Abstract
Calmodulin-like proteins (CMLs) represent a large family of plant calcium sensor proteins involved in the regulation of plant responses to environmental cues and developmental processes. In the present work, we identified four alternatively spliced mRNA forms of the grapevine CML21 gene that encoded proteins with distinct N-terminal regions. We studied the transcript abundance of CML21v1, CML21v2, CML21v3, and CML21v4 in wild-growing grapevine Vitis amurensis Rupr. in response to desiccation, heat, cold, high salinity, and high mannitol stress using quantitative real-time RT-PCR. The levels of all four splice variants of VaCML21 were highly induced in response to cold stress. In addition, VaCML21v1 and VaCML21v2 forms were highly modulated by all other abiotic stress treatments. Constitutive expression of VaCML21v2 and VaCML21v4 improved biomass accumulation of V. amurensis callus cell cultures under prolonged low temperature stress. Heterologous expression of the grapevine CML21v2 and VaCML21v4 splice variants in Arabidopsis improved survival rates of the transgenic plants after freezing. The VaCML21v2 overexpression enhanced activation of the cold stress-responsive marker genes AtDREB1A and AtDREB2A, while VaCML21v4 overexpression—AtCOR47, AtRD29A, AtRD29B, and AtKIN1 genes after freezing stress in the transgenic Arabidopsis. The results indicate that the grapevine CML21 gene acts as a positive regulator in the plant response to cold stress. The detected variety of CML21 transcripts and their distinct transcriptional responses suggested that this expansion of mRNA variants could contribute to the diversity of grapevine adaptive reactions.
Keywords: calcium sensor protein, cold stress, mRNA splice variants, transgenic plants and cell cultures, Vitis amurensis
1. Introduction
Plants encode a remarkable variety of Ca2+-binding sensor proteins that are known to detect, decode, and transmit the changes in cellular Ca2+ concentrations to downstream events [1,2,3]. Calmodulin-like proteins (CMLs) constitute a large group of plant EF-hand containing Ca2+-binding proteins that have no enzymatic function but transmit information to downstream targets via Ca2+-dependent protein–protein interaction [4,5]. Therefore, CMLs were classified as sensor relay proteins [6,7]. While CMLs are closely related to calmodulin (CaM), a versatile calcium sensor relay protein in all eukaryotes, they are restricted only to higher plants and some protists [8]. CMLs considerably vary in their lengths containing only one to six predicted Ca2+-binding EF-hand motifs and lack any other known functional domains or intrinsic activities [4,5]. CaMs are more conserved proteins in structure harboring four EF-hands and encoded by a small gene family.
At present, CMLs remain the least studied family of plant Ca2+ sensor proteins. Much remains unknown about the molecular regulation and biological functions of individual CML family protein members. It is known that CMLs are implicated in plant responses to abiotic stresses [9], plant pathogen defense [10], growth and developmental processes [4,11]. For example, specific plant CMLs are known to regulate expression of stress/defense-related genes [12], the most abundant vacuolar Na+-proton exchanger, Na+/H+ exchanger 1 [13] or trichome branching [14]. All these molecular players and processes could contribute to plant adaptation to changing environment and unfavorable conditions. Constitutive expression of certain stress-responsive CMLs in transgenic plants revealed the involvement of the CMLs in plant responses to abiotic stresses as positive [15,16] and negative regulators [17,18]. Genome-wide expression analyses revealed that transcription activity of CMLs can be markedly affected in tea, apple, and grapevine plants by abiotic stimuli, including temperature stresses, high soil salinity, or drought [19,20,21,22]. Search for CML targets has shown that CMLs interact with diverse protein targets, e.g., transcription factors [23], DNA repair proteins [24] or ion transporters [13], which are known to be involved in plant adaptation to abiotic stress.
In a recent study, we identified and characterized the CaM and CML gene families in wild-growing grapevine Vitis amurensis Rupr. under the influence of abiotic stress conditions and found that most of the genes actively responded to abiotic stresses [22]. Wild-growing relatives of cultured plant species often exhibit higher tolerance to abiotic stresses. Therefore, regulatory genes of the wild plants represent an important source for improving abiotic stress tolerance of their cultivated counterparts. During previous work, we also found that several CML genes in V. amurensis, including VaCML21, undergo alternative splicing. Alternative splicing is the regulatory process where pre-mRNA fragments are rearranged by the spliceosomes generating several transcripts from one pre-mRNA molecule, which leads to the production of several protein isoforms. Alternative splicing is currently considered as a regulatory process expanding the genome capacity and protein diversity in response to the unfavorable environmental conditions [25].
Despite there being intron-rich CMLs and some other Ca2+ sensors in the plant genome [3], the existing literature present scarce information on alternative splicing of plant Ca2+ sensors in general and do not present the evidence on alternative splicing of plant CMLs. To the best of our knowledge, there are only three investigations reporting on alternatively spliced plant Ca2+ sensor proteins, presented by three genes of Ca2+-dependent protein kinases (CDPKs or CPKs) [26,27,28]. CDPKs are the prominent Ca2+ sensors combining both Ca2+-binding and kinase domains [1,7]. Nishiyama et al. [26] reported that the liverwort CDPK gene has two almost identical and contiguous exons that were alternatively spliced into two mRNAs. These results indicated that the intron between the two alternative exons was not independently spliced out. Recently, Almadanim et al. [28] identified two alternatively spliced forms (alternative 3′ splice site selection) of rice CPK17 gene that is required for a proper cold stress response. The OsCPK17 splice transcript forms encoded truncated versions of the protein missing both the CDPK autoregulatory (activation) and Ca2+-binding domain due to a premature stop codon. The protein forms exhibited distinct functions and distributions. Previously, we found that the CPK9 gene of V. amurensis undergoes unproductive alternative splicing mediated by canonical 5′GT and 3′AG splicing sites in V. amurensis to generate mRNA isoforms lacking key catalytic kinase subdomains [27]. In addition, Sanyal et al. [29] reported that the CBL-interacting protein kinase 3 (CIPK3) of Arabidopsis is regulated by alternative splicing. CIPKs are known to form functional complexes with calcineurin B-like proteins (CBLs), which are also important EF-hand containing Ca2+ sensors in plants [30].
Therefore, the purpose of the present study was to identify and describe the alternative spliced grapevine CML21 forms and determine whether the spliced CML21 variants play a role in grapevine responses to abiotic stress.
2. Results
2.1. VaCML21 Is Regulated by Alternative Splicing
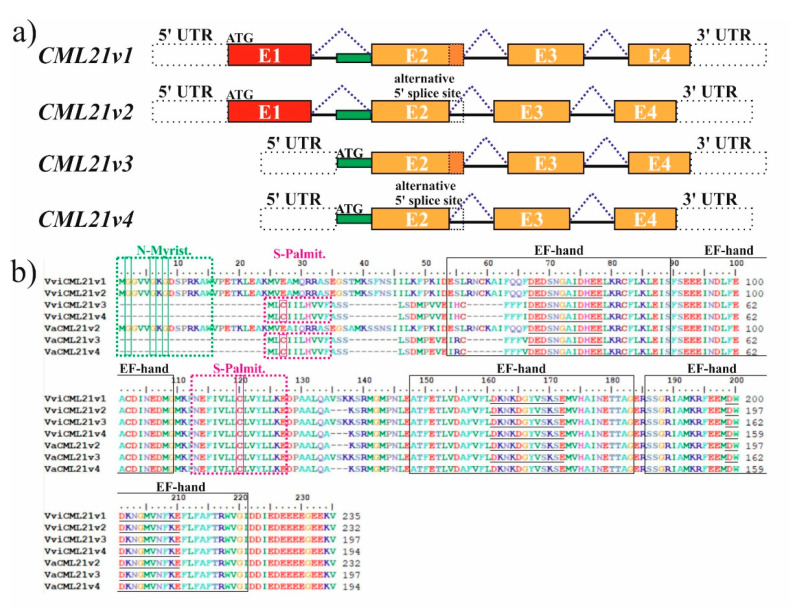
In addition to the VviCML21v1 mRNA, which was predicted previously for V. vinifera [19], our analysis of the V. vinifera CML transcript sequences computationally predicted and retrieved from the CRIBI and NCBI databases made it possible to identify three additional VviCML21 splice variants, i.e., VviCML21v2, VviCML21v3, and VviCML21v4 (Figure 1; Table 1). Our transcript sequence analysis revealed that the transcription start sites differed between the VviCML21v1/VviCML21v2 and VviCML21v3/VviCML21v4 pairs. As a result, the VviCML21v1 and VviCML21v2 splice variants contained exon 1 in contrast to the VviCML21v3 and VviCMLv4, where transcription was predicted to start later. This resulted in the loss of the exon 1 in VviCML21v3 and VviCMLv4 and a retention of the intron 1 partial sequence (Figure 1). Due to the partial intron retention, exon 2 was extended in the VviCML21v3 and VviCMLv4 splice variants (Figure 1). In addition, VviCML21v1 and VviCML21v3 transcript variants contained an identical insertion of nine nucleotides at the end of the exon 2 arising due to an alternative 5′ splice site selection (Figure 1). The boundaries of all putative introns in CML21 contained the highly conserved splice site sequence patterns (both the 5′ donor GT and the 3′ acceptor AG splice sites), supporting the alternative splicing hypothesis. None of the four splice variants of VviCML21 contained premature stop codons (no reading frame shift).
Figure 1.
The structure representation and protein sequence analysis of CML21 mRNA splice variants in grapevine. (a) Alternatively spliced CML21 transcript variants of Vitis amurensis identified in this work (VaCML21v2, VaCML21v3, VaCML21v4) and CML21 transcript variants of Vitis vinifera (VviCML21v1, VviCML21v2, VviCML21v3, VviCML21v4) predicted by genome sequence analysis and retrieved from the databases. Exons and introns are shown using boxes and lines, respectively, with white dashed boxes representing untranslated regions (UTRs). (b) Alignment and conserved motif analysis of CML21 deduced proteins in V. amurensis and V. vinifera. The EF hands were predicted by PROSITE scan [31,32] and the lipid modification sites were predicted using GPS-Lipid [33,34]. Multiple sequence alignment was conducted using BioEdit software [35].
Table 1.
Sequence characteristics of the CML21 mRNAs and their corresponding proteins in Vitis amurensis and Vitis vinifera.
|
Vitis amurensis (cDNA Clone) |
No. of EF Hands |
Predicted Lipid Modification Sites (Position) | MW (kDA) |
Vitis vinifera (mRNA Prediction) |
No. of EF Hands |
Predicted Lipid Modification Sites (Position) | MW (kDA) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcript Variant |
ID (NCBI GenBank) |
CDS (Amino Acids) |
Transcript Variant |
ID | CDS (Amino Acids) |
||||||
| nd | nd | nd | nd | nd | nd | VviCML21v.1 | VIT_219s0015g01200.1 | 708 (235) | 4 | N-Myrist(2,6,8) | 26.9 |
| VaCML21v.2 | MN540599 | 699 (232) | 4 | N-Myrist(2,6,8) N-Palmit(120) |
26.4 | VviCML21v.2 | VIT_219s0015g01200.2 | 699 (232) | 4 | N-Myrist(2,6,8) N-Palmit(120) |
26.5 |
| VaCML21v.3 | MW033207 | 594 (197) | 4 | N-Palmit(3) | 22.7 | VviCML21v.3 | VIT_219s0015g01200.7 | 594 (197) | 4 | N-Palmit(3) | 22.7 |
| VaCML21v.4 | MW033208 | 585 (194) | 4 | N-Palmit(3,82) | 22.4 | VviCML21v.4 | VIT_219s0015g01200.8 | 585 (194) | 4 | N-Palmit(3,82) | 22.4 |
Using RT-PCRs with specific primers designed for VviCML21v1, VviCML21v2, VviCML21v3, and VviCML21v4 splice variants, we cloned and sequenced cDNAs containing full-length coding DNA sequences of CML21 mRNAs from wild-growing V. amurensis using RNA samples isolated from the salt- (VaCML21v2) and cold-stressed (VaCML21v3, VaCML21v4) leaves of V. amurensis. The following sequencing analysis identified three splice variants of CML21 in V. amurensis, including VaCML21v2, VaCML21v3, and VaCML21v4 (Table 1). The transcripts VaCML21v2, VaCML21v3, and VaCML21v4 shared a high nucleotide sequence identity with the VviCML21v2, VviCML21v3, and VviCML21v4 (99%). We were not able to clone the VaCML21v1-like transcript variant using the RNA samples of V. amurensis. Table 1 shows a comparison of the cloned and sequenced CML21 transcript forms of V. amurensis with those computationally predicted for V. vinifera. The deduced amino acid sequences of the cloned and sequenced CML21s of V. amurensis shared high identities with the homologous CML21s of V. vinifera (Table 1).
Like all plant CMLs, the deduced amino acid sequences of the VviCML21 and VaCML21 transcript variants did not contain functional domains other than EF-hands (Table 1). The main structural features and motifs of the deduced CML21s in V. vinifera and V. amurensis are depicted in Figure 1b. Absence of the exon 1 in the VaCML21v3 and VaCML21v4 transcript forms shortened the first EF hand, although the central 12 amino acid residues forming the central Ca2+ binding loop were not changed (Figure 1b). Furthermore, the computational prediction of post-translational modifications in the grapevine CML21s showed the presence of putative N-myristoylation sites in the CML21v1 and CML21v2 protein isoforms, while S-palmitoylation sites were detected in CML21v2, CML21v3, and CML21v4 (Table 1). The differences in the pattern of putative lipid modification sites were due to the removal of exon 1 and partial intron 1 retention (Figure 1). When comparing the deduced amino acid sequences of grapevine CML21s with the Arabidopsis CMLs, the grapevine CML21 shared the highest homology with AtCML21. However, to the best of our knowledge, the current literature does not present reports on the functions and expression pattern of AtCML21.
2.2. CML21 Expression Profile under Different Abiotic Stresses
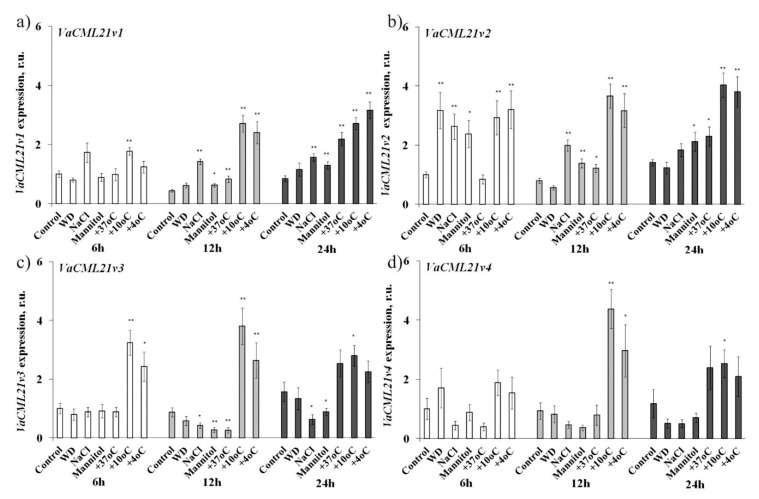
The transcript abundance of the VaCML21v1, VaCML21v2, VaCML21v3, and VaCML21v4 splice variants were analyzed under desiccation, high salinity, high mannitol, cold and heat stresses in the cuttings of wild-growing grape V. amurensis by quantitative real-time RT-PCR (qRT-PCR) (Figure 2). The levels of all four VaCML21 transcript forms were highly up-regulated under both +10 °C and +4 °C cold stress treatments at a minimum of two treatment intervals (Figure 2). Among the four splice variants, the level of VaCML21v4 showed a lower cold stress responsiveness being cold-regulated only 12 h and 24 h post-treatment. At the same time, the transcript pairs VaCML21v1/v2 and VaCML21v3/v4 exhibited distinct inducibility in respect of other abiotic stresses (Figure 2). While the structurally similar pair VaCML21v1 and VaCML21v2 was activated in response to salt, desiccation, mannitol, and heat stress, the pair of VaCML21v3 and VaCML21v4 either did not respond to these stresses or were even down-regulated under the treatments (Figure 2).
Figure 2.
Expression analysis of VaCML21v1 (a), VaCML21v2 (b), VaCML21v3 (c), and VaCML21v4 (d) splice variants 6 h, 12 h, and 24 h post-treatment in the leaves of V. amurensis exposed to control (Control, filtered water, +25 °C), water-deficit (WD, cuttings laid on a paper towel, +25 °C), high salt (0.4 M NaCl, +25 °C), osmoticum (0.4 M mannitol, +25 °C), low temperature (+10 °C and +4 °C), and high temperature (+37 °C) abiotic stress conditions. The expression of the CMLs was profiled by quantitative real-time RT-PCR. The data are presented as the mean ± SE (three independent experiments). *, **—significantly different from the Control for each time point at p ≤ 0.05 and 0.01 according to Student’s t-test.
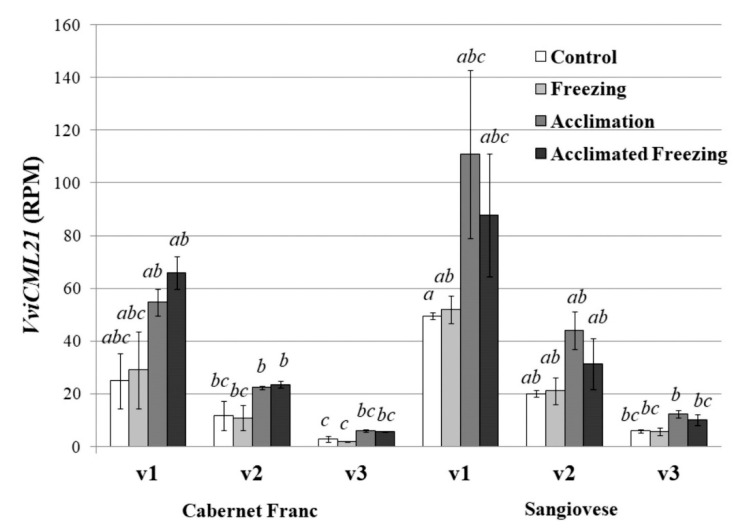
In the present work, we downloaded the RNAseq libraries of cold-treated V. vinifera by Londo et al. [37] from the Sequence Read Archive (SRA) and analyzed the content of the individual VviCML21v1, VviCML21v2, and VviCML21v3 transcripts under acclimation (chilling) and/or freezing stress (Figure 3). We did not find VviCML21v4 in the analyzed RNAseq libraries. Two different V. vinifera cultivars were chosen for this analysis: the most-damaged cultivar “Cabernet Franc” and the least-damaged cultivar “Sangiovese” (Figure 3). The data analysis revealed that the chilling acclimation, i.e., chilling stress, but not freezing itself, increased the transcript abundance of VviCML21v1, VviCML21v2, and VviCML21v3. Freezing without the chilling pretreatment did not have an effect on VaCML21 transcript abundance. Notably, transcript abundance of VviCML21v1, VviCML21v2, and VviCML21v3 was higher in the cold-resistant cultivar “Sangiovese” than that in the cold-sensitive cultivar “Cabernet Franc” by 1.4–3.1 times. In addition, we noted that the VviCML21v3 splice variant was produced at a much lower level than VviCML21v1 and VviCML21v2.
Figure 3.
Expression levels of the CML21v1, CML21v2, and CML21v3 splice variants in Vitis vinifera in the most-damaged cultivar “Cabernet Franc” and the least-damaged cultivar “Sangiovese” cultivated under low-temperature stress conditions. The RNAseq libraries were obtained by Londo et al. [37] and analyzed in the present study for VviCML21 abundance. The V. vinifera plants were exposed to control treatment, freezing (gradual freezing to −3 °C for 45 min), acclimation or chilling (+4 °C for 48 h), and acclimated freezing (+4 °C for 48 h and −3 °C for 45 min). RPM—reads per million mapped reads. The data are presented as the mean ± SE. Means followed by the same letter were not different using one-way analysis of variance (ANOVA), followed by the Tukey HSD multiple comparison test. A value of p ≤ 0.05 was considered significant.
2.3. Genetic Plant Transformation with the VaCML21v2 and VaCML21v4 Splice Variants and Selection of the Transformants
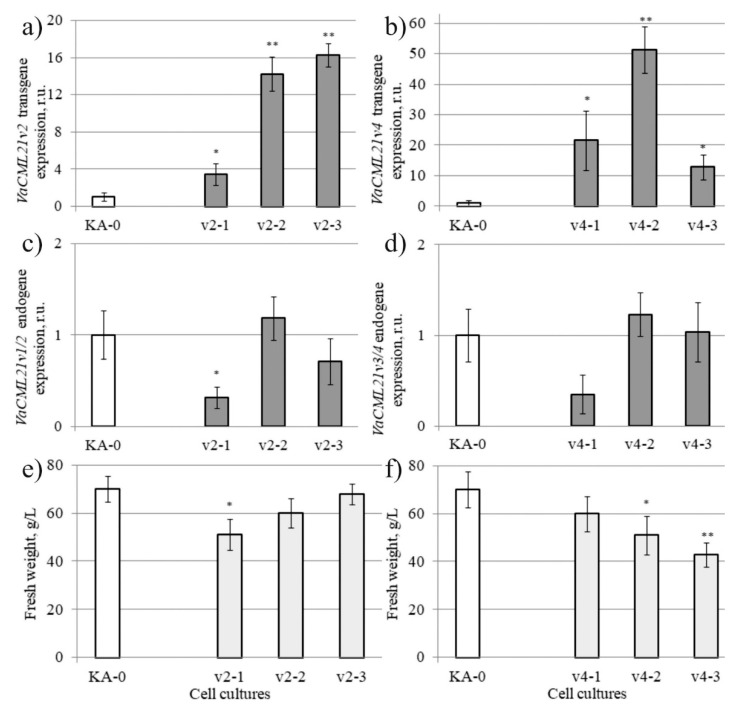
To gain further insight into the involvement of CML21 into the grapevine response to abiotic stresses, we transferred the 35s::VaCML21v2 and 35s::VaCML21v4 splice variants to cell suspension of V. amurensis and established independent transgenic callus cell lines. qRT-PCR revealed that the VaCML21v2-transformed (v2-1, v2-2, v2-3) and VaCML21v4-transformed (v4-1, v4-2, v4-3) callus cell cultures actively expressed the VaCML21v2 and VaCML21v4 transgenes (Figure 4a,b). qRT-PCR revealed that the levels of endogenous VaCML21v1/v2 and VaCML21v3/v4 mRNAs were not sufficiently changed, except for those in v2-1 cell line (Figure 4c,d). Under standard cultivation conditions, the v2-1, v4-2, and v4-3 cell lines accumulated biomass at a lower level than the KA0 control during the 30 d of cultivation (Figure 4e,f).
Figure 4.
Characterization of the grapevine callus cell cultures transformed with the VaCML21v2 and VaCMLv4 splice variants. (a,b) Quantification of the VaCML21v2 and VaCML21v4 transgene mRNAs; (c,d) Quantification of the endogenous VaCML21v1/v2 and VaCML21v3/v4 mRNAs. (e,f) Fresh biomass accumulation in the VaCML21v2-transgenic and VaCML21v4-transgenic cell cultures after 30-day cultivation. v2-1, v2-2, v2-3—VaCML21v2-transformed calli; v4-1, v4-2, v4-3—VaCML21v4-transformed calli. KA-0—control cell culture transformed with the “empty” vector. The data are presented as the mean ± SE (two independent experiments). * p ≤ 0.05; ** p ≤ 0.01 versus values of VaCML21 expression in the empty vector-transformed KA-0 cell culture according to Student’s t-test.
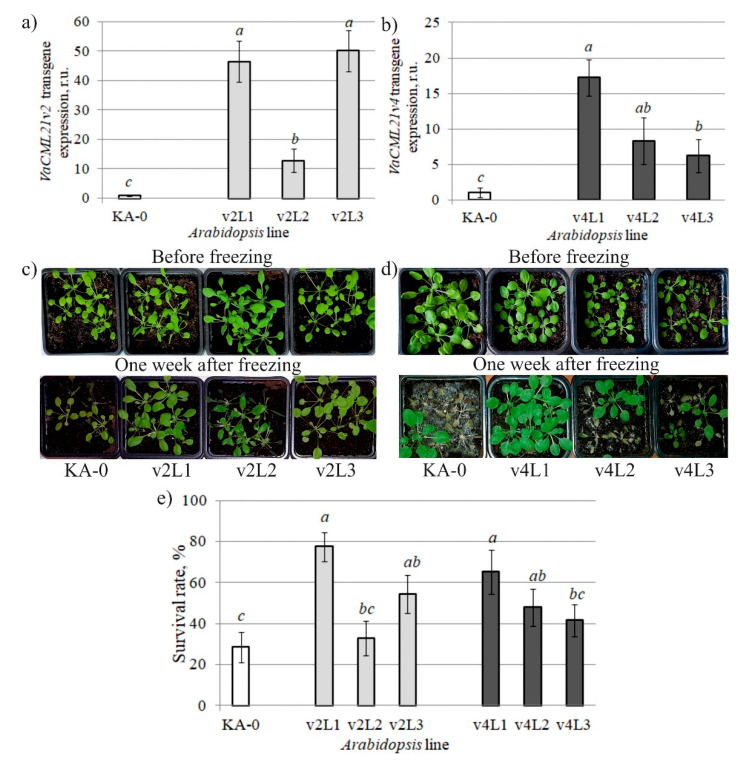
Then, the overexpression construct of VaCML21 or empty vector was transformed in Arabidopsis for generating the VaCML21v2-transgenic (v2L1, v2L2, v2L3), VaCML21v4-transgenic (v4L1, v4L2, v4L3), and control (KA-0) A. thaliana plant lines. The transgenic plant lines showed no morphological aberrations through the T4 generation. Quantification of the VaCML21v2 and VaCML21v4 mRNAs in the transgenic A. thaliana was performed by qRT-PCR (Figure 5a,b). As shown in Figure 5a,b, the transgenic 35S::VaCML21v2 and 35S::VaCML21v4 A. thaliana displayed different levels of VaCML21v2 and VaCML21v4 expression with the highest level of transgene mRNAs being detected in the v2L1, v2L3, and v4L1 lines. Expression of the VaCML21v2 transgene in v2L1 and v2L3 was considerably higher than that observed in v2L2 (by 3.6–3.9 times), while expression of VaCML21v4 transgene in v4L1 markedly exceeded the transgene levels in v4L2 and v4L3 (by 2.1–2.8 times).
Figure 5.
Characterization and responses to freezing of vector control (KA-0), VaCML21v2- (v2L1, v2L2, v2L3) and VaCML21v4-transgenic (v4L1, v4L2, v4L3) Arabidopsis. (a,b) Quantification of the VaCML21v2 and VaCML21v4 mRNAs in A. thaliana performed by qRT-PCR. (c,d) 4-week-old KA-0 and VaCML21-transgenic plants were stressed at −10 °C for 45 min and then cultured at +4 °C for 1 h for recovery. Photographs of representative seedlings were taken after 7 d of recovery. (e) Survival rates determined as the number of visibly green plants 7 d after freezing. Values are the mean ± SE. Six plants of each line were used in each of ten experiments. Means followed by the same letter were not different using one-way analysis of variance (ANOVA), followed by the Tukey HSD multiple comparison test. A value of p ≤ 0.05 was considered as significant.
2.4. Stress Tolerance of VaCML21-Overexpressing V. amurensis Cell Cultures and A. thaliana Plants
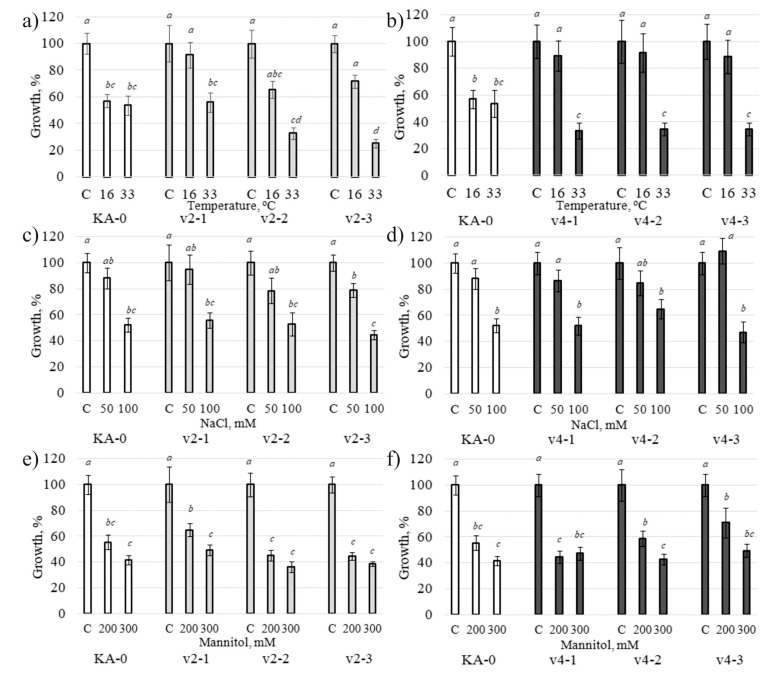
To assess the effect of cold and heat stress conditions on the cell culture biomass accumulation, we cultivated the VaCML21v2- and VaCML21v4-transgenic calli at 16 °C and 33 °C for 30 days (Figure 6a,b). To assess the effect of high salinity and mannitol-induced osmotic stress, the calli were also cultivated in the presence of NaCl or mannitol for 30 days (Figure 6c–f). Under high mannitol and salt stress conditions, the growth of the v2-1,2,3 and v4-1,2,3 cell lines was reduced to approximately the same level as the growth of the control KA-0 calli. However, the VaCML21v2- and VaCML21v4-transformed calli showed a higher biomass accumulation in response to cold stress in comparison with the growth of the KA-0 control (Figure 6a,b). Under cold stress conditions, growth of the v2-1,2,3 and v4-1,2,3 calli was reduced 1.1–1.5-fold and 1.1–1.2-fold, respectively, while the growth of KA-0 was reduced 1.8-fold. Under heat stress conditions, most of the VaCML21-transgenic lines exhibited a lowered biomass accumulation in comparison with that of KA0 control.
Figure 6.
Influence of temperature (a,b), salt (c,d) and osmotic (e,f) stresses on the fresh biomass accumulation in the transgenic grapevine callus cell lines overexpressing the VaCML21v2 and VaCML21v4 splice variants. The average growth rates were assessed after 30 days of cultivation in the dark under the control (24 °C), cold stress (+16 °C), heat stress (+37 °C), salt stress (NaCl 50 and 100 mM), and high-mannitol (200 and 300 mM) conditions. v2-1, v2-2, v2-3—VaCML21v2-transformed calli; v4-1, v4-2, v4-3—VaCML21v4-transformed calli. KA-0—control cell culture transformed with the “empty” vector. The data are presented as the mean ± SE (two independent experiments with ten replicates each). Means followed by the same letter were not different using one-way analysis of variance (ANOVA), followed by the Tukey HSD multiple comparison test. A value of p ≤ 0.05 was considered as significant.
The stress tolerance assays of the transgenic A. thaliana showed that the survival rates of the 35S::VaCML21 lines were higher than the survival rates of the KA-0 control plants (Figure 5c–e). The increase in cold stress tolerance was statistically significant only for those two transgenic lines (v2L1, v2L3, v4L1, v4L2) that expressed the VaCML21v2 and VaCML21v4 splice variants at a higher level (Figure 5a,b,d).
2.5. Transformation with VaCML21 Enhanced Transcription of Cold-Inducible Regulatory Genes in Stressed Arabidopsis
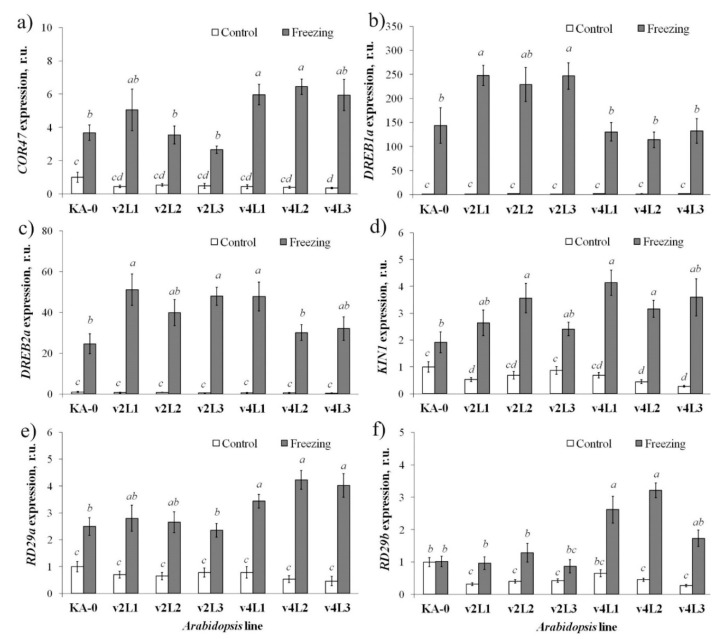
To further evaluate the involvement of VaCML21 to cold stress response, we analyzed expression of nine cold-inducible genes, including AtCOR47, AtRD29A, AtRD29B, AtDREB1A, AtDREB2A, AtKIN1, AtLEA, AtCBF1, and AtRAB18, which are known to function in plant cold stress protection and signaling, in the control and VaCML21-transgenic plants after freezing (Figure 7). The data revealed that the expression of the cold stress-associated genes did not differ between the KA-0 control and VaCML21-transgenic lines under normal conditions, except for lowered AtRD29B expression. However, in response to freezing, expression of six genes AtCOR47, AtRD29A, AtRD29B, AtDREB1A, AtDREB2A, AtKIN1 was up-regulated either in the VaCML21v2- or VaCML21v4-transgenic Arabidopsis lines to a considerably higher degree than in the control KA-0 plant line of A. thaliana (Figure 7). Overexpression of VaCML21v2 led to enhanced induction of AtDREB1A and AtDREB2A in response to freezing in at least two lines (Figure 7b,c), while overexpression of VaCML21v4—AtCOR47, AtKIN1, AtRD29A, and AtRD29B (Figure 7a,d,e,f) Transcription abundance of AtLEA, AtCBF1, and AtRAB18 was not essentially changed under either the control conditions or after freezing in all plant lines (data not shown).
Figure 7.
The expression of stress-responsive genes AtCOR47 (a), AtDREB1A (b), AtDREB2A (c), AtKIN1 (d), AtRD29A (e), and AtRD29B (f) in transgenic Arabidopsis transformed with the VaCML21v2 and VaCML21v4 splice variants in response to freezing stress. The 4-week-old control KA-0, 35S::VaCML21v2 (lines v2L1, v2L2, and v2L3), and 35S::VaCML21v4 (lines v4L1, v4L2, and v4L3) plants were exposed to control conditions (at 22 °C) or freezing (at −10 °C for 45 min and then cultured at +4 °C for 1 h for recovery). Total RNA was extracted from plants just before freezing (white bars) and 1 h after freezing (grey bars). The data are presented as the mean ± SE (two independent experiments). Means followed by the same letter were not different using one-way analysis of variance (ANOVA), followed by the Tukey HSD multiple comparison test. A value of p ≤ 0.05 was considered significant.
3. Discussion
The diverse and unique plant CML subfamily has been reported to serve as a set of Ca2+ sensor relay proteins with important functions in plant responses to environmental cues and developmental processes [6,7]. The presence of several introns in some plant CMLs makes the genes a target of alternative splicing, a process where generation of several mRNA splice variants could contribute to the diversity of plant reactions and stress adaptation. Despite there being intron-rich CMLs and some other Ca2+ sensors in the plant genome [3], the existent literature present little information about alternative splicing of Ca2+ protein sensors in plants. This study showed that the grapevine CML21 gene produced four splice variants arising from using an alternative transcription start site and a partial intron retention events combined with alternative splice site selection. The modifications affected CML21 structure, including the type of putative lipid modification sites and structure of the first EF-hands. These changes could potentially change localization and Ca2+- binding properties of CML21.
The present analysis revealed that all four splice variants of V. amurensis CML21 were highly activated in response to cold stress. The data on V. amurensis were in accordance with the chilling inducibility of the V. vinifera CML21. At the same time, VaCML21v1 and VaCML21v2 (but not VaCML21v3 and VaCMLv4) variants were highly activated in response to several other abiotic stress treatments, including desiccation, high mannitol, salinity, and heat stress. Overexpression of the VaCML21v2 and VaCML21v4 splice variants in the callus cultures of V. amurensis resulted in a higher biomass accumulation of the CML21v2- and CML21v4-transgenic cell lines during cultivation under lowered temperature, supporting the positive role of CML21 in cold stress adaptation. Furthermore, heterologous overexpression of the grapevine CML21v2 and CML21v4 splice variants in Arabidopsis led to improved survival rates of the transgenic plant lines after freezing and resulted in an enhanced transcriptional responsiveness of the cold stress-inducible genes AtCOR47, AtRD29A, AtRD29B, AtDREB1A, AtDREB2A, and AtKIN1 to freezing stress. The transcription factors DRE-BINDING PROTEIN 1A and 2A (DREB2A and DREB1A) have been reported to regulate dehydration-responsive element (DRE)-mediated transcription of target genes under cold stress, dehydration, and high salinity [38,39]. The expression of COR genes has been shown to be critical in plants for cold stress acclimation and tolerance [40]. The COR47, RD29A, RD29B, LEA, and some other COR genes known as group 2 LEA (LEA II) proteins, encode dehydrins, which are cold-inducible proteins that are supposed to sustain membrane stabilization and prevent protein aggregation [41]. The cold-inducible protein AtKIN1 has been suggested to mediate both cold and osmotic stress responses and possesses similarities with antifreeze proteins [42].
Thus, the VaCML21v2 and VaCML21v4 splice variants exhibited not only distinct expression patterns in response to abiotic stress treatments of V. amurensis, but also induced different cold stress-protective genes in response to freezing in A. thaliana. This indicates a distinct molecular mechanism for the VaCML21v1/v2- and VaCML21v3/v4-mediated regulation of cold stress response in grapevine. The mRNA transcript diversity of some Ca2+ sensor proteins and their distinct transcriptional properties detected in this and other works suggested that this splicing-induced expansion of mRNA variants could contribute to the diversity of plant adaptive reactions to unfavorable environmental conditions.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
For the abiotic stress treatments, we used young vines of wild-growing grapevine V. amurensis Rupr. (Vitaceae) sampled from a non-protected natural population near Vladivostok, Russia (Akademgorodok, the southern Primorsky region of the Russian Far East, longitude 43.2242327 and latitude 131.99112300) and identified at the Botany Department of the Federal Scientific Center of the Biodiversity FEBRAS. The freshly harvested V. amurensis vines were divided into cuttings (excised young stems 7–8 cm long with one healthy leaf) that were placed in individual beakers and used for the stress treatments. The cuttings were placed into the filtered water at 25 °C immediately after cutting. Then, the cuttings were acclimated to the “non-stress” condition for 30 min before they were treated with the stress treatments. For the control non-stress treatment, the V. amurensis cuttings were placed in filtered water at 25 °C. To induce water deficit stress, the cuttings were laid on a paper towel at 25 °C. To induce osmotic stress, the cuttings were placed in 400 mM NaCl (Himreaktivsnab, Ufa, Russia) and 400 mM D-mannitol (AppliChem, Darmstadt, Germani) solutions at 25 °C. To apply cold and heat stress, the V. amurensis cuttings were placed in filtered water in the growth chamber (Sanyo MLR-352, Panasonic, Osaka, Japan) at +4 °C, +10 °C, and +37 °C. The V. amurensis cuttings were grown under a 16/8 h light/dark photoperiod. We used similar experimental design and employed the same NaCl and mannitol concentrations as Chung et al. [43] and Dubrovina et al. [44] for studying calcium-dependent protein kinase gene expression in Capsicum annuum and V. amurensis. The experiments were repeated three times for each stress treatment time and for the control treatment.
Plants (Arabidopsis thaliana ecotype Columbia L., stored by our lab) were grown in pots filled with commercially available rich soil in a controlled environmental chamber at 22 °C (Sanyo MLR-352, Panasonic, Japan) kept on a 16/8 h day/night cycle at a light intensity of ~120 μmol m−2 s−1.
4.2. Isolation and Sequencing of VaCML21 Spliced Transcript Variants
Full-length cDNA coding sequences of VaCML21v2, VaCML21v3, and VaCML21v4 were amplified using RNA samples extracted from stressed leaves of V. amurensis, the high-fidelity Tersus polymerase (Evrogen, Moscow, Russia) and primers designed to the predicted coding sequences of VvCML21 mRNAs of V. vinifera PN20024 genotype (V2 mRNA prediction) retrieved from the Grapevine Genome Database hosted at Centro Di Ricerca Interdipartimentale Per Le Biotecnologie Innovative (CRIBI) Biotech Centre (http://genomes.cribi.unipd.it/grape; Table 2). The RT-PCR products were subcloned into pJET1.2/blunt and sequenced as described previously [22]. The VaCML amino acid sequences were predicted using the Gene Runner program (http://www.generunner.net). We performed a domain analysis by PROSITE scan ([31]; http://prosite.expasy.org), calculated Compute pI/Mw tool (http://web.expasy.org/compute_pi) and prediction of myristoylation and palmitoylation motif numbers with GPS-Lipid ([32]; http://lipid.biocuckoo.org/webserver.php). Multiple sequence alignments were done with the BioEdit 7.0.8 program [35]. The full-length coding mRNA sequences of VaCML21 and VviCML21 splice variants are given in Table S1.
Table 2.
Primers used for amplification of grapevine CML21 cDNAs and the house-keeping genes.
| Transcript Name (GenBankID) | Primers, 5′–3′ |
|---|---|
| Cloning and sequencing full-length cDNA coding sequences of VaCML21, 5′–3′ | |
|
CML21v1/v2 (no sequence/MN540599) |
F1:ATGGGAGGCGTGGTGGG R:TCAAACTTTCTCTTCACCTTC |
|
CML21v3/v4 (MW033207/ MW033208) |
F2:ATGCTGTGTATCATCCTTCATG R:TCAAACTTTCTCTTCACCTTC |
| Cloning VaCML21v2 and VaCML21v4 for plant transformation by the BglII and SalI sites (underlined), 5’–3’ | |
|
VaCML21v2 (MN540599) |
F:GCTCAGATCTATGGGAGGCGTGGTGGGAAAA R:TCGAGTCGACTTAAACTTTCTCTTCACCTTC |
|
VaCML21v4 (MW033208) |
F:GCTCAGATCTATGCTGTGTATCATCCTTCAT R:TCGAGTCGACTCAAACTTTCTCTTCACCTT |
| Primers for real-time PCR, 5′–3′ | |
|
VaCML21v1 (no sequence) |
GAACTGCAAAGCTATTTTTCAGCA CCCATCCGTGATTTTTTGGATAC |
|
VaCML21v2 (MN540599) |
GAACTGCAAAGCTATTTTTCAGCA ATCCGTGATTTGGCCTGAAGG |
|
VaCML21v3 (MW033207) |
TGCCAGAGGTTGAAATTCGTTGTT CCCATCCGTGATTTTTTGGATAC |
|
VaCML21v4 (MW033208) |
TGCCAGAGGTTGAAATTCGTTGTT ATCCGTGATTTGGCCTGAAGG |
|
VaActin1 (DQ517935) |
GTATTGTGCTGGATTCTGGTGAT AGCAAGGTCAAGACGAAGGATAG |
|
VaGAPDH (XM_002263109) |
CACTGAAGATGATGTTGTTTCC GCTATTCCAGCCTTGGCAT |
|
AtGAPDH (NM_111283.4) |
TTGGTGACAACAGGTCAAGCA AAACTTGTCGCTCAATGCAAT |
|
AtEF1a (XM_002864638) |
TGAGCACGCTCTTCTTGCTTTCA GGTGGTGGCATCCATCTTGTTACA |
| Primers for cDNA check-up on DNA contamination, 5′–3′ | |
|
VaActin1 (DQ517935) |
TTGCCATTCAGGCTGTTCTTTCT AGGAGCTGCTCTTTGCAGTTTCC |
|
AtActin1 (NM_112764) |
GATTCAGATGCCCAGAAGTC TCTGTGAACGATTCCTGGA |
| Primers for qRT-PCR quantification of the VaCML21v2 and VaCML21v4 transgene and endogene mRNAs in transformed plant cells, 5′–3′ | |
|
VaCML21v2 transgene (MN540599) |
CAAGCATTCTACTTCTATTG ACCATTTTGGCCTCAAGC |
|
VaCML21v1/v2 endogene (no sequence/MN540599) |
CATACTTAGCACTTGTCCCTTTTCC ACCATTTTGGCCTCAAGC |
|
VaCML21v4 transgene (MW033208) |
CAAGCATTCTACTTCTATTG TCTTCATAGATGAGGATTCAAATG |
|
VaCML21v3/v4 endogene (MW033207/ MW033208) |
TGAACCAGTTTTCATTTTAATTCAACGTG TCTTCATAGATGAGGATTCAAATG |
4.3. Generation of Transgenic Grapevine Cell Cultures
The overexpression constructs of VaCML21 (pZP-RCS2-VaCML21v2-nptII; pZP-RCS2- VaCML21v4-nptII) or empty vector (pZP-RCS2-nptII) were obtained as described [45]. Briefly, the full-length cDNA of VaCML21v2 and VaCML21v4 were amplified by PCR using the primers listed in Table 2 from pJET1.2. The full-length cDNAs of VaCML21v2 and VaCML21v4 were cloned into the pZP-RCS2-nptII vector [46] under the control of the double cauliflower mosaic virus (CaMV 35S) promoter. The pZP-RCS2-nptII construction also carried the nptII gene under the control of the double CaMV 35S promoter. Plasmid DNA samples (pSAT1 and pZP-RCS2-nptII) were kindly provided by Professor Alexander Krichevsky (State University of New York, Stony Brook, USA). The overexpression constructs were introduced into the Agrobacterium tumefaciens strain GV3101:pMP90 and transformed into the suspension culture V7 of V. amurensis by co-cultivation as described [47]. Briefly, A. tumefaciens strains bearing pZP-RCS2-nptII or pZP-RCS2-VaCML21v2/v4-nptII constructs were inoculated in multiple separate flasks with cell suspensions of V. amurensis to establish the independently transformed KA-0 (empty vector), VaCML21v2-transgenic (v2-1, v2-2, v2-3), and VaCML21v4-transgenic (v4-1, v4-2, v4-3) callus cell cultures. The KA-0 cell culture was used as a control in all further experiments. The V7 callus culture were established in 2017 from young stems of the mature V. amurensis plants as described [48]. After the transformation, the calli were cultivated for a 4-month period in the presence of 10–20 mg/L of Km to select transgenic cells and for a 6-month period in the presence of 250 mg/L of Cf to suppress the bacteria. Transgenic callus cell cultures were selected as described [47]. Transgenic callus cell cultures were selected and confirmed by PCR as described [45]. To analyze the level of exogenous VaCML21v2 and VaCML21v4 (transgenes), we used primers designed to the CaMV 35S promoter and the 5’ end of the VaCPK21 protein coding region (Table 2). To analyze the level of endogenous VaCML21 splice variants, we used primers designed to the 5′ UTR and the 5′ end of the VaCPK21 protein coding region (Table 2).
4.4. Generation of Transgenic Arabidopsis Plants
To create A. thaliana lines overexpressing the VaCML21v2 and VaCML21v4 splice variants, we used the same plasmid construction as for overexpression of VaCPK21 in cell cultures of V. amurensis (described above). The overexpression construct of VaCPK21 or empty was introduced into the A. tumefaciens strain GV3101::pMP90 and transformed by floral dip method into wild-type A. thaliana [49]. Transgenic plants were selected and confirmed by PCR as described [50] using primers designed for the CaMV 35S promoter in the pSAT1 vector and the 5′ end of the VaCPK21 protein coding region (Table 2). The PCR products were verified by DNA sequencing. The transgenic lines used in this study were homozygous plants with a single copy insertion. Six representative independent T3 homozygous lines (v2L1, v2L2, v2L3, v4L1, v4L2, v4L3) with high mRNA levels of the VaCML21v2 and VaCML21v4 transgenes were chosen for further analysis. We also selected a T3 homozygous KA-0 line of A. thaliana transformed with the empty vector (pZP-RCS2-nptII).
4.5. RNA Isolation and cDNA Synthesis
Total RNA isolation was performed using the cetyltrimethylammonium bromide-based extraction [51]. Complementary DNAs were synthesized using 1.5 µg of total RNA by the MMLV RT Kit (Evrogen, Moscow, Russia). The reactions were performed in 20 µL aliquots of the reaction mixture, which contained the first strand buffer, 2 µL of dNTP mix (10 mM each), 2 µL of oligo-(dT)15 primer (20 µM), and 2 µL of MMLV reverse transcriptase (100 u/µL), at 37 °C for 1.5 h. The 1 µL samples of reverse transcription products were then amplified by PCR and verified on the absence for DNA contamination using primers listed in Table 2.
4.6. Gene Expression Analysis by qRT-PCR
The qRT-PCRs were performed with EvaGreen Real-Time PCR (Biotium, Hayward, CA, USA) as described in [52,53], using two internal controls (VaActin1, VaGAPDH, AtGAPDH, AtEF1a). The expression was calculated by the 2−ΔΔCT method [54]. All GenBank accession numbers and primers are listed in Table 2. The specific primers for qRT-PCR were designed for the regions specific to each CML21 splice variant (Table 2). The qRT-PCR data shown were obtained from at least two independent experiments and are averages of eight technical replicates for each independent experiment (four qPCR reactions normalized to Actin and four qPCR reactions normalized to GAPDH expression for each independent experiment).
4.7. V. vinifera RNAseq Library Analsis
The sequences of the VviCML21 four transcript variants were combined with the grapevine genome file (https://www.ncbi.nlm.nih.gov/genome/401?genome_assembly_id=214125). Bowtie2 (bowtie2-build genome_name.fna index_db) was used to construct index files Bowtie2 for the V. vinifera genome. Recently, Londo et al. 2018 [37] produced Illumina RNAseq libraries of to analyze the broad transcriptional response of V. vinifera to low temperature stress. The RNAseq libraries [37] were downloaded from the Sequence Read Archive (SRA) data available through NCBI (accession: PRJNA402079). The RNAseq library numbers and cultivation conditions are listed in Table 3. The SRA files in fastq format were aligned to the V. vinifera indexed genome using Bowtie2 (bowtie2-q -x index_db -U SAR_file.fastq -S output_file.sam --un-gz 1.un.fq.gz -N 1 –local). Samtools was used to convert the SAM files to BAM files (samtools view file.sam –b –o file.bam) and for further sortingBAM files (samtools sort –l 9 file.bam –o file.sorted.bam). The command “samtools view –c” (samtools view –c file.sorted.bam “sample number ID”) was used to quantitate the number of primary aligned reads in each sample for each treatment. All bioinformatics assays were performed “blind” and were-based solely on sample number ID. The resulting number of reads was recalculated per million reads of the SRA file.
Table 3.
RNAseq libraries of Vitis vinifera cultivated under cold stress [37].
| Sangiovese | Cabernet Franc | Cultivation Conditions a | ||
|---|---|---|---|---|
| SRR6026721 | SRR6026711 | SRR6026737 | SRR6026699 | Control |
| SRR6026722 | SRR6026712 | SRR6026736 | SRR6026700 | Freezing (−3) for 45 min |
| SRR6026716 | SRR6026735 | SRR6026738 | SRR6026698 | Chilling stress (+4) for 48 h |
| SRR6026715 | SRR6026734 | SRR6026739 | SRR6026697 | Acclimated freezing (+4 → −3) |
a A detailed description of the cultivation conditions was presented in [37].
4.8. Abiotic Stress Treatments of Transgenic Cell Cultures and Plants
The V. amurensis calli were cultivated in 150 × 16 mm test tubes containing 9 mL of agarized Murashige and Skoog (MS) modified WB/A medium [55] supplemented with 0.5 mg/L 6-benzylaminopurine and 2.0 mg/L α-naphthaleneacetic acid. Growth analysis and abiotic stress treatments of transgenic callus cell cultures were conducted as described [50,56]. Briefly, salt treatment was applied by adding 50 and 100 mM of NaCl to the WB/A culture media. Mannitol treatment was applied by adding 200 and 300 mM of mannitol to the medium. Cold and heat treatments were performed by culturing the transgenic cells at 16 °C and 33 °C in a growth chamber (TSO-1/80 SPU, SKTB, Smolensk, Russia). The average growth rates were assessed after 30 days of cultivation under the control and stress conditions in the dark at 24–25 °C. The data were obtained from two independent experiments with 10 replicates each.
The plants of A. thaliana were subjected to freezing as described [50,52]. Briefly, the sterilized transgenic seeds of Arabidopsis were germinated on plates and the seven-day-old seedlings were transferred to commercially available rich well-watered soil in a controlled environmental chamber at standard conditions. For the low-temperature stress, normally cultured four-week-old A. thaliana plants were stressed in a −10 °C freezer for 45 min and then cultured at +4 °C for 1 h before RNA isolation. Six plants of each line were used in each of ten experiments.
4.9. Statistical Analysis
The data are presented as mean ± standard error (SE) and were evaluated by Student’s t-test (expression analysis for the grapevine cuttings and callus cultures) or one-way analysis of variance (ANOVA) (all other analyses), followed by the Tukey HSD multiple comparison test. A value of p < 0.05 was considered significant.
Acknowledgments
The authors are grateful to Eugene Gladyshev for the technical assistance in bioinformatic analysis of RNAseq libraries.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/21/7939/s1. Table S1. The nucleotide sequences of CML21 splice variants of Vitis vinifera and Vitis amurensis.
Author Contributions
A.S.D. and K.V.K. performed the research design, data analysis, RNA isolations, and paper preparation. O.A.A. obtained the cell cultures, transgenic plants, qRT-PCR data, and performed RNA isolations. Z.V.O. performed the bioinformatic analysis of RNAseq libraries. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (grant number 18-04-00284).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.DeFalco T.A., Bender K.W., Snedden W.A. Breaking the code: Ca2+ sensors in plant signaling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 2.Batistič O., Kudla J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012;1820:1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Mohanta T.K., Yadav D., Khan A.L., Hashem A., Abd Allah E.F., Al-Harrasi A. Molecular players of EF-hand containing calcium signaling event in plants. Int. J. Mol. Sci. 2019;20:1476. doi: 10.3390/ijms20061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perochon A., Aldon D., Galaud J.P., Ranty B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie. 2011;93:2048–2053. doi: 10.1016/j.biochi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 5.La Verde V., Dominici P., Astegno A. Towards understanding plant calcium signaling through calmodulin-like proteins: A biochemical and structural perspective. Int. J. Mol. Sci. 2018;19:1331. doi: 10.3390/ijms19051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders D., Pelloux J., Brownlee C., Harper J.F. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto K., Kudla J. Calcium decoding mechanisms in plants. Biochimie. 2011;93:2054–2059. doi: 10.1016/j.biochi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X., Dunand C., Snedden W., Galaud J.P. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015;20:483–489. doi: 10.1016/j.tplants.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Zeng H., Xu L., Singh A., Wang H., Du L., Poovaiah B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015;6:600. doi: 10.3389/fpls.2015.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheval C., Aldon D., Galaud J.P., Ranty B. Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta. 2013;1833:1766–1771. doi: 10.1016/j.bbamcr.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Midhat U., Ting M.K.Y., Teresinski H.J., Snedden W.A. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol. Biol. 2018;96:375–392. doi: 10.1007/s11103-018-0703-3. [DOI] [PubMed] [Google Scholar]
- 12.Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi T., Aharon G.S., Sottosanto J.B., Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+-and pH-dependent manner. Proc. Natl. Acad. Sci. USA. 2005;102:16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobney S., Chiasson D., Lam P., Smith S.P., Snedden W.A. The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 2009;284:31647–31657. doi: 10.1074/jbc.M109.056770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munir S., Liu H., Xing Y., Hussain S., Ouyang B., Zhang Y., Li H., Ye Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016;6:31772. doi: 10.1038/srep31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S., Xiong X., Arif S., Gao L., Zhao L., Shah I.H., Zhang Y. A calmodulin-like CmCML13 from Cucumis melo improved transgenic Arabidopsis salt tolerance through reduced shoot’s Na(+), and also improved drought resistance. Plant Physiol. Biochem. 2020;155:271–283. doi: 10.1016/j.plaphy.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Wu X., Qiao Z., Liu H., Acharya B.R., Li C., Zhang W. CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 2017;8:824. doi: 10.3389/fpls.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X.X., Wang T.Z., Liu M., Sun W., Zhang W.H. Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ. Exp. Bot. 2019;157:79–90. doi: 10.1016/j.envexpbot.2018.09.022. [DOI] [Google Scholar]
- 19.Vandelle E., Vannozzi A., Wong D., Danzi D., Digby A.M., Dal Santo S., Astegno A. Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiol. Biochem. 2018;129:221–237. doi: 10.1016/j.plaphy.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Li C., Meng D., Zhang J., Cheng L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malus × domestica) Plant Physiol. Biochem. 2019;139:600–612. doi: 10.1016/j.plaphy.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Ma Q.P., Zhou Q.Q., Chen C.M., Cui Q.Y., Zhao Y.X., Wang K., Arkorful E., Chen X., Sun K., Li X.H. Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant (Camellia sinensis) Sci. Rep. 2019;9:8211. doi: 10.1038/s41598-019-44681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubrovina A.S., Aleynova O.A., Ogneva Z.V., Suprun A.R., Ananev A.A., Kiselev K.V. The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis. Plants. 2019;8:602. doi: 10.3390/plants8120602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasani E., DalCorso G., Costa A., Zenoni S., Furini A. The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol. Biol. 2019;99:517–534. doi: 10.1007/s11103-019-00833-x. [DOI] [PubMed] [Google Scholar]
- 24.La Verde V., Trande M., D’Onofrio M., Dominici P., Astegno A. Binding of calcium and target peptide to calmodulin-like protein CML19, the centrin 2 of Arabidopsis thaliana. Int. J. Biol. Macromol. 2018;108:1289–1299. doi: 10.1016/j.ijbiomac.2017.11.044. [DOI] [PubMed] [Google Scholar]
- 25.Shang X.D., Cao Y., Ma L.G. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017;18:432. doi: 10.3390/ijms18020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiyama R., Mizuno H., Okada S., Yamaguchi T., Takenaka M., Fukuzawa H., Ohyama K. Two mRNA species encoding calcium-dependent protein kinases are differentially expressed in sexual organs of Marchantia polymorpha through alternative splicing. Plant Cell Physiol. 1999;40:205–212. doi: 10.1093/oxfordjournals.pcp.a029529. [DOI] [PubMed] [Google Scholar]
- 27.Dubrovina A.S., Aleynova O.A., Kiselev K.V., Novikova G.V. True and false alternative transcripts of calcium-dependent protein kinase CPK9 and CPK3a genes in Vitis amurensis. Acta Physiol Plant. 2014;36:1727–1737. doi: 10.1007/s11738-014-1547-3. [DOI] [Google Scholar]
- 28.Almadanim M.C., Gonçalves N.M., Rosa M.T.G., Alexandre B.M., Cordeiro A.M., Rodrigues M., Saibo N.J.M., Soares C.M., Romão C.V., Oliveira M.M., et al. The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:231–246. doi: 10.1016/j.bbamcr.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Sanyal S.K., Kanwar P., Samtani H., Kaur K., Jha S.K., Pandey G.K. Alternative splicing of CIPK3 results in distinct target selection to propagate ABA signaling in Arabidopsis. Front Plant Sci. 2017;8:1924. doi: 10.3389/fpls.2017.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma X., Li Q.H., Yu Y.N., Qiao Y.M., Haq S.U., Gong Z.H. The CBL-CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 2020;21:5668. doi: 10.3390/ijms21165668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C.J., Hofmann K., Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Database of Protein Domains, Families and Functional Sites. [(accessed on 18 September 2020)]; Available online: http://prosite.expasy.org/
- 33.Xie Y., Zheng Y., Li H., Luo X., He Z., Cao S., Shi Y., Zhao Q., Xue Y., Zuo Z., et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016;16:28249. doi: 10.1038/srep28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prediction of Lipid Modification Sites. [(accessed on 18 September 2020)]; Available online: http://lipid.biocuckoo.org/webserver.php.
- 35.Alzohairy A.M. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
- 36.Compute pI/Mw Tool. [(accessed on 18 September 2020)]; Available online: http://web.expasy.org/compute_pi/
- 37.Londo J.P., Kovaleski A.P., Lillis J.A. Divergence in the transcriptional landscape between low temperature and freeze shock in cultivated grapevine (Vitis vinifera) Hortic Res. 2018;5:10. doi: 10.1038/s41438-018-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruyama K., Takeda M., Kidokoro S., Yamada K., Sakuma Y., Urano K., Fujita M., Yoshiwara K., Matsukura S., Morishita Y., et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150:1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miura K., Furumoto T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013;14:5312–5337. doi: 10.3390/ijms14035312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Z., Wang X., Zhang L. Structural and functional dynamics of dehydrins: A plant protector protein under abiotic stress. Int. J. Mol. Sci. 2018;19:3420. doi: 10.3390/ijms19113420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurkela S., Franck M. Cloning and characterization of a cold-and ABA-inducible Arabidopsis gene. Plant Mol. Biol. 1990;15:137–144. doi: 10.1007/BF00017731. [DOI] [PubMed] [Google Scholar]
- 43.Chung E., Park J.M., Oh S.K., Joung Y.H., Lee S., Choi D. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta. 2004;220:286–295. doi: 10.1007/s00425-004-1372-9. [DOI] [PubMed] [Google Scholar]
- 44.Dubrovina A.S., Kiselev K.V., Khristenko V.S. Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis. J. Plant Physiol. 2013;170:1491–1500. doi: 10.1016/j.jplph.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Aleynova-Shumakova O.A., Dubrovina A.S., Manyakhin A.Y., Karetin Y.A., Kiselev K.V. VaCPK20 gene overexpression significantly increased resveratrol content and expression of stilbene synthase genes in cell cultures of Vitis amurensis Rupr. Appl. Microbiol. Biotechnol. 2014;98:5541–5549. doi: 10.1007/s00253-014-5625-7. [DOI] [PubMed] [Google Scholar]
- 46.Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 47.Kiselev K.V., Dubrovina A.S., Shumakova O.A., Karetin Y.A., Manyakhin A.Y. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013;32:431–442. doi: 10.1007/s00299-012-1375-0. [DOI] [PubMed] [Google Scholar]
- 48.Tyunin A.P., Suprun A.R., Nityagovsky N.N., Manyakhin A.Y., Karetin Y.A., Dubrovina A.S., Kiselev K.V. The effect of explant origin and collection season on stilbene biosynthesis in cell cultures of Vitis amurensis Rupr. Plant Cell Tiss. Organ Cult. 2019;136:189–196. doi: 10.1007/s11240-018-1490-x. [DOI] [Google Scholar]
- 49.Zhang X.R., Henriques R., Lin S.S., Niu Q.W., Chua N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 50.Dubrovina A.S., Kiselev K.V., Khristenko V.S., Aleynova O.A. VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr, mediates cold and drought stress tolerance. J. Plant Physiol. 2015;185:1–12. doi: 10.1016/j.jplph.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 51.Kiselev K.V., Shumakova O.A., Manyakhin A.Y., Mazeika A.N. Influence of calcium influx induced by the calcium ionophore, A23187, on resveratrol content and the expression of CDPK and STS genes in the cell cultures of Vitis amurensis. Plant Growth Regul. 2012;68:371–381. doi: 10.1007/s10725-012-9725-z. [DOI] [Google Scholar]
- 52.Dubrovina A.S., Kiselev K.V., Khristenko V.S., Aleynova O.A. The calcium-dependent protein kinase gene VaCPK29 is involved in grapevine responses to heat and osmotic stresses. Plant Growth Regul. 2017;82:79–89. doi: 10.1007/s10725-016-0240-5. [DOI] [Google Scholar]
- 53.Dubrovina A.S., Aleynova O.A., Manyakhin A.Y., Kiselev K.V. The role of calcium-dependent protein kinase genes CPK16, CPK25, CPK30, and CPK32 in stilbene biosynthesis and the stress resistance of grapevine Vitis amurensis Rupr. Appl. Biochem. Microbiol. 2018;54:410–417. doi: 10.1134/S0003683818040051. [DOI] [Google Scholar]
- 54.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Kiselev K.V., Dubrovina A.S., Bulgakov V.P. Phenylalanine ammonia-lyase and stilbene synthase gene expression in rolB transgenic cell cultures of Vitis amurensis. Appl. Microbiol. Biotechnol. 2009;82:647–655. doi: 10.1007/s00253-008-1788-4. [DOI] [PubMed] [Google Scholar]
- 56.Dubrovina A.S., Kiselev K.V., Aleynova O.A. Influence of overexpression of the true and false alternative transcripts of calcium-dependent protein kinase CPK9 and CPK3a genes on the growth, stress tolerance, and resveratrol content in Vitis amurensis cell cultures. Acta Physiol. Plant. 2016;38:78. doi: 10.1007/s11738-016-2093-y. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.