Abstract
Autistic spectrum disorder (ASD) is a complex neurodevelopmental disability with a genetic basis, and several studies have suggested a potential role of the reelin gene (RELN) in ASD susceptibility. Accordingly, genetic association studies have explored this potential association, but the results have been controversial thus far. For this reason, we assessed the association of four genetic variants of RELN (the 5′UTR CGG triplet repeat and polymorphisms rs736707, rs362691, and rs2229864) with ASD by means of a systematic review and meta-analysis. We retrieved studies comparing the distribution of the above-mentioned genetic variants between ASD patients and healthy controls. A meta-analysis was conducted using a random effects model, and calculations of the odds ratios (ORs) and confidence intervals (CIs) were performed. A sensitivity analysis and tests to determine the heterogeneity of the results were also performed. Eleven previous studies fulfilled the inclusion criteria and analyzed the association of the above-mentioned genetic variants and ASD. We did not find any significant association between the allele or genotype frequencies of the analyzed polymorphisms and ASD, and large heterogeneity was found for the rs736707 polymorphism. Moreover, no significant differences were found between the 5′UTR triplet repeat and this disorder. In light of current evidence, no single genetic variant within this gene is clearly associated with the development of ASD, and ethnic differences may explain part of the observed heterogeneity. Larger studies among different ethnic groups are needed to establish the role of specific genetic variants within RELN in the etiology of this disorder.
Keywords: reelin, autistic spectrum disorder, polymorphism, genetics, meta-analysis
1. Introduction
Autistic spectrum disorder (ASD) is a group of complex neurodevelopment disorders, including autism, pervasive developmental disorder not otherwise specified, Asperger syndrome, and other related conditions [1,2]. This disorder is characterized by impairments in social interactions and communication, with stereotypical patterns of behaviors and activities [3,4]. The prevalence of this disorder is at least approximately 1.5% in developed countries. Specifically, the prevalence is 18.5 per 1000 (one in 54) children aged eight years in the United States of America [5]. In other countries, such as the United Kingdom and Italy, the prevalence of ASD is 15.7 per 1000 [6] children aged 5–9 years and 11.5 per 1000 children aged 7–9 years [7], respectively. In 2010, a review of 23 studies found that the estimated prevalence of ASD across Asian countries/territories (Japan, China, Iran, Taiwan, Israel, and Indonesia) varied from 1.1 to 21.8 per 10,000 [8]. The prevalence of ASD is increasing in Asia. Recently, it was estimated that the prevalence of ASD in East Asia (Korea, India, and China) is 0.51%, 0.31% in South Asia (Nepal and Sri Lanka), and 0.35% in West Asia (Israel, Lebanon, Bangladesh, and Iran). In particular, ASD prevalence ranges between 0.06% in Iran and 2.64% in Korea [9]. Other authors have described an ASD prevalence of 785 per 100,000 children younger than five years in North Africa [10]. Thus, there are discrepancies in the prevalence rates across cultures, although differences in the prevalence of ASD obtained in such studies may also be justified, in part, by differences in the diagnostic criteria or epidemiological sampling methods used [9].
ASD affects all ethnic and socioeconomic groups. It is often associated with pronounced personal suffering and a heavy burden of care to families and society [11]. Moreover, children with ASD are substantially afflicted by ASD-related outcomes, including co-existing disorders [12] and bullying [13]. Moreover, one out of six children with ASD has several degrees of development disability, which may lead to intellectual disability [3,4,5].
The etiology of ASD is far from completely elucidated, but twin and family studies strongly support a genetic component [3,14,15,16]. Some single nucleotide polymorphisms (SNPs), such as rs10099100 on chromosome 8 and rs1000177 on chromosome 20, have been associated with ASD risk [16]. Moreover, several independent genome-wide scans have highlighted loci within the long arm of chromosome 7 as potential candidate genes explaining ASD susceptibility [17,18]. Among the potential loci of interest, the reelin gene (RELN) maps to 7q22 and encodes a signaling glycoprotein considered to play a key role in the migration of several neuronal cell types and the development of neural connections. Furthermore, Fatemi et al. [19] showed decreased levels of reelin protein in autistic patients. The role of RELN in ASD is thus supported by functional studies, as well as genome scans showing ASD linkage peaks in the region that contains this gene.
Several genetic association studies have been performed to analyze the association between genetic variants within the RELN gene and ASD, but the results have been conflicting. Some studies have found significant associations between ASD and longer triplet repeats in the 5′UTR region of this gene [20] or certain SNPs. Specifically, attention has been mostly focused on the polymorphisms rs736707, rs362691, and rs2229864. However, while some authors have found positive associations [21,22], other researchers have published negative findings [23,24]. Given these controversial results, the aim of this study is to analyze the association between ASD and genetic variants within the RELN gene by means of a systematic review and meta-analysis.
2. Materials and Methods
2.1. Inclusion Criteria
In this review, we included case-control studies that analyzed the relationship between ASD and 3 single nucleotide polymorphisms (SNPs) located within RELN (rs736707 in intron 59, rs362691 in exon 22, and rs2229864 in exon 50 of RELN). We also analyzed the potential association of ASD with a polymorphic trinucleotide repeat (CGG/GCC) within the 5′UTR region of RELN. The included reports had to include patients with ASD as cases alongside a control group comprised of healthy unrelated subjects. ASD had to be defined by the use of accepted diagnostic criteria (according to the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM–IV); Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM–5); the International Classification of Diseases–10 (ICD–10); the Autism Diagnostic Interview; the Autism Diagnostic Interview-Revised; or the Autism Diagnostic Observation Schedule).
2.2. Bibliographic Search and Data Extraction
Reports published before 3 1July 2020 that fulfilled the inclusion criteria were then identified. For this process, a bibliographic search was undertaken in the following databases: Medline (PubMed), Embase, and Web of Science. The terms used to carry out the search were “autism”, “autistic”, “polymorphism”, “genetic variant”, “reelin”, “reln”, “Polymorphism, Genetic”, and “Autistic Disorder”. There were no language restrictions for this study. The search was complemented by reviewing the references of the included articles. Additional reports were retrieved using the PubMed option "Related Articles" and the Web of Science option “Times cited”.
The search and data extraction were independently carried out by three of the authors (C–AJ, HG–I, and TV–HG).
Each author imported their search results into a reference manager software (Endnote 6) and removed duplicate references at import. The titles and abstracts were screened independently by the authors (C–AJ, HG–I, and TV–HG) for inclusion and exclusion. Disagreements were resolved by consensus. We recorded the following information: authors’ name(s), year of publication, and country. Moreover, allele and genotype frequencies were extracted or calculated from the raw data.
2.3. Statistical Analysis
Our meta-analysis compared the presence of the above-mentioned allelic variants among patients with ASD as cases versus healthy unrelated controls. The odds ratio (OR) and its 95% confidence interval (CI) were estimated for each paper. The pooled results are reported as the OR with a 95% CI and p-values using a random effects model [25]. A p-value < 0.05 was considered statistically significant. Cochran’s Q–statistic was used to assess heterogeneity: a significant Q–statistic (p = 0.10) indicated heterogeneity across studies. The I2 statistic was used for estimation of the inconsistency in the meta-analyses (percentage of the observed between-study variability due to heterogeneity rather than chance). The following cut-off points were used: I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; and I2 = 75–100%, extreme heterogeneity [26,27]. In cases with significant results, a sensitivity analysis was carried out to analyze the effect when excluding individual studies in the results.
This meta-analysis was performed using the computer software packages Review Manager 5.4 and MIX v.1.7. [28,29]. As in other meta-analyses [30], the information available for other studies was assessed; for this reason, ethical approval was not required.
3. Results
3.1. Study Identification and Selection
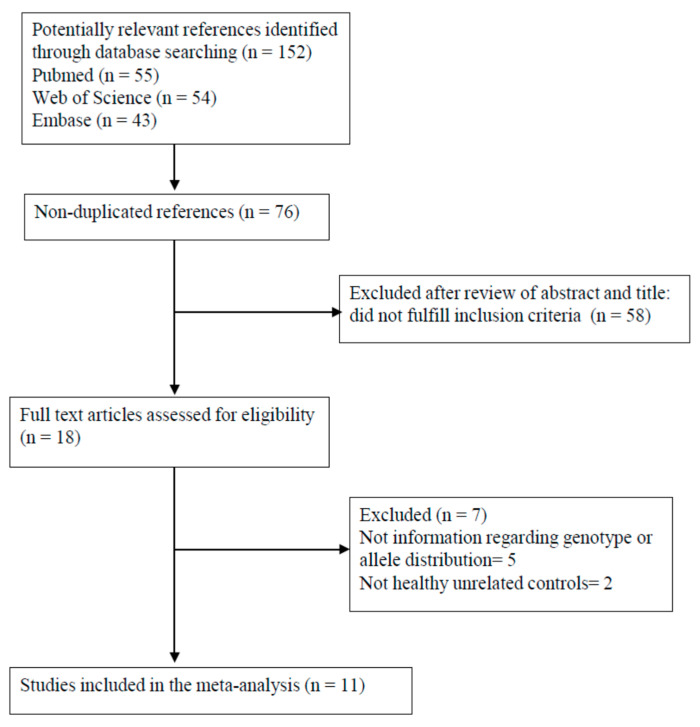
The flow of study identification and selection is shown in Figure 1. Our search strategy detected 152 potentially relevant papers, 18 of which were selected for further analysis. Five of the 18 studies [21,31,32,33,34] were excluded because they did not provide the distribution of SNPs or the distribution of the polymorphic trinucleotide repeat (CGG/GCC) within the RELN in the cases and controls.
Figure 1.
Flowchart of the selection of studies for inclusion in the meta-analysis.
Moreover, the studies by Krebs et al. [35] and Li et al. [36] were excluded since they only considered cases and parents of cases but not healthy unrelated controls. Therefore, 11 studies [20,22,23,24,37,38,39,40,41,42,43] were ultimately included in our meta-analysis. The allele and genotype distribution among the cases and controls is summarized in Table 1, Table 2 and Table 3.
Table 1.
Distribution of RELN CGG repeat genotypes.
| First Author, Year | N | Number of CGG Repeats | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3/10 | 4/8 | 4/10 | 6/10 | 7/8 | 7/10 | 8/8 | 8/9 | 8/10 | 8/11 | 8/11-15 | 8/12 | 8/13 | 8/14 | 9/10 | 9/13 | 10/10 | ||||
| (Persico et al., 2001) [20] | 1. Patients with ASD a | 95 | 1 | 1 | 16 | 44 | 0 | 5 | 2 | 0 | 16 | |||||||||
| 2. Healthy controls | 186 | 0 | 0 | 36 | 85 | 3 | 1 | 3 | 1 | 48 | ||||||||||
| (Zhang et al., 2002) [37] | 1. Patients with ASD 2. Healthy controls |
126 347 |
0 1 |
1 0 |
0 1 |
0 1 |
1 0 |
16 60 |
0 2 |
44 138 |
8 16 |
1 1 |
1 0 |
40 97 |
||||||
| (Dutta el al., 2007) [38] | 1. Patients with ASD 2. Healthy controls |
55 80 |
|
|
0 1 |
10 12 |
31 42 |
|||||||||||||
| First Author, Year | N | Number of CGG Repeats | ||||||||||||||||||
| 10/11 | 10/11-16 | 10/12 | 10/13 | 10/23 | 12/10 | 12/12 | 12/13 | 13/8 | 13/10 | 13/13 | 14/10 | 15/10 | 16/10 | |||||||
| (Persico et al., 2001) [20] | 1. Patients with ASD a | 95 | 0 | 5 | 3 | 1 | 1 | |||||||||||||
| 2. Healthy controls | 186 | 1 | 2 | 6 | 0 | 0 | ||||||||||||||
| (Zhang et al., 2002) [37] | 1. Patients with ASD 2. Healthy controls |
126 347 |
14 28 |
0 2 |
||||||||||||||||
| (Dutta el al., 2007) [38] | 1. Patients with ASD 2. Healthy controls |
55 80 |
0 3 |
2 4 |
11 14 |
1 1 |
0 1 |
0 1 |
0 1 |
|||||||||||
a ASD: Autistic spectrum disorder.
Table 2.
Distribution of RELN CGG repeat alleles.
| First Author, Year | N | Number of CGG Repeats | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 23 | |||
| (Persico et al., 2001) [20] | 1. Patients with ASD a | 95 | 2 | 84 | 86 | 0 | 11 | 6 | 0 | 1 | |||||||
| 2. Healthy controls | 186 | 0 | 165 | 190 | 4 | 3 | 9 | 1 | 0 | ||||||||
| (Zhang et al., 2002) [37] | 1. Patients with ASD | 126 | 0 | 1 | 0 | 1 | 85 | 2 | 140 | 4 | 5 | 13 | 0 | 0 | 1 | ||
| 2. Healthy controls | 347 | 1 | 0 | 1 | 1 | 277 | 3 | 363 | 7 | 12 | 26 | 1 | 2 | 0 | |||
| (Dutta et al., 2007) [38] | 1. Patients with ASD | 55 | 12 | 83 | 0 | 15 | 0 | 0 | 0 | ||||||||
| 2. Healthy controls | 80 | 18 | 116 | 3 | 20 | 1 | 1 | 1 | |||||||||
a ASD: Autistic spectrum disorder.
Table 3.
Genotype and allele distribution: intron 59, exon 22, and exon 50.
| First Author, Year | N | rs736707 (intron 59) | rs362691(exon22) L997V | rs2229864 (exon 50) | |||||||||||||
| Genotype distribution | Allele distribution | Genotype distribution | Allele distribution | Genotype distribution | Allele distribution | ||||||||||||
| CC | CT | TT | C | T | CC | CG | GG | C | G | TT | CT | CC | T | C | |||
| (Dutta et al., 2007) [38] | 1. Patients with ASD a | 55 | 4 | 24 | 27 | 32 | 78 | ||||||||||
| 2. Healthy controls | 80 | 8 | 31 | 41 | 47 | 113 | |||||||||||
| (Dutta et al., 2008) [23] | 1. Patients with ASD | 77 | 11 | 34 | 32 | 56 | 98 | 56 | 20 | 1 | 132 | 22 | |||||
| 2. Healthy controls (exon 22 N= 100) | 101 | 19 | 49 | 33 | 87 | 115 | 76 | 23 | 1 | 175 | 25 | ||||||
| (Li et al., 2008) [39] | 1. Patients with ASD (intron 59 N= 210) |
213 | 52 | 108 | 50 | 212 | 208 | 159 | 47 | 7 | 365 | 61 | 8 | 76 | 129 | 92 | 334 |
| 2. Healthy controls | 160 | 29 | 78 | 53 | 136 | 184 | 125 | 30 | 5 | 280 | 40 | 7 | 53 | 100 | 67 | 253 | |
| (He et al., 2011) [24] | 1. Patients with ASD (exon 22 N= 219) |
221 | 50 | 116 | 55 | 216 | 226 | 180 | 36 | 3 | 396 | 42 | 9 | 73 | 139 | 92 | 350 |
| 2. Healthy controls (exon 22 N= 277) (exon 50 N= 278) |
282 | 48 | 146 | 88 | 242 | 322 | 216 | 53 | 8 | 485 | 69 | 13 | 87 | 178 | 113 | 443 | |
| (Sharma et al., 2013) [22] | 1. Patients with ASD (intron 59 N= 129) |
136 | 14 | 50 | 65 | 78 | 180 | 3 | 16 | 117 | 22 | 250 | |||||
| 2. Healthy controls (intron 59 N= 208) |
193 | 35 | 94 | 79 | 164 | 252 | 2 | 34 | 157 | 38 | 348 | ||||||
| (Mehdizadeh et al., 2015) [40] | 1. Patients with ASD | 74 | 41 | 26 | 7 | 108 | 40 | ||||||||||
| 2. Healthy controls | 86 | 52 | 28 | 6 | 132 | 40 | |||||||||||
| (Mehdizadeh et al., 2016) [41] | 1. Patients with ASD | 74 | 0 | 16 | 58 | 16 | 132 | ||||||||||
| 2. Healthy controls | 88 | 0 | 28 | 60 | 28 | 148 | |||||||||||
| (Wang et al., 2018) [43] | 1. Patients with ASD | 157 | 33 | 78 | 46 | 144 | 170 | 19 | 70 | 68 | 108 | 206 | |||||
| 2. Healthy controls | 256 | 54 | 126 | 76 | 234 | 278 | 13 | 76 | 167 | 102 | 410 | ||||||
| (Şahin et al., 2018) [42] | 1. Patients with ASD | 61 | 0 | 10 | 51 | 10 | 112 | ||||||||||
| 2. Healthy controls | 64 | 0 | 8 | 56 | 8 | 120 | |||||||||||
a ASD: Autistic spectrum disorder.
Regarding the country of origin, three studies were carried out in China [24,39,43], two studies were performed in India [23,38], two studies were performed in Iran [40,41], and the other studies were performed in Canada [37], South Africa [22], Italy [20], and Turkey [42]. Other demographic characteristics are shown in Table 4.
Table 4.
Characteristics of the studies included in the meta-analysis.
| First Author, Year |
Country | Criteria for ASD a Definition | Ethnicity | Female/Male Ratio | Age (Mean [SD]) | ||
|---|---|---|---|---|---|---|---|
| Patients with ASD | Healthy Controls | Patients with ASD | Healthy Controls | ||||
| Persico et al., 2001 [20] |
Italy | DSM–IV b criteria for Autistic disorder | Caucasian | 6/89 | 89/97 | 6.25 (2.8) | 51.7 (19.6) |
| Zhang et al., 2002 [37] |
Canada | ADI–R c algorithm / ADOS d | N/A e | N/A | 170/177 | N/A | N/A |
| Dutta et al., 2007 [38] |
India | DSM–IV criteria for Autistic disorder | Indian | N/A | N/A | N/A | N/A |
| Dutta et al., 2008 [23] |
India | DSM–IV criteria for Autistic disorder | Indian | 13/64 | N/A | 5.8 (2.9) | N/A |
| Li et al., 2008 [39] |
China | DSM–IV criteria for Autistic disorder or ICD-10 f | Chinese Han | 32/181 | 25/135 | 5.3 (N/A) | 6.7 (N/A) |
| He et al., 2011 [24] |
China | DSM–IV criteria for Autistic disorder | Chinese Han | 35/197 | 43/240 | N/A | 32.8 (10.5) |
| Sharma et al., 2013 [22] |
South Africa | DSM–IV criteria for Autistic disorder | Black, white, and mixed ancestry | N/A | N/A | N/A | N/A |
| Mehdizadeh et al., 2015 [40] | Iran | DSM–IV criteria for Autistic disorder | Caucasian | 18/53 | 65/21 | 8.57 (N/A) | N/A |
| Mehdizadeh et al., 2016 [41] | Iran | DSM–IV criteria for Autistic disorder | Caucasian | 18/53 | 66/22 | 8.57 (0.07) | 7.79 (0.14) |
| Wang et al., 2018 [43] | China | DSM–IV criteria for Autistic disorder | Chinese Han | 21/108 | 72/184 | 8.4 (3.9) | 8.3 (3.9) |
| Şahin et al., 2018 [42] | Turkey | g DSM–5 criteria for Autistic disorder | N/A | 5/56 | 12/52 | 5.54 (3.1) | 6.43 (4.0) |
a ASD: Autistic Spectrum Disorder. b DSM–IV: Diagnostic and Statistical Manual of Mental Disorders, 4th edition. c ADI–R: Autism Diagnostic Interview-Revised. d ADOS: Autistic Diagnostic Observation Schedule. e N/A: not available. f ICD–10: International Classification of Diseases–10. g DSM–5: Diagnostic and Statistical Manual of Mental Disorders, 5th edition.
All studies used genomic DNA extracted from nucleated peripheral blood cells and carried out genotyping using polymerase chain reaction.
3.2. Relationship of Reelin Gene Polymorphisms with ASD
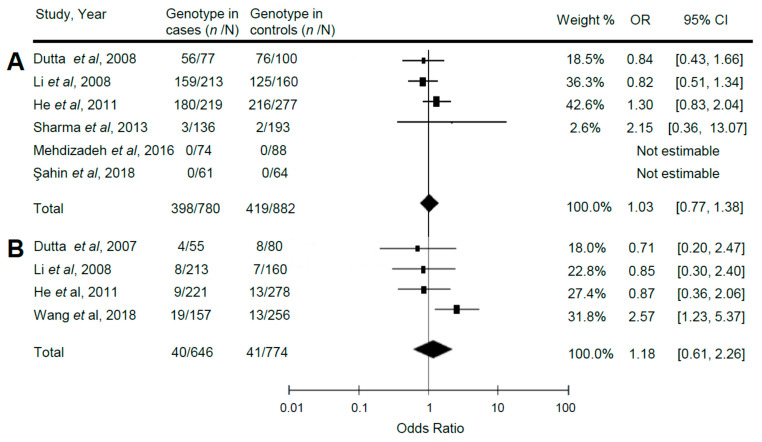
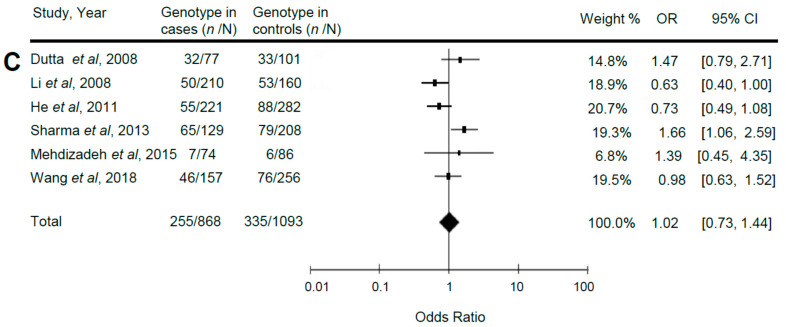
The summary and statistics for the association of reelin gene polymorphisms with ASD are shown in Table 5 and Figure 2 (A: Distribution of the genotype CC of the exon 22 polymorphism (rs362691) compared between patients with ASD (cases) and healthy controls under a random effects model. Test for overall effect: Z = 0.21 (p = 0.83). Test for heterogeneity: χ2 = 2.85 (p = 0.42); I2 = 0%. B: Distribution of the genotype TT of the exon 50 polymorphism (rs2229864) compared between patients with ASD (cases) and healthy controls under a random effects model. Test for overall effect: Z = 0.49 (p = 0.63). Test for heterogeneity: χ2 = 5.70 (p = 0.13); I2 = 47%. C: Distribution of the genotype TT of the intron 59 polymorphism (rs736707) compared between patients with ASD (cases) and healthy controls under a random effects model. Test for overall effect: Z = 0.12 (p = 0.90). Test for heterogeneity: χ2 = 13.08 (p = 0.02); I2= 62%).
Table 5.
Comparison of the allele and genotype frequencies of genetic variants in patients with autistic spectrum disorder (cases) versus healthy controls under a random effects model.
| Polymorphisms | OR | 95% CI | P overall effect | Q | P heterogeneity |
|---|---|---|---|---|---|
| Exon 22 | |||||
| C vs. G | 0.95 | 0.76, 1.20 | 0.68 | 5.32 | 0.38 |
| CC vs. CG + GG | 1.03 | 0.77, 1.38 | 0.83 | 2.85 | 0.42 |
| GG vs. CG + CC | 1.20 | 0.83, 1.75 | 0.34 | 4.08 | 0.54 |
| Exon 50 | |||||
| C vs. T | 0.81 | 0.55, 1.19 | 0.28 | 13.56 | 0.004 |
| CC vs. CT + TT | 0.75 | 0.48, 1.16 | 0.19 | 11.50 | 0.009 |
| TT vs. CT + CC | 1.18 | 0.61, 2.26 | 0.63 | 5.70 | 0.13 |
| Intron 59 | |||||
| C vs. T | 0.98 | 0.77, 1.24 | 0.84 | 16.05 | 0.007 |
| CC vs. CT + TT | 1.02 | 0.76, 1.37 | 0.88 | 7.84 | 0.17 |
| TT vs. CT + CC | 1.02 | 0.73, 1.44 | 0.90 | 13.08 | 0.02 |
| Triplet repeat number |
|||||
| 4 | 9.09 | 1.00, 82.50 | 0.05 | 0.01 | 0.94 |
| 8 | 0.86 | 0.69, 1.08 | 0.19 | 1.30 | 0.52 |
| 10 | 1.00 | 0.78, 1.29 | 0.98 | 2.77 | 0.25 |
| 11 | 0.89 | 0.14, 5.59 | 0.90 | 1.63 | 0.20 |
| 12 | 1.68 | 0.30, 9.50 | 0.56 | 7.41 | 0.02 |
| 13 | 1.26 | 0.81, 1.97 | 0.31 | 0.22 | 0.89 |
| 14 | 0.66 | 0.10, 4.20 | 0.66 | 0.08 | 0.96 |
| 15 | 0.52 | 0.06, 4.69 | 0.56 | 0.00 | 0.95 |
| 16 | 2.00 | 0.12, 32.62 | 0.63 | 1.52 | 0.22 |
Figure 2.
Meta-analysis of the association of reelin gene polymorphisms with autistic spectrum disorder (ASD). (A): Distribution of the genotype CC of the exon 22 polymorphism (rs362691) compared between patients with ASD (cases) and healthy controls under a random effects model. (B): Distribution of the genotype TT of the exon 50 polymorphism (rs2229864) compared between patients with ASD (cases) and healthy controls under a random effects model. (C): Distribution of the genotype TT of the intron 59 polymorphism (rs736707) compared between patients with ASD (cases) and healthy controls under a random effects model.
We did not find any significant association between possession of the CC genotype of exon 22 (rs362691), TT of exon 50 (rs2229864), or TT of intron 59 (rs736707) and the presence of ASD (Figure 2A: OR = 1.03; 95% CI: 0.77, 1.38; p = 0.83; Figure 2B: OR = 1.18; 95% CI: 0.61, 2.26; p = 0.63; Figure 2C: OR = 1.02; 95% CI: 0.73, 1.44; p = 0.90, respectively). Large heterogeneity was found (I2 = 62%, p = 0.02) when analyzing the rs736707 polymorphism. The comparison of allele frequencies and other models of inheritance also did not yield any significant results.
3.3. Relationship of Polymorphic Trinucleotide Repeat (CGG/GCC) within the Reelin Gene with ASD
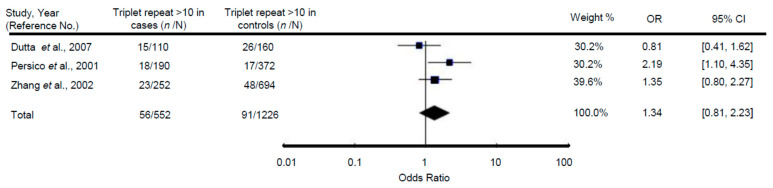
Meta-analysis regarding the association of the presence of >10 repeats of this triplet and ASD did not find any significant association (OR = 1.34; 95% CI: 0.81, 2.23; p = 0.26) (Figure 3: The presence of >10 repeats compared between patients with ASD (cases) and healthy controls under a random effects model. Test for the overall effect: Z = 1.13 (p = 0.26). Test for heterogeneity: χ2 = 3.97 (p = 0.14); I2 = 50%). The comparison of specific genotype frequencies also did not show any significant relationship (Table 5).
Figure 3.
Meta-analysis of the association between the polymorphic trinucleotide repeat (CGG/GCC) within the reelin gene and ASD.
4. Discussion
In our meta-analysis, we found no significant relationship between ASD and the distribution of the analyzed SNPs (rs736707, rs362691, and rs2229864) or the presence of longer triplet repeats in the 5′UTR region of the RELN gene. It is clear, however, that functional studies [19,44,45] and the results from genome wide scans [17,18] strongly suggest a role for this gene and the reelin protein in the development of ASD.
To explain our negative results, our meta-analysis indicates that previous studies with different ethnic groups found diverse and even contradictory results when analyzing these allelic variants. For instance, Li et al. [39] found that the TT genotype of the rs736707 polymorphism was significantly less prevalent among autistic patients in China compared to the controls, whereas Sharma et al. [22] reported a significantly higher prevalence of the T allele of this SNP among autistic patients of mixed ethnicity from South Africa. This may explain the large heterogeneity found for this polymorphism in the meta-analysis. Further, the G allele of the rs362691 polymorphism has been alternatively identified as the minor [31] or the major allele [22], although similar frequencies have been observed in the same ethnic group. All these results point towards genetic heterogeneity of ASD according to ethnicity, which may be partially responsible for the disparity of the results. Regrettably, an ethnicity analysis was not performed because a low number of studies were finally available for inclusion in this meta-analysis. This was, at least in part, because we were not able to fully combine the results from candidate gene association studies with those from family-based association studies since these studies differed in their design and statistical methods. The inclusion of a small number of studies, and their small sample sizes, are potentially associated with the lack of power to detect a small effect of common variants, and we acknowledge that this may be one of the major shortcomings of our work.
Apart from the risks of false negative results due to ethnicity as a confusion factor and/or low sample sizes, we cannot discount other explanations for our findings. Genetic susceptibility to ASD is probably mediated by many loci, each with a small to moderate effect [46]. Therefore, other genetic factors within this gene may have a relevant role in the development of ASD, such as different polymorphisms or haplotypes within RELN [33]. It has been also suggested that a specific paternal 5′UTR-CGG repeat allele effect may be of relevance [38]. Moreover, it is possible that the SNPs of the reelin gene that have been significantly associated with ASD are not directly involved with this disorder but exist in linkage disequilibrium with other functional polymorphisms in a nearby locus. Finally, gene–environment interactions or epigenetic factors may be more important than individual SNPs [47]. All these hypotheses are very difficult to test [48], and we admit that it may be challenging to determine the specific genetic factors associated with ASD, particularly since the relevant genetic bases may be subtle [32,46].
For our study, we selected the four genetic variants which had been more frequently analyzed in previous studies in order to be able to conduct a meta-analysis [20,21,22,23,24,31,32,33,34,35,36,37,38,39,40,41,42,43]. Due to the low sample sizes of previous studies and the low number of studies, it would be of interest to conduct more original gene candidate association studies (in addition to genome-wide association studies, such as the one recently developed by Matoba [16], but, unlike that paper, including healthy unrelated controls) to combine these studies with the meta-analysis. Otherwise, it will remain difficult to detect small or medium effects. Furthermore, the criteria for ASD definitions should be standardized between different studies to decrease heterogeneity.
5. Conclusions
In summary, our meta-analysis represents a comprehensive and up-to-date revision of the association of RELN genetic variants and ASD, including data from 1289 patients and 1858 controls. In light of the current evidence, no single genetic variant within this gene is clearly associated with the development of ASD. Larger studies in different ethnic groups are needed to establish the role of specific genetic variants within RELN in the etiology of this disorder.
Author Contributions
Conceptualization, M.M. and J.-A.M.-C.; methodology, I.H.-G., M.M. and A.-J.C.; formal analysis, I.H.-G., A.-J.C., and C.C.; data curation, I.H.-G., H.G.T.-d.l.V., A.-J.C., and C.C.; writing—original draft preparation, I.H.-G., A.-J.C., H.G.T.-d.l.V., and M.M.; writing—review and editing, I.H.-G., C.C., J.-A.M.-C., and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly funded by the program I3SNS, Instituto de Salud Carlos III (Spain) for Miguel Marcos.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elsabbagh M., Divan G., Koh Y.J., Kim Y.S., Kauchali S., Marcín C., Montiel-Nava C., Patel V., Paula C.S., Wang C., et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning-Courtney P., Murray D., Currans K., Johnson H., Bing N., Kroeger-Geoppinger K., Sorensen R., Bass J., Reinhold J., Johnson A., et al. Autism spectrum disorders. Curr. Probl. Pediatr. Adolesc. Health Care. 2013;43:2–11. doi: 10.1016/j.cppeds.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Lyall K., Croen L., Daniels J., Fallin M.D., Ladd-Acosta C., Lee B.K., Park B.Y., Snyder N.W., Schendel D., Volk H., et al. The Changing Epidemiology of Autism Spectrum Disorders. Annu Rev. Public Health. 2017;38:81–102. doi: 10.1146/annurev-publhealth-031816-044318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen A., Pettygrove S., Meaney F.J., Mancilla K., Gotschall K., Kessler D.B., Grebe T.A., Cunniff C. Prevalence of autism spectrum disorders in Hispanic and non-Hispanic white children. Pediatrics. 2012;129:e629–e635. doi: 10.1542/peds.2011-1145. [DOI] [PubMed] [Google Scholar]
- 5.Maenner M.J., Shaw K.A., Baio J., Washington A., Patrick M., DiRienzo M., Christensen D.L., Wiggins L.D., Pettygrove S., Andrews J.G., et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020;69:1. doi: 10.15585/mmwr.ss6904a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron-Cohen S., Scott F.J., Allison C., Williams J., Bolton P., Matthews F.E., Brayne C. Prevalence of autism-spectrum conditions: UK school-based population study. Br. J. Psychiatry. 2009;194:500–509. doi: 10.1192/bjp.bp.108.059345. [DOI] [PubMed] [Google Scholar]
- 7.Narzisi A., Posada M., Barbieri F., Chericoni N., Ciuffolini D., Pinzino M., Romano R., Scattoni M.L., Tancredi R., Calderoni S., et al. Prevalence of Autism Spectrum Disorder in a large Italian catchment area: A school-based population study within the ASDEU project. Epidemiol. Psychiatr. Sci. 2020;29:e5. doi: 10.1017/S2045796018000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X., Allison C. A review of the prevalence of Autism Spectrum Disorder in Asia. Res. Autism Spectr. Disord. 2010;4:156–167. doi: 10.1016/j.rasd.2009.10.003. [DOI] [Google Scholar]
- 9.Qiu S., Lu Y., Li Y., Shi J., Cui H., Gu Y., Li Y., Zhong W., Zhu X., Liu Y., et al. Prevalence of autism spectrum disorder in Asia: A systematic review and meta-analysis. Psychiatry Res. 2020;284:112679. doi: 10.1016/j.psychres.2019.112679. [DOI] [PubMed] [Google Scholar]
- 10.Olusanya B.O., Davis A.C., Wertlieb D., Boo N.Y., Nair M.K.C., Halpern R., Kuper H., Breinbauer C., De Vries P.J., Gladstone M., et al. Developmental disabilities among children younger than 5 years in 195 countries and territories, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Glob. Health. 2018;6:e1100–e1121. doi: 10.1016/S2214-109X(18)30309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howlin P., Goode S., Hutton J., Rutter M. Adult outcome for children with autism. J. Child. Psychol. Psychiatry. 2004;45:212–229. doi: 10.1111/j.1469-7610.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 12.Gordon-Lipkin E., Marvin A.R., Law J.K., Lipkin P.H. Anxiety and Mood Disorder in Children with Autism Spectrum Disorder and ADHD. Pediatrics. 2018;141:e20171377. doi: 10.1542/peds.2017-1377. [DOI] [PubMed] [Google Scholar]
- 13.Lung F.W., Shu B.C., Chiang T.L., Lin S.J. Prevalence of bullying and perceived happiness in adolescents with learning disability, intellectual disability, ADHD, and autism spectrum disorder: In the Taiwan Birth Cohort Pilot Study. Medicine (Baltimore) 2019;98:e14483. doi: 10.1097/MD.0000000000014483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg R.E., Law J.K., Yenokyan G., McGready J., Kaufmann W.E., Law P.A. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. pediatrics Adolesc. Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- 15.Lamb J.A., Parr J.R., Bailey A.J., Monaco A.P. Autism: In search of susceptibility genes. Neuromolecular Med. 2002;2:11–28. doi: 10.1385/NMM:2:1:11. [DOI] [PubMed] [Google Scholar]
- 16.Matoba N., Liang D., Sun H., Aygün N., McAfee J.C., Davis J.E., Raffield L.M., Qian H., Piven J., Li Y., et al. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl. Psychiatry. 2020;10:265. doi: 10.1038/s41398-020-00953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonan A.L., Alarcon M., Cheng R., Magnusson P.K., Spence S.J., Palmer A.A., Grunn A., Juo S.H.H., Terwilliger J.D., Liu J., et al. A genomewide screen of 345 families for autism-susceptibility loci. Am. J. Hum. Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett S., Beck J.C., Bernier R., Bisson E., Braun T.A., Casavant T.L., Childress D., Folstein S.E., Garcia M., Gardiner M.B., et al. An autosomal genomic screen for autism. Am. J. Med. Genet.—Neuropsychiatr. Genet. 2001;105:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Fatemi S.H., Snow A.V., Stary J.M., Araghi-Niknam M., Reutiman T.J., Lee S., Brooks A.I., Pearce D.A. Reelin signaling is impaired in autism. Biol. Psychiatry. 2005;57:777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Persico A.M., D’Agruma L., Maiorano N., Totaro A., Militerni R., Bravaccio C., Wassink T.H., Schneider C., Melmed R., Trillo S., et al. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol. Psychiatry. 2001;6:150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 21.Serajee F.J., Zhong H., Mahbubul Huq A.H. Association of Reelin gene polymorphisms with autism. Genomics. 2006;87:75–83. doi: 10.1016/j.ygeno.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Sharma J.R., Arieff Z., Gameeldien H., Davids M., Kaur M., van der Merwe L. Association analysis of two single-nucleotide polymorphisms of the RELN gene with autism in the South African population. Genet. Test. Mol. Biomark. 2013;17:93–98. doi: 10.1089/gtmb.2012.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta S., Sinha S., Ghosh S., Chatterjee A., Ahmed S., Usha R. Genetic analysis of reelin gene (RELN) SNPs: No association with autism spectrum disorder in the Indian population. Neurosci. Lett. 2008;441:56–60. doi: 10.1016/j.neulet.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 24.He Y., Xun G., Xia K., Hu Z., Lv L., Deng Z., Zhao J. No significant association between RELN polymorphism and autism in case-control and familybased association study in Chinese Han population. Psychiatry Res. 2011;187:462–464. doi: 10.1016/j.psychres.2010.04.051. [DOI] [PubMed] [Google Scholar]
- 25.Cochran W.G. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 26.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cochrane Training: Review Manager (RevMan) [(accessed on 16 August 2020)]; Available online: https://training.cochrane.org/online-learning/core-software-cochrane-reviews/revman.
- 29.Bax L., Yu L.M., Ikeda N., Tsuruta H., Moons K.G. Development and validation of MIX: Comprehensive free software for meta-analysis of causal research data. BMC Med. Res. Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Wang C., Yu J., Fan Y., Liu D., Zhou W., Shi T. Residential Radon and Histological Types of Lung Cancer: A Meta-Analysis of Case‒Control Studies. Int. J. Environ. Res. Public Health. 2020;17:1457. doi: 10.3390/ijerph17041457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonora E., Beyer K.S., Lamb J.A., Parr J.R., Klauck S.M., Benner A., Paolucci M., Abbott A., Ragoussis I., Poustka A., et al. Analysis of reelin as a candidate gene for autism. Mol. Psychiatry. 2003;8:885–892. doi: 10.1038/sj.mp.4001310. [DOI] [PubMed] [Google Scholar]
- 32.Devlin B., Bennett P., Dawson G., Figlewicz D.A., Grigorenko E.L., McMahon W., Minshew N., Pauls D., Smith M., Spence M.A., et al. Alleles of a reelin CGG repeat do not convey liability to autism in a sample from the CPEA network. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2004;126B:46–50. doi: 10.1002/ajmg.b.20125. [DOI] [PubMed] [Google Scholar]
- 33.Skaar D.A., Shao Y., Haines J.L., Stenger J.E., Jaworski J., Martin E.R., DeLong G.R., Moore J.H., McCauley J.L., Sutcliffe J.S., et al. Analysis of the RELN gene as a genetic risk factor for autism. Mol. Psychiatry. 2005;10:563–571. doi: 10.1038/sj.mp.4001614. [DOI] [PubMed] [Google Scholar]
- 34.Kelemenova S., Schmidtova E., Ficek A., Celec P., Kubranska A., Ostatnikova D. Polymorphisms of candidate genes in Slovak autistic patients. Psychiatr. Genet. 2010;20:137–139. doi: 10.1097/YPG.0b013e32833a1eb3. [DOI] [PubMed] [Google Scholar]
- 35.Krebs M.O., Betancur C., Leroy S., Bourdel M.C., Gillberg C., Leboyer M. Absence of association between a polymorphic GGC repeat in the 5’ untranslated region of the reelin gene and autism. Mol. Psychiatry. 2002;7:801–804. doi: 10.1038/sj.mp.4001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Nguyen L., Gleason C., Lotspeich L., Spiker D., Risch N., Myers R.M. Lack of evidence for an association between WNT2 and RELN polymorphisms and autism. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2004;126B:51–57. doi: 10.1002/ajmg.b.20122. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H., Liu X., Zhang C., Mundo E., Macciardi F., Grayson D.R., Guidotti A.R., Holden J.J.A. Reelin gene alleles and susceptibility to autism spectrum disorders. Mol. Psychiatry. 2002;7:1012–1017. doi: 10.1038/sj.mp.4001124. [DOI] [PubMed] [Google Scholar]
- 38.Dutta S., Guhathakurta S., Sinha S., Chatterjee A., Ahmed S., Ghosh S., Gangopadhyay P.K., Singh M., Usha R. Reelin gene polymorphisms in the Indian population: A possible paternal 5’UTR-CGG-repeat-allele effect on autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:106–112. doi: 10.1002/ajmg.b.30419. [DOI] [PubMed] [Google Scholar]
- 39.Li H., Li Y., Shao J., Li R., Qin Y., Xie C., Zhao Z. The association analysis of RELN and GRM8 genes with autistic spectrum disorder in Chinese Han population. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008;147:194–200. doi: 10.1002/ajmg.b.30584. [DOI] [PubMed] [Google Scholar]
- 40.Mehdizadeh L., Shahrokhi H., Adampourezare M., Hosseinpour M.A., Bonyadi M., Eslami A. Association between Common Single-nucleotide Polymorphism of Reelin Gene, rs736707 (C/T) with Autism Spectrum Disorder in Iranian-Azeri Patients. Int. J. Pediatrics. 2015;3:1065–1071. [Google Scholar]
- 41.Mehdizadeh L., Hosseinpour M.A., Zare M.A., Shahrokhi H. An Association Analysis of Reelin Gene (RELN) Exon 22 (G/C), Rs.362691, Polymorphism with Autism Spectrum Disorder among Iranian-Azeri Population. Int. J. Pediatrics. 2016;4:2027–2033. [Google Scholar]
- 42.Şahin N., Kara M., Kara B., Topal H. Evaluation of RELN gene polymorphism in children with autism spectrum disorder. Anadolu Psikiyatri Dergisi. 2018;19:599–606. doi: 10.5455/apd.292500. [DOI] [Google Scholar]
- 43.Wang G.F., Ye S., Gao L., Han Y., Guo X., Dong X.P., Su Y.Y., Zhang X. Two Single-Nucleotide Polymorphisms of the RELN Gene and Symptom-Based and Developmental Deficits Among Children and Adolescents with Autistic Spectrum Disorders in the Tianjin, China. Behav. Brain Res. 2018;350:1–5. doi: 10.1016/j.bbr.2018.04.048. [DOI] [PubMed] [Google Scholar]
- 44.Fatemi S.H., Stary J.M., Egan E.A. Reduced blood levels of reelin as a vulnerability factor in pathophysiology of autistic disorder. Cell. Mol. Neurobiol. 2002;22:139–152. doi: 10.1023/A:1019857620251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey A., Luthert P., Dean A., Harding B., Janota I., Montgomery M., Rutter M., Lantos P. A clinicopathological study of autism. Brain. 1998;121:889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- 46.Risch N., Spiker D., Lotspeich L., Nouri N., Hinds D., Hallmayer J., Kalaydjieva L., McCague P., Dimiceli S., Pitts T., et al. A genomic screen of autism: Evidence for a multilocus etiology. Am. J. Hum. Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu C.E., Dawson G., Munson J., D’Souza I., Osterling J., Estes A., Leutenegger A.L., Flodman P., Smith M., Raskind W.H., et al. Presence of large deletions in kindreds with autism. Am. J. Hum. Genet. 2002;71:100–115. doi: 10.1086/341291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daly A.K., Day C.P. Candidate gene case-control association studies: Advantages and potential pitfalls. Br. J. Clin. Pharmacol. 2001;52:489–499. doi: 10.1046/j.0306-5251.2001.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]