Abstract
Objectives
Aberrant Wnt signaling cascade is a hallmark of the triple-negative breast cancer (TNBC) that is linked with the increased proliferation, invasion, and poor overall survival. many genes are post-transcriptionally regulated by microRNAs (miRNAs) therefore; it is indisputable that the dysregulation of the miRNAs is an explanation for the aberrant signaling cascades. Thus, the present study was conducted to find the putative miRNA targeting the key players of Wnt/β -catenin cascade in the TNBC.
Methods
The miR-130a-3p was found as a potential regulator of the Wnt signaling cascade by applying several bioinformatic algorithms. Quantitative real-time PCR (qRT-PCR) was used to analyze the expression levels of miR-130a-3p and Wnt cascade genes in the TNBC cells. Afterward, TNBC cells were transiently transfected with the miR-130a-3p to investigate its effects on the expression of Wnt cascade genes. Subsequently, MTT, soft agar colony formation, scratch, transwell cell migration, and transwell cell invasion assays were used to determine the behavior of the TNBC cells in response to miR-130a-3p restoration.
Results
Results of the qRT-PCR showed downregulation of miR-130a-3p and upregulation of the Wnt cascade genes in the TNBC cells compared to the normal cells. Transient overexpression of miR-130a-3p decreased the expression levels of Wnt cascade genes significantly in the TNBC cells. Moreover, following the miR-130a-3p overexpression, the proliferation, anchorage-independent growth, and migration of the TNBC cells were reduced.
Conclusion
Overall, our findings provided an evidence for the significant role of miR-130a-3p in the regulation of Wnt/β-catenin cascade, and also introduced the miR-130a-3p as a new therapeutic target for the patients with TNBC.
Keywords: Cell biology, Bioinformatics, Biotechnology, Biochemistry, Molecular biology, Cancer research, Triple-negative breast cancer, miR-130a-3p, Wnt/β-catenin, ZEB1, FZD6, LRP6
Cell biology; Bioinformatics; Biotechnology; Biochemistry; Molecular biology; Cancer research; Triple-negative breast cancer; miR-130a-3p; Wnt/β-catenin; ZEB1; FZD6; LRP6.
1. Introduction
Breast cancer (BC) is the most common type of cancer among the women accounting for 25% of the cancer diagnosed cases [1]. Pathophysiology of BC is incredibly complex and therefore, it is divided into various subtypes with different therapeutic outcomes [2]. Triple-negative breast cancer (TNBC) is an invasive molecular subtype of BC widely overlapping with the basal-like phenotype [3]. TNBC tumor cells do not express the estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 (HER2) hence, the novel targeted drugs are unsuccessful, and therapeutic options are limited to the chemotherapy to enhance the survival of the patients [4].
Large-scale studies have pointed to the important role of Wnt/β-catenin signaling cascade in the genesis and development of BC. Besides, aberrant Wnt cascade is a hallmark of the TNBC that is linked with many tumor characteristics, such as the increased proliferation and metastasis [5, 6, 7]. The Wnt cascade is generally classified as canonical and non-canonical depending on the involvement of β-catenin [8]. The canonical cascade consists of the components located in different parts of the cell including the ligands, receptors, co-receptors, cytoplasmic proteins, and nuclear transcription factors. During the malignancy, oncogenic Wnt ligands bind to the Frizzled (FZD) receptors and/or LRP6 or LRP5 co-receptors (low-density lipoprotein receptor-related protein 5/6). Then, β-catenin, a key cytoplasmic protein, is separated from the β-catenin's “destruction complex”, relocates to the nucleus, and interacts with the transcription factors to activate the expression of downstream oncogenes associated with higher proliferation and metastasis [9, 10]. Despite having a significant role in the development of many types of cancer, molecular alterations leading to the aberrant Wnt cascade have not been well understood [11].
MicroRNAs (miRNAs) appear as the negative regulators of gene expression, and are involved in the regulation of almost every signaling pathway [12]. These small endogenous RNAs are capable of identifying messenger RNAs (mRNAs) through the sequence-specific interactions, which are common in the mRNA's 3′-UTR (untranslated region), and arrested protein synthesis by translational inhibition or mRNA cleavage [13]. Numerous studies have already shown the abnormal expression of miRNAs in the human cancers, where they can influence the characteristics of the cancer cells as a tumor suppressor or oncogene [14]. Since, each miRNA can target hundreds of mRNAs; it would be really interesting to identify targets of the miRNAs and their biological functions. Meanwhile, transcriptomic, proteomic, and computer-based approaches have provided valuable insights into the identification of miRNA targets [15, 16]. Among these approaches, computer-based methods are not only very simple, time-saving, and cost-effective, but also are important as an initial step in studying the miRNAs and target genes [17].
Accordingly, in the present study, some of the most precise miRNA target prediction algorithms were used to find the putative miRNAs targeting the key players of canonical Wnt cascade at multiple points including the ligands, receptors, cytoplasmic proteins, and nuclear transcription factors with major emphasis on Wnt2B, FZD6, LRP6, CTNNB1, and ZEB1 genes. Consequently, miR-130a-3p was predicted as a potential regulator of Wnt cascade. Then, it was transfected into the highly aggressive human triple-negative breast cancer cell lines, namely MDA-MB-468 and MDA-MB-231 to assess the effects of miR-130a-3p overexpression on (i) the expression of target genes by applying the quantitative real-time polymerase chain reaction (qRT-PCR) (ii) cell proliferation and anchorage-independent growth by performing the MTT and soft agar colony formation assays, respectively, and (iii) cell migration by performing the scratch, transwell cell migration, and transwell cell invasion assays.
2. Material and methods
2.1. In silico analysis
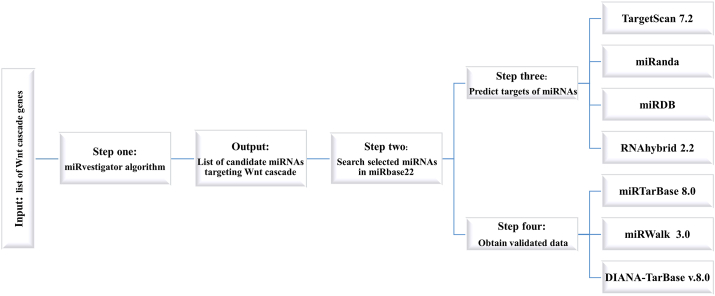
Figure 1 shows the schematic overview of the computational methods used for identification of the miRNA targeting the Wnt cascade genes. Prior to the analysis, a list of Wnt/β-catenin cascade genes over-expressed in the human cancers was prepared as reported by the previous studies and the Kyoto encyclopedia of the genes and genomes (KEGG) pathway enrichment analysis (http://www.genome.jp/kegg/). Then, the key genes with therapeutic potential were selected. The ultimate list of genes included the Wnt2B, FZD6, LRP6, CTNNB1, and ZEB1. Then, the following search direction was used to predict the miRNAs targeting the above-mentioned genes: (i) Gene IDs were imported into the miRvestigator algorithm to precisely associate the 3′-UTR patterns of the genes with the complementary region of miRNA based on the hidden Markov model (HMM). The resulting miRNAs are potentially capable of targeting the co-expressed genes. (ii) Predicted miRNAs were explored in the miRBase and national center for biotechnology information (NCBI) databases to obtain the basic information and to assess the novelty of the study. (iii) TargetScan, miRanda, miRDB, and RNAhybrid were applied as the prediction tools to predict the targets of miRNAs based on some combination of base pairing, conservation of interspecies, and thermodynamic behaviors. (iiii) miRTarBase, miRWalk and DIANA-TarBase databases were applied to obtain the experimentally validated data.
Figure 1.
Schematic overview of bioinformatic software and programs used for predicting miRNA and target genes.
2.2. Cell lines and culture
MDA-MB-468, MDA-MB-231 (human TNBC cell lines), and MCF-10A (normal breast epithelial cells) were purchased from the National Cell Bank of Iran (NCBI, Tehran, Iran). The cells were authenticated through appropriate Short-tandem repeat (STR) profiling, supplied by the NCBI, and verified to be mycoplasma-free using the PCR test. Dulbecco's Modified Eagle Medium (DMEM)-high glucose combined with 10% fetal bovine serum (FBS-Gibco, USA) and 1% antibiotic solution (100 IU/mL penicillin and 100 μg/mL streptomycin) was used for culturing TNBC cell lines. MCF-10A cell line was grown in the same way as other cell lines, except that it was supplemented with 10% horse serum (HS-Gibco, USA) instead of FBS. The cultured cells were maintained at 37 °C, 95% air, 5% CO2 and 100% humidity. Prior to performing any experiment, the number of cells and their viability were determined using trypan blue dye. Cell viability was higher than 90% in all experiments.
2.3. Transient transfection
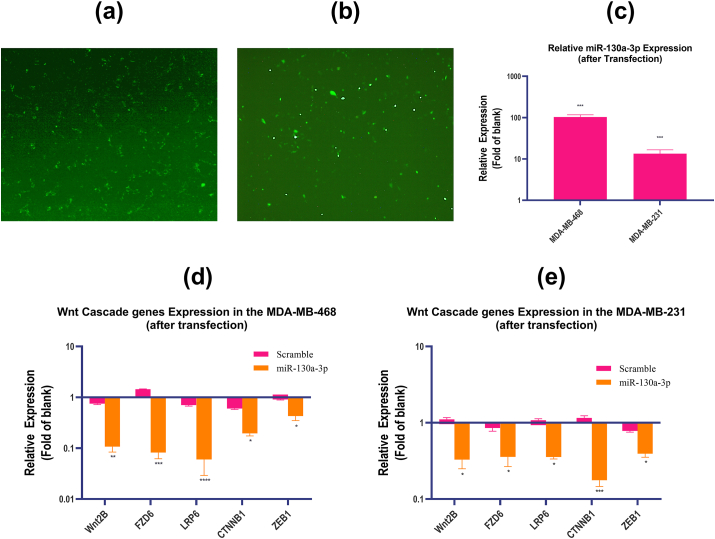
Introduction of the miR-130a-3p miRNA Mimic or scrambled miRNA (Exiqon-Qiagen, Germany) into the TNBC cells was conducted using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as detailed in the manufacturer's protocol. Cells from passages 3–5 were seeded in the 24-well plates and were grown to 90% of confluency at 37 °C in 5% CO2 to improve the transfection efficiency. After 24 h, the cells were transfected by addition of 5nM mimetic or scrambled miRNA solution, which were prepared in combination with the Lipofectamine. The medium was prepared to be FBS-free in this step. Non-transfected cells were used as the controls (blank). Cells were incubated for 6–12 h, followed by replacing the transfection medium with fresh medium containing 10% FBS and antibiotics. Transfection efficiency was evaluated by the fluorescence microscope (Nikon TE2000) and qRT-PCR analysis (Figure 5(a-c)). All the transfection tests were done three times.
Figure 5.
The expression level of miR-130a-3p and Wnt cascade genes after transfection. (a–b) The transfection efficiency of miR-130a-3p under fluorescence microscope in the TNBC cell lines, (a) MDA-MB-468 cell line; (b) MDA-MB-231 cell line. (c) The relative expression of miR-130a-3p in the TNBC cells after transfection with miR-130a-3p determined by qRT-PCR. (d–e) The relative expression of Wnt cascade genes in the TNBC cell lines determined by qRT-PCR, (d) MDA-MB-468 cell line; (e) MDA-MB-231 cell line. miR-130a-3p: cells transfected with miR-130a-3p miRNA Mimic; Scramble: cells transfected with scramble miRNA; Blank: non-transfected cells; The data are presented as mean ± SD for three independent experiments (∗p < 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001 compared to blank).
2.4. RNA extraction
The cellular RNA enriched for miRNAs was isolated from cells using hybrid-R™ RNA extraction kit (GeneAll, Korea) as detailed in the manufacturer's protocol. Briefly, 1ml of RiboEx™ lysis buffer was directly added to the culture flask, and mixed thoroughly by pipetting for 5 min. The mixtures were then transferred to the microtubes and 0.2 ml of chloroform was added to each tube. Mixtures were vigorously shaken and centrifuged for 15 min at 12 000 × g. The supernatant was transferred to a mini spin column and centrifuged for 30 s at around 10 000 × g. Next, the passed-through was discarded, and the spin column washed out two times with wash solutions. Purified RNA was then eluted with RNase-free water. Total RNA isolated from MCF-10A cells was utilized as a corresponding control sample. The concentration of purified RNAs were determined using Nanodrop (Aosheng, China). The integrity of RNAs was assessed by performing denaturing agarose gel electrophoresis. For long term storage, purified RNAs were aliquoted and maintained at -80 °C.
2.5. cDNA synthesis and qRT-PCR
Reverse transcription was conducted for mRNAs or miRNA using RevertAid® RT enzyme (Thermo Scientific, USA). The RT stem loop primers, specific for miR-130a-3p and SNORD47 (U47, as internal control), were designed according to our previously published protocol [18]. Random hexamer primers were used for reverse transcription of the genes and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), as an internal control. qRT-PCR was conducted using the SYBR GreenI detection system by a StepOne plus™ thermal cycler (ABI, USA). The cycling conditions were set at 10 min of 95 °C for first denaturation, followed by 40 rounds of a three-step temperature (95 °C 15 s- 60 °C 40 s- 72 °C 50 s). Melting curves were also analyzed at the end of the reaction. The expression data for genes and miR-130a-3p were normalized with respective internal controls (GAPDH and U47, respectively), and represented as relative expression using 2–ΔΔCt method. The entire experiment was repeated twice for biological replicates, and the measurements were carried out in triplicate as technical replicates. The sequences of qRT-PCR primers are listed in Table1.
Table 1.
Primers used for qRT-PCR.
| Gene | Sense | Anti-sense |
|---|---|---|
| Wnt2B | TCAGAAGTAGCCGAGAGG | ACCGTAGTGGATGTTGTC |
| FZD6 | GGAGTGAAGGAAGGATTAGTC | CAAGCAGAGATGTGGAACC |
| LRP6 | TCAACCCAGAGCTATTGCCTT | TAACCACTGCCTGCCGATTT |
| CTNNB1 | CAACTAAACAGGAAGGGATGG | ACAGTACGCACAAGAGCC |
| ZEB1 | TTACCAGGGAGGAGCAGTGA | CCTTCCTTTCCTGTGTCATCCT |
| GAPDH | CTCTGACTTCAACAGCGAC | CGTTGTCATACCAGGAAATGAG |
| miR-130a-3p | GACCCAGTGCAATGTTAAAAGG | GAGCAGGGTCCGAGGT |
| SNORD47 | ATCACTGTAAAACCGTTCCA | GAGCAGGGTCCGAGGT |
2.6. Proliferation assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed for evaluating cell proliferation. In brief, cells (3 × 103) were seeded in 96-well plate and placed in 37 °C and 5% CO2 incubator. After 24 h incubation, miR-130a-3p miRNA mimic or scrambled miRNA were transfected into cells and cell proliferation was followed for 24, 48, and 72 h. Then after, cells were rinsed twice with PBS, exposed to the MTT solution (0.5mg/ml in PBS) and incubated for 3h at 37 °C in a humidified atmosphere with 5% CO2. The medium was subsequently aspirated and formazan crystals were thoroughly resolved by adding 150 μl of dimethyl sulfoxide (DMSO) per well. After 5–10 min shaking, the absorbance was measured at 570 nm using Microplate Reader (Bio-Rad, Hercules, CA, USA). Non-transfected cells were considered as the controls (blank).
2.7. Soft agar colony formation assay
Anchorage-independent growth ability of cells was determined by performing the soft agar colony formation assay as reported by Borowicz et al. [19] with slight modifications. In brief, the synthetic sequences of miR-130a-3p or scramble-miRNA were transfected into the TNBC cells prior to performing the experiment. The 6-well plate was used to form two layers of agar with different concentrations. To do so, 1.5 ml of a mix of 0.8% noble agar and 2X DMEM medium (containing 10% FBS and 1% penicillin/streptomycin solution) were added to each well to form the bottom layer of agar. Then, the plates were covered and agar mixture was allowed to be solidified at 25 °C in the cell culture hood. The upper layer of agar containing cells was prepared by mixing cells solution (5 × 103 cells per well) with 0.5% noble agar in a 1:1 ratio, and adding 1.5 ml of the mixture into each well. The plate was then covered and allowed agar mixture to solidify before transferring it into a 37 °C and 5% CO2 incubator. After 21 days, the cells were stained by adding 200 μl of nitroblue tetrazolium chloride solution (Sigma, Germany) per well, and incubated for 24 h. Subsequently, colonies with a diameter greater than 50μm were counted. The experiment was repeated in three independent assays. Non-transfected cells were considered as the controls (blank).
2.8. Scratch/wound healing assay
The migratory capacity of whole-cell masses was assessed by performing scratch or wound closure assay according to Grada et al. protocol [20]. Briefly, 1.2 × 105 cells were seeded in 24-well plate, and grown within to reach a 90% confluent cell monolayer. Then, the cells were transfected as described above, and a vertical scratch was generated by a pipette tip through the cell monolayer. The medium along with detached cells was removed, wells were washed twice with PBS, and fresh culture medium containing 0.2–0.5% FBS was added to each well. Following wound formation and observing under inverted microscope, an initial image of the representative area was taken. The next images (in exactly the same area) were captured at multiple time points up to 48h to follow subsequent scratch closure. Non transfected cells were considered as the controls (blank), and the experiment was repeated three times. To analyze scratch closure, snapshots were imported into the ImageJ v1.8.0 (NIH, USA) program, and the scratch width was quantified. The migration area was calculated using the following formula:
| Migration Area = A(t=0)−A(t=Δh) |
A(t=0h) is the area of the wound measured immediately after scratching (t = 0h).
A(t=Δh) is the area of the wound measured hours after the scratch is performed.
2.9. Transwell migration assay
The migratory behavior of cells was further assessed using 8μm transwell inserts (SPL, Life Bioscience, Korea). At first, the synthetic sequence of miR-130a-3p or scramble miRNA was transfected into the TNBC cells. After transfection, on top of the filter membrane in transwell insert, 200 μl of cell solution (~5 × 104 cells per well in FBS-free medium) was added, and then incubated for 10 min with the purpose of settling the cells. 700 μl of the medium containing 10% FBS was added into the bottom of the lower chamber as a chemoattractant. After 24 h incubation at 37 °C and 5% CO2, using a cotton-tipped applicator, non-migrated cells were manually removed from the upper surface of the membrane filter, while migrated cells on the lower surface were fixed with 70% ethanol (Sigma, Germany). The fixed cells were then stained with 0.05% crystal violet (Merck-Darmstadt, Germany), and enumerated as cells per field of view (5 random fields) under an inverted microscope to calculate an average number of the cells. Non transfected cells were considered as the controls (blank).
2.10. Transwell invasion assay
Transwell cell invasion assay was conducted by adding of 100 μl of diluted Matrigel (Corning, USA) into the upper chamber of transwell membranes, and incubation for 2 h at 37 °C for gelling. Afterward, transfected or non-transfected cells (~5 × 104 cells/well, in FBS-free medium) were inoculated into the upper chamber, while 700 μl of DMEM medium containing 10% FBS was added into the lower chamber of the transwell. After 24 h incubation at 37 °C, non-migrated cells were manually removed from the upper surface of the membrane filter, while the invasive cells on the lower surface were fixed, stained, and counted as described in the transwell migration assay.
2.11. Statistical analysis
All data were analyzed with GraphPad Prism version 8.0.1 (GraphPad, La Jolla, CA) using ANOVA followed by Tukey or Bonferroni post hoc tests for multiple comparisons. Results were represented as mean ± SD for three independent experiments, and were considered significant if two-tailed-P- value < 0.05.
3. Results
3.1. In silico analysis showed that hsa-miR-130a-3p targets Wnt signaling cascade
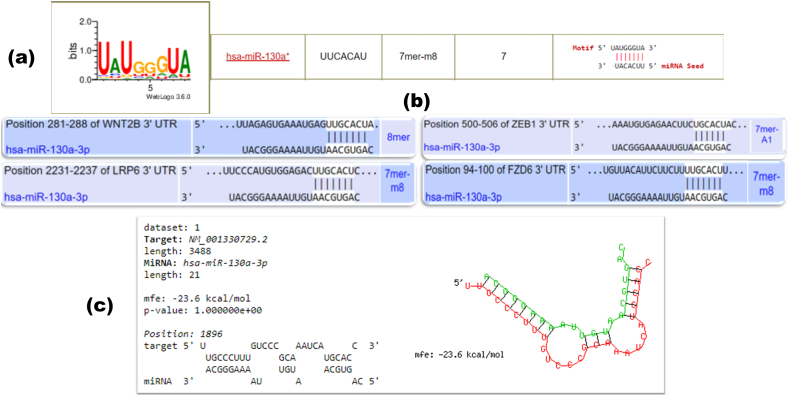
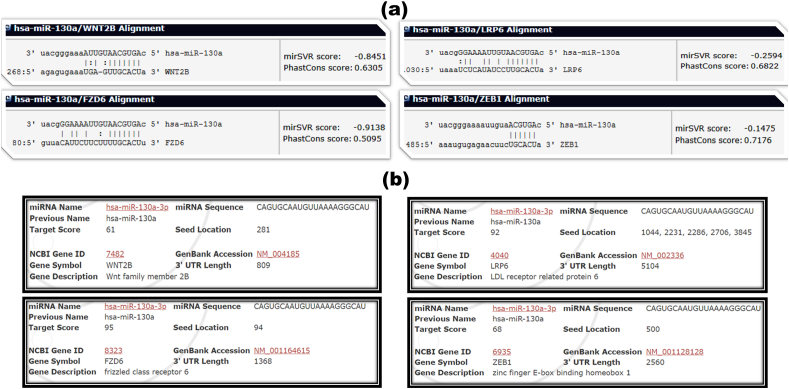
Different miRNA-target prediction tools were used to find a precise miRNA involved in the Wnt/β-catenin signaling cascade. The miRvestigator algorithm has been developed to predict the candidate miRNAs targeting the over-represented sequence pattern of the genes of interest. The gene IDs were submitted into the miRvestigator framework and a list of miRNAs that could potentially target the Wnt signaling cascade were predicted. Then, the predicted miRNAs were explored for more detail in the miRBase database. The predicted miRNAs were also scrutinized by reviewing the previously published papers in the NCBI. Subsequently, hsa-miR-130a-3p was chosen for further computational analyses. As shown in Figure 2(a), hsa-miR-130a-3p could target the Wnt cascade genes with a 7mer-m8 seed model and low P-value based on the miRvestigator results. In the further analysis, the results of miRNA-target prediction tools including TargetScan (Figure 2(b)), RNAhybrid (Figure 2(c)), miRanda (Figure 3(a)), and miRDB (Figure 3(b)) indicated that the miR-130a-3p has at least one matching site in the 3′-UTRs of target genes. Experimentally validated data were also collected from the miRWalk, DIANA-TarBase, and miRTarBase databases, which confirmed that the genes are novel candidates for miR-130a-3p, apart from the FZD6, which is validated to be a target gene for miR-130a-3p based on the next generation sequencing (NGS) studies.
Figure 2.
Screenshots of the result page of miRvestigator, TargetScan, and RNAhybrid. (a) The result page of the miRvestigator framework (mirvestigator.systemsbiology.net). (b) The complementary sequences of miR-130a-3p and Wnt cascade genes are presented according to the TargetScan (www.targetscan.org). (c) The complementary sequences of miR-130a-3p and 3′-UTR mRNA of CTNNB1 is presented according to the RNAhybrid (bibiserv.cebitec.uni-bielefeld.de/rnahybrid).
Figure 3.
Screenshots of the result page of miRanda and miRDB (a) The complementary sequences of miR-130a-3p and Wnt cascade genes are presented according to the miRanda (www.mirna.org). (b) The complementary sequences of miR-130a-3p and Wnt cascade genes are presented according to the miRDB (www.mirdb.org).
3.2. miR-130a-3p is downregulated, while Wnt cascade genes are upregulated in the TNBC cell lines
For obtaining more evidence on the role of miR-130a-3p in the pathogenesis of TNBC, its expression level was determined in the MDA-MB-468 and MDA-MB-231 cells compared to the normal cells. As shown in Figure 4(a), the expression level of miR-130a-3p in the MDA-MB-468 and MDA-MB-231 cells was 45 and 13 fold lower than that of the MCF-10A as the normal cells, respectively. These findings support the possible role of miR-130a-3p in the pathogenesis of the breast epithelial cells. In addition, the expression levels of the components of the Wnt signaling cascade listed above were analyzed in the TNBC cell lines. As shown in Figure 4(b), the expression levels of Wnt2B, FZD6, LRP6, CTNNB1, and ZEB1 were higher in the MDA-MB-468 cells than those of the MCF-10A cells (p-value ˂ 0.01). The expression levels of Wnt2B, FZD6, LRP6, CTNNB1, and ZEB1 were also higher in the MDA-MB-231 cell line than those of the normal breast epithelial cells (p-value ˂ 0.05). The results provided an evidence for irregular activation of the Wnt signaling cascade in the TNBC cells.
Figure 4.
The expression level of miR-130a-3p and Wnt cascade genes determined by qRT-PCR. (a) The relative expression of miR-130a-3p in the MDA-MB-468 and MDA-MB-231 cells. (b–c) The relative expression of Wnt cascade genes in the TNBC cell lines, (b) MDA-MB-468 cell line; (c) MDA-MB-231 cell line. The data are presented as mean ± SD for three independent experiments (∗p < 0.05, ∗∗P ≤ 0.01 compared to MCF-10A cell line).
3.3. Transient transfection of miR-130a-3p downregulates Wnt cascade genes in the TNBC cell lines
For further analysis of the bioinformatic findings, the synthetic sequences of miR-130a-3p or scramble miRNA were transfected into the MDA-MB-468 and MDA-MB-231cells. Then, the transfection efficiency of miR-130a-3p was determined by fluorescent microscope and qRT-PCR. As shown in Figure 5(a-c), miR-130a-3p was highly expressed in the MDA-MB-468 and MDA-MB-231 cells compared to non-transfected cells. Furthermore, our results showed that the expression levels of the target genes were decreased markedly when the miR-130a-3p was overexpressed in the MDA-MB-468 cells compared to the non-transfected MDA-MB-468 cells (Figure 5(d)). Additionally, MDA-MB-231 cells transfected with miR-130a-3p showed significant decrease in the expression of Wnt2B, FZD6, LRP6, CTNNB1 and, ZEB1 compared to non-transfected cells (Figure 5(e)). In both cell lines, there was no significant difference between the scramble-transfected cells and non-transfected cells (p-value ˃ 0.05).
3.4. High expression level of miR-130a-3p reduces the proliferation and anchorage-independent growth of the TNBC cells
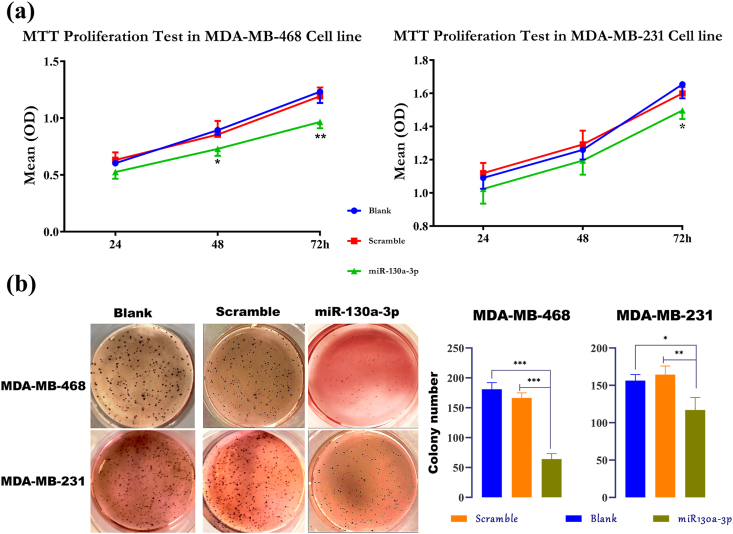
The MTT and colony formation assays were performed to assess the impact of miR-130a-3p overexpression on the proliferation and anchorage-independent growth of the MDA-MB-468 and MDA-MB-231 cells. In the MTT test, the absorbance values were considered for comparisons. As shown in Figure 6(a), after 48 and 72 h, the proliferation of the MDA-MB-468 cells transfected with the miR-130a-3p decreased substantially as compared with non-transfected cells (p-value ˂ 0.05). Moreover, while the influence of miR-130a-3p on the 24 h and 48 h proliferation of the MDA-MB-231 cells was not significant, the absorbance values were decreased significantly 72h after transfection with the miR-130a-3p in comparison with the non-transfected cells (p-value ˂ 0.05). In both cell lines, there was no significant difference between the scramble-transfected cells and non-transfected cells (p-value ˃ 0.05).
Figure 6.
Overexpression of miR-130a-3p decreases the proliferation and anchorage independent growth in the TNBC cells. (a) MTT test in the TNBC cells transfected with miR-130a-3p, scramble, or non-transfected cells. Data are presented as mean ± SD for three independent experiments; One-way ANOVA followed by Tukey's post-hoc test was used for comparison between groups; (∗p < 0.05, and ∗∗P ˂ 0.01 against blank). (b) The soft agar colony formation assay in the TNBC cells transfected with miR-130a-3p, scramble-miRNA, and non-transfected cells. Data are presented as mean ± SD for three independent experiments (∗p < 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001). miR-130a-3p: cells transfected with miR-130a-3p miRNA Mimic; Scramble: cells transfected with scramble miRNA; Blank: non-transfected cells.
The effect of miR-130a-3p overexpression on the anchorage-independent growth ability of the cells was evaluated by the soft agar colony formation assay. As shown in Figure 6(b), the formation of soft agar colonies was decreased markedly in the MDA-MB-468 cells transfected with the miR-130a-3p compared with scramble-transfected cells and non-transfected cells (p-value ˂ 0.0001). In line with MDA-MB-468 results, the number of soft agar colonies in MDA-MB-231 cells transfected with miR-130a-3p was decreased significantly compared to the scramble-transfected cells and non-transfected cells (p-value ˂ 0.05).
3.5. High expression level of miR-130a-3p inhibits the migration of TNBC cells
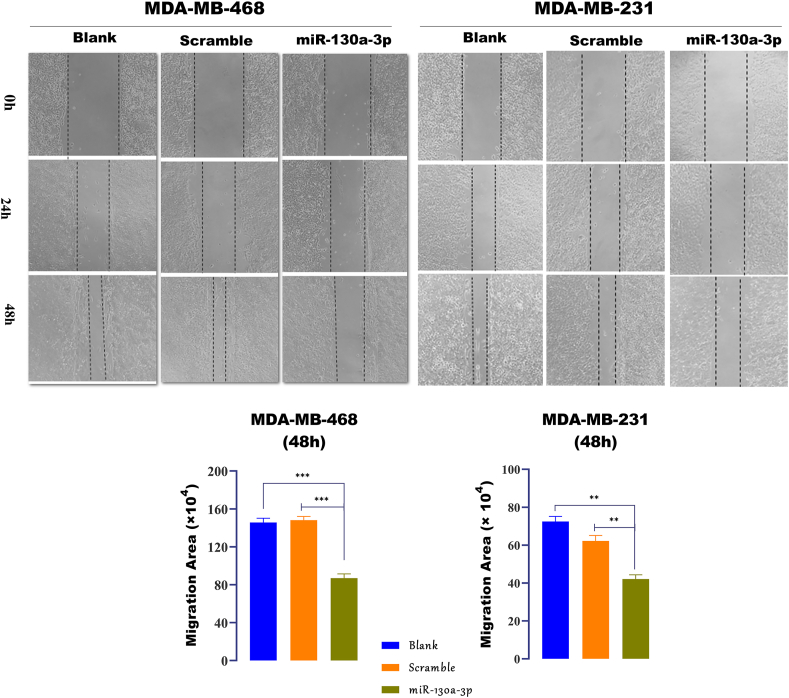
Migration is a feature of the metastatic cells, in which the cells can detach from the primary tumors and move to the surrounding area. The influence of miR-130a-3p on the cell migration was assessed by performing the scratch, transwell cell migration, and transwell cell invasion assays. Scratch or wound-healing assay is based on rendering a wound in a 90% confluent cell monolayer, and then, capturing the images at regular time points to track the wound closure. Quantification of the wound closure using the ImageJ software showed that in both cell lines, wound closure was decreased significantly within 24h and 48h after the restoration of miR-130a-3p expression in comparison with the scramble-transfected cells or non-transfected cells (Figure 7, p-value ˂ 0.0001 for MDA-MB-468 and p-value ˂ 0.001 for MDA-MB-231).
Figure 7.
Cell migration determined by the scratch assay. Wound healing assay and quantification of migration area (magnification, X 100) in the TNBC cells transfected with miR-130a-3p, scramble, and non-transfected cells after 24 h and 48 h. miR-130a-3p: cells transfected with miR-130a-3p miRNA Mimic; Scramble: cells transfected with scramble miRNA; Blank: non-transfected cells. Migrating and invasive cells were counted in ≥5 microscopic fields. Data in the bar charts are presented as the mean ± SD for three independent experiments (∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001).
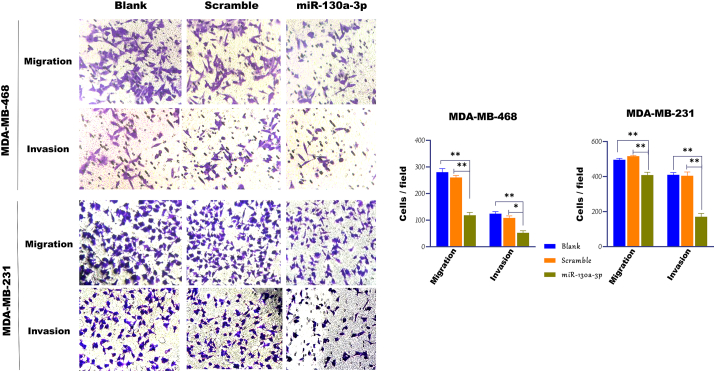
The effects of miR-130a-3p overexpression on the migration of the TNBC cells was further elucidated by the transwell cell migration and invasion assays. As indicated in Figure 8, high expression level of miR-130a-3p reduced the number of cells migrating through the transwell membrane in the MDA-MB-468 cells compared to the scramble-transfected cells, and non-transfected cells (p-value ˂ 0.01). Similar results were observed in the MDA-MB-231 cells, where the miR-130a-3p overexpression induced a significant decrease in the number of migrating cells compared to the scramble-transfected cells, and non-transfected cells (p-value ˂ 0.01). Regarding transwell invasion assay, the number of cells invasive through the transwell membrane, in both cell lines, decreased significantly after transfection with the miR-130a-3p compared to scramble-transfected cells or non-transfected cells (p-value ˂ 0.05).
Figure 8.
Cell migration determined by the transwell assays. Transwell cell migration and invasion assays, and quantification of migrating cells (magnification, X 200) in the TNBC cells transfected with miR-130a-3p, scramble, and non-transfected cells. miR-130a-3p: cells transfected with miR-130a-3p miRNA Mimic; Scramble: cells transfected with scramble miRNA; Blank: non-transfected cells. Migrating and invasive cells were counted in ≥5 microscopic fields. Data in the bar charts are presented as the mean ± SD for three independent experiments (∗p < 0.05, ∗∗P ≤ 0.01).
4. Discussion
The lack of effective treatments and poor prognosis of the patients with TNBC have prompted the significant attempts to find the reliable molecular targets in order to improve the patients’ survival [21]. Wnt signaling cascade is an example of such molecular targets that is dysregulated in more than half of the breast tumors, and is linked with poor overall survival of the patients [22]. MiRNAs are the main regulators of the gene expression and signaling pathways that are differentially expressed in the cancer cells and play a significant role in the development of cancer; thereby emerging as the potential therapeutic agents [23, 24]. Herein, we focused on the miRNA-based therapeutic intervention of breast cancer by applying the bioinformatic approaches. The results of bioinformatic analysis showed that the miR-130a-3p can block the Wnt/β -catenin cascade at multiple points with major emphasis on the Wnt2B, FZD6, LRP6, CTNNB1, and ZEB1 genes. Then, the role of miR-130a-3p was determined in the proliferation, anchorage-independent growth, and migration of the TNBC cells.
The biological significance of each miRNA depends on its targets however, the role of many of them has not been determined yet. Meanwhile, a wide range of computational methods has been developed to recognize the targets of miRNAs [25]. While these methods are important as an initial step to study the miRNAs and target genes, their outcomes are variable [17]. Hence, there is a need for the credible approaches to identify the targets of miRNA with admissible accuracy and sensitivity. Therefore, in this study, a direct, simple, and effective approach of bioinformatic analyses was used to find the miRNA targeting the Wnt/β -catenin signaling cascade. Using the miRvestigator algorithm as a first step to find the candidate miRNA is noticeable, because the purpose of this study is finding the miRNAs targeting the co-expressed genes. The results of bioinformatic prediction led us to select the miR-130a-3p as a potential regulator of the Wnt cascade. We also focused on the key players of the pathway under study, in which Wnt2B, LRP6, and CTNNB1 were novel candidate targets for the miR-130a-3p. Although, CTNNB1 was not listed as a target gene for the miR-130a-3p in most databases, the results of RNAhybrid indicated that the miR-130a-3p has a complementary site in the 3′-UTR of CTNNB1. Similar finding was observed with regards to another gene and miRNA in our previous study as the RNAhybrid predicted the target gene, and the experiments confirmed this interaction hence, the RNAhybrid is suggested as a precise miRNA-target prediction tool [26].
The previous studies have indicated that the miR-130a is involved in the development of different types of cancer, such as liver, cervical, ovarian, glioma, and prostate cancers [27]. In the current study, the expression level of miR-130a-3p was lower in the TNBC cells than the normal cells, suggesting that it is a tumor suppressor miRNA. The results are in agreement with the study by Pan et al. [28] in which the expression level of miR-130a was lower in the BC tissues and cell lines (MCF-7 and MDA-MB-435). In addition, the restoration of miR-130a expression in these cell lines reduced the cell proliferation, migration, and invasion by blocking the RAB5A. The results of another study revealed that overexpression of the miR-130a decreased the viability and colony numbers of doxorubicin-resistant MCF-7 cell line; and consequently, it was concluded that the miR-130a can decrease the drug resistance of human BC cells [29]. No more studies were found concerning the role of miR-130a-3p in the pathogenesis of breast cancer. However, our results were obtained from the TNBC cell lines, and, to the best of our knowledge, this study is the first report in this context.
Wnt ligands are differentially expressed in the BC tissues and cell lines [30]. Previous studies have indicated that the Wnt2B is overexpressed in the non-small cell lung cancer and polycystic ovarian syndrome and its inhibition by the miRNAs decreases the proliferation and invasion of cancer cells [31, 32]. Other studies have indicated that the suppression of Wnt2B decreases the resistance against the chemotherapeutic drugs in the ovarian and nasopharyngeal cancers [33, 34]. Such studies have demonstrated the role of Wnt2B in the development of human cancers however; the role of Wnt2B in the pathogenesis of BC remains as an open question. In the present study, it was shown that the Wnt2B is upregulated in the TNBC cells. Several bioinformatic prediction tools indicated that the Wnt2B is a target for miR-130a-3p. Accordingly, overexpression of miR-130a-3p significantly reduced the expression of Wnt2B in the TNBC cells compared to the control cells.
It has been demonstrated that the FZD6 is involved in both canonical and non-canonical Wnt signaling cascade. However, most studies have suggested an important role for the non-canonical cascade [35]. In the present study, the expression level of FZD6 was shown to be upregulated in the TNBC cells compared to the normal cells. Ectopic expression of the miR-130a-3p decreased the mRNA levels of FZD6 in both TNBC cell lines compared to the control cells. Although, FZD6 has been previously validated as a target for the miR-130a-3p based on the NGS studies that are not a strong evidence but, our validated method was qRT-PCR, which is a strong evidence. The roles of miR-130a-3p and FZD6 were also investigated jointly in the proliferation and migration of the TNBC cells. In line with our results, Corda et al. [36] reported that among 10 human FZD receptors altered in the breast tumors, FZD6 had the highest expression level in the TNBC samples than the normal samples, and its inhibition by the siRNA decreased the metastatic behavior of cancer cells in-vitro and in-vivo. It is important to note that, the Vantictumab, as a monoclonal antibody is produced to block 5 out of 10 FZD receptors. Its efficiency and safety have been tested alone or in combination with the chemotherapeutic drugs in some solid tumors, such as BC. However, FZD6 is not among the targets of Vantictumab, while it has a strong therapeutic potential in the patients with TNBC [22].
The results of the current study showed that the LRP6, as an important Wnt co-receptor is upregulated in the TNBC cells. In line with our results, it has been reported that the LRP6 is not only upregulated in the tissues and cell lines of BC, but also increases more significantly in the TNBC samples than the other subtypes. Moreover, blocking the LRP6 with an antagonist (a specialized chaperone) effectively prevents the tumor growth in-vivo, indicating its importance as a drug target in the human cancers [37, 38]. In the current study, bioinformatic analysis indicated that the LRP6 is a target gene for the miR-130a-3p. Besides, qRT-PCR results showed that the miR-130a-3p can suppress the expression of LRP6.
The β -catenin (CTNNB1 gene) is an important target gene of the Wnt/β -catenin signaling cascade. The expression of β -catenin in the BC is not only high but also is correlated with the histological grade of the tumors, and is negatively correlated with the survival of the patients [39]. As previously indicated, in this study, the expression level of CTNNB1was higher in the TNBC cells than the normal cells. In addition, in our previous study, it was shown that the suppression of the key genes of the Wnt signaling cascade, such as CTNNB1 with miR-381 decreases the invasion of cancer cells, while improving the overall survival in a mouse model of TNBC [40]. The high expression level of miR-130a-3p also decreased the expression level of CTNNB1 in the current study. As a result, miR-130a-3p can be considered as another regulator of CTNNB1.
Zinc finger E-box-binding homeobox 1 (ZEB1) is a major transcription factor regulating the epithelial to mesenchymal transition (EMT), which is a feature of the metastatic cells [41]. The abnormal expression of ZEB1 has been indicated in a number of human cancers, where it facilitates the migration, invasion, and metastasis [42]. In the present study, the expression level of ZEB1 was higher in the TNBC cells than the normal cells. In line with our results, it has been reported that the patients with TNBC express high levels of ZEB1 mRNA and display the features of EMT and metastasis [43]. The expression of ZEB1 and Snail1 in the lung cancer has been reported to be regulated by the Wnt/β -catenin cascade. Besides, inhibition of β -catenin induces the downregulation of ZEB1 [44]. In the study by Chi et al. [45] the results of the luciferase reporter assay, qRT-PCR, and western blot analysis showed that the ZEB1 is a direct target of miR-130a-3p in the cisplatin-resistant cervical cancer cells. Therefore, it could be assumed that the expression of ZEB1 was directly suppressed by the miR-130a-3p in the current study. In addition, inhibition of the β -catenin may provide a further explanation for this effect.
For corroborating these results, MTT, soft agar colony formation, scratch, transwell cell migration, and invasion assays were applied to investigate the proliferation, anchorage-independent growth, and migration capacity of the TNBC cells in response to miR-130a-3p restoration. Cells can migrate in several migratory phenotypes; unicellular form, as seen in mesenchymal or ameboid-like motion or by multicellular form, namely whole-cell mass migration [46]. In the present study, both whole-cell mass migration and single-cell migration were explored by performing wound closure assay and transwell assays, respectively, and the results showed that miR-130a-3p can decrease the migratory abilities of the TNBC cells in both phenotypes. Suppression of the Wnt signaling cascade by the miR-130a-3p could explain the resulting reduction in the proliferation and migration of the cancerous cells. It could also be assumed that this decrease in the cell migration may especially be attributed to the direct influence of the miR-130a-3p on the expression of ZEB1, FZD6, and LRP6, which has been reported to be involved in the EMT and metastasis [36, 43, 47].
5. Conclusion
This report puts a spotlight on deciphering the processes underlying the proliferation and migration of the triple-negative breast cancer. Our findings revealed that the miR-130a-3p can target up to five components of the Wnt signaling cascade at the same time. The bioinformatic approaches proposed in the present study can be extended for studies with similar purposes. Although there were some limitations to perform more experiments especially the luciferase reporter assay or analysis of the expression of the target genes at protein level, qRT-PCR assay, as a strong evidence (based on the miRTarBase database), revealed that the selected genes from Wnt/β -catenin cascade are the targets of the miR-130a-3p. Taken as a whole, our findings can be of widespread importance, but further scientific studies are required to confirm these data, which may lead to the development of new therapeutic strategies for the patients with triple-negative breast cancer.
Acknowledgements
This work was financially assisted by a dissertation grant (Ph.D. thesis of J.P, No: 239), and in part by fund from School of Advanced Technologies in Medicine from Shahid Beheshti University of Medical Sciences, Tehran, Iran (No: 15230), and also was supported in part by fund from the Iran National Science Foundation (No: 98003102).
Declarations
Author contribution statement
J. Poodineh: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
M. Sirati-Sabet: Analyzed and interpreted the data.
M. Rajabibazl: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
S. Mohammadi-Yeganeh: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was supported by the School of Advanced Technologies in Medicine from Shahid Beheshti University of Medical Sciences, Tehran, Iran (No: 15230), and the Iran National Science Foundation (No: 98003102).
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Masoumeh Rajabibazl, Email: rajabi_m@sbmu.ac.ir.
Samira Mohammadi-Yeganeh, Email: s.mohammadiyeganeh@sbmu.ac.ir.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Prado-Vázquez G., Gámez-Pozo A., Trilla-Fuertes L., Arevalillo J.M., Zapater-Moros A., Ferrer-Gómez M., Díaz-Almirón M., López-Vacas R., Navarro H., Maín P., Feliú J., Zamora P., Espinosa E., Fresno Vara J.Á. A novel approach to triple-negative breast cancer molecular classification reveals a luminal immune-positive subgroup with good prognoses. Sci. Rep. 2019;9:1538. doi: 10.1038/s41598-018-38364-y. 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 4.Schmid P., Adams S., Rugo H.S., Schneeweiss A., Barrios C.H., Iwata H., Dieras V., Hegg R., Im S.A., Shaw Wright G., Henschel V., Molinero L., Chui S.Y., Funke R., Husain A., Winer E.P., Loi S., Emens L.A. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 5.Arnold K.M., Pohlig R.T., Sims-Mourtada J. Co-activation of Hedgehog and Wnt signaling pathways is associated with poor outcomes in triple negative breast cancer. Oncol. Lett. 2017;14:5285–5292. doi: 10.3892/ol.2017.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey N., Barwick B.G., Moreno C.S., Ordanic-Kodani M., Chen Z., Oprea-Ilies G., Tang W., Catzavelos C., Kerstann K.F., Sledge G.W., Abramovitz M., Bouzyk M., De P., Leyland-Jones B.R. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Canc. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer F.C., Lacroix-Triki M., Savage K., Arnedos M., Lambros M.B., MacKay A., Natrajan R., Reis-Filho J.S. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol. 2011;24:209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 8.Yin P., Wang W., Zhang Z., Bai Y., Gao J., Zhao C. Wnt signaling in human and mouse breast cancer: focusing on Wnt ligands, receptors and antagonists. Canc. Sci. 2018;109:3368–3375. doi: 10.1111/cas.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koval A., Katanaev V.L. Dramatic dysbalancing of the Wnt pathway in breast cancers. Sci. Rep. 2018;8:7329. doi: 10.1038/s41598-018-25672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taciak B., Pruszynska I., Kiraga L., Bialasek M., Krol M. Wnt signaling pathway in development and cancer. J. Physiol. Pharmacol. 2018;69 doi: 10.26402/jpp.2018.2.07. [DOI] [PubMed] [Google Scholar]
- 12.Onyido E.K., Sweeney E., Nateri A.S. Wnt-signalling pathways and microRNAs network in carcinogenesis: experimental and bioinformatics approaches. Mol. Canc. 2016;15:56. doi: 10.1186/s12943-016-0541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avraham R., Yarden Y. Regulation of signalling by microRNAs. Biochem. Soc. Trans. 2012;40:26–30. doi: 10.1042/BST20110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa P.M., Pedroso de Lima M.C. MicroRNAs as molecular targets for cancer therapy: on the modulation of MicroRNA expression. Pharmacy (Basel) 2013;6:1195–1220. doi: 10.3390/ph6101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quillet A., Saad C., Ferry G., Anouar Y., Vergne N., Lecroq T., Dubessy C. Improving bioinformatics prediction of microRNA targets by ranks aggregation. Front. Genet. 2019;10:1330. doi: 10.3389/fgene.2019.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammadi-Yeganeh S., Paryan M., Mirab Samiee S., Soleimani M., Arefian E., Azadmanesh K., Mostafavi E., Mahdian R., Karimipoor M. Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Mol. Biol. Rep. 2013;40:3665–3674. doi: 10.1007/s11033-012-2442-x. [DOI] [PubMed] [Google Scholar]
- 19.Borowicz S., Van Scoyk M., Avasarala S., Karuppusamy Rathinam M.K., Tauler J., Bikkavilli R.K., Winn R.A. The soft agar colony formation assay. J. Vis. Exp. 2014 doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grada A., Otero-Vinas M., Prieto-Castrillo F., Obagi Z., Falanga V. Research techniques made simple: analysis of collective cell migration using the wound healing assay. J. Invest. Dermatol. 2017;137:e11–e16. doi: 10.1016/j.jid.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Canc. Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naidu S., Magee P., Garofalo M. MiRNA-based therapeutic intervention of cancer. J. Hematol. Oncol. 2015;8:68. doi: 10.1186/s13045-015-0162-0. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pohl S.G., Brook N., Agostino M., Arfuso F., Kumar A.P., Dharmarajan A. Wnt signaling in triple-negative breast cancer. Oncogenesis. 2017;6 doi: 10.1038/oncsis.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn D.E., Martin M.M., Feldman D.S., Terry A.V., Jr., Nuovo G.J., Elton T.S. Experimental validation of miRNA targets. Methods (San Diego, Calif.) 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammadi-Yeganeh S., Mansouri A., Paryan M. Targeting of miR9/NOTCH1 interaction reduces metastatic behavior in triple-negative breast cancer. Chem. Biol. Drug Des. 2015;86:1185–1191. doi: 10.1111/cbdd.12584. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H.D., Jiang L.H., Sun D.W., Li J., Ji Z.L. The role of miR-130a in cancer. Breast Cancer. 2017;24:521–527. doi: 10.1007/s12282-017-0776-x. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y., Wang R., Zhang F., Chen Y., Lv Q., Long G., Yang K. MicroRNA-130a inhibits cell proliferation, invasion and migration in human breast cancer by targeting the RAB5A. Int. J. Clin. Exp. Pathol. 2015;8:384–393. [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J., Zhao M., Hu H., Wang J., Ang L., Zheng L. MicroRNA-130a reduces drug resistance in breast cancer. Int. J. Clin. Exp. Pathol. 2019;12:2699–2705. [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh M. Differential regulation of WNT2 and WNT2B expression in human cancer. Int. J. Mol. Med. 2001;8:657–660. doi: 10.3892/ijmm.8.6.657. [DOI] [PubMed] [Google Scholar]
- 31.Wang B., Sun L., Li J., Jiang R. miR-577 suppresses cell proliferation and epithelial-mesenchymal transition by regulating the WNT2B mediated Wnt/beta-catenin pathway in non-small cell lung cancer. Mol. Med. Rep. 2018;18:2753–2761. doi: 10.3892/mmr.2018.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuanyuan Z., Zeqin W., Xiaojie S., Liping L., Yun X., Jieqiong Z. Proliferation of ovarian granulosa cells in polycystic ovarian syndrome is regulated by MicroRNA-24 by targeting wingless-type family member 2B (WNT2B) Med. Sci. Mon. 2019;25:4553–4559. doi: 10.12659/MSM.915320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G., Liu Y., Su Z., Ren S., Zhu G., Tian Y., Qiu Y. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. Eur. J. Canc. 2013;49:2596–2607. doi: 10.1016/j.ejca.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Wang H., Fan L., Xia X., Rao Y., Ma Q., Yang J., Lu Y., Wang C., Ma D., Huang X. Silencing Wnt2B by siRNA interference inhibits metastasis and enhances chemotherapy sensitivity in ovarian cancer. Int. J. Gynecol. Canc. 2012;22:755–761. doi: 10.1097/IGC.0b013e3182540284. [DOI] [PubMed] [Google Scholar]
- 35.Corda G., Sala A. Non-canonical WNT/PCP signalling in cancer: fzd6 takes centre stage. Oncogenesis. 2017;6:e364. doi: 10.1038/oncsis.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corda G., Sala G., Lattanzio R., Iezzi M., Sallese M., Fragassi G., Lamolinara A., Mirza H., Barcaroli D., Ermler S., Silva E., Yasaei H., Newbold R.F., Vagnarelli P., Mottolese M., Natali P.G., Perracchio L., Quist J., Grigoriadis A., Marra P., Tutt A.N., Piantelli M., Iacobelli S., De Laurenzi V., Sala A. Functional and prognostic significance of the genomic amplification of frizzled 6 (FZD6) in breast cancer. J. Pathol. 2017;241:350–361. doi: 10.1002/path.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C.C., Prior J., Piwnica-Worms D., Bu G. LRP6 overexpression defines a class of breast cancer subtype and is a target for therapy. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5136–5141. doi: 10.1073/pnas.0911220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J., Lu W., Chen D., Xu B., Li Y. Role of Wnt Co-receptor LRP6 in triple negative breast cancer cell migration and invasion. J. Cell. Biochem. 2017;118:2968–2976. doi: 10.1002/jcb.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., Zhang H., Hou J., Niu J., Ma Z., Zhao H., Liu C. Clinical implications of β-catenin protein expression in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:14989–14994. [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammadi-Yeganeh S., Hosseini V., Paryan M. Wnt pathway targeting reduces triple-negative breast cancer aggressiveness through miRNA regulation in vitro and in vivo. J. Cell. Physiol. 2019;234:18317–18328. doi: 10.1002/jcp.28465. [DOI] [PubMed] [Google Scholar]
- 41.Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caramel J., Ligier M., Puisieux A. Pleiotropic roles for ZEB1 in cancer. Canc. Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 43.Maturi V., Enroth S., Heldin C.H., Moustakas A. Genome-wide binding of transcription factor ZEB1 in triple-negative breast cancer cells. J. Cell. Physiol. 2018;233:7113–7127. doi: 10.1002/jcp.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X., Li L., Huang Q., Xu W., Cai X., Zhang J., Yan W., Song D., Liu T., Zhou W., Li Z., Yang C., Dang Y., Xiao J. Wnt signaling through Snail1 and Zeb1 regulates bone metastasis in lung cancer. Am. J. Canc. Res. 2015;5:748–755. [PMC free article] [PubMed] [Google Scholar]
- 45.Chi C., Mao M., Shen Z., Chen Y., Chen J., Hou W. HOXD-AS1 exerts oncogenic functions and promotes chemoresistance in cisplatin-resistant cervical cancer cells. Hum. Gene Ther. 2018 doi: 10.1089/hum.2017.256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Justus C.R., Leffler N., Ruiz-Echevarria M., Yang L.V. In vitro cell migration and invasion assays. JoVE. 2014:51046. doi: 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ibrahim S.A., Hassan H., Vilardo L., Kumar S.K., Kumar A.V., Kelsch R., Schneider C., Kiesel L., Eich H.T., Zucchi I., Reinbold R., Greve B., Götte M. Syndecan-1 (CD138) modulates triple-negative breast cancer stem cell properties via regulation of LRP-6 and IL-6-mediated STAT3 signaling. PloS One. 2014;8 doi: 10.1371/journal.pone.0085737. [DOI] [PMC free article] [PubMed] [Google Scholar]