Abstract
In this work, a sialic acid (SA)-imprinted thermo-responsive hydrogel layer was prepared for selective capture and release of cancer cells. The SA-imprinting process was performed at 37 °C using thermo-responsive functional monomer, thus generating switchable SA-recognition sites with potent SA binding at 37 °C and weak binding at a lower temperature (e.g., 25 °C). Since SA is often overexpressed at the glycan terminals of cell membrane proteins or lipids, the SA-imprinted hydrogel layer could be used for selective cancer cell recognition. Our results confirmed that the hydrogel layer could efficiently capture cancer cells from not only the culture medium but also the real blood samples. In addition, the captured cells could be non-invasively released by lowing the temperature. Considering the non-invasive processing mode, considerable capture efficiency, good cell selectivity, as well as the more stable and durable SA-imprinted sites compared to natural antibodies or receptors, this thermo-responsive hydrogel layer could be used as a promising and general platform for cell-based cancer diagnosis.
Keywords: Molecular imprinting, Cell capture and release, Thermo-responsive hydrogels, Molecular recognition, Sialic acid
Graphical abstract
Highlights
-
•
Thermo-responsive sialic acid (SA)-imprinted hydrogel layer was prepared.
-
•
The hydrogel layer could efficiently and selective capture cancer cells at 37 °C.
-
•
The captured cancer cells could be released at a lower temperature (e.g., 25 °C).
-
•
The hydrogel layer could be used for capture and release cancer cells from blood.
1. Introduction
Molecular recognition in living systems represents a series of fundamental events that are crucial to a variety of biological processes in molecular and cellular levels [[1], [2], [3]]. As a consequence of external or internal biological stimuli, reversible changes in these natural molecular recognition systems like the receptor-ligand or antibody-antigen interactions will occur. These dynamic interactions then trigger specific cell-signaling and intracellular cascades, which subsequently result in normal or abnormal cellular processes [4,5]. In other words, the molecular recognition-mediated cellular processes are central to both physiology and pathology in biosystems [6,7]. In this context, molecular recognition has attracted increasing interest in the development of advanced biomaterials and biomedical devices [5]. For biomaterial design and bioengineering, selective integration of natural molecular recognition (e.g., ligand conjugations, antibody modifications, growth factor loadings, and so forth) into non-biogenic materials have proven to be an efficient strategy to enhance their initiative to communicate with cells; and the increased vitality could greatly promote the biological performance of materials or devices, e.g., improving biocompatibility, accelerating tissue regeneration, or increasing the sensitivity of biological detections [[8], [9], [10], [11], [12], [13], [14]].
Despite tremendous progress in biomaterial development, these natural molecular recognition systems (e.g., receptor-ligand or antibody-antigen interactions) still have inherent drawbacks in practical uses [15,16]. Apart from the physicochemical instability and short shelf life, their isolation and purification are often time-consuming and high-cost. These drawbacks thus necessitate scientists to seek more durable and robust substitutions with receptor or antibody-like molecular specificity. Currently, various synthetic strategies to create artificial molecular recognition have been developed for mimicking the molecular specificity of receptors or antibodies [17,18]. Amongst, molecular imprinting technology has aroused great concern due to the molecularly tunable reversibility and specificity of the resultant molecularly imprinted polymers (MIPs) in analogy with natural receptors and antibodies [3,[19], [20], [21]]. The idea of molecular imprinting was inspired by the mechanism of enzyme catalysis and antibody formation. Molecular recognition sites in MIPs are generally created by the self-assembling and imprinting process of molecular templates with functional monomers and crosslinkers. Similar to natural molecular recognition systems, these imprinted sites are spatially complementary to the shape and functionality of the template molecules. In view of the high physicochemical stability, customizable molecular specificity, low cost, and simplicity in synthesis, MIPs have found potentials over wide areas including separation, immunoassays, catalysis, drug delivery, sensing, bio-imaging and so forth [3,19].
As known, the original intention of MIPs is to mimic natural molecular recognition that commonly occurs in cellular processes [22]. The applications of current MIPs in cellular levels, however, are still in its fancy. A sharp contrast is that the molecular separation and extraction using MIPs have achieved great success [[23], [24], [25]], while cell separation based on the molecular specificity of MIPs have rarely been explored [3,[26], [27], [28]]. In fact, cell separations are very important in both fundamental cell biology and modern medical diagnosis [29]. For example, selective isolation of cancer cells from blood stream (i.e., circulating tumor cells, CTCs) has shown great promise in cancer theranostics. This is because CTCs analysis will provide important information for cancer early diagnosis and prognosis [7,30]. In view of this, fabrication of a MIPs-based platform for selective cancer cell isolation is of great significance in not only the functioning replicates of natural molecular recognition in biosystems but also the development of advanced medical theranostics.
To create MIPs with cell recognition, one of the rational methods is imprinting of a specific biomarker that is attached on the out layer of cell membrane [3]. Sialic acid (SA) has been found to be a key constituent in the glycans of cell membrane proteins or lipids, and it has proven to correlate with several pathological processes, in particular, the cancers [31,32]. The overexpression of SA residues on cancer cell surface thus provides the opportunity to generate cancer affinity by tailoring MIPs with specific SA-recognition, and this mechanism has been used for targeted cancer cell imaging using the SA-imprinted nanoparticles [[33], [34], [35]]. These precedents indicated the SA-imprinted sites in MIPs are crucial to specific cancer cell recognition, and theoretically, they may also act as a selective driving force for cancer cell separation. In view of this, we take one step further in this study and intent to employ the specific SA-recognition of MIPs for selective cancer cell isolation (Fig. 1). This work will expend the applicability of artificial molecular recognition (i.e., MIPs) from simple molecular separation or extraction to selective cellular isolation, and this is promising in cell-based cancer early diagnosis and prognosis. In our design, we will prepare a layer of SA-imprinted hydrogel and investigate SA-recognition-induced cancer cell attachment on the surface. To facilitate subsequent cell detachment and harvesting, thermo-responsiveness is involved in this system in order to generate switchable SA-recognition through a temperature transition manner. When the thermo-responsive hydrogel layer is incubated with cancer cells at 37 °C, surface SA-recognition will trigger specific cell capture. Upon reduction of the temperature to 25 °C, the deteriorative SA-recognition caused by changes of the SA-imprinted sites in shape and functionality will lead to rapid release of the bound cancer cells. Compared with its natural counterparts, MIPs-based molecular recognition features low-cost and durable, and especially in this study, the molecular specificity towards SA could be tailored with thermo-switchability. Therefore, we believe that the work may give a paragon for the fabrication of cell isolation platforms through chemically designed molecular recognition. Moreover, the physiologically acceptable thermo-responsiveness enables the low- or non-invasive cell harvesting, implying the superiority of this study in accurate cancer diagnosis that is highly depended on the state of captured cancer cells.
Fig. 1.
Thermo-responsive SA-imprinted hydrogel layer enables selective capture and release of cancer cells (e.g., HepG-2, human liver carcinoma cells) through surface molecular specificity towards the overexpressed SA on the glycan terminal of cell membrane proteins.
2. Materials and methods
2.1. Materials
Sialic acid (SA, 98%, HEOWNS), 3-methacrylamidophenylboronic acid (MAPBA, 98%, HEOWNS), N-(3-aminopropyl)methacrylamide hydrochloride (NAPMAAm, 98%, Aladdin), ammonium persulphate (APS, 98%, Sinopharm), N,N,N′,N′-tetramethylethylenediamine (TEMED, 99.5%, Aladdin), 3-(trimethoxysilyl)propyl methacrylate (MPTS, 97%, Aladdin) was used as received. N-isopropylacrylamide (NIPAAm, 98%, Aladdin) was recrystallized from n-hexane, acrylamide (AAm, 99%, Aladdin) and N,N′-methylenebisacrylamide (MBAAm, 99%, Aladdin) were used after recrystallization from acetone. Cover glass with a size of 24 mm × 32 mm and quartz slides with diameters of 15 mm were purchased from Citotest. By using the Spring laboratory pure water system based on RO-DI step-by-step purification technology, the minimum resistivity of deionized water (dH2O) can reach 18.2 MΩ cm. Trypsin/EDTA (ethylenediaminetetraacetic acid) solution (0.25%), streptomycin, penicillin, RPMI (Roswell Park Memorial Institute) 1640 medium, MEM (minimum essential medium) medium and FBS (fetal bovine serum) were purchased from Gibco BRL (USA). Phosphate-buffered saline solution (PBS, 10 mmol/L, pH = 7.4) was purchased from HyClone (USA). Acridine orange (AO) and propidium iodide (PI) were purchased from Beijing Solarbio Science & Technology Co., Ltd. Phycoerythrin (PE)-anti-CD45 and FITC-anti-Asialoglycoprotein receptor 1 (ASGPR1) were purchased from Santa Cruz Biotechnology (USA), Cell Counting Kit-8 (CCK-8), 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO), 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) were purchased from Beyotime Biotechnology (Shanghai, China) and were used as received. The brands and sources of all other biochemical reagent will be mentioned in the experimental methods as described below.
2.2. Preparation of thermo-responsive SA-imprinted and non-imprinted hydrogel layer
-
a)
Surface silanization of quartz slides
Quartz slides were firstly activated by using piranha solutions, a mixture of concentrated H2SO4 to 30% H2O2 solutions (2:1, v/v), at 90 °C for 1 h. Then, the slide was washed with ultrapure water and dried with argon. This step was repeated 3 times. After that, the slide was treated for 1 h in a silanization solution that was prepared by diluting MPTS (0.5 mL) in the mixture of ethanol (50 mL) and diluted glacial acetic acid (1.5 mL, 10% v/v). The double bond functionalized quartz slides were washed with ethanol, dried in the N2 for further use.
-
b)
SA-imprinting on the silanized quartz slides
The schematic illustration of SA-imprinted hydrogel layer was shown in Fig. 2. Typically, SA (9.3 mg, 0.03 mmol), MAPBA (5.7 mg, 0.03 mmol), NAPMAAm (5.3 mg, 0.03 mmol), NIPAAm (40.5 mg, 0.36 mmol), AAm (12.7 mg, 0.18 mmol), MBAAm (3.9 mg, 0.025 mmol) were dissolved in 1 mL PBS (10 mmol/L, pH = 7.4). After filtered with a 0.22 μm membrane, the solution was purged with N2 for 30 min to remove oxygen in the reaction system. Subsequently, APS (15 μL, 10%, m/v) and TEMED (30 μL, 10%, v/v) were added. Then, 10 μL of the solution was immediately placed between the cover glass slide and the MPTS-functionalized quartz slide, the reaction was conducted in an incubator at 37 °C for 3 h. After the reaction, the cover glasses were removed with forceps and the resultant SA-imprinted hydrogels was coated on the support quartz substrate (i.e., SA-imprinted hydrogel, SIH). SIH was washed with HCl solution (pH = 5) to remove the SA template and equilibrated in PBS solution for further use. The non-imprinted hydrogel (denoted as NIH) was also prepared and washed in the same way without the addition of SA template molecules.
Fig. 2.
Strategy to create SA-imprinted sites (i.e., SA-recognition) on thermo-responsive hydrogel layer by means of molecular imprinting on quartz substrate.
2.3. Preparation of SA-imprinted bulk hydrogel
SA-imprinted bulk hydrogel with the same composition was prepared for binding test. The preparation procedure was described as follows: 37.2 mg (0.12 mmol) of SA, 22.8 mg (0.12 mmol) of MAPBA, 21.6 mg (0.12 mmol) of NAPMAAm, 162 mg (1.432 mmol) of NIPAAm, 50.8 mg (0.716 mmol) of AAm, 15.6 mg (0.1 mmol) of MBAAm were dissolved in 4 mL PBS (10 mmol/L, pH = 7.4), and left to rest at 4 °C for 12 h. After taking out, purge with N2 for 30 min to remove oxygen in the reaction system. Subsequently, APS (60 μL, 10%, m/v) and TEMED (120 μL, 10%, v/v) were added. After joining the initiation system, immediately added the mixed prepolymer into the gel electrophoresis scaffold and react at 37 °C for 3 h. After the reaction, the gel-like bulk hydrogel polymer was taken out, and a uniform disk was punched out with a 3 mm diameter gel punch. After removal of the template with HCl solution (pH = 5), and the hydrogel was equilibrated in PBS for later use. As a control, non-imprinted bulk hydrogel was prepared by the same method without the addition of SA during the imprinting process.
2.4. Binding tests
-
a)
Equilibrium binding
Different hydrogel solutions (2, 4, 8, 12, 24 mg/mL in PBS) and SA (10 μg/mL in PBS) (C0) were mixed and incubated at 37 °C for 12 h. Upon reaching the binding equilibrium, the concentration of SA in the supernatant (Ceq) was analyzed by HPLC (high performance liquid chromatography), and the amount of SA bound (B%) onto the hydrogels was calculated. HPLC analysis was performed using 0.1% H3PO4 as mobile phase, a RP-AQUA column (Sunniest RP-AQUA, 250 × 4.6 mm), a flow rate of 0.8 mL min−1, an injection volume of 20 μL, and the detection wavelength for SA is 205 nm. The equation for the calculation of SA binding is:
-
b)
Isothermal binding
Binding isotherms were further studied to determine their association constants toward the SA through Scatchard analysis. Typically, hydrogel (24 mg/mL) was incubated with a series of SA solutions (C0 = 2–40 μg/mL, 1 mL) at 37 °C for 12 h. The amounts of SA bound onto the hydrogels were determined by HPLC. The Scatchard equation is:
where Ka and Bmax represent the binding association constant and apparent maximum number of the binding sites, respectively; F refers to the amount of SA template remaining in the supernatants after equilibrium binding.
-
c)
Thermo-responsive binding
Temperature-sensitive binding properties of the hydrogels is studied by measuring the SA adsorption capacity at a lower temperature 25 °C. The different adsorption data at 37 and 25 °C will indicate the thermo-responsive binging property.
2.5. Cell culture
Human liver cancer cells (HepG-2, with overexpressed SA on the cell surface) (obtained from Shanghai Cell Center) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Fibroblasts (L929 cells) (obtained from Shanghai Cell Center) were cultured in MEM medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. All cells were cultured at 37 °C with 5% CO2 atmosphere. The medium was changed three times weekly, and the cells were harvested using 0.25% trypsin and 0.26 mM EDTA in PBS after reaching sub-confluency. For cell capture, HepG-2 cells were pre-stained by DiO (green), and L929 cells were prestained by DiI (red) before use.
2.6. Cell capture and release
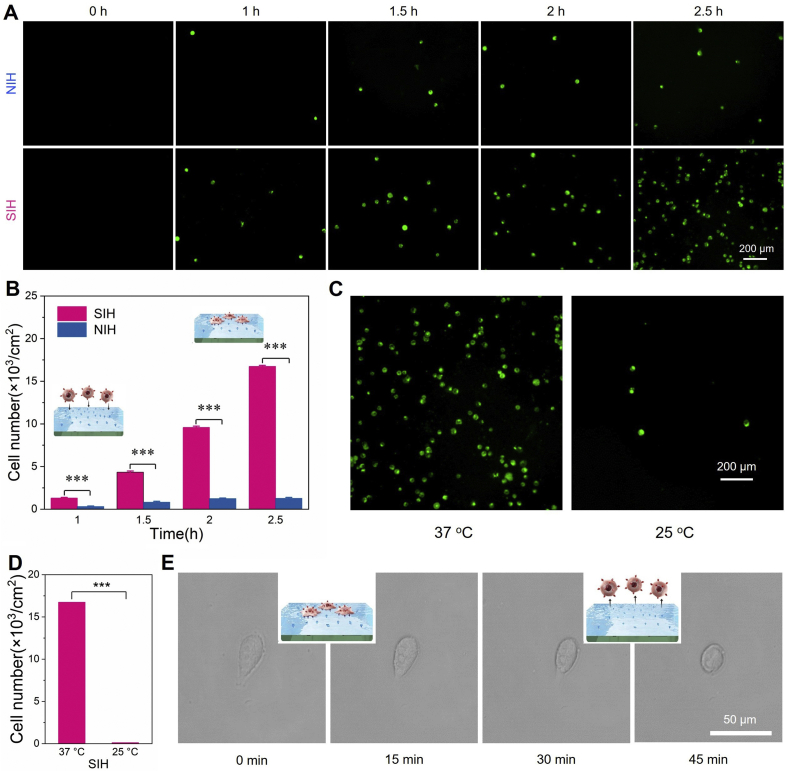
The hydrogel layer SIH and NIH were placed in a 24-well plate and stored with 1 mL PBS. Then removed PBS and added 1 mL of DiO-pre-stained HepG-2 cells (5 × 104 cells/mL) to each well, then incubated at 37 °C for 1, 1.5, 2, and 2.5 h respectively. After that, the medium was removed, and the weakly adsorbed cells were mildly washed with PBS. The captured cells on the SIH and NIH were then counted, respectively. For the cell release experiment, the captured cells were then cooled at 25 °C for 30 min and then mildly washed with PBS, followed by fluorescence microscope analysis and cell counting. Randomly shot with a digital camera was used to quantify the captured cells, and then scaled up to calculate the total amount of captured cells.
2.7. Cell viability
The original and recovered cells were first assessed by a live/dead assay by staining the live and dead cells with AO (green, 20 μg/mL) and PI (red, 15 μg/mL), respectively. The stained cells were examined and evaluated under fluorescence microscope. The original cells and recovered cells were sub-cultured in a 6-well plate (1 × 104 cells/well). After incubation for 1, 3, or 5 days in the medium at 37 °C, the cell adhesion status was observed using a microscope and cell proliferation was evaluated through CCK-8 assay. CCK-8 solution (100 μL) was added to each well, and the cells were incubated at 37 °C for 24 h. The absorbance was measured at 450 nm using a microplate spectrophotometer (BioTek, Winooski, VT, USA). Each sample was tested 6 times in parallel.
2.8. Selective capture in cell mixtures
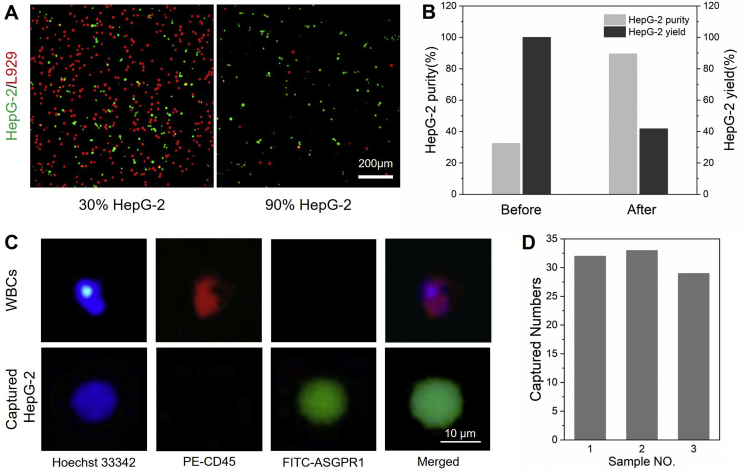
In order to study the selectivity of SIH towards SA-overexpressed cancer cells, HepG-2 cells and normal fibroblast cell line (L929 cells) were both introduced as interfering cells. Briefly, DiI-pre-stained L929 cells were added to the culture medium that containing DiO-pre-stained HepG-2 cells. The cell mixture was at a ratio of 2:1 (L929/HepG-2), and the total cell density was 5 × 104 cells/mL. The purity and yield of HepG-2 cells on hydrogel layer were studied by a fluorescence microscope.
2.9. Cell capture in artificial CTC blood samples
Rat blood sample was acquired from experimental animal center of Jiangsu university. 200 HepG-2 cells were added in 1 mL of whole blood to obtain artificial CTC blood sample. The blood sample was added to SIH and incubated at 37 °C for 120 min. Then, the non-specific adsorbed cells from the blood sample washed with PBS for 3 times. The captured cells were fixed and permeabilized, and then stained with 10 μL of PE-anti-CD45 and FITC-anti-ASGPR1 solutions and 10 μL Hoechst 33,342 at 4 °C for 12 h. The capture efficiency of HepG-2 cells was calculated according to the fluorescence images taken under an inverted fluorescence microscope.
2.10. Statistical analysis
Statistical analyses were performed using one-way analysis of variance (ANOVA) followed by Tukey's test. Two-way analysis of variance was only used when comparisons were made with two or more interconnected variables. The differences between two groups are considered significant when the p-value is less than 0.05.
3. Results and discussion
3.1. Preparation of SA-imprinted hydrogel
The procedure to prepare SA-imprinted hydrogel layer was presented in Fig. 2. The imprinting process was carried out by redox-initiated polymerization (ASP/TEMED) at 37 °C in PBS using NIPAAm as the thermo-sensitive monomer and MAPBA, NAPMAAm, AAm, and MBAAm as the diol-binding [[36], [37], [38]], amine-coordinated [39], hydrogen-bonding monomers and crosslinker, respectively. Polymerization was initiated between a MPTS-functionalized quartz slide and a cover glass, subsequently leading to a thin layer of SA-imprinted hydrogel on the quartz slide. After that, the SA template molecules, adsorbed oligomers, and unreacted monomers on the hydrogel layer were removed by alternately rinsing with deionized water and diluted solution of HCl (pH = 5), and finally was stored in PBS.
The SA-imprinted hydrogel (SIH) layer and its non-imprinted control hydrogel (NIH) layer were first characterized using Fourier transform infrared spectroscopy (FT-IR) to study their chemical composition (Fig. 3A). Clearly, the characteristic peaks of poly (NIPAAm) such as the amide I band (1644 cm−1, C O stretching) and amide II band (1541 cm−1, N–H stretching) were observed [40]. Note that the characteristic peak of B–O stretching around 1340 cm−1 in the hydrogels was not found, probably due to low content of poly (MAPBA) in the hydrogel network (less than 5% in the repeated units). Nevertheless, the result confirmed the successful grafting of a layer of polyacrylamide-based hydrogel on quartz slide. The SIH and NIH hydrogel layer were further studied using scanning electron microscope (SEM) (Fig. 3B). Both SIH and NIH showed flat surfaces owing to the cover glass during polymerization. According to the surface scratch caused by a syringe needle, the thickness of hydrogel layer was estimated to be 1–3 μm.
Fig. 3.
(A) FT-IR spectrum of the SIH and NIH hydrogels on quartz substrates. (B) The SEM images of SIH and NIH on quartz substrates. The scratch on SIH was caused by slicing the surface with a syringe needle. (C) the surface wettability of SIH and NIH at different temperature. Inset show the water drop profiles (4 μL) on the surfaces.
Since poly (NIPAAm) is sensitive to environmental temperature [26,41], we then investigated the thermo-responsiveness of SIH and NIH. Different surface wettability at different temperature represents one of the thermo-responsive properties of poly (NIPAAm). Thus, we checked the surface hydrophilicity/hydrophobicity through water contact angle measurement (Fig. 3C). The SIH and NIH both showed more spreading water drops on the surface at 25 °C as compared to that of a high temperature at 37 °C. The water contact angles of SIH and NIH both could increase approximately 6 ~ 7° when the temperature reduced from 37 to 25 °C. In addition to the temperature-dependent surface wettability, we also investigated the thermo-responsive swelling and shrinking properties of two hydrogels. The SA-imprinted and non-imprinted hydrogels without quartz as supports but with the same chemical compositions to SIH and NIH were also prepared. Changes of their sizes in PBS at different temperatures (i.e., 25 or 37 °C) were recorded upon achieving the swelling equilibria (in 30 min). As shown in Fig. 3D, the resultant two hydrogel both exhibited great volume increases (i.e., marked swelling) when the environment temperature reduced from 37 °C to 25 °C. This result, together with the temperature-dependent surface wettability, jointly suggested the thermo-responsiveness of the PNIPAAm-containing SIH and NIH, which may affect greatly on the SA binding properties.
3.2. SA rebinding properties of the thermo-responsive imprinted hydrogel
The imprinting mechanism was then confirmed by testing the SA rebinding properties of SIH and NIH. Equilibrium binding experiments were first performed by incubating the SIH or NIH hydrogels in fixed concentration of SA in PBS solution at 37 °C. As shown in Fig. 4A, the SIH hydrogel could bind more SA than their corresponding NIH over the testing concentration ranges. For example, in PBS with 10 μg/mL of SA, 24 mg of SIH hydrogel could bind 50.1% of SA, while the equivalent binding amount for the control NIH was only 21.3%, preliminarily suggesting the presence of SA binding sites in SIH and meanwhile verifying the success of SA imprinting process. To get more information about the SA binding characteristics, isothermal adsorption experiments were further performed and studied with Scatchard analysis (Fig. 4B and C) [42]. After incubating the two types of hydrogels in different SA solutions at 37 °C for 12 h (to ensure binding equilibrium), the amount of bound SA on SIH and NIH was quantified. Likewise, SIH showed significantly higher SA binding capacity as compared to NIH over a wide range of SA concentrations (Fig. 4B). Scatchard analysis of the isothermal binding data showed that the plots for SIH could be fitted into two straight lines, indicating the heterogeneous binding sites towards SA in SIH, i.e., the high-affinity and low-affinity sites. The binding association constant (Ka) of high-affinity sites was determined to be 4.75 × 104 M−1, which is significantly stronger than that of the low-affinity sites (1.12 × 104 M−1) (Fig. 4C). The presence of such kind of high specific binding sites in SIH further confirmed the successful imprinting of SA in the hydrogel network.
Fig. 4.
(A) Equilibrium bindings of SA in PBS (10 μg/mL) on different amount of SIH or NIH. (B) Binding isotherms of SA onto SIH or NIH (24 mg/mL) in PBS over a SA concentration range of 0–40 μg/mL. (C) Scatchard analysis of the binding isotherms. (D) Thermo-responsiveness of the binding capacity.
Since these poly (NIPAAm)-based hydrogels possessed thermo-responsive surface wettability and inner structural changing properties, their temperature-dependent SA rebinding property was further determined (Fig. 4D). At 37 °C, the SIH hydrogel show 2.4-fold higher SA binding capacity than that of the NIH hydrogel. When the temperature decreased into 25 °C, both SIH and NIH showed significantly reduced SA binding, and the SIH showed a much larger reduction than NIH. In addition, the difference of binding capacity between SIH and NIH at 25 °C became less significant. Such kind of reduction in the binding ability at 25 °C is probably due to the swelling-induced inner structural changing and subsequently the loss or even disappearance of SA imprinting sites in SIH. Conceivably, the thermo-responsive SA-imprinted sites in SIH hydrogel might be used for not only selective recognition of cancer cells with overexpressed SA on the membrane glycans at 37 °C, but also rapid release of the bound cells upon cooling down the temperature, such as 25 °C or lower.
3.3. Cancer cell capture and release
With this thermo-responsive and SA-specific platform in hand, we then investigated the capture and release properties of SIH and NIH towards a model cancer cell line HepG-2, which has proven to overexpress SA at the glycan terminal of membrane glycoproteins [43]. Both SIH and its control NIH were incubated with DiO-pre-stained HepG-2 cells (green) in 1 mL of cell culture medium (5 × 104 cells/mL). The specifically captured cells (i.e., the non-specific cell capture was mildly washed away) on SIH and NIH at different time intervals were recorded with the assistance of a fluorescence microscope. As shown in Fig. 5A, the number of captured HepG-2 cells on SIH and NIH both exhibited a time-dependent manner, and the SIH with SA-imprinted sites was found to be able to capture an abundance of cells, which was much more than that on the non-imprinted control NIH. Quantitative analysis showed that, after incubation for 2.5 h, the SIH could capture 1.67 × 104 cells with a capture efficiency around 33.4% (Fig. 5B). In contrast, only 0.13 × 104 HepG-2 cells (capture efficiency: 2.5%) were found on the control group NIH, indicating the low non-specificity of NIH for cell attachment. Note that the low and non-specific cell binding on NIH was mainly due to the relatively hydrophilic polymer backbone of poly (NIPAAm), which has proven, in our previous study [40], to be cell repellent in absent of surface tethered cell binding actors. In other words, the results confirmed that the SA-recognition sites in SIH could significantly enhance HepG-2 recognition and meanwhile improve the capture capacity in spite of its inherent cell repelling surface.
Fig. 5.
Cancer cell capture and release. (A) Time-dependent cell capture by the SIH and NIH hydrogel layer (HepG-2 cell, DiO-pre-staining, green). (B) The numbers of captured HepG-2 at different time intervals. (C) Temperature-induced cell release. (D) Cell release efficiency. (E) Details of the captured cells during the capture and release process.
Since the SA-recognition sites in SIH was thermo-sensitive and the SIH surface would loss its SA binding capacity at low temperature (e.g., 25 °C) (Fig. 4D), the SIH layer with capture cells was then incubated in cell culture medium at 25 °C to examine its thermo-responsive cell release property. As shown in Fig. 5C, a rapid cell detachment from the SIH surface could be observed in 30 min. Quantitative analysis revealed that more than 99% of the captured HepG-2 cells could be released, and the residual cells were rarely found on the hydrogel surface (Fig. 5D). This phenomenon demonstrated that the SIH hydrogel layer with thermo-responsive SA binding ability could efficiently capture SA-overexpressed cancer cells and rapidly release them through a biologically acceptable temperature transition method. To get insight into the mechanism of cell attachment and detachment, we then observed the states of captured cells on SIH during the releasing process (Fig. 5E). We found that the captured HepG-2 cells on SIH after 2.5 h of incubation showed a slightly spreading state, implying that the SA-binding sites of SIH facilitated cell attachment. Upon lowing the temperature (e.g., 25 °C), a gradual change of the binding morphology of HepG-2 cells from spreading to round shape was clearly observed in 30 min. Therefore, we deduced that the low-temperature-induced loss of SA-binding, decreasing cell recognition on the surface, and subsequently the cell shrinking behavior contributed to the rapid and efficient release of captured HepG-2 cells. This result, together with the high capture efficiency and rapid release rate, implied the potential of this MIPs-based thermo-responsive platform for cancer cell collection.
3.4. Cell viability after capture/release process
To verify the non-invasive cell capture and release process, we then evaluated the viability of released cells from SIH during the temperature transition process. After one capture/release cycle, the cells were collected and re-seeded on tissue culture polystyrene (TCPS) plates. Adhesion and growth profiles of the recovered and original HepG-2 cells on the TCPS plate were recorded after 1, 3, and 5 days of culture (Fig. 6A). Clearly, the recovered HepG-2 cells could re-adhere on TCPS, and the cell growth behaviors was found to be comparable to that of untreated original ones. This result indicated that the recovered cancer cells was not negatively affected by temperature transition process. Live/dead cell-staining assay was further employed to evaluate the viabilities of recovered HepG-2 cells. Fluorescent images of the original and recovered cells in Fig. 6B both indicated an overall vitality with only very few or negligible damaged cells. Quantitative analysis, by counting the live and dead cells on the surface, revealed that the 98.3% of the recovered cells were alive, further confirming the non-invasive, highly biocompatible properties of the thermo-responsive hydrogel layer for cell harvesting and isolation.
Fig. 6.
The biocompatibility of thermo-responsive cell harvesting process. (A) The proliferation profiles of the original and recovered HepG-2 cells in 5 days. (B) The fluorescent images of HepG-2 cell after live/dead staining. (C) Quantitative result of live/dead cell-staining assay.
3.5. Selectivity of cell capture
The ability to selectively capture cancer cells from the mixture of multiple cells is highly desired, in particular, in the practical isolation of CTCs from blood. Thus, the specific cancer cell capture was further carried out in a cell mixture with both the SA-overexpressed HepG-2 and normal mouse fibroblast L929 cells. Such cell-purification assay was performed using an initial ratio of HepG-2 and L929 cells around 1:3. We intended to applied one capture/release cycle to improve the purity of HepG-2 cells. The two type of cells were pre-stained with DiI (red, for L929) and DiO (green, for HepG-2), respectively. Fluorescent microscope was first used to analyze the cell purity. After incubation of SIH in the cell mixture at 37 °C for 2 h, the SIH layer was taken out, mildly washed with PBS, and then placed at 25 °C followed by collecting the released cells. As shown in Fig. 7A, the fluorescent images of cell mixture after one cycle of capture/release process showed a significant increase in the numbers of HepG-2 cells. Quantitative analysis revealed that the purities of HepG-2 cells were increased from 32.5% to 89.6%, and more than 40% the initial HepG-2 cells could be finally recovered (Fig. 7B). The result indicated the potential of this thermo-responsive platform for selective cancer cell isolation in complex cell samples.
Fig. 7.
(A) and (B) Selective cancer cell capture. (A) The fluorescent images of cell mixture of HepG-2 cells (green, DiO) and L929 cells (red, Dil) before (left) and after (right) on capture/release cycle. (C) Enumeration of the captured HepG-2 cells from artificial CTC blood sample spiked with 200 HepG-2 cells per mL. (D) The numbers of captured HepG-2 cells in three parallel trials.
3.6. Identification and analysis of artificial CTC blood samples
In view of the above positive results, we finally proceed to use the thermo-responsive hydrogel layer for isolating rare cancer cells (e.g., HepG-2 cells) from blood samples. To mimic the CTCs-containing blood of cancer patients, we employed mouse whole blood as the substitute and spiked with 200 HepG-2 cells per mL. This artificial CTC blood sample was then incubated with the SIH hydrogel layer for cell capture. After 2 h of incubation, the captured cells on SIH were identified and enumerated through a three-color immunocytochemistry assay, in which PE-labeled anti-CD45 (PE-CD45), FITC-labeled anti-asialoglycoprotein receptor 1 (FITC-ASGPR1), and Hoechst 33,342 were used to recognize the white blood cells (WBCs), HepG-2 cells, and the cell nuclei, respectively [44,45]. Fluorescent images indicated that the HepG-2 cells in blood could be captured by SIH with non-specific attachment of WBCs (Fig. 7C). Quantitative analysis of three parallel trials indicated that the thermo-responsive cell capture platform could capture more than 15% of the spiked HepG-2 cells from the artificial CTCs blood samples, suggesting a considerable cancer cell capture efficiency (Fig. 7D). It was also worth mentioning that, by lowing the temperature, all the bound cells could be the released from SIH surface, implying a simple and convenient process for cell harvesting. In a word, the above results jointly demonstrated that the MIPs-based platform with switchable SA-recognition would be a promising tool for CTC isolation and purification, and the thermo-responsive mechanism in this work could also provide a low- or non-invasive strategy that is highly desired in accurate cancer diagnosis.
4. Conclusions
In summary, we reported here a thermo-responsive hydrogel layer with chemically designed molecular recognition sites for selective cancer cell isolation. Molecular imprinting technique was employed to generate the molecular affinity towards SA, a specific biomarker that is commonly overexpressed at the glycan chains of glycoproteins on cancer cells. By introducing thermo-responsive poly (NIPAAm) as the hydrogel backbone, switchable SA-recognition could be achieved on the hydrogel layer and shown up in a temperature-dependent manner. Due to the cancer-specificity of SA, the thermo-responsive SA-recognition possessed controlled cancer cell-targeting activity, which enabled selective capture and release of cancer cells (e.g., HepG-2) by changing the temperature. Our results showed that the thermo-responsive hydrogel layer could efficiently capture cancer cells from both the cell culture medium and real blood samples at 37 °C, while the captured cells could be non-invasively released and harvested at 25 °C. We anticipate that, the non-invasive processing mode, considerable capture efficiency, good cell selectivity, as well as the more durable molecular recognition sites compared to natural antibodies or receptors, would make this MIPs-based platform a useful and general tool for CTCs detection and collection.
Declaration of competing interest
The authors declare that no conflict of interest exists in this paper.
CRediT authorship contribution statement
Yue Ma: Methodology, Investigation, Data curation, Writing - original draft. Yimei Yin: Methodology, Investigation, Data curation, Writing - original draft. Li Ni: Methodology, Investigation, Writing - original draft. Haohan Miao: Methodology, Investigation. Yingjia Wang: Investigation. Cheng Pan: Investigation. Xiaohua Tian: Investigation, Validation. Jianming Pan: Investigation, Writing - review & editing. Tianyan You: Resources, Writing - review & editing. Bin Li: Conceptualization, Resources, Supervision, Writing - review & editing. Guoqing Pan: Conceptualization, Resources, Supervision, Writing - review & editing.
Acknowledgement
This work was supported by the National Key Research and Development Program of China (2019YFA0112000 and 2016YFC1100203), the National Natural Science Foundation of China (21875092, 81925027, 21706099 and 81471790), the China Postdoctoral Science Foundation (2016M601745), the Innovation and Entrepreneurship Program of Jiangsu Province, the “Six Talent Peaks” program of Jiangsu Province (2018-XCL-013), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Bin Li, Email: binli@suda.edu.cn.
Guoqing Pan, Email: panguoqing@ujs.edu.cn.
References
- 1.Frederick K.K., Marlow M.S., Valentine K.G., Wand A.J. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persch E., Dumele O., Diederich F. Molecular recognition in chemical and biological systems. Angew. Chem. Int. Ed. 2015;54:3290–3327. doi: 10.1002/anie.201408487. [DOI] [PubMed] [Google Scholar]
- 3.Pan J., Chen W., Ma Y., Pan G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018;47:5574–5587. doi: 10.1039/c7cs00854f. [DOI] [PubMed] [Google Scholar]
- 4.Robertus J., Browne W.R., Feringa B.L. Dynamic control over cell adhesive properties using molecular-based surface engineering strategies. Chem. Soc. Rev. 2010;39:354–378. doi: 10.1039/b906608j. [DOI] [PubMed] [Google Scholar]
- 5.Rosales A.M., Anseth K.S. The design of reversible hydrogels to capture extracellular matrix dynamics. Nature Reviews Materials. 2016;1:15012. doi: 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Midwood K.S., Williams L.V., Schwarzbauer J.E. Tissue repair and the dynamics of the extracellular matrix. Int. J. Biochem. Cell Biol. 2004;36:1031–1037. doi: 10.1016/j.biocel.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Fidler I.J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Canc. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 8.Wang W., Yeung K.W.K. Bone grafts and biomaterials substitutes for bone defect repair: a review. Bioactive Materials. 2017;2:224–247. doi: 10.1016/j.bioactmat.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng X., Shen Z., Mernaugh R. Recombinant antibodies and their use in biosensors. Anal. Bioanal. Chem. 2012;402:3027–3038. doi: 10.1007/s00216-011-5569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y., Zhang L., Yang L., Zhu F., Ding M., Lin F., et al. “Click” chemistry in polymeric scaffolds: Bioactive materials for tissue engineering. J. Contr. Release. 2018;273:160–179. doi: 10.1016/j.jconrel.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Yang T., Du Z., Qiu H., Gao P., Zhao X., Wang H., et al. From surface to bulk modification: plasma polymerization of amine-bearing coating by synergic strategy of biomolecule grafting and nitric oxide loading. Bioactive Materials. 2020;5:17–25. doi: 10.1016/j.bioactmat.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jabbari E. Bioconjugation of hydrogels for tissue engineering. Curr. Opin. Biotechnol. 2011;22:655–660. doi: 10.1016/j.copbio.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song S., Wang L., Li J., Fan C., Zhao J. Aptamer-based biosensors. Trac. Trends Anal. Chem. 2008;27:108–117. [Google Scholar]
- 14.Guo Z., Liu H., Dai W., Lei Y. Responsive principles and applications of smart materials in biosensing. Smart Materials in Medicine. 2020;1:54–65. doi: 10.1016/j.smaim.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subrahmanyam S., Piletsky S.A., Turner A.P.F. Application of natural receptors in sensors and assays. Anal. Chem. 2002;74:3942–3951. doi: 10.1021/ac025673+. [DOI] [PubMed] [Google Scholar]
- 16.Ruigrok Vincent J.B., Levisson M., Eppink Michel H.M., Smidt H., van der Oost J. Alternative affinity tools: more attractive than antibodies? Biochem. J. 2011;436:1–13. doi: 10.1042/BJ20101860. [DOI] [PubMed] [Google Scholar]
- 17.Mahon C.S., Fulton D.A. Mimicking nature with synthetic macromolecules capable of recognition. Nat. Chem. 2014;6:665. doi: 10.1038/nchem.1994. [DOI] [PubMed] [Google Scholar]
- 18.Chen W., Tian X., He W., Li J., Feng Y., Pan G. Emerging functional materials based on chemically designed molecular recognition. BMC Materials. 2020;2:1. [Google Scholar]
- 19.Chen L., Wang X., Lu W., Wu X., Li J. Molecular imprinting: perspectives and applications. Chem. Soc. Rev. 2016;45:2137–2211. doi: 10.1039/c6cs00061d. [DOI] [PubMed] [Google Scholar]
- 20.Xu J., Miao H., Wang J., Pan G. Molecularly imprinted synthetic antibodies: from chemical design to biomedical applications. Small. 2020;16:1906644. doi: 10.1002/smll.201906644. [DOI] [PubMed] [Google Scholar]
- 21.Haupt K., Medina Rangel P.X., Bui B.T.S. Molecularly imprinted polymers: antibody mimics for Bioimaging and therapy. Chem. Rev. 2020;120:9554–9582. doi: 10.1021/acs.chemrev.0c00428. [DOI] [PubMed] [Google Scholar]
- 22.Vlatakis G., Andersson L.I., Müller R., Mosbach K. Drug assay using antibody mimics made by molecular imprinting. Nature. 1993;361:645–647. doi: 10.1038/361645a0. [DOI] [PubMed] [Google Scholar]
- 23.Kubo T., Otsuka K. Recent progress in molecularly imprinted media by new preparation concepts and methodological approaches for selective separation of targeting compounds. Trac. Trends Anal. Chem. 2016;81:102–109. [Google Scholar]
- 24.Arabi M., Ostovan A., Bagheri A.R., Guo X., Wang L., Li J., et al. Strategies of molecular imprinting-based solid-phase extraction prior to chromatographic analysis. Trac. Trends Anal. Chem. 2020;128:115923. [Google Scholar]
- 25.Cui Y., Li M., Hong X., Du D., Ma Y. Solid-phase interfacial synthesis of dual-imprinted colloid particles for multifunctional nanomedicine development. Colloid and Interface Science Communications. 2020;36:100267. [Google Scholar]
- 26.Pan G., Guo Q., Ma Y., Yang H., Li B. Thermo-responsive hydrogel layers imprinted with rgds peptide: a system for harvesting cell sheets. Angew. Chem. Int. Ed. 2013;52:6907–6911. doi: 10.1002/anie.201300733. [DOI] [PubMed] [Google Scholar]
- 27.Pan G., Shinde S., Yeung S.Y., Jakštaitė M., Li Q., Wingren A.G., et al. An epitope-imprinted Biointerface with dynamic Bioactivity for modulating cell–biomaterial interactions. Angew. Chem. Int. Ed. 2017;56:15959–15963. doi: 10.1002/anie.201708635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv S.-W., Liu Y., Xie M., Wang J., Yan X.-W., Li Z., et al. Near-infrared light-responsive hydrogel for specific recognition and photothermal site-release of circulating tumor cells. ACS Nano. 2016;10:6201–6210. doi: 10.1021/acsnano.6b02208. [DOI] [PubMed] [Google Scholar]
- 29.Witek M.A., Freed I.M., Soper S.A. Cell separations and sorting. Anal. Chem. 2020;92:105–131. doi: 10.1021/acs.analchem.9b05357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorin M.A., Verdone J.E., van der Toom E., Bivalacqua T.J., Allaf M.E., Pienta K.J. Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat. Rev. Urol. 2017;14:90–97. doi: 10.1038/nrurol.2016.224. [DOI] [PubMed] [Google Scholar]
- 31.Pearce O.M.T., Läubli H. Sialic acids in cancer biology and immunity. Glycobiology. 2015;26:111–128. doi: 10.1093/glycob/cwv097. [DOI] [PubMed] [Google Scholar]
- 32.Teoh S.T., Ogrodzinski M.P., Ross C., Hunter K.W., Lunt S.Y. Sialic acid metabolism: a key player in Breast cancer metastasis revealed by metabolomics. Frontiers in Oncology. 2018;8 doi: 10.3389/fonc.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinde S., El-Schich Z., Malakpour A., Wan W., Dizeyi N., Mohammadi R., et al. Sialic acid-imprinted fluorescent core–shell particles for selective labeling of cell surface glycans. J. Am. Chem. Soc. 2015;137:13908–13912. doi: 10.1021/jacs.5b08482. [DOI] [PubMed] [Google Scholar]
- 34.Liu R., Cui Q., Wang C., Wang X., Yang Y., Li L. Preparation of sialic acid-imprinted fluorescent conjugated nanoparticles and their application for targeted cancer cell imaging. ACS Appl. Mater. Interfaces. 2017;9:3006–3015. doi: 10.1021/acsami.6b14320. [DOI] [PubMed] [Google Scholar]
- 35.Yin D., Li X., Ma Y., Liu Z. Targeted cancer imaging and photothermal therapy via monosaccharide-imprinted gold nanorods. Chem. Commun. 2017;53:6716–6719. doi: 10.1039/c7cc02247f. [DOI] [PubMed] [Google Scholar]
- 36.Liu S., Pan J., Liu J., Ma Y., Qiu F., Mei L., et al. Dynamically PEGylated and Borate-coordination-polymer-coated polydopamine nanoparticles for synergetic tumor-targeted, chemo-photothermal combination therapy. Small. 2018;14:1703968. doi: 10.1002/smll.201703968. [DOI] [PubMed] [Google Scholar]
- 37.Tian X., Chen X., Feng Y., Duan Y., Dong M., Pan G., et al. Biomimetic fabrication of dynamic biointerfaces with optional and diversified bioactivities through reversible covalent and bioorthogonal chemistry. Chem. Eng. J. 2020;398:125620. [Google Scholar]
- 38.Liu L., Tian X., Ma Y., Duan Y., Zhao X., Pan G. A versatile dynamic mussel-inspired Biointerface: from specific cell behavior modulation to selective cell isolation. Angew. Chem. Int. Ed. 2018;57:7878–7882. doi: 10.1002/anie.201804802. [DOI] [PubMed] [Google Scholar]
- 39.Shno D., Kubo A., Murata Y., Koyama Y., Kataoka K., Kikuchi A., et al. Amine effect on phenylboronic acid complex with glucose under physiological pH in aqueous solution. J. Biomater. Sci. Polym. Ed. 1996;7:697–705. doi: 10.1163/156856296x00462. [DOI] [PubMed] [Google Scholar]
- 40.Guo B., Pan G., Guo Q., Zhu C., Cui W., Li B., et al. Saccharides and temperature dual-responsive hydrogel layers for harvesting cell sheets. Chem. Commun. 2015;51:644–647. doi: 10.1039/c4cc08183h. [DOI] [PubMed] [Google Scholar]
- 41.Schild H.G. Poly (N-isopropylacrylamide): experiment, theory and application. Prog. Polym. Sci. 1992;17:163–249. [Google Scholar]
- 42.Pan G., Zu B., Guo X., Zhang Y., Li C., Zhang H. Preparation of molecularly imprinted polymer microspheres via reversible addition–fragmentation chain transfer precipitation polymerization. Polymer. 2009;50:2819–2825. [Google Scholar]
- 43.Millar J.S. The sialylation of plasma lipoproteins. Atherosclerosis. 2001;154:1–13. doi: 10.1016/s0021-9150(00)00697-3. [DOI] [PubMed] [Google Scholar]
- 44.Varillas J.I., Zhang J., Chen K., Barnes, Liu C., George T.J., et al. Microfluidic isolation of circulating tumor cells and cancer stem-like cells from patients with pancreatic ductal adenocarcinoma. Theranostics. 2019;9:1417–1425. doi: 10.7150/thno.28745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Chen B., He M., Wang H., Xiao G., Yang B., et al. Simultaneous detection of MCF-7 and HepG2 cells in blood by ICP-MS with gold nanoparticles and quantum dots as elemental tags. Biosens. Bioelectron. 2017;90:343–348. doi: 10.1016/j.bios.2016.11.030. [DOI] [PubMed] [Google Scholar]