Abstract
Rhizoctonia solani (Rs) is a soil-borne pathogen with a broad host range. This pathogen incites a wide range of disease symptoms. Knowledge regarding its infection process is fragmented, a typical feature for basidiomycetes. In this study, we aimed at identifying potential fungal effectors and their function. From a group of 11 predicted single gene effectors, a rare lipoprotein A (RsRlpA), from a strain attacking sugar beet was analyzed. The RsRlpA gene was highly induced upon early-stage infection of sugar beet seedlings, and heterologous expression in Cercospora beticola demonstrated involvement in virulence. It was also able to suppress the hypersensitive response (HR) induced by the Avr4/Cf4 complex in transgenic Nicotiana benthamiana plants and functioned as an active protease inhibitor able to suppress Reactive Oxygen Species (ROS) burst. This effector contains a double-psi beta-barrel (DPBB) fold domain, and a conserved serine at position 120 in the DPBB fold domain was found to be crucial for HR suppression. Overall, R. solani seems to be capable of inducing an initial biotrophic stage upon infection, suppressing basal immune responses, followed by a switch to necrotrophic growth. However, regulatory mechanisms between the different lifestyles are still unknown.
Keywords: basidiomycete, biotrophy, effector, cathepsin, hypersensitive response, papain inhibitor, protease, soil-borne pathogen, sugar beet
1. Introduction
Plants are constantly exposed to vast numbers of microorganisms and insects. Survival depends on efficient recognition of the intruders and the timely activation of the local and systemic defense machinery. To succeed, plants have evolved two main strategies to detect pathogens [1,2]. The first defense level resides on the external face of a plant cell and includes conserved secreted microbial elicitors, called pathogen-associated molecular patterns (PAMPs), which are recognized by plant receptor proteins or pattern recognition receptors (PRRs) [3]. PAMPs are typically essential components of all classes of pathogens, such as the bacterial flagellin or the fungal chitin. Plants also respond to endogenous molecules released by pathogen invasion, such as cell wall fragments. Altogether, stimulation of PRRs leads to PAMP-triggered immunity (PTI). The second level of defense involves recognition of pathogen virulence molecules, termed effectors, by intracellular nucleotide-binding–leucine-rich repeat receptor genes, and most of them function as resistance (R) genes. This recognition induces effector-triggered plant immunity (ETI). ETI has co-evolved together with specific pathogen genotypes leading to strain or race-specific interactions, which culminates in local and strong hypersensitive response (HR) of plant-host cells restricting pathogen spread. Diversification of ETI receptors and pathogen effectors both within and between species is the norm, whereas the function of some PRRs is conserved widely across plant families [4]. Generally, PTI and ETI give similar overall responses, such as HR, although ETI is qualitatively stronger and much faster [5].
Soil contains millions of organisms and the nutrient-rich root exudates region attracts a myriad of invaders, including pathogens. Soil-borne pathogens represent an understudied category of plant pathogens, much due to their choice of the environment which is complex to monitor and control. The fungus Rhizoctonia solani (in Greek “root killer”) has a wide host range and causes the damping-off disease in seedlings, although plants in all ages can be infected [6]. The disease was first reported in 1858 by Kühn and has a growing impact due to warmer climate conditions. Despite knowledge of R. solani for more than 160 years many questions remain to be answered on its lifecycle and infection processes. This basidiomycete does not form any asexual spores, while the sexual stage (teleomorph: Thanatephorus cucumeris) is extremely rare. Restring structures (microsclerotia) can be formed, which can survive in the soil or plant debris for long periods before new rounds of infection [7,8]. Rhizoctonia solani isolates vary in morphology and genetics and they are grouped in different anastomosis groups (AGs) based on hyphal anastomosis reactions [9]. Isolates that belong to the AG2-2IIIB group are more aggressive against sugar beets and cause root and crown rot, damping off or foliar blight [10].
The majority of effectors, which have been characterized in R. solani so far, are necrosis-inducing ones [11,12,13,14]. In this work, we aimed to identify novel effectors for the R. solani-sugar beet interaction. We prioritized a predicted singleton from the genome dataset of the R. solani AG2-2IIIB strain [15] encoding a rare lipoprotein A (RlpA) gene, henceforth RsRlpA. This protein previously has been reported in bacterial strains, but its exact function is still unknown [16]. We found that RsRlpA was able to suppress PTI-related HR induced by the Avr4/Cf4 complex. Avr4 is a chitin-binding effector recognized by the Cf4 receptor in resistant tomato plants leading to strong HR [17,18]. It is also an active protease inhibitor, interacting with a plant cathepsin. Taken together, this study shows that R. solani deploys effectors other than necrosis-inducing ones, indicating that R. solani relies on an initial biotrophic stage to establish a successful infection, followed by extensive necrotrophic growth when plant tissue is heavily colonized.
2. Results
2.1. The RsRlpA Contains a DPBB Fold and Is Highly Induced upon Early Infection
Genome analysis of R. solani AG2-2IIIB (Rs) revealed 126 predicted, secreted and cysteine-rich proteins [15]. Among them, only 61 were shorter than 400 amino acids and cysteine-rich, and thus considered as candidate effectors [19,20]. Eleven remaining single-gene candidates were predicted, including the RsRlpA (RSOLAG22IIIB_08473) gene. We first monitored the transcript levels in a time-course of sugar beet plantlets grown in R. solani infested soil. The RsRlpA gene was highly induced 2 days post inoculation (dpi) compared to 4 and 6 dpi, indicating a role in the early infection process (Figure 1a). Next, the protein structure of RsRplA was investigated. This gene encodes a putative rare lipoprotein A (RlpA)-like protein containing a conserved region that has a double-psi beta-barrel (DPBB) fold (IPR009009) (Figure S1). Structure analyses revealed 37% homology to a papain inhibitor from Streptomyces species, 35% to a cellulose-binding protein from Clavibacter species and 29% to kiwellin, a protein present in plant cell walls.
Figure 1.
Analysis of the RsRlpA candidate effector. (a) Transcription patterns of the RsRplA gene upon sugar beet infection. Rhizoctonia solani mycelia was grown in potato dextrose broth (PDB) medium and used as control for comparison. The G3PDH gene was used as internal standard. Error bars represent SD based on at least three biological replicates. Different letters (a,b) indicate statistically significant differences according to Tukey’s test (p value < 0.05). (b) Fungal biomass quantification upon infection of sugar beet leaves with Cercospora beticola wild type, empty vector and RsRlpA+ overexpression strains, while inoculation with H20 (mock) was used a negative control. For qPCR, the C. beticola actin (act) gene was used. Data were normalized with the elongation factor gene (elf-1) from Beta vulgaris. Data show the average of three independent overexpression strains and three biological replicates. Different letters (a,b) indicate significant differences according to Tukey’s test (p < 0.05) and error bars represent SD.
2.2. Heterologous Expression of the RsRlpA Promotes Fungal Infection
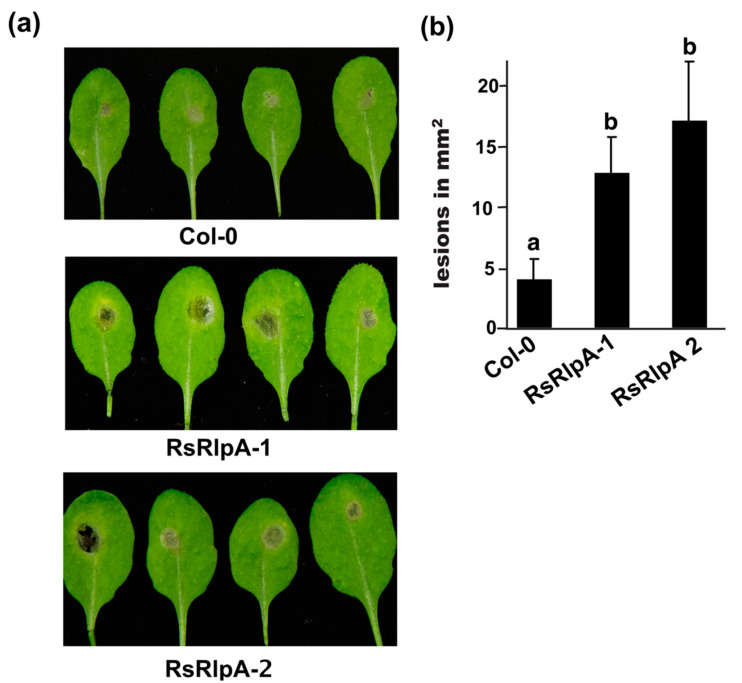
To investigate the role of RsRlpA in virulence, we chose the sugar beet ascomycete pathogen Cercospora beticola for heterologous gene analysis, since R. solani is not amenable to any kind of molecular editing. Reverse transcription (RT) PCR analysis confirmed the expression of RsRlpA+ strains (Figure S2a). Confocal microscopy demonstrated active GFP in overexpression and empty vector strains (Figure S3). The fungal biomass in RsRlpA+ infected plant tissues was significantly increased further supporting the importance of RsRlpA in the infection process (Figure 1b). No differences in the size of necrotic lesions were observed among C. beticola wild type (WT) and the RsRplA+ strains. (Figure S4). The role of RsRlpA in virulence was also investigated in transgenic Arabidopsis thaliana plants (Col-0) harboring a 35S:RsRplA construct. Phenotypic analysis of these lines displayed no differences as compared to WT plants, despite the high expression of RsRplA (Figure S2b). Potential responses of these overexpression lines to Botrytis cinerea were tested. Infected leaves showed more severe symptoms compared to WT (Figure 2a,b) suggesting a non-specific recognition of the RsRplA in plant-fungal interactions.
Figure 2.
Expression of RsRlpA in Arabidopsis thaliana promotes fungal infection. (a) Symptoms of A. thaliana leaves from wild type (Col-0) and overexpression RsRplA lines after infection with Botrytis cinerea conidia. Images taken 3 dpi. (b) Lesion area on A. thaliana leaves infected with B. cinerea 3 dpi. Different letters (a,b) indicate significant differences according to Tukey’s test (p < 0.05). Error bars represent SD of four biological replicates, containing four infected leaves from four plants.
2.3. The RsRplA is Localized to the Cell Periphery and Suppresses Hypersensitive Response Induced by the Avr4/Cf4 Complex
To monitor the localization of RsRlpA in plant cells, a construct was made with GFP at the C-terminus, keeping the signal peptide intact, and transiently expressed in Nicotiana benthamiana plants. The RsRplA protein was accumulated in the cell periphery and nucleoplasm (Figure 3a). The RsRlpA was also tagged with RFP at the C-terminus. Our data showed localization in the cell periphery similar to the GFP-tagged variants, but no localization to the nucleus was observed (Figure 3b), indicating that it was most likely free GFP that localized to the nucleoplasm. Treatment of leaves with mannitol (plasmolysis) confirmed the localization to the cell periphery (Figure 3c).
Figure 3.
The RsRplA effector is localized to plant cell periphery. Live-cell imaging of (a) C-terminal GFP-tagged or (b) tagRFP-tagged RsRlpA in Agro-infiltrated N. benthamiana leaves. The localization was monitored with a laser-scanning confocal microscope with a sequential scanning mode 48 h post infiltration. The GFP and the chlorophyll were excited at 488 nm. GFP (green) and chlorophyll (red) fluorescent signals were collected at 505–525 and 680–700 nm, respectively. (c) Plasmolysis was incited by using 1M mannitol for 30 min. The tagRFP was excited at 558 nm and collected at 545–620 nm. c: cytoplasm, ER: endoplasmic reticulum, n: nucleoplasm, nc: nucleolus.
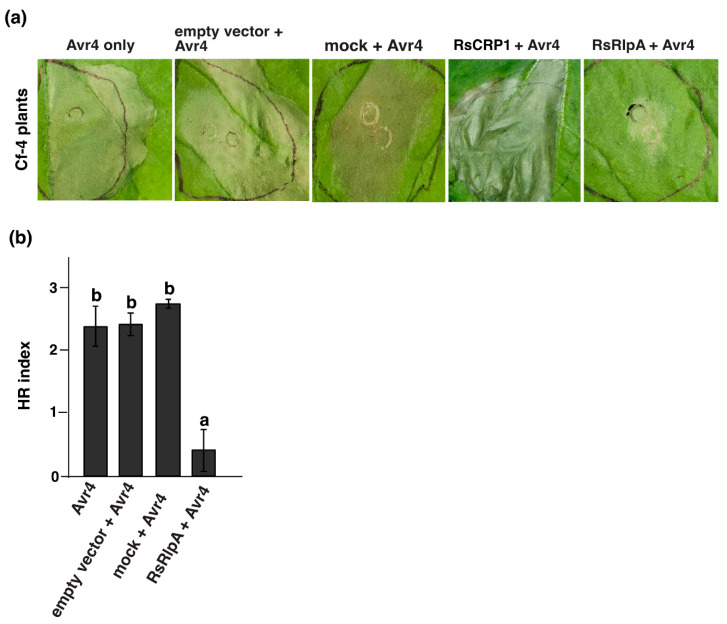
Firstly, we checked whether RsRplA could induce necrosis. In contrast to earlier studies [11,12,13,14], transient expression of this protein in N. benthamiana did not induce cell death (Figure S5). Furthermore, it is known that many pathogens secrete effectors function as HR suppressors [19,20]. For that reason, the ability of RsRlpA to suppress HR was analyzed in the Avr4/Cf4 complex [17,18]. In leaf areas where the RsRlpA had been previously Agro-infiltrated, significantly reduced HR was observed compared to the area where only the Avr4 was expressed (Figure 4a,b). In parallel, potential suppression of ETI-triggered HR was checked in N. bethamiana infected with Pseudomonas syringae pv. tomato DC3000. This bacterial strain secretes effector proteins to the hosts through the type III secretion system, recognized by N. benthamiana R proteins, leading to a strong HR [21]. We found that RsRplA was not able to suppress HR induced by this bacterial strain (Figure S5). Taken together, these data suggest that RsRlpA is only involved in PTI responses.
Figure 4.
The RsRplA effector suppresses the Avr4-mediated hypersensitive response (HR). (a) HR suppression assay by the RsRplA on N. benthamiana expressing the Avr4/Cf4 complex. (b) HR index-scale from 0–3 with “0” indicates no symptoms and “3” severe symptoms on N. benthamiana. Leaves were Agro-infiltrated first with the RsRlpA effector driven by the 35S promoter and HR challenged 24hpi with the Avr4 effector. Agro-infiltration with: empty vector, induction buffer (mock) or the RsCRP1 candidate effector protein were used as negative controls. Images taken 3dpi. Different letters (a,b) indicate significant differences according to Tukey’s test (p < 0.05). Error bars represent SD of samples of two Agro-infiltrated leaves from eight plants.
2.4. The RsRlpA is an Active Protease Inhibitor Suppressing Oxidative Burst
As mentioned before, the predicted 3D structure of RsRlpA displayed homology to papain-like inhibitors. Thus, we investigated whether this protein was able to block proteolytic activity. To this end, we expressed RsRplA in E. coli cells and nanoLC-MS/MS analysis confirmed the correct amino acid sequence of this protein. The inhibitory activity of RsRlpA was examined against papain, a common plant protease from the papaya plant. The Z-Phe-Arg-NMec substrate was used, which upon cleavage emits fluorescence which can be quantified fluorometrically. The RsRlpA was able to block papain proteolytic activity, similar to the N[N-L-3-trans-carboxyoxirane-2-carbonyl-L-leucyl]-agmatine (E-64) inhibitor, a well-known protease inhibitor, used as a positive control (Figure 5a). To clarify whether RsRlpA was also able to suppress oxidative burst, a luminol-based assay was used. ROS production was significantly reduced on leaves which previously had been treated with the RsRplA protein compared to non-treated leaves (Figure 5b). These data suggest that suppression of HR is possibly attributed to the reduction in ROS burst.
Figure 5.
The RsRplA is a protease inhibitor suppressing ROS burst. (a) Protease inhibition assay using 10 μM of purified RsRlpA protein mixed with the Z-Phe-Arg-NMec substrate and 5 μM papain. The protease inhibitor E64 and BSA protein were used as positive and negative controls, respectively. (b) Chitin-induced oxidative (ROS) burst assay in N. benthamiana leaves. Production of ROS was determined using luminol-dependent chemiluminescence. Leaf discs were treated with chitin, 10 μΜ RsRplA protein, while only buffer was used as a negative control. Eight biological replicates were run in both assays.
2.5. The RsRplA Associates with a Plant Cathepsin
To study whether RsRlpA interacts with plant proteases, this protein was tagged with GFP, immunopurified from N. benthamiana and interacting proteins were analyzed by MS/MS. Among the candidate proteases, six were selected for pairwise yeast-two-hybrid analysis (Table S1). Using RsRlpA as a prey and plant proteases as baits, we identified two proteases potentially interacting with the RsRlpA; protease II (NbS00008029g0007.1) and cathepsin B-like cysteine proteinase (NbS00025385g0005.1) (Figure 6a). Structure analysis of cathepsin B-like revealed that it is a papain-like C1A cysteine protease, containing a signal peptide and a cathepsin pro-peptide inhibitor domain (I29).
Figure 6.
The RsRplA interacts with a plant cathepsin. (a) Pairwise yeast-two-hybrid analysis between RsRlpA (used as a prey in pGADT7 vector) and potentially interacting plant proteases (used as baits in pGBKT7 vector). Growth of yeast cells on SD-4 (-His, -Ade, -Leu, -Trp) selective media represents protein–protein interaction and growth on SD-2 (-Leu, -Trp) media confirms yeast transformation. Yeasts transformed with the empty vectors were used as negative controls. (b) Co-immunoprecipitation (co-IP) assay between the GFP-tagged RsRlpA and HA-tagged cathepsin, transiently co-expressed in N. benthamiana and pull-downed using the GFP-trap agarose magnetic beads.
Previously, plant cathepsins have been reported to be involved in different processes such as programmed cell death and autophagy [22,23]. To test RsRlpA-cathepsin B interaction a co-immunoprecipitation assay was conducted. Cathepsin B was tagged with the HA epitope (either in C or N terminus) and transiently co-expressed with the GFP-tagged RsRlpA in N. benthamiana. Our results demonstrated that the C-HA tagged cathepsin B-like could co-immunoprecipitate with RsRlpA (Figure 6b), indicating that this interaction leads to HR suppression.
2.6. The Ser120 is Involved in RsRplA Ability to Suppress Cell Death
Amino acid sequences among known effector proteins function as protease inhibitors were then compared. The EPIC1 and EPIC2 from Phytophthora infestans, which target the Phytophthora Inhibited Protease 1 (PIP1), a papain-like cysteine protease (PLCP) from tomato [24] and the Avr2 effector from the hemibiotrophic pathogen C. fulvum, which inhibits the tomato Rcr3 protease [25] were aligned with RsRlpA. We found that S120, Y122, G129 and C141 were identical between these four effectors (Figure 7a). To evaluate the importance of the conserved amino acids, point mutations were created. In the case of RsRlpA, these four amino acids were located in the DPBB fold. When monitoring the impact of the RsRlpAS120T mutant, it failed to suppress HR induced by the Avr4/Cf4 complex (Figure 7b,c). No visible effects were seen from the other three mutants (Figure 7b,c). The in-planta localization and protein stability of the four RsRlpA mutants were unaltered (Figure 7d).
Figure 7.
Serine 120 in the RsRplA protein is essential for suppression of Avr4-mediated HR. (a) Alignment among the RsRplA amino acid sequence and protease inhibitors from Phytophthora infestans (EPIC1 and EPIC2) and Cladosporium fulvum (Avr2) conducted by Clustal X algorithm. Asterisks indicate identical amino acids. (b) HR suppression assay by the RsRplA mutants on N. benthamiana plants expressing the Cf4/Avr4 complex. (c). HR scale from 0–3 where ‘’0‘’ indicates no symptoms and “3” severe symptoms in N. benthamiana. Leaves were Agro-infiltrated first with the RsRlpA or mutants and HR challenged 24hpi with the Avr4 effector. Empty vector was used as a negative control. Images taken 3dpi. Different letters (a,b) indicate significant differences according to Tukey’s test (p < 0.05). Error bars represent SD of eight plants, each containing two Agro-infiltrated leaves. (d) Live-cell imaging of C-terminal GFP-tagged RsRlpA mutants in Agro-infiltrated N. benthamiana leaves. The localization was monitored with a laser-scanning with a sequential scanning mode 48 hrs post infiltration. The GFP was excited at 488 nm and collected at 505–525 nm.
3. Discussion
Rhizoctonia solani is a soil-borne plant pathogen with an almost unknown infection cycle. Here we have presented data suggesting that R. solani is a potential hemibiotrophic pathogen since it has capacity to generate features of a short biotrophic stage. Mechanistic studies of selected genes in R. solani is complicated since this species is not amenable to any genetic modification. Thus, the employment of tools and strategies from other plant-pathogen systems is the only way forward to generate new information.
Here we followed-up data generated in the genome sequencing of the sugar beet R. solani isolate AG2–2IIIB, known also to cause disease in maize [26]. A predicted unigene effector candidate encoding a rare lipoprotein A (RlpA)-like protein was chosen for further analysis using multiple approaches. The RsRlpA protein contains a DPBB fold with six β-barrels identified previously in various protein super-families, such as expansins, dehydrogenases, endo-glucanases, and in papain-like inhibitors [27,28]. For example, in the rice blast pathogen Magnaporthe oryzae, induction of a RlpA-like gene (MGG_07556.6) was identified at the early biotrophic stage of rice infection [29]. These data are in agreement with the RsRplA transcription patterns, indicating an involvement of this effector at the early stages of the infection. RlpAs have also been identified in the secretomes of virulent Pyrenophora teres f. teres isolates and in ectomycorrhizal species causing brown and white rot [30,31]. In bacteria, the RlpAs are involved in morphogenesis, cell division and peptidoglycan metabolism [32,33]. Despite the rather frequent incidence among various microorganisms, not much is known about their function in a plant defense context. Cysteine protease inhibitors are also present in plants, for example CaPR4c in pepper that interacts with the AvrBsT effector from Xanthomonas campestris [34]. CaPR4c is required for HR and is localized at the plant plasma membrane during cell death. Thus, this type of responses seems to be a common trans-kingdom event.
The predicted protein structure of RsRplA showed homology to papain-like inhibitors. Papain-like cysteine proteases (PLCPs) are involved in numerous plant immune responses against invading pathogens, such as induction of cell death or function as co-receptors for R proteins [35,36,37]. Their precise roles in plant immunity remain to be elucidated. It is speculated that they release damage-associated molecular patterns (DAMPs) which can induce defense mechanisms [38]. Hence, PLCPs are obvious targets for particularly biotrophic pathogens to suppress such responses. Here we found that RsRplA is an active protease inhibitor suppressing papain proteolytic activity and induction of HR by interacting with a plant cathepsin. Effectors inhibiting PLCPs have been previously reported in biotrophic and hemibiotrophic plant pathogens [39,40,41] and the roles of these inhibitors in the suppression of HR have been demonstrated [42,43].
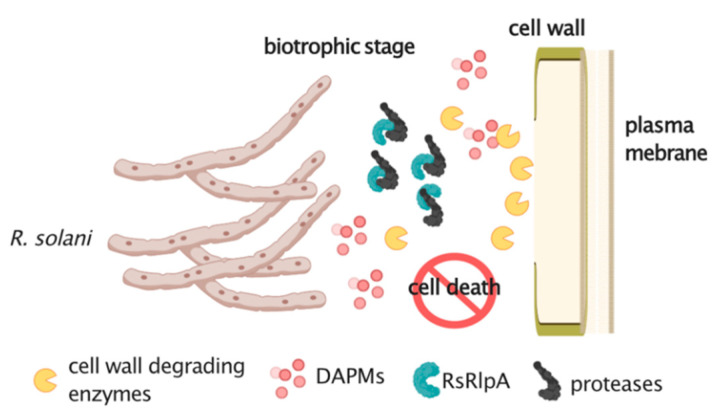
Lifestyles are rarely a strict phenomenon. Changes between different forms or ways to invade or somehow associate with a host plant are far from rare [44]. Based on our studies on the R. solani sugar beet strain AG2-2IIIB, we propose the following model of its different modes of lifestyle (Figure 8). This basidiomycete spends most of its time in soil and infected debris. It has a large repertoire of carbohydrate-active enzyme (CAZy) encoding genes in its genome suitable for cell wall degradation and possible saprophytic survival [25]. These enzymes can generate DAMPs able to induce PTI. At the same time, R. solani possibly deploys effectors to suppress basal host defense responses, such as suppression of programmed cell death, in order to establish a successful infection. These results, in combination with our previous studies, showing that R. solani produces a chitin-binding LysM effector perturbing chitin-induced immunity, supporting the preference of this pathogen to living cells (biotrophic state) at least at the early stages of infection [45]. Intriguingly, in sugar beet, both major latex proteins (MLPs) and NBS-LRR encoding R genes contribute to R. solani defense, suggesting that multiple fungal invasion and defense strategies are employed [46].
Figure 8.
Proposed function of the RsRplA effector during plant infection. Carbohydrate active enzymes degrade the plant cell wall and produce damage-associated molecular patterns (DAMPs) able to induce pattern-triggered immunity (PTI). The RsRplA effector could be deployed by R. solani to perturb protease activity suppressing hypersensitive response during an initial biotrophic stage. This is followed by activity of cell wall degrading enzymes leading to the onset of the necrotrophic stage. Figure created by the Biorender software.
Progression of the pathogen growth and host colonization leads to the sugar beet root and crown rot. This late necrotrophic phase of the infection is most likely dominated by the CAZYs and necrosis-inducing effectors encoded by the fungus. Signals or environmental conditions that determine the switches between the different growth and infection strategies of R. solani remain to be identified.
4. Materials and Methods
4.1. Fungal Isolates and Growth Conditions
The R. solani AG2-2IIIB isolate BBA 69,670 (DSM 101808), originating from sugar beet plants grown in Bavaria, Germany, was used in the current study [26]. Soil was infested with R. solani using media containing perlite, corn flour and water in 1:1:5 ratio. Cercospora beticola isolate Ty1, isolated from infected sugar beet leaves grown in Germany, was kept on potato dextrose agar (PDA, DB Difco, Franklin Lakes, NJ, USA) plates at 22 °C in darkness and sporulation was induced on tomato growth extract medium [47]. Botrytis cinerea strain B05.10 [48] was kept on PDA (DB Difco, Franklin Lakes, NJ, USA) and sporulation was induced by 0.5 M NaCl under continuous blue light.
4.2. RNA Isolation and qRT-PCR Analysis
Three-week-old sugar beet plantlets were transplanted into soil infested by the R. solani as described previously [45]. RNA from plantlets 2, 4, and 6 days post infestation (dpi) and R. solani mycelia, grown in potato dextrose broth (PDB, DB Difco, Franklin Lakes, NJ, USA) and used as a control, was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. qRT-PCR was carried out as previously described [49]. Primers are listed in Table S2. Data were normalized to R. solani G3PDH (RSOLAG22IIIB_07022) transcript levels [50] and relative expression values were calculated according to the 2−ΔΔCt method [51]. At least three biological replicates were used.
4.3. Construction of Cercospora Beticola Transgenic Strains and Infection Assays
For the construction of C. beticola transgenic overexpression strains, the In-Fusion cloning technology was used (Takara, Kusatsu, Japan). The RsRplA gene from R. solani cDNA was amplified using the primers listed in Table S2. The gene fragment was ligated to the pRFHUE-eGFP vector [52] and transformed to C. beticola using an Agrobacterium-mediated protocol [53]. The expression of RsRlpA gene in transgenic strains was investigated by reverse transcription PCR and confocal microscopy.
Leaves of three-week-old sugar beet plantlets were infected with 106 conidia/mL derived from C. beticola (WT), overexpression strains (RsRlpA+) and strains where only the empty vector was expressed as previously described [45]. Mock inoculation with H20 was used as a negative control. The fungal DNA biomass was quantified in infected leaves 7 dpi using quantitative real-time PCR. Total genomic DNA was extracted from infected leaves using a CTAB-mediated protocol [54]. Fungal DNA was quantified using the C. beticola actin (act) reference gene and normalized with B. vulgaris elongation factor (elf-1) gene. Primers are listed in Table S2.
4.4. Transgenic Arabidopsis Plants and Pathogenicity Assays
The RsRlpA effector sequence was amplified from R. solani cDNA using the Phusion Taq polymerase (Thermo Fisher Scientific, Waltham, MA, USA), cloned into pENTR/D-TOPO Gateway vector (Thermo Fisher Scientific, Waltham, MA, USA). The orientation and integrity of the insertion were confirmed by DNA sequencing (Macrogen Inc., Seoul, South Korea). Clones entered the pGWB602 binary vector and transformed to Agrobacterium tumefaciens strain C58 cells, followed by transformation to Arabidopsis Col-0 using the floral dip method as previously described [55]. Expression of the RsRlpA in transgenic lines was confirmed by RT-PCR. Rosette leaves of three-week-old Arabidopsis plants grown on short-day conditions (8 h light/16 h dark) at 22 °C/17 °C, were inoculated with 106/mL conidia of B. cinerea and symptoms were evaluated after three days. Four biological replicates for each transgenic line were used. Each sample comprised four plants and four infected leaves per plant.
4.5. Effector Subcellular Localization and Hypersensitive Response Assays
Using the RsRplA:pENTR/D-TOPO clone, the RsRplA gene entered to the pGWB605 or pGWB660 binary vectors, tagged with C-terminus GFP or tagRFP fluorescence proteins, respectively. Overnight cultures of A. tumefaciens (C58) harboring the 35S:RsRplA constructs were used for Agro-infiltration on 4-week old N. benthamiana leaves [56]. The subcellular localization of the RsRlpA effector was monitored by using a Zeiss LSM 800 confocal microscope. The GFP was excited at 488 nm and collected at 505–525 nm, and tagRFP was excited at 558 nm and collected at 545–620 nm.
For the HR suppression assay, the RsRlpA gene was entered to the pGWB602 binary vector and transiently expressed in N. benthamiana plants (grown under 17 h light/7 h dark at 23 °C) harboring the Cf-4 receptor protein from tomato plants. The condition was: OD600 = 0.5 in 10 mM MES, 10 mM MgCl2 and 150 μM acetosyringone. The HR was triggered 24 hrs after RsRplA Agro-infiltration with the Cladosporium fulvum Avr4 effector protein at OD600 = 0.03. The HR suppression ability of RsRlpA was also investigated in N. benthamiana wild type plants upon infection with P. syringae pv. tomato DC3000 as described previously [57]. Agro-infiltration with empty vector, only induction buffer (mock) and the R. solani RsCRP1 (RSOLAG22IIIB_02432) effector candidate were used as negative controls.
For single point mutation, the procedure by Zheng et al. [58] was applied using primers for site-directed mutagenesis listed in Table S2. PCR products treated with the DpnI restriction enzyme, ligated to pGWB602 vector and transformed to A. tumefaciens C58 cells. The HR suppression assay for the four mutants (RsRlpAS120T, RsRlpAY122F, RsRlpAG129A and RsRplAC141A) was conducted as described above. In total, eight four-week-old plants were Agro-infiltrated and two upper leaves in each plant were used. HR response was evaluated using a scale from 0 to 3, ranging from no symptoms (0) to severe symptoms (3).
4.6. Prediction of Protein 3D Structure, Production and Purification
The presence of conserved domains in the RsRplA effector was tested using the SMART 6.0 protein analysis tool and InterProScan 5.0 [59,60]. Its 3D structure was predicted using the SWISS-MODEL [61] and the RaptorX servers [62]. For protein production, the entire RsRlpA gene was amplified from R. solani cDNA using primers in Table S2. PCR products were digested with NdeI and XhoI restriction enzymes (New England Biolabs, Ipswich, MA, USA), ligated to the pET26b(+) vector and transformed to E. coli SHuffle® T7 cells (New England Biolabs, Ipswich, MA, USA). The orientation and integrity of the insertion were confirmed by DNA sequencing (Macrogen Inc., Seoul, South Korea). Protein production and purification were done as described previously [63]. Transformed cells were grown overnight in LB medium and protein production was induced by 1 mM IPTG (Thermo Fisher Scientific, Waltham, MA, USA) for 18 hrs at 18 °C. Then bacterial cells were precipitated and resuspended in 50 mL PBS buffer and homogenized using a cell press disruptor at 1.93 kbar. The supernatant was loaded onto HisPur cobalt chromatography cartridges (Thermo Fisher Scientific, Waltham, MA, USA) and eluted from the column using 150–300 mM imidazole. Protein identification was performed by denaturation with 8 M urea and MS/MS analysis (Proteome Factory AG, Berlin, Gernany). Before use, excess salt and imidazole were removed by eluting the protein through desalting spin columns (Thermo Fisher Scientific, Waltham, MA, USA).
4.7. Protease Inhibition and ROS Burst Suppression Assays
The protease inhibition assay was done as described previously [64] by measuring the enzymatic activity of Z-Phe-Arg-NMec. 10 μM of purified RsRlpA protein was mixed with 5 μM Z-Phe-Arg-NMec (Merck, Kenilworth, NJ, USA) and 5 μM papain (Sigma-Aldrich, St. Louis, MO, USA) in 850 μL of total reaction buffer (0.1 M buffer KH2PO4/Na2HPO4, pH 6.8; 4 mM cysteine; 1 mM Na2EDTA; 200 mM NaCl; 0.05% Brij 35). 10 μM of cysteine protease inhibitor E-64 (Sigma-Aldrich, St. Louis, MO, USA) or BSA protein were used as a positive and negative control, respectively. For the suppression of the oxidative burst of reactive oxygen species (ROS), a luminol-based protocol was used [65]. Briefly, leaf discs from N. benthamiana plants, treated with chitin (100 μL/mL), luminol (200 μM) (Sigma-Aldrich, St. Louis, MO, USA) and 10 μg/mL horseradish peroxidase (Sigma-Aldrich, St. Louis, MO, USA). The suppression of ROS was analyzed by using 10 μM RsRlpA protein and measuring the chemiluminescence levels.
4.8. Pull-Down and Mass Spectrometry
The RsRlpA protein, tagged with GFP at the C-terminal, was transiently expressed in N. benthamiana leaves, while free GFP was used as a negative control. Proteins were extracted using an extraction buffer containing 20 mM HEPES pH 6.8, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Tween 20, 1 mM PMFS and proteases inhibitor cocktail (Roche) and pull-downed using the GFP-trap agarose magnetic beads (Chromotek, Munich, Germany). Samples proceeded for LC-ESI-MS/MS analysis at the Clinical Proteomics Mass Spectrometry facility, Karolinska Institute/Karolinska University Hospital/Science for Life Laboratory, Stockholm.
On-bead reduction, alkylation and digestion (trypsin, sequencing grade modified, Pierce) was performed, followed by SP3 peptide clean-up of the resulting supernatant [66]. Each sample was separated using a Thermo Scientific Dionex nano LC-system in a 3 hr 5–40% ACN gradient coupled to Thermo Scientific High Field QExactive. The software Proteome Discoverer vs. 1.4 including Sequest-Percolator for improved identification was used to search the N. benthamiana proteome database for protein identification, limited to a false discovery rate of 1%.
4.9. Protein-Protein Interaction
For the yeast-two-hybrid assay, full-length cDNA from the RsRlpA gene was ligated to the pGADT7 prey vector (Takara, Kusatsu, Japan) and plant proteases to the pGBKT7 bait plasmid (Takara, Kusatsu, Japan) and simultaneously transformed to the Saccharomyces cerevisiae AH109 strain. Transformations with empty vectors were used as negative controls. For co-immunoprecipitation assays, the RsRplA GFP-tagged protein and HA-tagged plant protease were transiently co-expressed in N. benthamiana and pull-downed as described above. GFP-tagged protein was detected using the B2 anti-GFP HRP-conjugated antibody (Santa Cruz Biotechnology, Dallas, TX, USA), and HA-tagged proteases were detected using the anti-HA peroxidase-conjugated antibody (Sigma-Aldrich, St. Louis, MO, USA), according to manufacturers’ instructions.
4.10. Statistical Analysis
Analysis of variance (ANOVA, one way) was conducted on gene expression and phenotypic data using a General Linear Model implemented in SPSS version 24 (IBM, Armonk, NY, USA). Pairwise comparisons were made using the Tukey’s test at the 95% significance level.
Acknowledgments
The authors would like to thank Wageningen University for providing Cf-4 transgenic N. benthamiana plants, and the Clinical Proteomics Mass Spectrometry facility, Karolinska Institutet/Karolinska University Hospital/Science for Life Laboratory, for the MS/MS analysis.
Abbreviations
| AG | Anastomosis group |
| DAMP | Damage-associated molecular patterns |
| CAZy | Carbohydrate-Active enZYmes |
| DPBB | Double psi beta barrel |
| DPI | Days post inoculation |
| ETI | Effector-triggered immunity |
| HR | Hypersensitive response |
| PAMPs | Pathogen-associated molecular patterns |
| PLCP | Papain-like cysteine protease |
| PRR | Pattern recognition receptors |
| PTI | Pattern-triggered immunity |
| RlpA | Rare lipoprotein A |
| ROS | Reactive oxygen species |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/21/8070/s1.
Author Contributions
Conceptualization, S.N.C., L.H., P.N.M., C.D. and G.T.; Funding acquisition, C.D., G.T. and P.N.M.; Methodology, S.N.C., F.D., P.N.M. and G.T.; Writing—original draft, L.H., G.T. and C.D.; Writing—review & editing, P.N.M., C.D., F.D., L.H., S.N.C. and G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been funded by Helge Ax:son Johnson’s foundation (G.T.), the Swedish Research Council Formas (grant number 2015-00368) (G.T.), the Swedish Research Council VR (grant number 621-2012-3982) (C.D.), MariboHilleshög Research AB (L.H.), the Swedish University of Agricultural Sciences (F.D.) and an IMBB-FORTH startup fund (P.N.M.). The APC was funded by Stiftelsen Knut och Alice Wallenbergs forskarstipendiefond (grant number GFS2019-0018) (G.T.).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Boller T., Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 4.Giraldo M.C., Valent B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013;11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Tyler B.M., Wang Y. Defense and counter defense during plant-pathogenic oomycete infection. Annu. Rev. Microbiol. 2019;8:667–696. doi: 10.1146/annurev-micro-020518-120022. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay A.N. Handbook on Diseases of Sugar Beet. 1st ed. CRC Press; Boca Raton, FL, USA: 1987. [Google Scholar]
- 7.Cubeta M.A., Vilgalys R. Population Biology of the Rhizoctonia solani complex. Phytopathology. 1997;87:480–484. doi: 10.1094/PHYTO.1997.87.4.480. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez M., Pujol M., Metraux J.P., Gonzalez-Garcia V., Bolton M.D., Borras-Hidalgo O. Tobacco leaf spot and root rot caused by Rhizoctonia solani Kuhn. Mol. Plant Pathol. 2011;12:209–216. doi: 10.1111/j.1364-3703.2010.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sneh B., Burpee L., Ogoshi A. Identification of Rhizoctonia Species. APS Press Inc.; St Paul, MI, USA: 1991. [Google Scholar]
- 10.Bolton M.D., Panella L., Campbell L., Khan M.F. Temperature, moisture, and fungicide effects in managing Rhizoctonia root and crown rot of sugar beet. Phytopathology. 2010;100:689–697. doi: 10.1094/PHYTO-100-7-0689. [DOI] [PubMed] [Google Scholar]
- 11.Zheng A., Lin R., Zhang D., Qin P., Xu L., Ai P., Ding L., Wang Y., Chen Y., Liu Y., et al. The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 2013;4:1424. doi: 10.1038/ncomms2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson J.P., Sperschneider J., Win J., Kidd B., Yoshida K., Hane J., Saunders D., Singh K.B. Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 2017;7:10410. doi: 10.1038/s41598-017-10405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., Peng X., Wang Y., Hua K., Xing F., Zheng Y., Liu W., Sun W., Wei S. The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front. Microbiol. 2019;10:2228. doi: 10.3389/fmicb.2019.02228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei M., Wang A.J., Liu Y., Ma L., Niu X., Zheng A. Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front. Plant Sci. 2020;11:01115. doi: 10.3389/fmicb.2020.01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wibberg D., Andersson L., Rupp O., Goesmann A., Pühler A., Varrelmann M., Dixelius C., Schlüter A. Draft genome sequence of the sugar beet pathogen Rhizoctonia solani AG2-2IIIB strain BBA69670. J. Biotechnol. 2016;222:11–12. doi: 10.1016/j.jbiotec.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Gerding M.A., Liu B., Bendezu F.O., Hale C.A., Bernhardt T.G., de Boer P.A. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosten M.H., Vogelsang R., Cozijnsen T.J., Verberne M., de Wit P.J. The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell. 1997;9:367–379. doi: 10.1105/tpc.9.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Burg H.A., Harrison S.J., Joosten M.H., Vervoort J., de Wit P.J. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 2006;19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 19.Stergiopoulos I., de Wit P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 20.Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L., Tollot M., Zuccaro A., Reissmann S., Kahmann R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015;66:513–545. doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- 21.Collmer A., Lindeberg M., Petnicki-Ocwieja T., Schneider D.J., Alfano J.R. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 2002;10:462–469. doi: 10.1016/S0966-842X(02)02451-4. [DOI] [PubMed] [Google Scholar]
- 22.Martínez M., Cambra I., González-Melendi P., Santamaría M.E., Díaz I. C1A cysteine-proteases and their inhibitors in plants. Physiol. Plant. 2012;145:85–94. doi: 10.1111/j.1399-3054.2012.01569.x. [DOI] [PubMed] [Google Scholar]
- 23.Bárány I., Berenguer E., Solís M.T., Pérez-Pérez Y., Santamaría M.E., Crespo J.L., Risueño M.C., Díaz I., Testillano P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018;69:1387–1402. doi: 10.1093/jxb/erx455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney H.C., van’t Klooster J.W., van der Hoorn R.A., Joosten M.H., Jones J.D., de Wit P.J. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 26.Wibberg D., Andersson L., Tzelepis G., Rupp O., Blom J., Jelonek L., Pühler A., Fogelqvist J., Varrelmann M., Schlüter A., et al. Genome analysis of the sugar beet pathogen Rhizoctonia solani AG2-2IIIB revealed high numbers in secreted proteins and cell wall degrading enzymes. BMC Genom. 2016;17:245. doi: 10.1186/s12864-016-2561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castillo R.M., Mizuguchi K., Dhanaraj V., Albert A., Blundell T.L., Murzin A.G. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure. 1999;7:227–236. doi: 10.1016/S0969-2126(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 28.Quiroz-Castaneda R.E., Martinez-Anaya C., Cuervo-Soto L.I., Segovia L., Folch-Mallol J.L. Loosenin, a novel protein with cellulose-disrupting activity from Bjerkandera adusta. Microb. Cell Fact. 2011;10:8. doi: 10.1186/1475-2859-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21:1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellegrin C., Morin E., Martin F.M., Veneault-Fourrey C. Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 2015;6:1278. doi: 10.3389/fmicb.2015.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ismail I.A., Able A.J. Secretome analysis of virulent Pyrenophora teres f. teres isolates. Proteomics. 2016;16:2625–2636. doi: 10.1002/pmic.201500498. [DOI] [PubMed] [Google Scholar]
- 32.Arends S.J., Williams K., Scott R.J., Rolong S., Popham D.L., Weiss D.S. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J. Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorgenson M.A., Chen Y., Yahashiri A., Popham D.L., Weiss D.S. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol. Microbiol. 2014;93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim N.H., Hwang B.K. Pepper pathogenesis-related protein 4c is a plasma membrane-localized cysteine protease inhibitor that is required for plant cell death and defense signaling. Plant J. 2015;81:81–94. doi: 10.1111/tpj.12709. [DOI] [PubMed] [Google Scholar]
- 35.Krüger J., Thomas C.M., Golstein C., Dixon M.S., Smoker M., Tang S., Mulder L., Jones J.D. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;296:744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- 36.Xu Q.F., Cheng W.S., Li S.S., Li W., Zhang Z.X., Xu Y.P., Zhou X.P., Cai X.Z. Identification of genes required for Cf-dependent hypersensitive cell death by combined proteomic and RNA interfering analyses. J. Exp. Bot. 2012;63:2421–2435. doi: 10.1093/jxb/err397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misas-Villamil J.C., van der Hoorn R.A., Doehlemann G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016;212:902–907. doi: 10.1111/nph.14117. [DOI] [PubMed] [Google Scholar]
- 38.Pearce G., Yamaguchi Y., Barona G., Ryan C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA. 2010;107:14921–14925. doi: 10.1073/pnas.1007568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L.M., Jones A.M.E., Huitema E., van der Hoorn R.A.L., et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müeller A.N., Ziemann S., Treitschke S., Assmann D., Doehlemann G. Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 2013;9:e1003177. doi: 10.1371/journal.ppat.1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark K., Franco J.Y., Schwizer S., Pang Z., Hawara E., Liebrand T.W.H., Pagliaccia D., Zeng L., Gurung F.B., Wang P., et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018;9:1718. doi: 10.1038/s41467-018-04140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Silva I., Poirier G.G., Heath M.C. Activation of cysteine proteases in cowpea plants during the hypersensitive response: A form of programmed cell death. Exp. Cell Res. 1998;245:389–399. doi: 10.1006/excr.1998.4256. [DOI] [PubMed] [Google Scholar]
- 43.Lozano-Torres J.L., Wilbers R.H., Gawronski P., Boshoven J.C., Finkers-Tomczak A., Cordewener J.H., America A.H., Overmars H.A., Van’t Klooster J.W., Baranowski L., et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA. 2012;109:10119–10124. doi: 10.1073/pnas.1202867109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spanu P.D., Panastruga R. Editorial: Biotrophic plant-microbe interactions. Front. Plant Sci. 2017;8:192. doi: 10.3389/fpls.2017.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dölfors F., Holmquist L., Dixelius C., Tzelepis G. A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol. Genet. Genom. 2019;294:1211–1218. doi: 10.1007/s00438-019-01573-9. [DOI] [PubMed] [Google Scholar]
- 46.Holmquist L., Dölfors F., Fogelqvist J., Cohn J., Kraft T., Dixelius C. Major latex protein-like encoding genes contribute to Rhizoctonia solani defense responses in sugar beet. Mol. Genet. Genom. 2020:accepted. doi: 10.1007/s00438-020-01735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcuzzo L.L., Haveroth R., Nascimento A. Induction technique of sporulation in vitro of Cercospora beticola. Summa Phytopathol. 2015;41:74. doi: 10.1590/0100-5405/2054. [DOI] [Google Scholar]
- 48.Staats M., van Kan J.A. Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot Cell. 2012;11:1413–1414. doi: 10.1128/EC.00164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tzelepis G., Melin P., Jensen-Funck D., Stenlid J., Karlsson M. Functional analysis of glycoside hydrolase family 18 and 20 genes in Neurospora crassa. Fungal Genet. Biol. 2012;49:717–730. doi: 10.1016/j.fgb.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Chamoun R., Samsatly J., Pakala S.B., Cubeta M.A., Jabaji S. Suppression subtractive hybridization and comparative expression of a pore-forming toxin and glycosyl hydrolase genes in Rhizoctonia solani during potato sprout infection. Mol. Genet. Genom. 2015;290:877–900. doi: 10.1007/s00438-014-0962-x. [DOI] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Crespo-Sempere A., Lopez-Perez M., Martinez-Culebras P.V., Gonzalez-Candelas L. Development of a green fluorescent tagged strain of Aspergillus carbonarius to monitor fungal colonization in grapes. Int. J. Food Microbiol. 2011;148:135–140. doi: 10.1016/j.ijfoodmicro.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 53.Utermark J., Karlovsky P. Genetic transformation of filamentous fungi by Agrobacterium tumefaciens. [(accessed on 1 March 2017)];Protocol Exchange. 2008 Available online: http://www.nature.com/protocolexchange/protocols/427.
- 54.Möller E.M., Bahnweg G., Sandermann H., Geiger H.H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20:6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods. 2009;5:3. doi: 10.1186/1746-4811-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X. Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protocol. 2011;1:e95. [Google Scholar]
- 57.Gimenez-Ibanez S., Hann D.R., Chang J.H., Segonzac C., Boller T., Rathjen J.P. Differential suppression of Nicotiana benthamiana innate immune responses by transiently expressed Pseudomonas syringae Type III effectors. Front. Plant Sci. 2018;9:688. doi: 10.3389/fpls.2018.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I., Doerks T., Bork P. SMART 6, Recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5, Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S., Li W., Liu S., Xu J. RaptorX-Property: A web server for protein structure property prediction. Nucleic Acids Res. 2016;44:W430–W435. doi: 10.1093/nar/gkw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charova S.N., Gazi A.D., Mylonas E., Pozidis C., Sabarit B., Anagnostou D., Psatha K., Aivaliotis M., Beuzon C.R., Panopoulos N.J., et al. Migration of type III secretion system transcriptional regulators links gene expression to secretion. mBio. 2018;9:e01096-18. doi: 10.1128/mBio.01096-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gawlik K., Gutowicz J. Inhibitory activity against papain, a CA1 cysteine peptidase, in Saccharomycetaceae. Microbiol. Res. 2008;163:545–555. doi: 10.1016/j.micres.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Albert M., Butenko M.A., Aalen R.B., Felix G., Wildhagen M. Chemiluminescence detection of the oxidative burst in plant leaf pieces. Bioprotocol. 2015;5:e1423. doi: 10.21769/BioProtoc.1423. [DOI] [Google Scholar]
- 66.Moggridge S., Sorensen P.H., Morin G.B., Hughes C.S. Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. J. Proteome Res. 2018;17:1730–1740. doi: 10.1021/acs.jproteome.7b00913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.