlight the diverse mechanisms by which RGS protein complexes control plasticity in response to opioid and psychostimulant drug exposure; we further discuss how the understanding of these pathways may lead to new opportunities for therapeutic interventions in G protein pathways.
SIGNIFICANCE STATEMENT
Regulator of G protein signaling (RGS) proteins are signal transduction modulators, expressed widely in various tissues, including brain regions mediating addiction and analgesia. Evidence from preclinical work suggests that members of the RGS family act by unique mechanisms in specific brain regions to control drug-induced plasticity. This review highlights interesting findings on the regulation and function of RGS proteins in models of analgesia and addiction.
Introduction
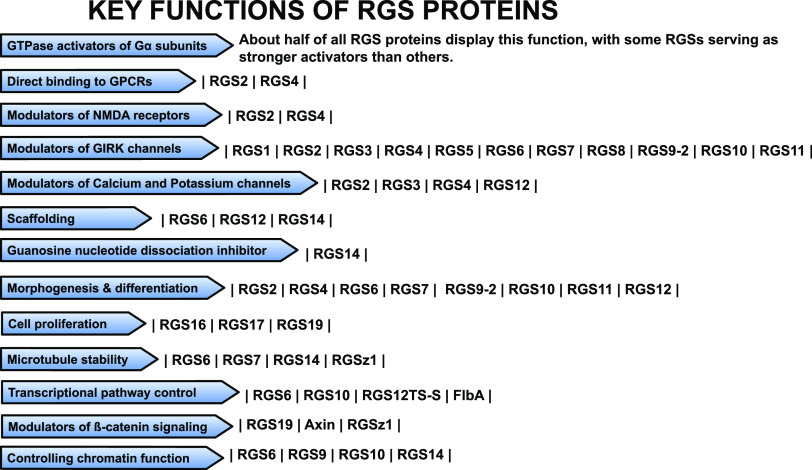
Regulator of G protein signaling (RGS) proteins are multifunctional signal transduction molecules playing dynamic roles in physiologic processes such as cardiac function, responses to stress, and cognition (Traynor and Neubig, 2005; Wieland et al., 2007; Terzi et al., 2009; Senesee et al., 2018; Squires et al., 2018). RGS proteins modulate dopamine, noradrenergic, serotonin, opioid, muscarinic, metabotropic glutamate, adenosine, and other G-protein coupled receptors (GPCRs) in central and peripheral cells. Human studies link RGS proteins to pathologic conditions including Parkinson’s disease (Greenbaum et al., 2009; Dusonchet et al., 2014), schizophrenia (Levitt et al., 2006; Rivero et al., 2013), hypertension (Kvehaugen et al., 2014; Chang et al., 2015), posttraumatic stress disorders (Amstadter et al., 2009), and pain (Smith et al., 2012). Members of the RGS family contain an RGS domain as well as other domains that control their enzymatic activity, protein interactions, stability, and cellular localization (Hepler, 1999; Neubig and Siderovski, 2002; Siderovski et al., 2002; Talbot et al., 2010). Binding of the RGS domain to activated Gα subunits enhances their GTPase activity and accelerates their return to the inactive Gα-GDP state and termination of signaling (Berman et al., 1996; Hunt et al., 1996; Mukhopadhyay and Ross, 1999). Moreover, preclinical studies describe additional mechanisms by which members of the RGS family control the duration and direction of signal transduction. About half of the proteins categorized as RGS show significant GTPase activity, and several members of the RGS family may bind to activated Gα subunits to prevent activation of effectors (Abramow-Newerly et al., 2006). Additional functions attributed to RGS protein subtypes include direct interactions with GPCRs (Georgoussi et al., 2006; Langer et al., 2009), modulation of G protein–activated inwardly rectifying K+ (GIRK) channels (Labouebe et al., 2007; Zhou et al., 2012), and modulation of calcium channels (Mark et al., 2000; Clark et al., 2003; Richman et al., 2005). RGS proteins may act as scaffolds (Willard et al., 2007; Shu et al., 2010; Huang et al., 2014) and they may also control N-methyl-d-aspartate receptor function (Bouhamdan et al., 2006; Liu et al., 2006), guanosine nucleotide dissociation (Mittal and Linder, 2004), mitosis (Martin-McCaffrey et al., 2004), cytoskeletal processes (Liu et al., 2002; Nixon et al., 2002; Martin-McCaffrey et al., 2005; Dave et al., 2009; Sivori et al., 2019), and cell proliferation (Willard et al., 2007; Yang and Li, 2007; Tso et al., 2010; Vivot et al., 2016; Chi et al., 2017; Madrigal et al., 2017; Scherer et al., 2017). Members of the RGS family may additionally regulate transcriptional pathways (Ikeda et al., 1998; Shimizu et al., 2003; Liu and Fisher, 2004; Feigin and Malbon, 2007; Alqinyah et al., 2017; Gaspari et al., 2018) and chromatin function (Liu and Fisher, 2004; Mitsi et al., 2015; Alqinyah et al., 2017; Branch and Hepler, 2017). Figure 1 displays some of the key functions of RGS proteins. Overall, the 40 mammalian RGS proteins show distinct tissue distribution, cellular localization, and preference for Gα subunit and GPCR subtypes (Hepler, 1999; Hollinger and Hepler, 2002).
Fig. 1.
The schematic lists some of the key functions of RGS proteins. NMDA, N-methyl-d-aspartate receptor.
Several members of the RGS family are expressed in brain circuits mediating drug addiction and analgesia (Gold et al., 1997). The rise in the incidence of addiction-related disorders (Skolnick and Volkow, 2016; Volkow and Boyle, 2018) and the need for safer, more efficacious treatment of chronic pain call for a deeper understanding of the circuits and intracellular pathways mediating addiction, physical dependence, and analgesia. Over the last two decades, preclinical studies have used genetic tools along with brain biochemistry to understand the impact of RGS proteins on the modulation of GPCR responses in the brain. Such approaches have been used to globally or conditionally inactivate a specific RGS protein and to generate RGS insensitive Gα subunits (Clark et al., 2003; Lamberts et al., 2013). Studies from several research groups have documented the powerful role of RGS family members in the modulation of GPCR signaling in brain circuits mediating drug addiction and pain relief. There is also strong evidence that acute or chronic exposure to drugs of abuse triggers dynamic and unique changes in RGS protein expression; this result suggests critical roles of RGS family members in the rewarding and reinforcing actions of drugs of abuse as well as in processes controlling GPCR desensitization. This review will highlight preclinical evidence of the role of RGS family members in the actions of opioids and psychostimulants.
RGS Proteins Modulate the Actions of Opioids
Synthetic, semi-synthetic, and endogenous opioids activate the mu opioid receptor (MOR) to promote analgesia, addiction, sedation, and several other undesired effects (Le Merrer et al., 2009). Opioids have been prescribed for their treatment of acute, subacute, and chronic pain, but their prolonged use may lead to physical dependence and progression to addiction (Raehal et al., 2011; Ehrlich et al., 2019). Long-term use of morphine and other opioids for the management of chronic pain often leads to analgesic tolerance. This is a major concern in therapeutics as patients need significantly higher drug doses for pain relief and, as a consequence, are at higher risk for addiction or respiratory depression (Skolnick and Volkow, 2016; Volkow et al., 2019). MORs are present in the nucleus accumbens (NAc) and ventral tegmental area (VTA), which are part of the mesolimbic dopamine pathway, a circuit orchestrating reward, reinforcement, and pain perception (Serafini et al., 2020). Sudden cessation of opioid treatment leads to withdrawal, which is a result of aberrant activation of MOR and noradrenergic receptors in several brain regions (Kreek and Koob, 1998; Weinshenker and Schroeder, 2007; Burma et al., 2017). RGS proteins also control a number of other GPCRs implicated in opioid addiction, including monoamine and metabotropic glutamate receptors (Terzi et al., 2009). In the next segment, we will discuss findings from rodent models on how exposure to opioids affects the expression of RGS proteins in the brain, as well as findings on the functional role of specific RGS in opioid-induced behaviors.
Role of RGS Proteins in Opioid Analgesia
MOR agonists such as morphine, oxycodone, and fentanyl have been used for the alleviation of acute and chronic pain (Busse et al., 2018; Klimas et al., 2019). Studies in the rodent brain have demonstrated that morphine and other opioids alter the association of certain RGS proteins with the MOR (Garzon et al., 2005; Han et al., 2010; Psifogeorgou et al., 2011). Evidence from several groups documents that opioids trigger brain region-specific changes in the expression of RGS proteins. Notably, some members of the RGS family, such as RGS9-2, operate in an agonist-dependent manner, whereas other proteins, such as RGSz1, act solely as negative modulators of MOR function. In the next section, we discuss in detail evidence on the role of specific RGS family members in the modulation of opioid actions.
RGS9-2.
RGS9-2, a member of the R7 subfamily, is enriched in the striatum (Rahman et al., 1999). It is found as part of multiprotein complexes containing the Gβ5 protein, the adaptor protein R7-binding protein (R7BP) (Masuho et al., 2013; Muntean and Martemyanov, 2016), and the scaffold spinophilin (Charlton et al., 2008; Bonsi et al., 2019). R7 subfamily members (RGS9-1, RGS9-2, RGS7, RGS6, and RGS11) bind to Gβ5 G protein through a G gamma-like domain, and this interaction is crucial for their stability and half-life (Sondek and Siderovski, 2001; Chen et al., 2003). In the brain, the interaction of R7 subfamily proteins with R7BP mediates their membrane localization (Masuho et al., 2013). Studies in striatal homogenates have revealed the potent role of RGS9-2 in MOR-agonist–induced inhibition of cyclic adenosine monophosphate (cAMP) formation (Xie et al., 2012).
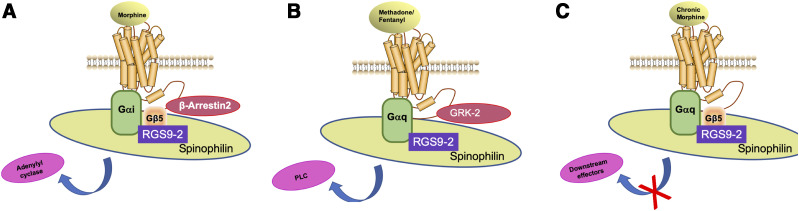
Research using genetically modified mice has demonstrated that RGS9-2 modulates the analgesic efficacy of opioids in an agonist-dependent mode. Specifically, behavioral analysis of RGS9 knockout (KO) mice in the hot plate assay has revealed that RGS9-2 negatively controls the analgesic efficacy of morphine (Zachariou et al., 2003). In contrast, deletion of RGS9-2 decreases the analgesic efficacy of methadone and fentanyl in the hot plate test (Psifogeorgou et al., 2011). Moreover, genetic inactivation of RGS9 does not affect the efficacy of acute oxycodone in the hot plate assay (Gaspari et al., 2017). This agonist-biased phenotype results from unique complexes formed between RGS9-2 and Gα subunits in the striatum (Fig. 2). Acute morphine treatment promotes the formation of complexes between RGS9-2, Gαi3 (and other Gαi subunits), Gβ5 (which stabilizes RGS9-2), and β-arrestin2 to eventually trigger inhibition of adenylyl cyclase (Fig. 2A) (Psifogeorgou et al., 2011). In contrast, chronic morphine promotes the formation of complexes between RGS9-2, Gαq, and Gβ5 that prohibit actions on downstream effectors (Fig. 2C) (Psifogeorgou et al., 2011). Notably, administration of either methadone or fentanyl favors the formation of complexes between RGS9-2, Gαq, and G protein–coupled receptor kinase 2 and the activation of phospholipase C (Fig. 2B) (Psifogeorgou et al., 2011). The above studies described in Psifogeorgou et al. (2011) and illustrated in Figure 2 assessed RGS9-2 complexes at doses that produced maximum analgesia in the hot plate assay and at a time point (30 minutes) at which maximal analgesia was observed (Psifogeorgou et al., 2011).
Fig. 2.
RGS9-2 forms distinct complexes in the striatum depending on the MOR agonist administered and the duration of treatment. (A) RGS9-2 complexes in the striatum following treatment with acute morphine. (B) RGS9-2 complexes in the striatum following treatment with methadone or fentanyl. (C) RGS9-2 complexes in the striatum following chronic morphine treatment (four consecutive days). GRK-2, G protein–coupled receptor kinase 2; PLC, phospholipase C. Figure summarizes findings from Psifogeorgou et al. (2011).
Notably, viral overexpression of RGS9-2 in the NAc attenuates the analgesic effects of morphine (Gaspari et al., 2014), suggesting a role of the brain reward pathway in the efficacy of opioid analgesics. Studies in rats using i.c.v administration of antisense oligodeoxynucleotides against RGS9 have also demonstrated increased antinociceptive potency of morphine and Ala2-MePhe4-Glyol5-enkephalin as well as prolonged duration of the analgesic response (Garzon, Rodriguez-Diaz et al., 2001).
RGS7.
RGS7 is present in several components of the reward pathway and shows wider overall distribution in the brain relative to RGS9-2. It is abundantly expressed in the locus coeruleus (LC), a noradrenergic nucleus with a prominent role in physical dependence, anxiety, and pain modulation (Gold et al., 1997, 2003). The functional role of RGS7 in opioid analgesia has been demonstrated using RGS7 KO mice. Constitutive deletion of the RGS7 gene increased morphine analgesia (Sutton et al., 2016). This phenotype involved RGS7 actions outside the striatum, as conditional deletion of RGS7 in striatal neurons did not affect the efficacy of morphine in the hot plate assay. There is no information on the role of RGS7 in analgesic responses to other opioids.
RGS4.
In vitro studies have demonstrated the role of the ubiquitin pathway in opioid-mediated actions and have provided evidence for crosstalk between opioid receptors and other GPCRs. Chronic treatment with either MOR or delta opioid receptor (DOR) agonists increases the breakdown of RGS4 protein by the ubiquitin-proteasome pathway. This opioid-mediated reduction in RGS4 expression selectively increases DOR and muscarinic receptor subtype three signaling (Wang and Traynor, 2011). Similar to RGS9-2, RGS4 modulates responses to opioid analgesics in an agonist-dependent fashion. Although knockout of the RGS4 gene does not affect the analgesic efficacy of morphine or the trajectory of morphine tolerance, prevention of RGS4 action decreases the analgesic efficacy of fentanyl and methadone in the hot plate assay. Several mechanisms may contribute to these agonist-dependent phenotypes, including the formation of GPCR dimers and the activation of distinct intracellular pathways. Coimmunoprecipitation assays have demonstrated that the abundance of RGS4-MOR complexes is decreased following fentanyl treatment and unaffected following morphine administration (Han et al., 2010). These findings further support the hypothesis that opioids trigger the formation of distinct MOR complexes with G proteins and other signal transduction components in the brain. It is also important to mention that pharmacological inhibition of RGS4 activity in certain brain regions may also modulate the antinociceptive actions of opioids. Recent studies have reported that microinjection of compound CCG-63802 in the ventrolateral periaqueductal gray (vlPAG) enhances the antinociceptive potency of morphine but has no effect on fentanyl’s actions in the same paradigm (Morgan et al., 2020). Although CCG-63802 is not a highly selective compound (Senese et al., 2020), these data suggest an agonist-dependent role of RGS4 in the modulation of opioid actions in the periaqueductal gray (PAG).
RGSz1.
RGSz1 is expressed in low amounts throughout the brain (Wang et al., 1998). Past in vitro work has identified a potent negative modulatory role of RGSz1 in MOR functional responses (Ajit et al., 2007). These studies have reported that RGSz1 may bind to Gαz or other inhibitory Gα subunits to negatively modulate pathways downstream of MOR, including cAMP and protein kinase C signaling. Our group demonstrated that prevention of RGSz1 action enhances the analgesic efficacy of opioids, such as morphine, methadone and fentanyl, in the hot plate assay (Gaspari et al., 2018). Thus, unlike RGS9-2 and RGS4 that act in an agonist-dependent manner, RGSz1 negatively regulates the analgesic efficacy of clinically prescribed opioid analgesics.
RGS19.
RGS19 [also known as Gα-interacting protein (GAIP)] negatively modulates MOR signaling and regulates several other intracellular events by forming complexes with GAIP-interacting protein N terminus and GAIP-interacting protein C terminus. Chronic treatment of SH-SY5Y cells with either MOR or DOR agonists increases RGS19 and GAIP-interacting protein C terminus protein levels via the protein kinase C/mitogen-activated protein kinase (MAPK) pathway, suggesting that these complexes may modulate the actions of several opioid receptors upon prolonged exposure to opioid analgesics (Wang and Traynor, 2013).
R7BP.
R7BP is localized in several analgesia and addiction circuits including the dorsal striatum, the NAc, the LC, and the hippocampus. Although R7 family proteins are regulated by morphine, the expression of R7BP remains unchanged after acute and chronic morphine administration. In the hot plate assay, R7BP KO mice show a greater analgesic response to low morphine doses compared with their wild-type controls (Terzi et al., 2012). Notably, knockout of R7BP affects the stability and expression of RGS9-2 in the striatum (Anderson et al., 2007).
RGS Insensitive Gαo Subunits.
RGS-insensitive Gαo subunits were created by a point mutation of G184S at the interface binding to RGS domains (Clark et al., 2003). Initial studies using knock-in mice expressing RGS-insensitive Gαo subunits have demonstrated that prevention of the interaction between RGS proteins and Gαo leads to enhanced endogenous opioid peptide activity (Lamberts et al., 2011). Using knock-in mice expressing RGS insensitive Gαo subunits, Lamberts et al. demonstrate that prevention of RGS modulation of Gαo enhances opioid supraspinal antinociception, whereas it attenuates spinal antinociceptive responses of opioid analgesics such as morphine and methadone (Lamberts et al., 2013). GIRK channels have been shown to play a role in modulating morphine antinociception. Recent studies have demonstrated reduced MOR-GIRK coupling in the vlPAG and LC as well as decreased MOR activation by Ala2-MePhe4-Glyol5-enkephalin and fentanyl in RGS-insensitive mice (McPherson et al., 2018). These experiments provide important insight on the consequences of loss of RGS modulation in Gαo signaling in responses to opioids.
A Role of RGS Proteins in Analgesic Tolerance
Evidence from in vitro and in vivo studies highlights the potent and selective role of RGS proteins in MOR desensitization and in mechanisms that contribute to analgesic tolerance to opioids. In vitro work in C6 cells has shown that endogenous RGS proteins inhibit opioid-induced desensitization and MOR downregulation, thereby controlling the actions of chronic agonist exposure (Clark et al., 2004; Clark and Traynor, 2005).
In accord with observations in models of analgesia, there is a brain region-specific role for RGS proteins in modulating analgesic tolerance. For example, RGS9-2 promotes morphine tolerance by actions in the NAc (Gaspari et al., 2014), whereas RGSz1 promotes morphine tolerance by actions in the vlPAG (Gaspari et al., 2018). These two RGS proteins act not only on distinct anatomic sites but also by distinct mechanisms, since RGS9-2 negatively modulates cAMP signaling whereas RGSz1 controls the transcriptional activity of beta-catenin. What is more noteworthy is that certain RGS proteins can have differing actions on the development of tolerance based on the opioid administered. For example, recent work from our group shows that prevention of RGS9 action delays the development of morphine tolerance, whereas it accelerates tolerance to oxycodone (Zachariou et al., 2003; Gaspari et al., 2017). Notably, studies using constitutive knockout mice suggest that RGS4 does not impact morphine tolerance (Han et al., 2010). Most behavioral studies on opioid tolerance focus on morphine as this drug promotes MOR desensitization and analgesic tolerance much faster than other clinically available opioids. However, it is still important to address the impact of RGS proteins in analgesic tolerance to other opioids.
RGS9-2.
Studies in striatal homogenates have documented that RGS9-2 controls cAMP signaling in response to dopamine or opioid receptor activation as well as sensitization and signaling kinetics of adenylyl cyclase 5 (Xie et al., 2012). In HEK237 cells, exposure to opioid results in the migration of RGS9-2 toward the cell membrane and increases the colocalization with MOR and β-arrestin-2. In support of the hypothesis stating that delayed MOR internalization contributes to analgesic tolerance, overexpression of RGS9-2 delays MOR endocytosis in response to agonist treatment (Psifogeorgou et al., 2007). Earlier work has also demonstrated the dynamic and region-specific regulation of R7 and RZ subfamily proteins as well as the role of Gβ5 and R7 proteins in opioid tolerance (Sanchez-Blazquez et al., 2003; Lopez-Fando et al., 2005). Consistent with findings on analgesic efficacy, RGS9-2 modulates opioid tolerance in an agonist-dependent manner. RGS9 KO or viral overexpression of a dominant-negative form of the protein in the NAc significantly delays analgesic tolerance to morphine (Zachariou et al., 2003; Gaspari et al., 2014). Conversely, overexpression of RGS9-2 in the NAc accelerates morphine tolerance, suggesting that maladaptive plasticity within the brain reward center is sufficient to decrease responsiveness to opioid analgesics (Gaspari et al., 2014). On the contrary, RGS9-2 actions protect from analgesic tolerance to oxycodone, as RGS9 KO mice develop tolerance during time points at which their wild-type counterparts still respond to the drug. This phenotype has been observed in the hot plate assay and in models on neuropathic pain in which oxycodone was used to alleviate mechanical allodynia (Gaspari et al., 2017).
RGS7.
The role of RGS7 in analgesic tolerance to morphine was similar to that of RGS9-2. Sutton and colleagues used the hot plate assay to demonstrate that constitutive RGS7 KO mice show delayed tolerance to morphine compared with their wild-type counterparts (Sutton et al., 2016). Striatal deletion of RGS7 was not sufficient to delay tolerance to morphine in the hot plate assay. Similar to the phenotypes observed in the analysis of RGS9 KO and RGS4 KO mice, RGS7 KO mice exhibit a more severe naloxone precipitated withdrawal compared with the control wild-type mice (Sutton et al., 2016). This phenotype is likely related to RGS7 actions in the LC, as striatal knockout of RGS7 did not affect the severity of precipitated naloxone withdrawal.
RGS4.
In vitro studies using lentiviral delivery of small hairpin RNA (shRNA) to specifically downregulate RGS4 protein in human neuroblastoma SH-SY5Y cells demonstrate that RGS4 differentially regulates MOR and DOR signaling. Indeed, although downregulation of RGS4 has no effect on forskolin-stimulated cAMP accumulation and MAPK activity in response to MOR activation, it facilitates inhibition of cAMP accumulation and MAPK activity following treatment with DOR agonists (Wang et al., 2009). In accord with these findings, RGS4 KO does not affect morphine tolerance. In spite of the abundant expression of RGS4 in several analgesia circuits including the spinal cord, the LC, and other midbrain and thalamic structures, studies using a 4-day hot plate assay show that knockout of RGS4 does not affect tolerance to morphine (Han et al., 2010). The impact of RGS4 in tolerance to other opioids used for pain management remains unknown.
RGSz1.
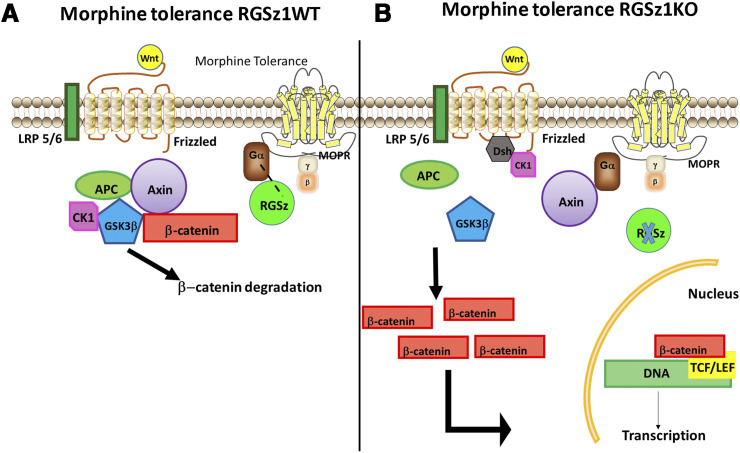
RGSz1 protein levels were downregulated in the mouse PAG 30 minutes after acute morphine administration and upregulated following a 4-day administration regimen that leads to analgesic tolerance. A similar pattern of regulation was observed for the Gαz subunit (Gaspari et al., 2018). The levels of RGSz1 protein remain unaffected by morphine tolerance in the NAc, the dorsal striatum, and the thalamus, highlighting the regional specificity of RGSz1 actions. Using a 5-day hot plate tolerance paradigm, we showed that constitutive inactivation of the RGSz1 gene delays the development of tolerance in both male and female mice. A similar role of RGSz1 in analgesic tolerance has been observed when morphine was used to alleviate thermal hypersensitivity in mice suffering from prolonged peripheral inflammation (Gaspari et al., 2018). RGSz1 actions in the vlPAG greatly contribute to this phenotype, as conditional downregulation of RGSz1 (by use of Cre-expressing adeno-associated vectors and a floxed RGSz1 mouse line) is sufficient to delay morphine tolerance. By combining brain biochemistry, next-generation RNA sequencing, and bioinformatic analysis, we found that RGSz1 in the mouse vlPAG promotes the development of morphine tolerance by antagonizing the transcriptional activity of β-catenin. β-catenin may promote the expression of genes necessary to restore MOR signaling and synaptic maladaptations associated with analgesic tolerance. The nuclear translocation of β-catenin is controlled by cytoplasmic multiprotein complexes, which contain the protein Axin-2 (Huang and He, 2008). Axin-2 also contains an RGS domain and may bind to activated Gα subunits (Castellone et al., 2005; Stemmle et al., 2006; Egger-Adam and Katanaev, 2010; Gaspari et al., 2018). Under states of tolerance, RGSz1 levels increase in the PAG as does the abundance of complexes between Gαz and MOR (Fig. 3). Knockout of RGSz1 permits the association of Axin-2 with Gαz, an effect that leads to dissociation of the destruction complex and the translocation of β-catenin to the nucleus (Gaspari et al., 2018) (Fig. 3). Future work will determine whether this mechanism is observed in other analgesia or addiction circuits and will further elucidate the role of RGSz1 and Gαz actions in MOR-expressing circuits.
Fig. 3.
Schematic summarizing the role of RGSz1 in modulating Axin2-controlled β-catenin complexes in models of morphine tolerance. (A) Morphine tolerance promotes the association of Gαz with RGSz1 in the PAG of RGSz1 wild-type mice. Axin2 complexes (destruction complexes) prevent the nuclear translocation of β-catenin and the transcription of genes counteracting morphine tolerance. (B) Knockout of RGSz1 permits the formation of Axin2-Gαz complexes, the dissociation of destruction complexes and the translocation of β-catenin to the nucleus. APC, XXXX; CK1, XXXX; LEF, XXXX; LRP, XXXX; MOPR, XXXX; RGSz1WT, RGSz1 wild type; TCF, XXXX. Figure summarizes findings from Gaspari et al. (2018).
R7BP.
The interacting partner of R7 RGS protein members, R7BP, mediates the docking of RGS proteins to the cell membrane (Muntean and Martemyanov, 2016). Inactivation of R7BP affects the function of both RGS7 and RGS9-2 in the striatum (Anderson et al., 2009). Consistent with the observations on mice lacking the RGS9 or the RGS7 genes, R7BP KO mice show delayed development of morphine tolerance (Terzi et al., 2012).
A Role of RGS Proteins in Opioid Reward and Addiction-Related Behaviors
As mentioned earlier, several RGS proteins are expressed in the mesolimbic dopamine system, controlling functional responses of GPCRs mediating reward, motivation, habit formation, and impulsivity. Among them, RGS9-2, RGS7, and RGS4 have documented roles in the intracellular signaling of dopamine and opioid receptors and have been the focus of many laboratories studying drug addiction. Most of the information on the functional role of RGS family members on opioid addiction-related behaviors derives from rodent studies using local overexpression, constitutive, and conditional gene knockdown approaches.
RGS9-2.
RGS9-2 complexes in the striatum are dynamically regulated by psychoactive drugs, antiparkinsonian agents, and pathologic conditions (Rahman et al., 2003; Gold et al., 2007).
Constitutive inactivation of the RGS9 gene results in a 10-fold increase in the rewarding sensitivity of morphine, as assessed by the place conditioning paradigm (Zachariou et al., 2003). Conversely, viral overexpression of RGS9-2 in the NAc decreases the rewarding efficacy of morphine (Gaspari et al., 2014). The agonist-dependent modulatory role of RGS9-2 reported in models of analgesia has also been observed in reward-assessing behaviors. Thus, RGS9 KO mice are less sensitive to the rewarding effects of oxycodone in the conditioned place preference paradigm, and they do not develop locomotor sensitization in response to repeated oxycodone treatment (Gaspari et al., 2017). Moreover, RGS9-2 positively regulates the extinction and reinstatement of oxycodone place preference (Gaspari et al., 2017). RGS9-2 has a similar function under neuropathic pain states, as RGS9 KO mice suffering from peripheral nerve injury show decreased sensitivity to the rewarding actions of oxycodone (Gaspari et al., 2017).
RGS7.
RGS7 KO mice show increased sensitivity to the rewarding and reinforcing effects of morphine. These actions are mediated by striatal medium spiny neurons, as conditional inactivation of RGS7 in striatal neurons recapitulates this phenotype. RGS7 KO mice also show enhanced locomotor activation in response to morphine treatment (Sutton et al., 2016). In accord with the findings from the analysis of RGS9 KO and RGS7 KO mice, R7BP KO mice show increased sensitivity to the locomotor stimulating effects of morphine, as they respond to lower doses than those required to promote locomotion in their wild-type littermates (Terzi et al., 2012).
RGSz1.
RGSz1 KO mice show decreased sensitivity to the rewarding effects of morphine in the conditioned place preference test compared with their wild type counterparts. Furthermore, RGSz1 KO mice do not develop locomotor sensitization in response to repeated morphine treatment (Gaspari et al., 2018). We recently found that downregulation of RGSz1 in the vlPAG recapitulates the decreased morphine reward sensitivity phenotype, whereas downregulation of RGSz1 in the NAc does not affect the rewarding efficacy of morphine (Sakloth et al., 2019). Since RGSz1 contraregulates the rewarding versus the analgesic efficacy of opioids, interventions in RGSz1 pathways activity may be applied to optimize the actions of opioids.
RGS4.
Global knockout or conditional knockdown of RGS4 in the adult mouse NAc leads to a small but significant increase in the rewarding efficacy morphine in the place preference paradigm (Han et al., 2010; Kim et al., 2018). Furthermore, RGS4 KO mice show enhanced locomotor sensitization in response to morphine (Han et al., 2010). More recently, Kim et al. applied shRNA methodology to downregulate RGS4 in the mouse NAc to investigate the mechanisms by which RGS4 modulates responses to opioids. This work demonstrates that RGS4 modulates morphine reward by controlling the phosphorylation of ionotropic glutamate receptors in the NAc (Kim et al., 2018).
R7BP.
The role of R7BP in modulating opioid rewarding actions has not been fully examined. Terzi and colleagues reported that knockout of R7BP enhances the locomotor sensitizing effects to morphine (Terzi et al., 2012), similar to what was observed with RGS9 and RGS7 knockout mice. It will be interesting to investigate the regional effects of R7BP in the rewarding actions of synthetic opioids.
A Role of RGS Proteins in Opioid Physical Dependence
There is a wealth of evidence on the regulation of RGS proteins in the brain by prolonged exposure to opioids. RGS2 and RGS4 are present in several brain regions implicated in physical dependence including the LC, the VTA, the prefrontal cortex, and the amygdala (Gold et al., 1997; Terzi et al., 2009). Studies by Gold et al. in the LC of morphine-dependent rats have demonstrated that RGS2 and RGS4 mRNA levels increased 2–3-fold at 6 hours following naloxone treatment, suggesting that these molecules control the aberrant firing of noradrenergic neurons upon precipitated opioid withdrawal. Notably, RGS3, 5, 7, 8, and 11 remain unchanged in the LC following chronic morphine administration (Gold et al., 2003). The levels of RGS4 protein are also increased 2-fold in the LC following chronic morphine treatment, and this effect may play a protective role against physical dependence (Gold et al., 2003). The analysis of RGS4 KO mice supports the hypothesis that RGS4 negatively controls the firing of LC neurons, as RGS4 KO mice show exacerbated signs of morphine withdrawal compared with their wild-type controls. Patch clamp recordings from slices of drug-naive and morphine-dependent mice reveal that inactivation of the RGS4 gene increases the firing sensitivity of LC neurons only in morphine dependence states (Han et al., 2010).
In addition to RGS4, the striatal enriched RGS9-2 also plays a prominent role in the modulation of physical dependence to morphine. Prevention of RGS9-2 action exacerbates behavioral and biochemical manifestations of morphine withdrawal (Zachariou et al., 2003). Unexpectedly, although precipitated oxycodone withdrawal produces similar symptoms to those observed with morphine withdrawal, knockout of the RGS9 gene did not affect the severity of oxycodone withdrawal symptoms (Gaspari et al., 2017). Notably, knockout of R7BP has no effect on morphine withdrawal (Terzi et al., 2012). In an in vitro model of adenylyl cyclase super sensitization that corresponds to withdrawal in animals, cells expressing RGS insensitive Gαo subunits demonstrated increased supersensitivity to adenylyl cyclase (Clark et al., 2004). These differences further highlight the distinct intracellular pathways triggered by opioid analgesics with high abuse potential. Thus, patients carrying polymorphisms affecting specific signaling pathways may be more vulnerable to certain opioids. Interestingly, downregulation of RGSz1 in the mouse vlPAG did not affect symptoms of naloxone-precipitated withdrawal. This is important information, as inhibition of RGSz1 pathways may provide a way to enhance the analgesic efficacy of opioids without impacting physical dependence (Gaspari et al., 2018). Table 1 summarizes the opioid-related phenotypes associated with genetic interventions in the expression of RGS proteins.
TABLE 1.
Summary of reports on opioid behavioral phenotypes observed with different RGS mutant lines in models of analgesia and addiction
| RGS subtype | Opioid | Genetic intervention | Phenotype | References | ||
|---|---|---|---|---|---|---|
| Analgesia and tolerance | Reward | Withdrawal | ||||
| RGS9-2 | Morphine | Constitutive KO | Increased analgesia, delayed tolerance (hot plate assay) | Increased (CPP) | Exacerbated naloxone precipitated withdrawal | Zachariou et al., 2003 |
| RGS9-2 | Morphine | AAV-RGS9-2 NAc overexpression | Decreased analgesia, accelerated tolerance (hot plate assay) | Decreased (CPP) | Ameliorated naloxone withdrawal | Gaspari et al., 2014 |
| RGS9-2 | Morphine | AAV-DEPless RGS9-2 NAc overexpression | Delayed tolerance (hot plate assay) | — | Exacerbated naloxone precipitated withdrawal | Gaspari et al., 2014 |
| RGS9-2 | Methadone, fentanyl | Constitutive KO | Decreased analgesia (hot plate assay) | — | — | Psifogeorgou et al., 2011 |
| RGS9-2 | Oxycodone | Constitutive KO | No change in analgesic efficacy, accelerated tolerance (hot plate assay) | Decreased (CPP) | No effect on naloxone precipitated withdrawal | Gaspari et al., 2017 |
| RGS7 | Morphine | Constitutive KO | Increased analgesia, delayed tolerance (hot plate assay) | Increased (CPP) | Exacerbated naloxone precipitated withdrawal | Sutton et al., 2016 |
| RGS7 | Morphine | Conditional striatal KD | No change in analgesic efficacy or tolerance (hot plate assay) | Increased (CPP) | No effect on naloxone precipitated withdrawal | Sutton et al., 2016 |
| RGS4 | Methadone, Fentanyl | Constitutive KO | Decreased analgesia (hot plate assay) | — | — | Han et al., 2010 |
| RGS4 | Morphine | Constitutive KO and conditional NAc KD | No change in analgesic efficacy or tolerance (hot plate assay) | Increased (CPP) | Exacerbated naloxone precipitated withdrawal | Han et al., 2010 |
| RGS4 | Fentanyl | RGS4 antagonist CCG-63802 in vlPAG | No change in analgesic efficacy (hot plate assay) | — | — | Morgan et al., 2019 |
| RGS4 | Morphine | RGS4 antagonist CCG-63802 in vlPAG | Increased analgesia (hot plate assay) | — | — | Morgan et al., 2019 |
| RGSz1 | Morphine | Constitutive KO | Increased analgesia, delayed tolerance (hot plate assay) | Decreased (CPP) | Gaspari et al., 2018 | |
| RGSz1 | Methadone, Fentanyl | Constitutive KO | Increased analgesia (hot plate assay) | — | — | Gaspari et al., 2018 |
| RGSz1 | Morphine | Conditional vlPAG KD | Delayed tolerance (hot plate assay) | Decreased (CPP) | No effect on naloxone precipitated withdrawal | Gaspari et al., 2018 |
| R7BP | Morphine | Constitutive KO | Increased analgesia, delayed tolerance (hot plate assay) | Increased locomotor sensitizing effects | No effect on naloxone precipitated withdrawal | Terzi et al., 2012 |
| RGS-insensitive Gαo | Morphine | Constitutive knock-in | Increased analgesia (supraspinal, hot plate assay) | — | Lamberts et al., 2013 | |
| RGS-insensitive Gαo | Morphine | Constitutive knock-in | Decreased analgesia (spinal, tail-withdrawal) | — | — | Lamberts et al., 2013 |
AAV, XXXX; CPP, XXXX; DEPless, XXXX; KD, knockdown.
So far, only a subset of the RGS proteins expressed in nociceptive circuits have been investigated for their role in opioid actions. Based on evidence on the unique role RGS9-2, RGS7, RGSz1, and RGS4 in opioid actions (Table 1) and the wide expression of MORs and RGS proteins in the spinal cord and brain subregions, it is essential to complete our understanding on how various members of the RGS family affect MOR signaling and desensitization. It will also be important to further characterize the RGS/Gα-protein complexes with MOR that are formed in response to acute and repeated opioid administration in central and in peripheral sites and to determine the cell-type specificity of these events. For example, coimmunoprecipitation assays highlight changes in the abundance of MOR complexes with both RGS9-2 and RGS4 in response to fentanyl administration; however, it is unknown if these events happen in parallel, if they occur in distinct cellular populations, or if they take place in dendritic versus somatic sites of the neurons. Such information will complement our knowledge on signal transduction events associated with short-term versus prolonged exposure to opioids. Another gap in our knowledge involves the signal transduction triggered by opioids under states of chronic pain, major depression, and other chronic debilitating disorders. This knowledge will guide drug development efforts toward more targeted interventions for pain management. Most studies have focused on evaluating RGS’s roles in morphine actions, and we still need a better understanding of how RGS proteins modulate the actions of commonly prescribed opioids, such as fentanyl, methadone, and oxycodone, as well as the actions of highly abused drugs such as heroin.
RGS Proteins and Psychostimulant Actions
Similar to observations with opioids, addiction to psychostimulants has been a health burden worldwide (Degenhardt et al., 2014). Psychostimulants trigger an increase in monoamine levels in several brain circuits including the brain reward pathway. Changes in dopamine and other monoamine receptor activity in response to psychostimulant exposure contribute to maladaptive plasticity associated with addiction (Kreek et al., 2012). Over the years, a number of studies documented changes in RGS protein expression in models of psychostimulant addiction, whereas genetic mouse models reveal a powerful role of specific RGS protein members in sensitivity to psychostimulant-induced behaviors.
In fact, psychostimulants may differentially regulate RGS transcripts depending on the brain region and the drug administration regimen. These studies indicate that members of the RGS family primarily control the actions of psychostimulants by modulating functional responses to D1 and D2 dopamine receptors in the striatum (Burchett et al., 1999; Rahman et al., 2003; Stanwood et al., 2006). As psychostimulant addiction involves multiple circuits and neurochemical pathways, it is expected that RGS proteins affect several other GPCRs in addiction circuitry. In fact, evidence from rat models of addiction shows that RGS proteins also affect the actions of psychostimulants by modulating the activity of metabotropic glutamate receptor 5 (mGluR5) (Schwendt and McGinty, 2007; Schwendt et al., 2012).
Cocaine-Induced Changes in RGS Expression
Preclinical evidence from various laboratories highlights the dynamic regulation of RGS proteins in response to psychostimulant exposure. Studies in rodents using acute and chronic cocaine administration regimens reveal a treatment- and brain region– specific regulation of RGS family members. Members of the R4 family appear to show distinct and often opposite patterns of expression upon exposure to cocaine. Acute cocaine administration triggers an increase in RGS4 mRNA levels in the NAc and in the dorsal central gray, as well as a reduction of RGS4 expression in the LC and in the reticulotegmental nucleus (Bishop et al., 2002). Similar to RGS4, RGS2 mRNA levels increase by 2-fold in the rat striatum following acute administration of cocaine (Burchett et al., 1999). On the other hand, chronic cocaine intake leads to a reduction in RGS4 expression in the striatum. RGS4 expression levels were downregulated in the rat caudate putamen following binge cocaine administration (Yuferov et al., 2003). Later studies also documented that cocaine self-administration increased RGS4 protein levels in the striatum, and this effect was reversed when rats were exposed to the cocaine-paired environment (McGinty et al., 2008). Notably, chronic cocaine treatment followed by abstinence and then administration of a challenge cocaine dose promotes the expression of RGS4 transcript in the LC and in the dorsal central gray; however, it decreases in RGS4 mRNA levels in the red nucleus and in the reticulotegmental nucleus (Bishop et al., 2002). Cocaine abstinence in rodents leads to a reduction in RGS4 and RGS2 expression in the dorsal striatum (Schwendt et al., 2007; Bilodeau and Schwendt, 2016). Cocaine withdrawal does not alter RGS4 or RGS7 levels in the amygdala, the frontal cortex, or the hypothalamic paraventricular nucleus of rats (Carrasco et al., 2003). Repeated exposure to cocaine has no effect on RGSz1 transcript in the rat hypothalamic paraventricular nucleus (Carrasco et al., 2004). As expected, the striatal enriched RGS9-2 protein is highly implicated in processes mediating cocaine addiction. Repeated exposure to cocaine for 7 days produces a significant increase in RGS9-2 protein levels in the NAc (Rahman et al., 2003).
A Role of RGS Proteins in Cocaine Addiction-Related Behaviors
Studies from several laboratories have addressed the functional role of RGS proteins in behavioral responses to cocaine. So far studies using genetically modified rodents have investigated the impact of RGS4, RGS2, RGS9-2, RGS12, and R7BP in behavioral responses to cocaine. RGS4 KO mice show decreased sensitivity to the rewarding effects of cocaine in the place preference assay in a sex-dependent manner (Rorabaugh et al., 2018). The mechanism of RGS4 action involves modulation of D2-receptor signaling in the NAc and other regions in the reward pathway (Min et al., 2012; Rorabaugh et al., 2018). In contrast, RGS2 has no effect on the rewarding actions of cocaine.
Knockout of RGS9 heightened the rewarding effects of cocaine, whereas overexpression of RGS9-2 in the NAc diminished cocaine’s locomotor activating effects. These actions of RGS9-2 are, at least in part, mediated through the dopamine D2 receptor subtype (Rahman et al., 2003). Indeed, expression of RGS9-2 in Xenopus oocytes accelerated the off kinetics of the D2 receptor–induced GIRK currents (Rahman et al., 2003). Dopamine signaling in the striatum is regulated by both RGS7 and RGS9 and by their binding to R7BP. Surprisingly, R7BP KO mice do not show any cocaine locomotor stimulation or locomotor sensitization phenotype, suggesting that RGS7 and RGS9-2 have distinct modulatory roles in the brain reward pathway (Anderson et al., 2010).
Amphetamine-Induced Changes in RGS Expression
Studies using rat models of amphetamine intake reveal a brain region– and receptor subtype–dependent pattern of RGS protein regulation. Notably, changes in RGS protein expression correlate with amphetamine intake. Single injections of amphetamine and methamphetamine increase RGS2 mRNA levels in the rat striatum by 4-fold, and these effects are prevented by pretreatment with the D1 antagonist SCH-23390 (Burchett et al., 1999). Five days of amphetamine self-administration promote RGS2 transcript expression in the midbrain, increase trafficking of RGS2 to the membrane, and disrupt the interaction between D2/D3 dopamine receptor subtypes and Gαi2 in the midbrain, ultimately leading to increased dopamine release (Calipari et al., 2014).
In ovariectomized rats, administration of 17-β-estradiol for 2 weeks enhances amphetamine-induced place preference and increases the levels of RGS9-2 protein in the NAc shell (Silverman and Koenig, 2007). Furthermore, RGS9-2 levels are decreased in the striatum of amphetamine-sensitized rats relative to their controls (Seeman et al., 2007).
In the NAc, RGS2 and RGS4 transcripts are upregulated after amphetamine self-administration, and their levels are positively correlated with amphetamine intake. Furthermore, there is a reduction of RGS7 and RGS8 in the NAc, but there was no correlation with amphetamine intake (Sun et al., 2015). Studies by Sun et al. show distinct patterns of RGS mRNA regulation in the rat VTA and NAc following amphetamine self-administration. Specifically, in the VTA, RGS2, RGS4, and RGS20 transcripts are upregulated, whereas RGS9, RGS10, and RGS17 mRNA levels are downregulated. Furthermore, the changes in RGS2, RGS4, RGS10, and RGS20 mRNA levels in the VTA are positively correlated with amphetamine intake (Sun et al., 2015).
Chronic amphetamine treatment followed by abstinence and then by an amphetamine challenge leads to a decrease in RGS4 protein levels in the dorsal striatum and in the NAc. Studies have additionally shown that RGS4 mRNA levels are downregulated in a time-dependent manner in rat forebrain regions following treatment with low doses of amphetamine (Schwendt et al., 2006). These actions of RGS4 involve interactions with the mGluR5 subtype in the striatum (Schwendt et al., 2007, 2012). Indeed, overexpression of RGS4 in the rat dorsal striatum, by use of herpes simplex virus–RGS4 viral vectors, augmented amphetamine-induced horizontal activity by modulating the activity of mGluR5 receptors.
A Role of RGS Proteins in Amphetamine Addiction-Related Behaviors
There is limited information on the functional role of RGS proteins in amphetamine addiction. So far, there is evidence on the role of RGS9-2 and RGS12 in mediating amphetamine’s actions. RGS9-2 is a negative modulator of the locomotor activating effects of several psychostimulants, including amphetamine (Rahman et al., 2003). RGS9 KO mice show increased locomotion in response to low doses of amphetamine compared with their wild-type littermates (Rahman et al., 2003; Walker et al., 2015). Walker and colleagues suggest that this phenotype is more prominent in female RGS9 KO mice, which show greater amphetamine-induced locomotion relative to male RGS9 KO mice (Walker et al., 2015).
RGS12 also appears to play a unique role in the actions of psychostimulants as RGS12 knockout mice show decreased amphetamine and cocaine-mediated hyperlocomotion; however, they do not show any deficits in locomotor sensitization and place preference (Gross et al., 2018). These actions of RGS12 were found to involve regulation of the dopamine transporter in the ventral striatum. It will be interesting to determine if RGS12 plays a role in other addiction-related behaviors or in the development of psychostimulant dependence.
In summary, evidence from multiple laboratories and addiction models highlights the highly specific and dynamic regulation of RGS protein/mRNA levels following exposure to cocaine or amphetamine. Furthermore, studies with constitutive knockout mice point to specific RGS subtypes as factors that affect vulnerability to psychostimulant addiction. Although it is speculated that the mechanism by which RGS proteins affect psychostimulant actions involves modulation of dopamine receptors in the striatum, additional receptors and intracellular pathways are likely involved. It will be essential to understand the role of RGS proteins in neuroanatomical circuits, cell types, and receptors associated with acute and repeated exposure to psychostimulants. It will also be important to delineate the functional role of additional RGS family members in psychostimulant addiction and to identify polymorphisms in patients suffering from drug abuse disorders.
Future Directions
As highlighted in this review, RGS proteins have distinct and nonoverlapping roles in addiction and analgesia. It will be important to understand how drugs of abuse modulate the expression and activity of RGS family members in a cell-type specific manner. Receptor signaling appears to differ in somatic versus axonal or dendritic segments and it is possible that RGS proteins have neuronal compartment-specific roles. It will also be essential to dissect the neuronal circuits affected by changes in the activity of RGS proteins. Regarding strategies to optimize the actions of opioid analgesics, targeting selective RGS pathways provides a way for interventions in analgesia circuits without impacting the actions of opioids in addiction hubs. On the other hand, interventions in RGS protein activity in the brain reward center may provide novel drug addiction treatment avenues. Given the critical role of several members of the RGS family in analgesia and addiction-related behaviors, an important next milestone involves the application of genetic studies for the identification of polymorphisms associated with addiction vulnerability.
These new insights from basic research complement our knowledge of the mechanisms underlying receptor signaling and desensitization, and they may guide pain and addiction therapeutics toward new targets. As indicated by findings discussed in this review, a therapeutic intervention may require inhibition or promotion of RGS function. Although the developments of highly selective RGS inhibitors remains a challenge (Senese et al., 2020), novel technologies (shRNA, clustered regularly interspaced short palindromic repeats) as well as detailed knowledge of the downstream pathways and protein complexes affected by RGS proteins may provide promising directions for therapeutic interventions.
Abbreviations
- DOR
delta opioid receptor
- GAIP
Gα-interacting protein
- GIRK
G protein–activated inwardly rectifying K+
- GPCR
G-protein coupled receptor
- KO
knockout
- LC
locus coeruleus
- MAPK
mitogen-activated protein kinase
- mGluR5
metabotropic glutamate receptor 5
- MOR
mu opioid receptor
- NAc
nucleus accumbens
- PAG
periaqueductal gray
- R7BP
R7-binding protein
- RGS
regulator of G protein signaling
- shRNA
small hairpin RNA
- vlPAG
entrolateral periaqueductal gray
- VTA
ventral tegumental area
Authorship Contributions
Collected literature, put together figures and tables and wrote the review: Sakloth, Polizu, Bertherat, Zachariou.
Footnotes
This study was supported by the National Institute of Neurologic Disorders and Stroke [NS086444 (V.Z.), NS111351 (V.Z.)] and the National Institute on Drug Abuse [PPG-POIDA08227 (V.Z.)]. The authors declare no competing financial interest.
References
- Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. (2006) RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal 18:579–591. [DOI] [PubMed] [Google Scholar]
- Ajit SK, Ramineni S, Edris W, Hunt RA, Hum WT, Hepler JR, Young KH. (2007) RGSZ1 interacts with protein kinase C interacting protein PKCI-1 and modulates mu opioid receptor signaling. Cell Signal 19:723–730. [DOI] [PubMed] [Google Scholar]
- Alqinyah M, Maganti N, Ali MW, Yadav R, Gao M, Cacan E, Weng HR, Greer SF, Hooks SB. (2017) Regulator of G Protein signaling 10 (Rgs10) expression is transcriptionally silenced in activated microglia by histone deacetylase activity. Mol Pharmacol 91:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstadter AB, Koenen KC, Ruggiero KJ, Acierno R, Galea S, Kilpatrick DG, Gelernter J. (2009) Variant in RGS2 moderates posttraumatic stress symptoms following potentially traumatic event exposure. J Anxiety Disord 23:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Cao Y, Davidson S, Truong HV, Pravetoni M, Thomas MJ, Wickman K, Giesler GJ, Jr, Martemyanov KA. (2010) R7BP complexes with RGS9-2 and RGS7 in the striatum differentially control motor learning and locomotor responses to cocaine. Neuropsychopharmacology 35:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Martemyanov KA. (2009) Changes in striatal signaling induce remodeling of RGS complexes containing Gbeta5 and R7BP subunits. Mol Cell Biol 29:3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Semenov A, Pravetoni M, Posokhova EN, Song JH, Uversky V, Chen CK, Wickman K, Martemyanov KA. (2007) Expression and localization of RGS9-2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci 27:14117–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 86:445–452. [DOI] [PubMed] [Google Scholar]
- Bilodeau J, Schwendt M. (2016) Post-cocaine changes in regulator of G-protein signaling (RGS) proteins in the dorsal striatum: relevance for cocaine-seeking and protein kinase C-mediated phosphorylation. Synapse 70:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GB, Cullinan WE, Curran E, Gutstein HB. (2002) Abused drugs modulate RGS4 mRNA levels in rat brain: comparison between acute drug treatment and a drug challenge after chronic treatment. Neurobiol Dis 10:334–343. [DOI] [PubMed] [Google Scholar]
- Bonsi P, Ponterio G, Vanni V, Tassone A, Sciamanna G, Migliarini S, Martella G, Meringolo M, Dehay B, Doudnikoff E, et al. (2019) RGS9-2 rescues dopamine D2 receptor levels and signaling in DYT1 dystonia mouse models. EMBO Mol Med 11:e9283 DOI: 10.15252/emmm.201809283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhamdan M, Yan HD, Yan XH, Bannon MJ, Andrade R. (2006) Brain-specific regulator of G-protein signaling 9-2 selectively interacts with alpha-actinin-2 to regulate calcium-dependent inactivation of NMDA receptors. J Neurosci 26:2522–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch MR, Hepler JR. (2017) Endogenous RGS14 is a cytoplasmic-nuclear shuttling protein that localizes to juxtanuclear membranes and chromatin-rich regions of the nucleus. PLoS One 12:e0184497 DOI: 10.1371/journal.pone.0184497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchett SA, Bannon MJ, Granneman JG. (1999) RGS mRNA expression in rat striatum: modulation by dopamine receptors and effects of repeated amphetamine administration. J Neurochem 72:1529–1533. [DOI] [PubMed] [Google Scholar]
- Burma NE, Kwok CH, Trang T. (2017) Therapies and mechanisms of opioid withdrawal. Pain Manag 7:455–459. [DOI] [PubMed] [Google Scholar]
- Busse JW, Wang L, Kamaleldin M, Craigie S, Riva JJ, Montoya L, Mulla SM, Lopes LC, Vogel N, Chen E, et al. (2018) Opioids for chronic noncancer pain: a systematic review and meta-analysis. JAMA 320:2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calipari ES, Sun H, Eldeeb K, Luessen DJ, Feng X, Howlett AC, Jones SR, Chen R. (2014) Amphetamine self-administration attenuates dopamine D2 autoreceptor function. Neuropsychopharmacology 39:1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco GA, Barker SA, Zhang Y, Damjanoska KJ, Sullivan NR, Garcia F, D’souza DN, Muma NA, van De Kar LD. (2004) Estrogen treatment increases the levels of regulator of G protein signaling-Z1 in the hypothalamic paraventricular nucleus: possible role in desensitization of 5-hydroxytryptamine1A receptors. Neuroscience 127:261–267. [DOI] [PubMed] [Google Scholar]
- Carrasco GA, Zhang Y, Damjanoska KJ, D’Souza DN, Garcia F, Battaglia G, Muma NA, Van de Kar LD. (2003) A region-specific increase in Galphaq and Galpha11 proteins in brains of rats during cocaine withdrawal. J Pharmacol Exp Ther 307:1012–1019. [DOI] [PubMed] [Google Scholar]
- Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. (2005) Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science 310:1504–1510. [DOI] [PubMed] [Google Scholar]
- Chang PY, Qin L, Zhao P, Liu ZY. (2015) Association of regulator of G protein signaling (RGS5) gene variants and essential hypertension in Mongolian and Han populations. Genet Mol Res 14:17641–17650. [DOI] [PubMed] [Google Scholar]
- Charlton JJ, Allen PB, Psifogeorgou K, Chakravarty S, Gomes I, Neve RL, Devi LA, Greengard P, Nestler EJ, Zachariou V. (2008) Multiple actions of spinophilin regulate mu opioid receptor function. Neuron 58:238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. (2003) Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci USA 100:6604–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Jin Q, Liu X, Xu L, He X, Shen Y, Zhou Q, Zhang J, Jin M. (2017) miR-203 inhibits cell proliferation, invasion, and migration of non-small-cell lung cancer by downregulating RGS17. Cancer Sci 108:2366–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Harrison C, Zhong H, Neubig RR, Traynor JR. (2003) Endogenous RGS protein action modulates mu-opioid signaling through Galphao. Effects on adenylyl cyclase, extracellular signal-regulated kinases, and intracellular calcium pathways. J Biol Chem 278:9418–9425. [DOI] [PubMed] [Google Scholar]
- Clark MJ, Neubig RR, Traynor JR. (2004) Endogenous regulator of G protein signaling proteins suppress Galphao-dependent, mu-opioid agonist-mediated adenylyl cyclase supersensitization. J Pharmacol Exp Ther 310:215–222. [DOI] [PubMed] [Google Scholar]
- Clark MJ, Traynor JR. (2005) Endogenous regulator of g protein signaling proteins reduce mu-opioid receptor desensitization and down-regulation and adenylyl cyclase tolerance in C6 cells. J Pharmacol Exp Ther 312:809–815. [DOI] [PubMed] [Google Scholar]
- Dave RH, Saengsawang W, Yu JZ, Donati R, Rasenick MM. (2009) Heterotrimeric G-proteins interact directly with cytoskeletal components to modify microtubule-dependent cellular processes. Neurosignals 17:100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt L, Baxter AJ, Lee YY, Hall W, Sara GE, Johns N, Flaxman A, Whiteford HA, Vos T. (2014) The global epidemiology and burden of psychostimulant dependence: findings from the Global Burden of Disease Study 2010. Drug Alcohol Depend 137:36–47. [DOI] [PubMed] [Google Scholar]
- Dusonchet J, Li H, Guillily M, Liu M, Stafa K, Derada Troletti C, Boon JY, Saha S, Glauser L, Mamais A, et al. (2014) A Parkinson’s disease gene regulatory network identifies the signaling protein RGS2 as a modulator of LRRK2 activity and neuronal toxicity. Hum Mol Genet 23:4887–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger-Adam D, Katanaev VL. (2010) The trimeric G protein Go inflicts a double impact on axin in the Wnt/frizzled signaling pathway. Dev Dyn 239:168–183. [DOI] [PubMed] [Google Scholar]
- Ehrlich AT, Kieffer BL, Darcq E. (2019) Current strategies toward safer mu opioid receptor drugs for pain management. Expert Opin Ther Targets 23:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin ME, Malbon CC. (2007) RGS19 regulates Wnt-beta-catenin signaling through inactivation of Galpha(o). J Cell Sci 120:3404–3414. [DOI] [PubMed] [Google Scholar]
- Garzón J, Rodríguez-Díaz M, López-Fando A, Sánchez-Blázquez P. (2001) RGS9 proteins facilitate acute tolerance to mu-opioid effects. Eur J Neurosci 13:801–811. [DOI] [PubMed] [Google Scholar]
- Garzón J, Rodríguez-Muñoz M, Sánchez-Blázquez P. (2005) Morphine alters the selective association between mu-opioid receptors and specific RGS proteins in mouse periaqueductal gray matter. Neuropharmacology 48:853–868. [DOI] [PubMed] [Google Scholar]
- Gaspari S, Cogliani V, Manouras L, Anderson EM, Mitsi V, Avrampou K, Carr FB, Zachariou V. (2017) RGS9-2 modulates responses to oxycodone in pain-free and chronic pain states. Neuropsychopharmacology 42:1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari S, Papachatzaki MM, Koo JW, Carr FB, Tsimpanouli ME, Stergiou E, Bagot RC, Ferguson D, Mouzon E, Chakravarty S, et al. (2014) Nucleus accumbens-specific interventions in RGS9-2 activity modulate responses to morphine. Neuropsychopharmacology 39:1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari S, Purushothaman I, Cogliani V, Sakloth F, Neve RL, Howland D, Ring RH, Ross EM, Shen L, Zachariou V. (2018) Suppression of RGSz1 function optimizes the actions of opioid analgesics by mechanisms that involve the Wnt/β-catenin pathway. Proc Natl Acad Sci USA 115:E2085–E2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoussi Z, Leontiadis L, Mazarakou G, Merkouris M, Hyde K, Hamm H. (2006) Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell Signal 18:771–782. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Han MH, Herman AE, Ni YG, Pudiak CM, Aghajanian GK, Liu RJ, Potts BW, Mumby SM, Nestler EJ. (2003) Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur J Neurosci 17:971–980. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Hoang CV, Potts BW, Porras G, Pioli E, Kim KW, Nadjar A, Qin C, LaHoste GJ, Li Q, et al. (2007) RGS9-2 negatively modulates L-3,4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson’s disease. J Neurosci 27:14338–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. (1997) Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci 17:8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum L, Smith RC, Rigbi A, Strous R, Teltsh O, Kanyas K, Korner M, Lancet D, Ben-Asher E, Lerer B. (2009) Further evidence for association of the RGS2 gene with antipsychotic-induced parkinsonism: protective role of a functional polymorphism in the 3′-untranslated region. Pharmacogenomics J 9:103–110. [DOI] [PubMed] [Google Scholar]
- Gross JD, Kaski SW, Schroer AB, Wix KA, Siderovski DP, Setola V. (2018) Regulator of G protein signaling-12 modulates the dopamine transporter in ventral striatum and locomotor responses to psychostimulants. J Psychopharmacol 32:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Renthal W, Ring RH, Rahman Z, Psifogeorgou K, Howland D, Birnbaum S, Young K, Neve R, Nestler EJ, et al. (2010) Brain region specific actions of regulator of G protein signaling 4 oppose morphine reward and dependence but promote analgesia. Biol Psychiatry 67:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler JR. (1999) Emerging roles for RGS proteins in cell signalling. Trends Pharmacol Sci 20:376–382. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 54:527–559. [DOI] [PubMed] [Google Scholar]
- Huang H, He X. (2008) Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol 20:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Stewart A, Maity B, Hagen J, Fagan RL, Yang J, Quelle DE, Brenner C, Fisher RA. (2014) RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated Dnmt1 degradation and promoting apoptosis. Oncogene 33:3604–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt TW, Fields TA, Casey PJ, Peralta EG. (1996) RGS10 is a selective activator of G alpha i GTPase activity. Nature 383:175–177. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J 17:1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Koob GF. (1998) Drug dependence: stress and dysregulation of brain reward pathways. Drug Alcohol Depend 51:23–47. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Levran O, Reed B, Schlussman SD, Zhou Y, Butelman ER. (2012) Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J Clin Invest 122:3387–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee S, Kang S, Jeon TI, Kang MJ, Lee TH, Kim YS, Kim KS, Im HI, Moon C. (2018) Regulator of G-Protein Signaling 4 (RGS4) controls morphine reward by glutamate receptor activation in the nucleus accumbens of mouse brain. Mol Cells 41:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimas J, Gorfinkel L, Fairbairn N, Amato L, Ahamad K, Nolan S, Simel DL, Wood E. (2019) Strategies to identify patient risks of prescription opioid addiction when initiating opioids for pain: a systematic review. JAMA Netw Open 2:e193365 DOI: 10.1001/jamanetworkopen.2019.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvehaugen AS, Melien Ø, Holmen OL, Laivuori H, Dechend R, Staff AC. (2014) Hypertension after preeclampsia and relation to the C1114G polymorphism (rs4606) in RGS2: data from the Norwegian HUNT2 study. BMC Med Genet 15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouèbe G, Lomazzi M, Cruz HG, Creton C, Luján R, Li M, Yanagawa Y, Obata K, Watanabe M, Wickman K, et al. (2007) RGS2 modulates coupling between GABAB receptors and GIRK channels in dopamine neurons of the ventral tegmental area. Nat Neurosci 10:1559–1568. [DOI] [PubMed] [Google Scholar]
- Lamberts JT, Jutkiewicz EM, Mortensen RM, Traynor JR. (2011) μ-Opioid receptor coupling to Gα(o) plays an important role in opioid antinociception. Neuropsychopharmacology 36:2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberts JT, Smith CE, Li MH, Ingram SL, Neubig RR, Traynor JR. (2013) Differential control of opioid antinociception to thermal stimuli in a knock-in mouse expressing regulator of G-protein signaling-insensitive Gαo protein. J Neurosci 33:4369–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer I, Tikhonova IG, Boulègue C, Estève JP, Vatinel S, Ferrand A, Moroder L, Robberecht P, Fourmy D. (2009) Evidence for a direct and functional interaction between the regulators of G protein signaling-2 and phosphorylated C terminus of cholecystokinin-2 receptor. Mol Pharmacol 75:502–513. [DOI] [PubMed] [Google Scholar]
- Le Merrer J, Becker JA, Befort K, Kieffer BL. (2009) Reward processing by the opioid system in the brain. Physiol Rev 89:1379–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. (2006) Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4). Biol Psychiatry 60:534–537. [DOI] [PubMed] [Google Scholar]
- Liu W, Yuen EY, Allen PB, Feng J, Greengard P, Yan Z. (2006) Adrenergic modulation of NMDA receptors in prefrontal cortex is differentially regulated by RGS proteins and spinophilin. Proc Natl Acad Sci USA 103:18338–18343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chatterjee TK, Fisher RA. (2002) RGS6 interacts with SCG10 and promotes neuronal differentiation. Role of the G gamma subunit-like (GGL) domain of RGS6. J Biol Chem 277:37832–37839. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fisher RA. (2004) RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J Biol Chem 279:14120–14128. [DOI] [PubMed] [Google Scholar]
- López-Fando A, Rodríguez-Muñoz M, Sánchez-Blázquez P, Garzón J. (2005) Expression of neural RGS-R7 and Gbeta5 proteins in response to acute and chronic morphine. Neuropsychopharmacology 30:99–110. [DOI] [PubMed] [Google Scholar]
- Madrigal A, Tan L, Zhao Y. (2017) Expression regulation and functional analysis of RGS2 and RGS4 in adipogenic and osteogenic differentiation of human mesenchymal stem cells. Biol Res 50:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark MD, Wittemann S, Herlitze S. (2000) G protein modulation of recombinant P/Q-type calcium channels by regulators of G protein signalling proteins. J Physiol 528:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-McCaffrey L, Willard FS, Oliveira-dos-Santos AJ, Natale DR, Snow BE, Kimple RJ, Pajak A, Watson AJ, Dagnino L, Penninger JM, et al. (2004) RGS14 is a mitotic spindle protein essential from the first division of the mammalian zygote. Dev Cell 7:763–769. [DOI] [PubMed] [Google Scholar]
- Martin-McCaffrey L, Willard FS, Pajak A, Dagnino L, Siderovski DP, D’Souza SJ. (2005) RGS14 is a microtubule-associated protein. Cell Cycle 4:953–960. [DOI] [PubMed] [Google Scholar]
- Masuho I, Xie K, Martemyanov KA. (2013) Macromolecular composition dictates receptor and G protein selectivity of regulator of G protein signaling (RGS) 7 and 9-2 protein complexes in living cells. J Biol Chem 288:25129–25142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Shi XD, Schwendt M, Saylor A, Toda S. (2008) Regulation of psychostimulant-induced signaling and gene expression in the striatum. J Neurochem 104:1440–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson KB, Leff ER, Li MH, Meurice C, Tai S, Traynor JR, Ingram SL. (2018) Regulators of G-protein signaling (RGS) proteins promote receptor coupling to G-protein-coupled inwardly rectifying potassium (GIRK) channels. J Neurosci 38:8737–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C, Cheong SY, Cheong SJ, Kim M, Cho DI, Kim KM. (2012) RGS4 exerts inhibitory activities on the signaling of dopamine D2 receptor and D3 receptor through the N-terminal region. Pharmacol Res 65:213–220. [DOI] [PubMed] [Google Scholar]
- Mitsi V, Terzi D, Purushothaman I, Manouras L, Gaspari S, Neve RL, Stratinaki M, Feng J, Shen L, Zachariou V. (2015) RGS9-2--controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states. Proc Natl Acad Sci USA 112:E5088–E5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V, Linder ME. (2004) The RGS14 GoLoco domain discriminates among Galphai isoforms. J Biol Chem 279:46772–46778. [DOI] [PubMed] [Google Scholar]
- Morgan MM, Tran A, Wescom RL, Bobeck EN. (2020) Differences in antinociceptive signalling mechanisms following morphine and fentanyl microinjections into the rat periaqueductal gray. Eur J Pain 24:617–624. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Ross EM. (1999) Rapid GTP binding and hydrolysis by G(q) promoted by receptor and GTPase-activating proteins. Proc Natl Acad Sci USA 96:9539–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean BS, Martemyanov KA. (2016) Association with the plasma membrane is sufficient for potentiating catalytic activity of regulators of G protein signaling (RGS) proteins of the R7 subfamily. J Biol Chem 291:7195–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubig RR, Siderovski DP. (2002) Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov 1:187–197. [DOI] [PubMed] [Google Scholar]
- Nixon AB, Grenningloh G, Casey PJ. (2002) The interaction of RGSZ1 with SCG10 attenuates the ability of SCG10 to promote microtubule disassembly. J Biol Chem 277:18127–18133. [DOI] [PubMed] [Google Scholar]
- Psifogeorgou K, Papakosta P, Russo SJ, Neve RL, Kardassis D, Gold SJ, Zachariou V. (2007) RGS9-2 is a negative modulator of mu-opioid receptor function. J Neurochem 103:617–625. [DOI] [PubMed] [Google Scholar]
- Psifogeorgou K, Terzi D, Papachatzaki MM, Varidaki A, Ferguson D, Gold SJ, Zachariou V. (2011) A unique role of RGS9-2 in the striatum as a positive or negative regulator of opiate analgesia [published correction appears in J Neurosci (2011) 31:7578]. J Neurosci 31:5617–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Gold SJ, Potenza MN, Cowan CW, Ni YG, He W, Wensel TG, Nestler EJ. (1999) Cloning and characterization of RGS9-2: a striatal-enriched alternatively spliced product of the RGS9 gene. J Neurosci 19:2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raehal KM, Schmid CL, Groer CE, Bohn LM. (2011) Functional selectivity at the μ-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev 63:1001–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, Kovoor A, Chen CK, DiLeone RJ, Schwarz SC, et al. (2003) RGS9 modulates dopamine signaling in the basal ganglia. Neuron 38:941–952. [DOI] [PubMed] [Google Scholar]
- Richman RW, Strock J, Hains MD, Cabanilla NJ, Lau KK, Siderovski DP, Diversé-Pierluissi M. (2005) RGS12 interacts with the SNARE-binding region of the Cav2.2 calcium channel. J Biol Chem 280:1521–1528. [DOI] [PubMed] [Google Scholar]
- Rivero G, Gabilondo AM, García-Sevilla JA, Callado LF, La Harpe R, Morentin B, Meana JJ. (2013) Brain RGS4 and RGS10 protein expression in schizophrenia and depression. Effect of drug treatment. Psychopharmacology (Berl) 226:177–188. [DOI] [PubMed] [Google Scholar]
- Rorabaugh BR, Rose MJ, Stoops TS, Stevens AA, Seeley SL, D’Souza MS. (2018) Regulators of G-protein signaling 2 and 4 differentially regulate cocaine-induced rewarding effects. Physiol Behav 195:9–19. [DOI] [PubMed] [Google Scholar]
- Sakloth F, Gaspari S, Singh M, Pryce K, Zachariou V. (2019) Targeting RGSz1 actions in the periaqueductal gray promotes opioid analgesia and decreases reward sensitivity (Abstract). FASEB J 33(suppl 1):498.7. [Google Scholar]
- Sánchez-Blázquez P, Rodríguez-Díaz M, López-Fando A, Rodríguez-Muñoz M, Garzón J. (2003) The GBeta5 subunit that associates with the R7 subfamily of RGS proteins regulates mu-opioid effects. Neuropharmacology 45:82–95. [DOI] [PubMed] [Google Scholar]
- Scherer SL, Cain MD, Kanai SM, Kaltenbronn KM, Blumer KJ. (2017) Regulation of neurite morphogenesis by interaction between R7 regulator of G protein signaling complexes and G protein subunit Gα13. J Biol Chem 292:9906–9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendt M, Gold SJ, McGinty JF. (2006) Acute amphetamine down-regulates RGS4 mRNA and protein expression in rat forebrain: distinct roles of D1 and D2 dopamine receptors. J Neurochem 96:1606–1615. [DOI] [PubMed] [Google Scholar]
- Schwendt M, Hearing MC, See RE, McGinty JF. (2007) Chronic cocaine reduces RGS4 mRNA in rat prefrontal cortex and dorsal striatum. Neuroreport 18:1261–1265. [DOI] [PubMed] [Google Scholar]
- Schwendt M, Sigmon SA, McGinty JF. (2012) RGS4 overexpression in the rat dorsal striatum modulates mGluR5- and amphetamine-mediated behavior and signaling. Psychopharmacology (Berl) 221:621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P, Ko F, Jack E, Greenstein R, Dean B. (2007) Consistent with dopamine supersensitivity, RGS9 expression is diminished in the amphetamine-treated animal model of schizophrenia and in postmortem schizophrenia brain. Synapse 61:303–309. [DOI] [PubMed] [Google Scholar]
- Senese NB, Kandasamy R, Kochan KE, Traynor JR. (2020) Regulator of G-protein signaling (RGS) protein modulation of opioid receptor signaling as a potential target for pain management. Front Mol Neurosci 13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese NB, Rasenick MM, Traynor JR. (2018) The role of G-proteins and G-protein regulating proteins in depressive disorders. Front Pharmacol 9:1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini RA, Pryce KD, Zachariou V. (2020) The mesolimbic dopamine system in chronic pain and associated affective comorbidities. Biol Psychiatry 87:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Hicks JK, Huang TP, Keller NP. (2003) Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 165:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu FJ, Ramineni S, Hepler JR. (2010) RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal 22:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderovski DP, Snow BE, Chung S, Brothers GM, Sondek J, Betts L. (2002) Assays of complex formation between RGS protein G gamma subunit-like domains and G beta subunits. Methods Enzymol 344:702–723. [DOI] [PubMed] [Google Scholar]
- Silverman JL, Koenig JI. (2007) Evidence for the involvement of ERbeta and RGS9-2 in 17-beta estradiol enhancement of amphetamine-induced place preference behavior. Horm Behav 52:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S, Meazza R, Quintarelli C, Carlomagno S, Della Chiesa M, Falco M, Moretta L, Locatelli F, Pende D. (2019) NK cell-based immunotherapy for hematological malignancies. J Clin Med 8:1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick P, Volkow ND. (2016) Re-energizing the development of pain therapeutics in light of the opioid epidemic. Neuron 92:294–297. [DOI] [PubMed] [Google Scholar]
- Smith SB, Maixner DW, Fillingim RB, Slade G, Gracely RH, Ambrose K, Zaykin DV, Hyde C, John S, Tan K, et al. (2012) Large candidate gene association study reveals genetic risk factors and therapeutic targets for fibromyalgia. Arthritis Rheum 64:584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Siderovski DP. (2001) Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharmacol 61:1329–1337. [DOI] [PubMed] [Google Scholar]
- Squires KE, Montañez-Miranda C, Pandya RR, Torres MP, Hepler JR. (2018) Genetic analysis of rare human variants of regulators of G protein signaling proteins and their role in human physiology and disease. Pharmacol Rev 70:446–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanwood GD, Parlaman JP, Levitt P. (2006) Genetic or pharmacological inactivation of the dopamine D1 receptor differentially alters the expression of regulator of G-protein signalling (Rgs) transcripts. Eur J Neurosci 24:806–818. [DOI] [PubMed] [Google Scholar]
- Stemmle LN, Fields TA, Casey PJ. (2006) The regulator of G protein signaling domain of axin selectively interacts with Galpha12 but not Galpha13. Mol Pharmacol 70:1461–1468. [DOI] [PubMed] [Google Scholar]
- Sun H, Calipari ES, Beveridge TJ, Jones SR, Chen R. (2015) The brain gene expression profile of dopamine D2/D3 receptors and associated signaling proteins following amphetamine self-administration. Neuroscience 307:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton LP, Ostrovskaya O, Dao M, Xie K, Orlandi C, Smith R, Wee S, Martemyanov KA. (2016) Regulator of G-protein signaling 7 regulates reward behavior by controlling opioid signaling in the striatum. Biol Psychiatry 80:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JN, Roman DL, Clark MJ, Roof RA, Tesmer JJ, Neubig RR, Traynor JR. (2010) Differential modulation of mu-opioid receptor signaling to adenylyl cyclase by regulators of G protein signaling proteins 4 or 8 and 7 in permeabilised C6 cells is Galpha subtype dependent. J Neurochem 112:1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi D, Cao Y, Agrimaki I, Martemyanov KA, Zachariou V. (2012) R7BP modulates opiate analgesia and tolerance but not withdrawal. Neuropsychopharmacology 37:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi D, Stergiou E, King SL, Zachariou V. (2009) Regulators of G protein signaling in neuropsychiatric disorders. Prog Mol Biol Transl Sci 86:299–333. [DOI] [PubMed] [Google Scholar]
- Traynor JR, Neubig RR. (2005) Regulators of G protein signaling & drugs of abuse. Mol Interv 5:30–41. [DOI] [PubMed] [Google Scholar]
- Tso PH, Wang Y, Wong SY, Poon LS, Chan AS, Wong YH. (2010) RGS19 enhances cell proliferation through its C-terminal PDZ motif. Cell Signal 22:1700–1707. [DOI] [PubMed] [Google Scholar]
- Vivot K, Moullé VS, Zarrouki B, Tremblay C, Mancini AD, Maachi H, Ghislain J, Poitout V. (2016) The regulator of G-protein signaling RGS16 promotes insulin secretion and β-cell proliferation in rodent and human islets. Mol Metab 5:988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Boyle M. (2018) Neuroscience of addiction: relevance to prevention and treatment. Am J Psychiatry 175:729–740. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Jones EB, Einstein EB, Wargo EM. (2019) Prevention and treatment of opioid misuse and addiction: a review. JAMA Psychiatry 76:208–216. [DOI] [PubMed] [Google Scholar]
- Walker PD, Jarosz PA, Bouhamdan M, MacKenzie RG. (2015) Effects of gender on locomotor sensitivity to amphetamine, body weight, and fat mass in regulator of G protein signaling 9 (RGS9) knockout mice. Physiol Behav 138:305–312. [DOI] [PubMed] [Google Scholar]
- Wang J, Ducret A, Tu Y, Kozasa T, Aebersold R, Ross EM. (1998) RGSZ1, a Gz-selective RGS protein in brain. Structure, membrane association, regulation by Galphaz phosphorylation, and relationship to a Gz gtpase-activating protein subfamily. J Biol Chem 273:26014–26025. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu-Chen LY, Traynor JR. (2009) Differential modulation of mu- and delta-opioid receptor agonists by endogenous RGS4 protein in SH-SY5Y cells. J Biol Chem 284:18357–18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Traynor JR. (2011) Opioid-induced down-regulation of RGS4: role of ubiquitination and implications for receptor cross-talk. J Biol Chem 286:7854–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Traynor JR. (2013) Modulation of μ-opioid receptor signaling by RGS19 in SH-SY5Y cells. Mol Pharmacol 83:512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker D, Schroeder JP. (2007) There and back again: a tale of norepinephrine and drug addiction. Neuropsychopharmacology 32:1433–1451. [DOI] [PubMed] [Google Scholar]
- Wieland T, Lutz S, Chidiac P. (2007) Regulators of G protein signalling: a spotlight on emerging functions in the cardiovascular system. Curr Opin Pharmacol 7:201–207. [DOI] [PubMed] [Google Scholar]
- Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP. (2007) Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation. EMBO J 26:2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Masuho I, Brand C, Dessauer CW, Martemyanov KA. (2012) The complex of G protein regulator RGS9-2 and Gβ(5) controls sensitization and signaling kinetics of type 5 adenylyl cyclase in the striatum. Sci Signal 5:ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li YP. (2007) RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev 21:1803–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V, Kroslak T, Laforge KS, Zhou Y, Ho A, Kreek MJ. (2003) Differential gene expression in the rat caudate putamen after “binge” cocaine administration: advantage of triplicate microarray analysis. Synapse 48:157–169. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, Neve RL, Sim-Selley LJ, Selley DE, Gold SJ, et al. (2003) Essential role for RGS9 in opiate action. Proc Natl Acad Sci USA 100:13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chisari M, Raehal KM, Kaltenbronn KM, Bohn LM, Mennerick SJ, Blumer KJ. (2012) GIRK channel modulation by assembly with allosterically regulated RGS proteins. Proc Natl Acad Sci USA 109:19977–19982. [DOI] [PMC free article] [PubMed] [Google Scholar]