Graphical abstract
Keywords: Flux balance analysis, Cell metabolism, Strain optimisation, Chinese hamster ovary cells, Hybrid modelling, Principal component analysis, Recombinant protein production
Abstract
With omics data being generated at an unprecedented rate, genome-scale modelling has become pivotal in its organisation and analysis. However, machine learning methods have been gaining ground in cases where knowledge is insufficient to represent the mechanisms underlying such data or as a means for data curation prior to attempting mechanistic modelling. We discuss the latest advances in genome-scale modelling and the development of optimisation algorithms for network and error reduction, intracellular constraining and applications to strain design. We further review applications of supervised and unsupervised machine learning methods to omics datasets from microbial and mammalian cell systems and present efforts to harness the potential of both modelling approaches through hybrid modelling.
1. Introduction
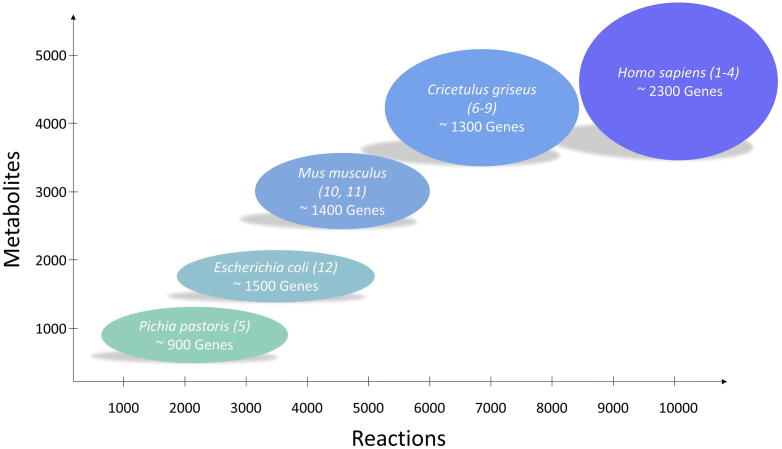
Genome-scale metabolic models (GeMs) are a database for all known information about an organism across multiple scales, including most of its known genes, the enzymes encoded by those genes and associated expression rules (gene-protein-reaction, GPR, rules), transport reactions and participating metabolites. There have been numerous GeMs published for a variety of organisms, most of which can be found in the BiGG database (http://bigg.ucsd.edu/). Relevant to the production of protein therapeutics are the human [1], [2], [3], [4], Pichia pastoris as described in Theron et al. [5], Chinese hamster ovary (CHO) cells [6], [7], [8], [9], murine cells [10], [11] and a plethora of GeMs for Escherichia coli [12], as summarised in Fig. 1.
Fig. 1.
Summary of genome-scale metabolic models for different organisms relevant to the production of recombinant proteins or valuable metabolic products.
Although GeMs provide valuable insight into biological networks, full integration of data across the genomic, transcriptomic, proteomic and metabolomic scales is yet to be effectively realised. Given the unprecedented rate of data generation, modelling efforts have evolved by, for example, developing hybrid kinetic/stoichiometric formulations to overcome the weaknesses of any individual approach whilst, at the same time, combining their strengths through alleviating the burden of parameter estimation [13], or including intracellular insight without loss of model tractability [14], [15]. The next logical step is the development of hybrid approaches that take advantage of known techniques that harness the information content of extensive datasets. Machine learning (ML) is the scientific study of algorithms applied to complex datasets for pattern recognition, classification, and prediction. The concept of automated learning was developed from the theory that machines learn without being pre-programmed on assignments of data patterns to classes. The iterative notion behind the learning theory relies on the independent adaptation of the ML model when presented to new data input. Such adaptation is based on the ability to recognise patterns in complex datasets to generate reliable, reproducible results from previous computations.
Herein, we review the latest advances in GeM development and application for process understanding and cell engineering and discuss efforts to hybridise FBA with dynamic kinetic models focusing on recombinant protein producing systems. These include CHO cells, which are the workhorse of industrial therapeutic glycoprotein production, but also microbial hosts such as Pichia pastoris and Escherichia coli. We then present the case for using ML to decipher the information carried in large omics datasets and review the main techniques for doing so, including supervised and unsupervised ML methods, as well as necessary data pre-treatment techniques prior to ML application. Given the limited number of studies on industrial protein production systems, we review the application of ML to omics more broadly, including microbial cell systems for metabolite or recombinant protein production as well as human disease models.
2. Advances in GeM development and application
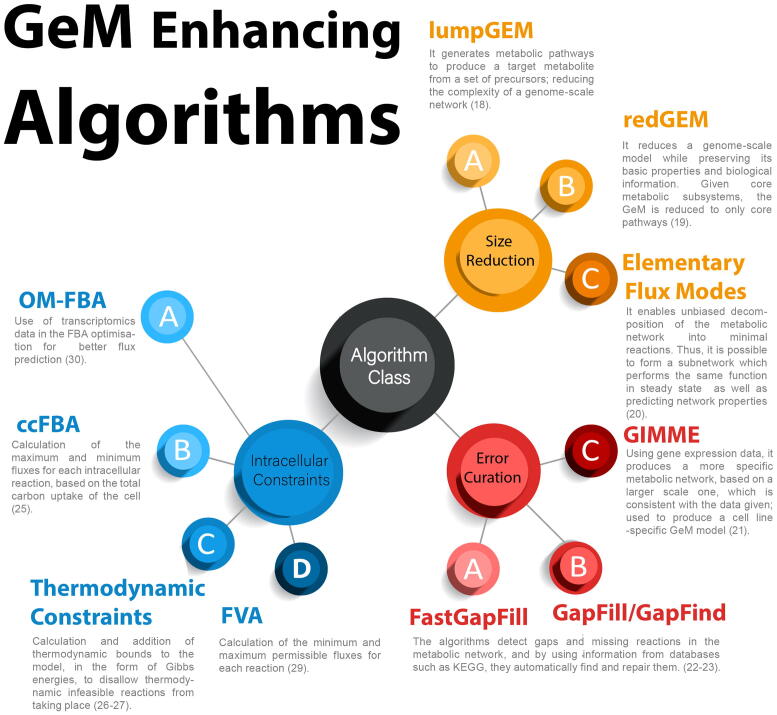
Research involving GeMs has largely focused on (a) the development of consensus models for commonly used organisms, (b) the advancement of computational algorithms and (c) the development of dynamic flux models. Recent key studies in these three areas are summarised in Fig. 2. Efforts to develop GeMs have yielded a new community-curated model of CHO cell metabolism developed by Hefzi et al. [6]. Their work includes cell line-specific models for CHO-S, CHO-K1 as well as the generic CHO GeM model, iCHO1766, which contains most of the known CHO genes, enzymes and metabolites. Calmels et al. have also developed a GeM specific to the DG-44 cell line [7]. The iCHO1766 GeM was recently expanded to include the secretory pathway in a study that paves the way for ascertaining the burden of individual recombinant protein molecules on metabolism and protein synthesis and secretion [16].
Fig. 2.
Algorithms used to improve the network and efficiency of genome-scale metabolic models.
A powerful tool for the analysis of GeMs and the calculation of fluxes is Flux Balance Analysis (FBA). FBA is an optimisation technique aiming to predict the flux distribution in a metabolic network. Cell metabolism can be represented by a stoichiometric matrix, S, whose columns represent the reactions, j, and rows the metabolites, i, of the metabolism. FBA assumes pseudo-steady state of the metabolite concentrations; so, if x represents the metabolite concentrations, thus , where v is a vector of the fluxes of the reactions. However, in any biological system the number of reactions is larger than the number of metabolites, so the number of degrees of freedom is greater than 1. Additionally, if we want to predict intracellular fluxes of a GeM using FBA we need to define the upper and lower bounds of fluxes [17].
Recent work on computational algorithms has focused on reducing model size [18], [19], [20] while keeping the core biological information intact as well as identifying missing links in the reaction network and filling them in with reactions from databases [21], [22], [23], such as the Kyoto Encyclopedia of Genes and Genomes or MetaCyC [24]. Lastly, considerable effort is being put into the development of computational techniques for the calculation of appropriate bounds for intracellular reactions [25], [26], [27], [28], [29], [30] and exchange reactions [31].
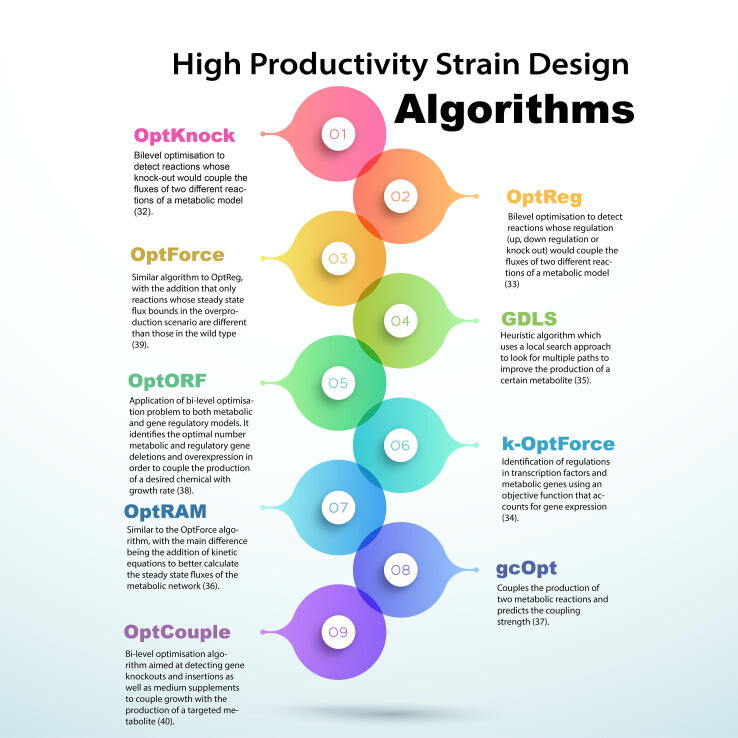
GeMs have found application in the optimisation-based design of genetic engineering strategies to alleviate the trade-off between cell growth and desired product formation. Key algorithms for this purpose [32], [33], [34], [35], [36], [37], [38], [39], [40] are summarised in Fig. 3. There are two distinct fields of optimisation techniques applied to metabolic models for this purpose: optimisation of the metabolic network or optimisation of the extracellular environment. The first is mainly composed of Mixed-Integer Linear Programming (MILP) algorithms, which identify reactions and therefore candidate genes for knockout, upregulation or downregulation. These algorithms have been applied to create engineered microbial strains as described in a recent review by Hendry et al. [41]. For example, Suástegui et al. applied the OptForce algorithm to increase shikimic acid production in Saccharomyces serevisiae [42], and Tan et al. were able to increase octanoic acid production in E. coli again with the use of OptForce [43]. Another example is the use of an E. coli GeM [44] to identify gene knock out strategies that improve glycan biosynthesis [45]. Saitua et al. employed a dynamic GeM of P. pastoris to predict system behaviour across batch and fed-batch cultivation under glucose-limited aerobic conditions, followed by the design of single knock-out genetic engineering strategies that can boost volumetric protein productivity [46]. The CHO GeM has also been used to identify burdensome host cell proteins for deletion to ease pressure on downstream processing [16], [47]. However, due to the size and complexity of mammalian cell GeMs, this kind of algorithms have not been widely applied to mammalian cell systems yet.
Fig. 3.
Algorithms used in strain design for the overproduction of metabolites.
The second category of optimisation applications is focused on improving the media formulation and/or feeding strategy of the culture. CHO cells have been the subject of numerous such studies due to their use for industrial antibody production. GeMs or smaller scale models have supported researchers to improve the design of culture media and feeds or develop more efficient CHO cell strains with the help of experimental data and FBA. The CHO-K1 GeM has been used to compare catabolism in the presence of different feeds and to optimise feed formulation using FBA [48]. Similarly, the CHO DG-44 GeM has been employed to predict cell phenotype when grown in different culture media and propose optimised formulations [7]. A reduced version of a CHO GeM was used by Xing et al. [49] to adjust the concentrations of certain amino acids in the culture media, by Junghans et al. [50] to propose changes to media and feed that could improve bioprocess efficiency and by Templeton et al. [51] to predict high producers based on intracellular flux distribution. The CHO GeM [6] has been used to study glucose and lactate metabolism using FBA coupled with dynamic equations to simulate the consumption and secretion rates of essential metabolites [52], and aid the design of new feeding strategies by analysing intracellular fluxes [53]. This approach has also been employed to model batch CHO cell culture conditions [54] and to study metabolic shifts as a result of switching from physiological temperature to mild hypothermic conditions [55].
Additionally, DFBA has been applied widely to microbial cell systems primarily for the prediction of biomass growth and production of metabolites. Topics of recent studies include the production of shikimic acid in E. coli [56], the prediction of growth rate and ethanol production in Saccharomyces cerevisiae [57] and the overproduction of secretory proteins in Streptomyces lividans [58]. In the context of recombinant protein production, Torres et al. applied a comprehensive GeM to chemostat cultures of P. pastoris conducted under different oxygenation levels to reveal high-order metabolic effects of different culture parameters on system performance [59]. An underpinning requirement for such applications is the availability of a high-fidelity GeM. To this end, a significant recent advancement was the refinement of P. pastoris GeM specifically for growth on non-glucose substrates such as glycerol and methanol, which are most relevant for recombinant protein production [60]. There are several methodologies for coupling FBA optimisation with kinetic equations to create a hybrid model as summarised in Table 1.
Table 1.
Techniques and algorithms used for steady-state and dynamic MFA and FBA.
| Type | Advantages | Disadvantages | Description | Ref. |
|---|---|---|---|---|
| Static Optimisation approach | Simple implementation Suitable for GeM models Fast |
Provides a simple, not very detailed solution Cannot predict metabolic shifts |
Separates culture period into intervals of pseudo steady-state and performs an FBA optimisation for each of them | [131] |
| [132] | ||||
| Dynamic Optimisation Approach | Detailed representation of metabolism Can describe metabolic shifts |
Accurate parameter estimation in differential equations necessary Need to avoid overfitting |
Performs optimization over the whole period of interest with the use of differential equations to describe biomass and media concentrations | [132] |
| [133] | ||||
| [54] | ||||
| DMFA | Calculates intracellular fluxes | Requires extracellular metabolite concentrations thus, cannot be used in underdetermined systems | Uses a linear spline function to calculate intracellular fluxes | [55] |
| Describes intracellular fluxes using linear changes of the fluxes though time | [134] | |||
| Multi-objective optimisation | Uses the duality theorem to achieve optimality | Numerical challenges arising from the DAE formulation | Uses logarithmic barrier functions on the constraints of the primal and dual problem. Converts them to a DAE with the dynamic balance equations for the substrates | [135] |
| Deals with LP infeasibility that can be caused during time integration | Requires careful objective function setting to achieve realistic solution | DFBA using lexicographic optimisation to deal with the LP feasibility problem | [136] |
3. Hybrid machine learning and constrained-based modelling approaches
Machine learning algorithms have found application in both the interpretation of high-dimensional metabolic data and the development of tools for the description of cellular metabolism. More specifically, unsupervised ML methods have been utilized for the identification of key metabolic parameters that accommodate model development, for the identification of sub-groups in the data and for the reduction of data complexity prior to downstream modelling applications. On the other hand, supervised algorithms have been used to both replace alternative approaches (kinetic and stoichiometric models) for metabolic modelling but also to synergistically work and improve the predictions of alternative models.
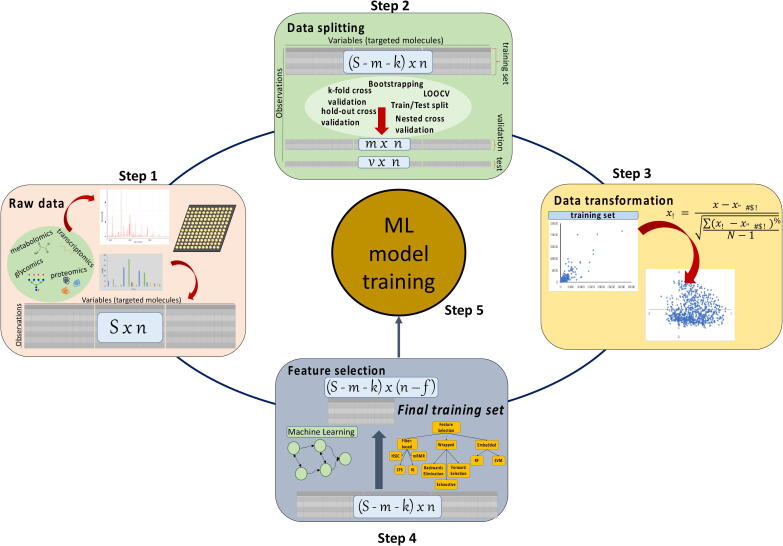
Prior to introducing the available dataset to the chosen ML configuration, data arising from different omics techniques requires pre-processing (Fig. 4) in order to increase model robustness and avoid overfitting. Typically, data pre-treatment is performed in three steps as described below. Whilst elaborating on data pre-treatment is not within the scope of this review, we invite interested readers to further explore the articles suggested herein.
Fig. 4.
Data pre-processing steps for improving the performance and ensuring the robustness of machine learning algorithms.
3.1. Data pre-treatment
3.1.1. Data splitting
As a first step, the dataset is sampled for the construction of (a) the training set, i.e. the data used for model training, (b) the validation set, the data used for tuning hyperparameters and (c) the test set, the set of data used to evaluate model’s predictive performance. Random sampling (RS) is typically applied for data splitting because it introduces low levels of bias and efficiently evaluates model generalization. RS methods can sample the same data point multiple times (RS with replacement) or only once (RS without replacement). Bootstrapping and cross validation (CV) are the most representative techniques of RS with and without replacement, respectively. Detailed comparisons of the data sampling methods can be found in Varoquaoux [61], Vabalas et al. [62], Kim [63], and Xu & Goodcare [64]. It is important to note that data splitting techniques such as CV, should not be used for the identification of optimum data splitting strategies but for the evaluation of model generalization. However, limitations arising from the underlying biological principles can introduce biases in the handling and splitting of the dataset, as the ML algorithms perform poorly when extrapolating. Thus, the user needs to ensure that the training set covers the space within which the validation and test sets lie. Hyperparameters are parameters of the ML algorithm that are set by the user in order to define the configuration of the model and remain constant during model training. The validation set is essential for tuning the hyperparameters (i.e. number of hidden layers and nodes) and should be separate to the training set. Grid or random search algorithms examine different combinations of hyperparameters to identify the top-performing set in an exhaustive or random manner, respectively. Whilst computationally intensive, nested or double cross validation, where the training set determined by the first CV is then further split to a new pair of training and validation sub-sets by a nested CV algorithm, is necessary in order to avoid overfitting of the estimated hyperparameters to the training set.
3.1.2. Data transformation (standardize/normalize)
Prior to any further data pre-processing, raw data are typically scaled in order to remove the bias towards the variables with the highest values. For example, large-scale discrepancies between the concentration of the targeted molecules are commonly observed in omics data and, if not appropriately scaled, the downstream ML algorithm will form strong dependencies to the most abundant targets. Whilst there is a plethora of available data transformation techniques that can be applied to omics data [65], [66] (Table 2), z-score normalization (also known as standardization or autoscaling) is the most widely used technique. Z-score normalization aims to equalize the variance of measured molecules by setting the mean of each variable equal to zero and the standard deviation equal to one. An excellent comparison between different transformation methods applied to metabolomics data prior to multivariate analysis is presented in van den Berg et al. [67]. Another pre-treatment step often applied in omics datasets and especially in single-cell RNA-seq data is the imputation for the estimation of missing points [68], [69], [70], [71], [72].
Table 2.
Summary of major data splitting, data transformation and feature selection techniques. Notation: and are the raw and normalized values of variable in observation , respectively. and are the mean and standard deviation of variable in the available dataset, respectively. and are the minimum and maximum values of variable observed in the available dataset.
| Class | Principle | Advantages | Disadvantages | Methods |
|---|---|---|---|---|
| Data splitting | ||||
| Random Sampling (RS) | The training, validation and test sets are randomly chosen from the population | Minimal levels of bias introduced during sampling When iteratively repeated can be used for model generalization evaluation |
Do not account for data distribution Model might perform poorly if requested to extrapolate Not appropriate for small sample sizes |
Train/Test split k-fold cross-validation (CV) Hold-out cross-validation Nested cross-validation Leave-one-out-cross-validation (LOOCV) Bootstrapping |
| Data Transformation | ||||
| z-score normalization | Accounts for both the mean and the variability of the dataset | Assumes normal distribution Could lead to over-amplification of small differences Increases the impact of measurement error |
||
| Pareto scaling | Reduction of large values effect on model training | Reduction of large variance in the data | ||
| Range scaling | Transformed features are equally important | Outliers can undermine the correct interpretation of data variation | ||
| min-max normalization | Most applicable when the data does not follow a normal distribution | Sensitive to outliers Does not account for the data dispersion |
||
| Mean centering | Mean of all features is zero Can partially alleviate multicollinearity |
Does not scale the data Usually applied in combination with other scaling methods |
||
| Log transformation | Can alleviate heteroskedasticity and impose normal distribution | Can be problematic when values reach transformation function boundaries | ||
| Feature Selection | ||||
| Filter | Features are selected based on their performance in statistical algorithms | High efficiency Independent of the predictor |
Do not interact with the predictor Results can be relatively poor |
Correlation based Feature Selection (CFS) Information Gain (IG) minimum Redundancy-Maximum Relevance (mRMR) Hilbert Schmidt Independence Criterion Lasso (HSIC-Lasso) |
| Wrapped | Evaluation of feature subsets based on ML model performance. They are composed of a search and an evaluation algorithm | Results in feature subsets with good performance | Computationally expensive (greedy) as they require multiple model simulations Biased towards the examined model Exhaustive search and fast evaluation necessary Danger of overfitting Optimization of evaluation method may be necessary |
Forward selection Backwards elimination Exhaustive search |
| Embedded | Feature selection is incorporated in model training | Non-contributing features are usually penalized Less computationally expensive than wrapped methods Robust to overfitting |
Selection is biased towards the model in use | Least absolute shrinkage and selection operator (LASSO) Support Vector Machine (SVM) SVM-Recursive feature elimination (SVM-RFE) Random Forest (RF) |
3.1.3. Feature selection
Implementation of ML algorithms usually requires the handling of large and high-dimensional datasets. Omics data typically include a vast number of measured variables (in the scale of thousands) that act as inputs for ML models, e.g. genes in RNA-seq or microarrays. Herein, the terms features and labels will be used to describe the inputs and outputs of ML models, respectively, following the convention used by the ML community. Feature selection is defined by the reduction of dataset dimensionality, meaning the selection of only a subset of features that maintain or even improve the accuracy of the model, as the importance of each feature towards labels prediction varies significantly [73]. Feature selection is a pre-processing step only performed on the training set to avoid information leaking from the test or validation sets to model training. Implementation of feature selection is necessary to avoid model overfitting and to ensure that the algorithm can correctly identify the dependencies and correlations between features and labels.
Feature selection methods pursue three possible objectives: (1) the identification and retention of the features that are (not necessarily linearly) strongly correlated with the labels and (2) the identification and exclusion of the features that are strongly correlated with each other and (3) the identification of feature combinations that improve model predictive capabilities. Feature selection, results in the designation of a subset of features that account for the maximum variance of the data or in a ranking list that reflects the importance of each feature toward labels variance [74]. Consequently, feature selection results in improved and faster model performance and deeper understanding of dependencies within the features and between the features and labels. Techniques for feature selection usually include unsupervised algorithms where the output is not known a priori. There are three popular classes of feature selection techniques: filter, wrapper and embedded methods [75], [76]. In filter methods, different statistical and ranking algorithms are applied to the available dataset to determine the highest scoring features [77], [78]. Wrapper methods include the development and training of several model configurations using different feature subsets [79]. Should the exclusion of the examined features not substantially decrease model performance, the features can be excluded from model construction [80], [81], [82], [83]. Finally, the ML models that include a dimensionality-reduction step and can be used simultaneously for feature selection and label prediction belong to the class of embedded algorithms. Ensembles that combine different modelling algorithms have also been proposed for feature selection [74]. More characteristics about these methods can be found in Table 2.
Although not typically included in any of the aforementioned feature selection classes, algorithms such as principal component analysis (PCA), hierarchical clustering, k-means clustering, k-nearest neighbours (k-NN) classification and autoencoders (unsupervised or self-supervised artificial neural networks) can contribute to dimensionality reduction. However, the dimensionality reduction techniques search for latent variables to explain the noise in data, whilst feature selection methods search within the available features of the input set. A further classification of dimensionality reduction to linear/non-linear methods and univariate/multivariate methods is also common. Unlike linear methods (i.e. least absolute shrinkage and selection operator – LASSO), algorithms that account for non-linear dependencies between the features and labels are more suitable when a non-linear predictor model is used [84]. Multivariate analysis involves the simultaneous evaluation of multiple variables for the better exploration of interdependencies between the examined variables.
3.2. Unsupervised machine learning applications
Omics data typically include thousands of measured variables. Unsupervised ML techniques have been widely used to process multidimensional datasets and expedite data analysis in biological and biomedical systems as summarised in Fig. 5. For example, autoencoders have been used as an initialization step in ML frameworks that classify breast cancer metabolomic data [13], while PCA of proteomic data has been utilized for the identification of metabolic engineering strategies for intensification of production in bacterial cultures [14]. Undoubtedly, PCA is the most widely used unsupervised tool for the interpretation of both experimental data and computational results. An overview of available methods and their application to cell systems is presented in Table 3.
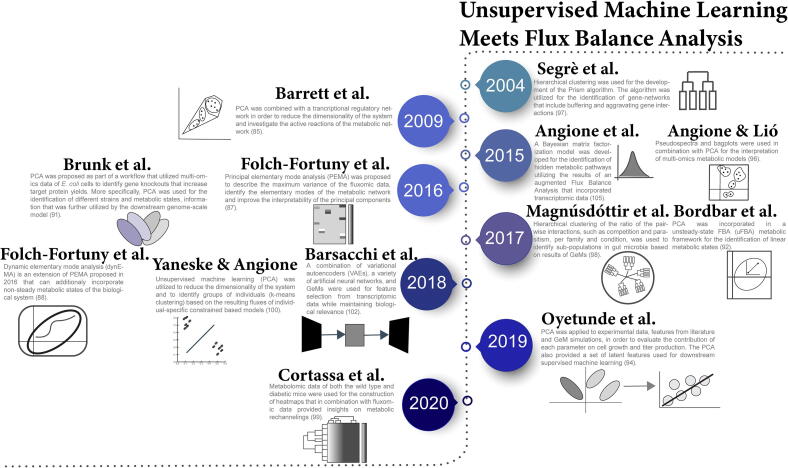
Fig. 5.
Timeline of unsupervised machine earning techniques and their application to constraint-based models.
Table 3.
Unsupervised machine learning methods used in combination with constraint-based models.
| Unsupervised ML method | Description | Applications |
|---|---|---|
| PCA | Dimensionality reduction Data interpretability Data simplification Identification of variation sources |
Dimensionality reduction of fluxomic data [85], [86] Can be applied to CBM results to identify central fluxes and pathways [85,86,95,96) Identification of metabolism active EMs [87], [88], [89], [90] Can be applied to experimental or simulation data to further inform downstream CBMs or ML algorithms [91], [92], [93], [94] |
| Clustering | Identification of sub-populations Clustering criteria, such as centroid- or distribution-based, vary depending on the chosen algorithm |
Hierarchical clustering for the identification of populations and the development of population specific CBMs [98], [100] Gene network reconstruction [97] Heatmaps of metabolomics data for studying metabolic alterations [99] k-means clustering to group CBM results [100] |
| Autoencoders | Unsupervised artificial neural networks Dimensionality reduction |
Variational autoencoders (VAEs) in combination with CBMs to identify biologically relevant features from microarray data [102] Autoencoders customized based on GeM models [104] |
| Bayesian factor model | Dimensionality reduction Identification of certain latent variables that account for data variation |
Metabolic pathways analysis from gene expression data [105] |
| MCR-ALS | Dimensionality reduction Application of custom constraints |
Pathways identification [106] |
3.2.1. Applications of PCA
PCA is a dimensionality reduction method that searches for artificial latent components (principal components) that maximize data variance and are linear functions of the original variables. PCA has been applied to statistically explain the variability of metabolic fluxes and identify important metabolic pathways that are related to cellular behaviour. Barrett et al. [85] combined a transcriptional regulatory network with PCA to first identify the active reactions of a metabolic network in a defined medium and further reduce system dimensionality. A similar framework that applies PCA to fluxomic data for analysing cellular behaviour has been also presented by Gonzalez-Martinez et al. [86].
Whilst PCA is a powerful tool for dimensionality reduction and simplification of data visualization, the resulting principal components lack biologically relevant meaning. In order to overcome this limitation, principal elementary mode analysis (PEMA) has been proposed for the identification of important fluxes and metabolic pathways in E. coli cells [87]. Part of PEMA is the implementation of PCA on the elementary modes (EMs) of the metabolic model, leading to the formation of principal components that represent groups of EMs on the same metabolic pathway. Dynamic elementary mode analysis (dynEMA) has been proposed as an extension of PEMA for non-steady state fluxes [88]. In a similar manner, von Stosch et al. [89] proposed the principal EM (PEM) analysis as a computationally efficient framework for the evaluation of the EMs using a branch and bound technique that enables the reduction of EM combinations under examination. Bhadra et al. [90] proposed a hybrid of PCA and stoichiometric flux analysis, termed principal metabolic flux analysis (PMFA), that accounts for non-steady state scenarios and can be expanded to the Sparse-PMFA version of the method (components can have sparse loadings) in order to capture the variance of genome-scale gene data. PMFA successfully identified the active EMs of a P. pastoris metabolic network and was found to outperform both PEMA and PCA in predicting data variance.
Brunk et al. [91] applied PCA to time-course metabolomic data to uncover metabolic fingerprints of strains and distinct metabolic states, which the authors later utilized to further investigate strain variation using GeM. Similarly, Bordbar et al. [92] proposed a framework of unsteady-state FBA (uFBA) that incorporated PCA of time-course metabolomic data for the identification of linearized metabolic states that could be included in downstream metabolic model construction. PCA has also been used as a pre-processing dimensionality reduction step in genome-scale ML for the prediction of metabolite concentrations based on enzymatic expression levels [93]. In a study that combined bioprocess and simulated data from GeM, PCA was applied to evaluate the effect of various bioprocess parameters on cell growth and productivity [94]. The authors also extracted the 40 first principal components and used them as features for the downstream ML algorithm. Occhipinti et al. [95] applied PCA to FBA results to identify pathways that when appropriately manipulated lead to rhamnolipids (biosurfactant) overproduction in Pseudomonas aeruginosa. PCA, in combination with pseudospectra and bagplots, has also been applied for the interpretation of multi-omics metabolic models that incorporate gene expression, metabolism and codon usage [96].
3.2.2. Clustering
Clustering algorithms can be used for the identification of sub-populations and groups of either cells, genes or metabolic pathways. Segrè et al. [97] developed the Prism algorithm, an unsupervised hierarchical clustering method for the formation of gene networks where the resulting clusters interact with each other in a single way, trying to mimic the monochromatic buffering and aggravating interactions of genes. Hierarchical clustering has also been applied to identify sub-populations in gut microbia based on results from constraint-based GeMs [98]. Heatmaps of metabolomic data in combination with fluxomic data have been utilized for studying the rechannelling of metabolic fluxes under the supplementation of palmitate in hearts of diabetic mice [99]. Yaneske & Angione [100] developed personalised constraint-based metabolic models of breast cancer patients by modifying the acceptable boundaries of fluxes according to the relevant gene expression levels from transcriptomic data [101] in an effort to identify correlations between the fluxomic data and patient age. As a second step, the authors used unsupervised learning to identify the fluxes that explain most of the data variation, reduce dimensionality (PCA) and create patient groupings (k-means clustering).
3.2.3. Other unsupervised techniques
Barsacchi et al. [102] developed a deep learning framework that utilized variational autoencoders (VAEs) and GeMs in order to extract biologically relevant features from transcriptomic data, extending previous work on forming meaningful latent space from gene expression levels [103]. Autoencoders have also been used as part of DeepMetabolism, a deep learning framework that utilizes GeMs to customize the connections between the layers of the model [104], where genes are included in the input layer, proteins in the first hidden layer and the phenotype in the second hidden layer. Bayesian matrix factorization assuming Gaussian-Markov random field properties has been applied for the identification of latent pathways in fluxomic data harvested from augmented FBA modelling of transcriptomic data [105]. The Multivariate Curve Resolution – Alternating Least Squares (MCR-ALS) technique [106] has also been proposed for the decomposition of fluxomic data of a constraint-based model of P. pastoris [107].
3.3. Supervised ML applications
Supervised ML algorithms (summarised in Table 4) have largely focused on inferring the relationship between multi-omics layers such as the identification of essential genes from an array of features extracted from complex biological data [108]. Yet it has become clear that ML could also benefit from exploring different omics layers of information generated from white-box mechanistic models [109]. These include GeM and flux analyses, which represent biomolecular interactions involved in networks of biochemical reactions [17], thus contributing, in principle, albeit with added complexity, to a fuller description of topological features arising from gene expression [110]. An overview of key studies is presented in Fig. 6.
Table 4.
Supervised machine learning methods and examples of their application to bioinformatics.
| Supervised ML method | Description | Strength | Weaknesses | Application examples |
|---|---|---|---|---|
| Linear Regression | Data Regression to its mean value Best Fit Line (Mean Pattern of the dataset) Gradient Descent Least Square Function Continuous Output Normality Assumption Linearity Assumption |
Computationally inexpensive Weighed Sum Prediction Reduces complex dataset to a singular function Less prone to overfitting |
Reduces larger complex dataset to a singular function Assumption of Linearity Relationship is seldom applicable Does not distinguish Outliers which might bias regression |
Prediction and Evolutionary info analysis of protein structure [137] Prediction and evolutionary information analysis of protein solvent accessibility [138] Genetic Expression inference [139] Genotype Prediction based on Single Nucleotide Polymorphism [140] Prediction of protein secondary structure [141] |
| Logistic Regression | Extension of Linear Regression Logistic line fitting Probability modelling Non-linearity acceptance |
Probability-based classification (rather than final classifications) Fast Training Extension to Multiclass Classifications Less prone to overfitting |
Complex Multiplicative weighted function Complete Separation of classes Unrepresentative for classes that highly overlap |
Cellular Phenotype classification based on gene expression profile [142] Gene Selection [143] Disease Classification from microarray data [144] Molecular Classification of Cancer [145] |
| Support Vector Machine | Hyperplane Data classification Classes separation on higher dimensionality Kernel Transformation |
Easy implementation to well defined classified categories Effective in high dimensional spaces Non-linear input acceptance |
Not suitable for overlapping classes Can be prone to overfitting when number of features exceeds the number of samples No probabilistic explanation for classification |
Classification on Gene functional annotations from a combination of protein sequence and structure data [146] Cancer Classification from genetic expression [147] Protein subcellular classification prediction [148] Structural Classification of proteins [149] |
| Naïve- Bayers | Probabilistic Classification Probabilistic Bayer’s theorem Conditional Independence between variables Most used for classification |
Reduced risk of overfitting on small datasets Probabilistic classification Fast training Computationally inexpensive Scales linearly |
Does not incorporate feature interactions Performance sensitive to skewed data Requires assumption that variables are conditionally independent |
MicroRNA target prediction [150] Prediction of Protein Interaction Sites [151] Prediction of Protein coupling specificity [152] |
| Decision Tree Classifier & Forest Tree |
Classification or Regression modelling Parameter based Data splitting of variable with highest information gain Data entropy Information Gain Theory Gini Coefficient |
Easy interpretation and analysis Valued on smaller datasets Multiclassification applicability White box model highlighting classification pattern Easily assembled |
Tendency to overfit Lack of linear smoothness |
Prediction of microorganism growth temperatures and enzyme catalytic optima [153] Protein Structure Prediction from enzymatic turnovers [129] Microbial Genome prediction [154] MS cancer data classification [155] Gene Selection for Cancer identification [156] Human protein function prediction [157] Prediction of Protein interaction [158] |
| k-NN | Classification and Regression modelling Clustering classification Instance-based learning, i.e. lazy learning Parameter selection on Kernel basis Higher dimensions for clustering |
Simple to implement Learn non-linear boundary Robust to noise in input data |
Inefficiency on training larger datasets Expensive computational cost K value evaluation based on mixed heuristics Unclear |
Gene selection for sample classification based on gene expression data [159] Classification for Cancer diagnosis [160] Prediction of Metabolic pathways dynamics [161] |
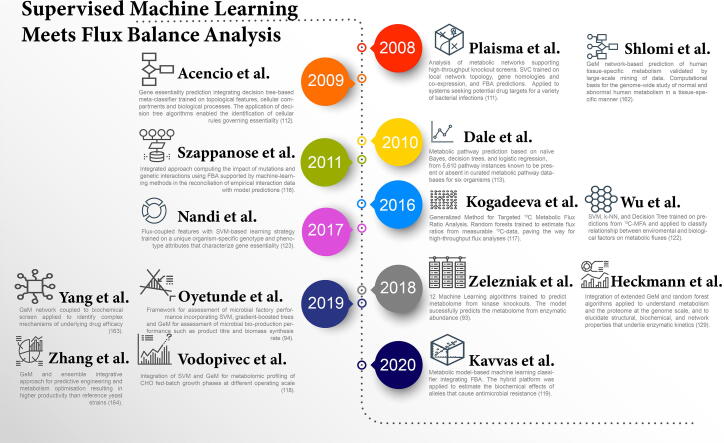
Fig. 6.
Timeline of efforts to integrate supervised machine learning with flux balance analysis. ([113], [117], [118], [119], [162], [163], [164].)
3.3.1. Gene essentiality analysis
Plaimas et al. [111] applied support vector machines (SVM) to an E. coli FBA network that incorporated both genomic and transcriptomic data to identify essential metabolic reactions and therefore essential enzymes. Acencio and Lemke [112] pushed the envelope further by training decision tree algorithms on several features including network topology, cellular compartments, and biological process information, such as cell cycle, metabolism, signal transduction, transcription, transport etc. Such a multi-faceted description not only aided the prediction of gene essentiality but also the identification of biological determinants of phenotypes. In addition, through the deconstruction of complex information by the application of decision tree classifiers, the study was able to identify cellular rules governing gene essentiality. In agreement with other studies [114], [115], the number of protein physical interactions, as the principal tree root node, was deemed the most important feature and therefore essential to algorithm performance.
Szappanos et al. [116] applied FBA to characterize gene interaction networks. In this case, a genetic algorithm was used to generate hypotheses that improved the prediction of gene interaction by reconciliating empirical interaction data with model predictions. Such a method is appealing, as it closes the gap between in silico and in vitro work. Nevertheless, and similar to previous studies [111], some of the predictions were compromised due to the inability of the FBA to capture the majority of experimentally determined genetic interactions, and the lack of regulation description at the gene expression and enzymatic reaction levels.
3.3.2. Integration with extracellular conditions
The integration of flux analysis and ML has also shown promising results in the context of analysing environmental impact on cell phenotype. Simple linear regression has been used to correctly predict E. coli growth from FBA data [120]. Zampieri et al. [121] applied supervised linear regression on gene expression profiling to estimate lactate production in CHO cells. The study successfully validated the hybrid modelling approach by comparing predicted lactate production with experimentally measured yields in a cross-validation setting. Other studies have investigated the extension of the hybrid approach to incorporate a wide description of factors ranging from the genome to the extracellular environment. For instance, Wu et al. [122] presented a web-based platform that applied an ensemble of ML techniques including SVM, k-nearest neighbours (k-NN), and decision tree, to literature data from nearly 100 13C-FMA studies on heterotrophic bacteria. The approach enabled the prediction of fluxomes as a function of bacterial species, substrate type, growth rate, oxygen conditions, and cultivation methods.
In 2017, Nandi et al. [123] extended the hybrid methodology utilising SVM-based implementation for binary classification of E. coli genes based on gene sequencing and expression, network topology and flux-based features. By also accounting for environmental factors, the model was able to capture the minimal set of genes that are essential in any given environment. The model was trained on 4094 metabolic reaction–gene pairs, out of which 384 were essential, 3120 were non-essential, and for around 590 reaction–gene pairs there was no phenotype information available. In addition, the model predicted the essentiality of 317 genes previously unidentified by exhaustive genome-scale knockout experiments. Such work further highlights that the appropriate choice of features arising from the genome-phenotype configuration and their correct description improves the performance of supervised algorithms.
3.3.3. Incorporation of regulatory system features
The complexity of cellular phenotypes stems from global transcription events and their perturbation due to changes in the extracellular environment [124], [125], resulting changes in enzyme capacities as well as changes of enzyme activities through metabolite-enzyme interactions [126], but also metabolic enzyme regulation due to post-translational events such as enzyme phosphorylation [127]. Last but not least, a small subset of metabolic reactions can occur spontaneously or are mediated by small molecules [128]. This knowledge underlines how fundamentally intertwined the relationships between the different levels of omics information and data are. Recently, Zelezniak et al. [93] mapped these regulatory patterns through the combination of ML with metabolic control analysis enabling quantitative prediction of entire cell metabolomes. First, flux analysis highlighted the importance of largely overlooked mechanisms in metabolic regulation. Second, ML captured the inherently complex multifactorial relationships at protein level. As a result, the study successfully quantified the role of enzyme abundance changes in metabolic regulation. Interestingly, all kinase deletions triggered global enzymatic expression changes. In fact, the detected variation at proteomic level led to wide metabolic control shifts between different sets of enzymes. While in earlier studies supervised algorithms were trained on a limited scope of sometimes overlooked omics features, Zelezniak et al. demonstrated that the incorporation of additional omics information yields robust classifications.
Since the incorporation of proteomics has been proved essential, Heckmann et al. [129] applied such a hybrid approach to elucidate structural, biochemical and network properties that underline enzymatic kinetics. The study integrated an ensemble methodology, including random forest and neural network algorithms, to predict catalytic turnover in E. coli from enzymatic biochemistry, protein structure and FBA computations. In a similar fashion, Amin et al. [130] applied a decision tree algorithm on enzymatic promiscuity data to predict hundreds of reactions and metabolites that may exist in E. coli but may not have been accounted for in databases.
4. Summary and outlook
GeMs are indispensable for organising and analysing omics datasets for a variety of cell systems. The development of GeMs for industrially-relevant organisms routinely used for the production of high-value chemicals or recombinant protein therapeutics together with the advancement of optimisation algorithms is enabling the design of improved cell factories and production processes. This is achieved by detecting cellular and process bottlenecks and genetic engineering strategies for overcoming them, better understanding cellular physiology and identifying alternative pathways to desired phenotypes. Although generic models of model organisms are a more accurate representation of cellular mechanisms, there is also tremendous value in methodologies that support tailoring GeMs to specific cell lines as well as reducing their size to serve a specific application. This is evident by the fact that the scale and complexity of, for example, CHO cell GeMs has so far limited the application of optimisation algorithms commonly used for strain design in microbial systems like E. coli or P. pastoris. Apart from the associated computational challenges, the genetic engineering strategies returned by optimisation algorithms are also often too complex to implement experimentally. In this regard, the introduction of thermodynamic constraints can decrease the solution space. However, the lack of Gibbs free energy data for a large number of metabolic reactions has so far hampered the applicability of such model reduction algorithms to mammalian cell systems. An additional drawback is the fact that most algorithms are used to solve a steady state model whereas the actual process is inherently dynamic.
Biological knowledge does not, however, advance as quickly as the current data generation rate. Although mechanistic approaches such as stoichiometric models have long been used for understanding cell phenotype and data integration, they cannot yet capture the full information content of available omics datasets due to lack of underpinning understanding necessary for model formulation. This has led to the emergence of ML methodologies, which have found application in identifying features of gene essentiality for guiding cell engineering. Another promising development is the creation of hybrid ML/FBA formulations for dynamicising GeMs, while maintaining a lower parameter estimation burden compared to hybrid kinetic/FBA approaches. However, although studies have successfully investigated the incorporation of multiple features across omics layers, they seem to heavily rely on public domain information for data curation, which may involve error and bias. Another caveat is the danger of overfitting ML models, which can significantly limit applicability and robustness.
Without a doubt, the ever-increasing capacity for high-throughput experimentation and sample analysis renders hybrid ML/FBA modelling a promising tool for harnessing the information content of multi-omics datasets and GeMs. Within the bioprocessing industry, this could support cell line classification, and process screening, leading to accelerated host selection and process development, respectively. Despite the aforementioned limitations, the studies reviewed herein showcase the advantages of hybrid methodologies in terms of the systematic representation of feature-vector relationship and decision boundaries, when compared to individual modelling approaches, be they stochastic, deterministic or data-driven.
CRediT authorship contribution statement
Athanasios Antonakoudis: Methodology, Visualization, Writing - original draft. Rodrigo Barbosa: Methodology, Visualization, Writing - original draft. Pavlos Kotidis: Methodology, Visualization, Writing - original draft. Cleo Kontoravdi: Supervision, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
AA thanks the UK Engineering and Physical Sciences Research Council for his studentship. RB thanks the UK Biotechnology and Biological Sciences Research Council and GlaxoSmithKline for his studentship. PK thanks the Department of Chemical Engineering, Imperial College London, for his scholarship.
References
- 1.Sigurdsson M.I., Jamshidi N., Steingrimsson E., Thiele I., Palsson B.Ø. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst Biol. 2010;4 doi: 10.1186/1752-0509-4-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swainston N. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunk E. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu J.Y., Kim H.U., Lee S.Y. Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism. Proc Natl Acad Sci. 2017;114:E9740. doi: 10.1073/pnas.1713050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theron C.W., Berrios J., Delvigne F., Fickers P. Integrating metabolic modeling and population heterogeneity analysis into optimizing recombinant protein production by Komagataella (Pichia) pastoris. Appl Microbiol Biotechnol. 2018;102:63–80. doi: 10.1007/s00253-017-8612-y. [DOI] [PubMed] [Google Scholar]
- 6.Hefzi H. A consensus genome-scale reconstruction of Chinese hamster ovary cell metabolism. Cell Systems. 2016;3:434–443.e438. doi: 10.1016/j.cels.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calmels C., McCann A., Malphettes L., Andersen M.R. Application of a curated genome-scale metabolic model of CHO DG44 to an industrial fed-batch process. Metab Eng. 2019;51:9–19. doi: 10.1016/j.ymben.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Fouladiha H, Marashi SA, Li S, Vaziri B, Lewis NE, Systematically gap-filling the genome-scale model of CHO cells. bioRxiv; 2020, 2020.2001.2027.921296. [DOI] [PubMed]
- 9.Yeo H.C., Hong J., Lakshmanan M., Lee D.-Y. Enzyme capacity-based genome scale modelling of CHO cells. Metab Eng. 2020;60:138–147. doi: 10.1016/j.ymben.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Sheikh K., Förster J., Nielsen L.K. Modeling Hybridoma Cell Metabolism Using a GenericGenome-Scale Metabolic Model ofMus musculus. Biotechnol Prog. 2008;21:112–121. doi: 10.1021/bp0498138. [DOI] [PubMed] [Google Scholar]
- 11.Khodaee S., Asgari Y., Totonchi M., Karimi-Jafari M.H. iMM1865: A New Reconstruction of Mouse Genome-Scale Metabolic Model. Sci Rep. 2020;10:6177. doi: 10.1038/s41598-020-63235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monk J.M. Genome-scale metabolic reconstructions of multiple <em>Escherichia coli</em> strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci. 2013;110:20338. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolan R.P., Lee K. Dynamic model for CHO cell engineering. J Biotechnol. 2012;158:24–33. doi: 10.1016/j.jbiotec.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Ahn W.S., Antoniewicz M.R. Towards dynamic metabolic flux analysis in CHO cell cultures. Biotechnol J. 2012;7:61–74. doi: 10.1002/biot.201100052. [DOI] [PubMed] [Google Scholar]
- 15.Robitaille J., Chen J., Jolicoeur M. A Single Dynamic Metabolic Model Can Describe mAb Producing CHO Cell Batch and Fed-Batch Cultures on Different Culture Media. PLoS ONE. 2015;10:e0136815. doi: 10.1371/journal.pone.0136815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez J.M. Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion. Nat Commun. 2020;11:68. doi: 10.1038/s41467-019-13867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orth J.D., Thiele I., Palsson B.O. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ataman M., Hatzimanikatis V. lumpGEM: Systematic generation of subnetworks and elementally balanced lumped reactions for the biosynthesis of target metabolites. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ataman M., Hernandez Gardiol D.F., Fengos G., V. Hatzimanikatis, redGEM: Systematic reduction and analysis of genome-scale metabolic reconstructions for development of consistent core metabolic models. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster S., Hilgetag C. On elementary flux modes in biochemical reaction systems at steady state. J Biol Syst. 1994;02:165–182. [Google Scholar]
- 21.Becker S.A., Palsson B.O. Context-Specific Metabolic Networks Are Consistent with Experiments. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satish Kumar V., Dasika M.S., Maranas C.D. Optimization based automated curation of metabolic reconstructions. BMC Bioinf. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiele I., Vlassis N., Fleming R.M.T. fastGapFill: efficient gap filling in metabolic networks. Bioinformatics (Oxford, England) 2014;30:2529–2531. doi: 10.1093/bioinformatics/btu321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caspi R. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lularevic M., Racher A.J., Jaques C., Kiparissides A. Improving the accuracy of flux balance analysis through the implementation of carbon availability constraints for intracellular reactions. Biotechnol Bioeng. 2019;116:2339–2352. doi: 10.1002/bit.27025. [DOI] [PubMed] [Google Scholar]
- 26.Pandey V., Hernandez Gardiol D., Chiappino Pepe A., Hatzimanikatis V., TEX-FBA, A constraint-based method for integrating gene expression, thermodynamics, and metabolomics data into genome-scale metabolic models. BioArchive. 2019 [Google Scholar]
- 27.Henry C.S., Broadbelt L.J., Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schellenberger J., Lewis N.E., Palsson B. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys J. 2011;100:544–553. doi: 10.1016/j.bpj.2010.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Guo W., Feng X. OM-FBA: Integrate Transcriptomics Data with Flux Balance Analysis to Decipher the Cell Metabolism. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0154188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y. An unconventional uptake rate objective function approach enhances applicability of genome-scale models for mammalian cells. NPJ Syst Biol Appl. 2019;5:25. doi: 10.1038/s41540-019-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 33.Pharkya P., Maranas C.D. An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng. 2006;8:1–13. doi: 10.1016/j.ymben.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Chowdhury A., Zomorrodi A.R., Maranas C.D. k-OptForce: Integrating Kinetics with Flux Balance Analysis for Strain Design. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lun D.S. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen F. OptRAM: In-silico strain design via integrative regulatory-metabolic network modeling. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alter T.B., Ebert B.E. Determination of growth-coupling strategies and their underlying principles. BMC Bioinf. 2019;20:447. doi: 10.1186/s12859-019-2946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Reed J.L. OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC Syst Biol. 2010;4 doi: 10.1186/1752-0509-4-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranganathan S., Suthers P.F., Maranas C.D. OptForce: An optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen K., Broeken V., Hansen A.S.L., Sonnenschein N., Herrgard M.J. OptCouple: Joint simulation of gene knockouts, insertions and medium modifications for prediction of growth-coupled strain designs. Metab Eng Commun. 2019;8 doi: 10.1016/j.mec.2019.e00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendry J.I., Bandyopadhyay A., Srinivasan S., Pakrasi H.B., Maranas C.D. Metabolic model guided strain design of cyanobacteria. Curr Opin Biotechnol. 2020;64:17–23. doi: 10.1016/j.copbio.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Suastegui M. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors. Metab Eng. 2017;42:134–144. doi: 10.1016/j.ymben.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Tan Z. Engineering of E. coli inherent fatty acid biosynthesis capacity to increase octanoic acid production. Biotechnol Biofuels. 2018;11:87. doi: 10.1186/s13068-018-1078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feist A.M. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wayman J.A., Glasscock C., Mansell T.J., DeLisa M.P., Varner J.D. Improving designer glycan production in Escherichia coli through model-guided metabolic engineering. Metab Eng Commun. 2019;9 doi: 10.1016/j.mec.2019.e00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitua F., Torres P., Pérez-Correa J.R., Agosin E. Dynamic genome-scale metabolic modeling of the yeast Pichia pastoris. BMC Syst Biol. 2017;11:27. doi: 10.1186/s12918-017-0408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kol S. Multiplex secretome engineering enhances recombinant protein production and purity. Nat Commun. 2020;11:1908. doi: 10.1038/s41467-020-15866-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z. CHO cell productivity improvement by genome-scale modeling and pathway analysis: Application to feed supplements. Biochem Eng J. 2020 [Google Scholar]
- 49.Xing Z. Optimizing amino acid composition of CHO cell culture media for a fusion protein production. Process Biochem. 2011;46:1423–1429. [Google Scholar]
- 50.Junghans L. From nutritional wealth to autophagy: In vivo metabolic dynamics in the cytosol, mitochondrion and shuttles of IgG producing CHO cells. Metab Eng. 2019;54:145–159. doi: 10.1016/j.ymben.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Templeton N. Application of13C flux analysis to identify high-productivity CHO metabolic phenotypes. Metab Eng. 2017;43:218–225. doi: 10.1016/j.ymben.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Martínez-Monge I. Concomitant consumption of glucose and lactate: A novel batch production process for CHO cells. Biochem Eng J. 2019;151 [Google Scholar]
- 53.Fouladiha H. A metabolic network-based approach for developing feeding strategies for CHO cells to increase monoclonal antibody production. Bioprocess Biosyst Eng. 2020;43:1381–1389. doi: 10.1007/s00449-020-02332-6. [DOI] [PubMed] [Google Scholar]
- 54.Zamorano F., Vande Wouwer A., Jungers R.M., Bastin G. Dynamic metabolic models of CHO cell cultures through minimal sets of elementary flux modes. J Biotechnol. 2013;164:409–422. doi: 10.1016/j.jbiotec.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Martínez V.S., Buchsteiner M., Gray P., Nielsen L.K., Quek L.E. Dynamic metabolic flux analysis using B-splines to study the effects of temperature shift on CHO cell metabolism. Metab Eng Commun. 2015;2:46–57. doi: 10.1016/j.meteno.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuriya Y., Araki M. Dynamic Flux Balance Analysis to Evaluate the Strain Production Performance on Shikimic Acid Production in Escherichia coli. Metabolites. 2020;10 doi: 10.3390/metabo10050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plaza J., Bogaerts P. Dynamic flux balance analysis for predicting biomass growth and ethanol production in yeast fed-batch cultures. IFAC-PapersOnLine. 2018;51:631–636. [Google Scholar]
- 58.Valverde J.R., Gullón S., García-Herrero C.A., Campoy I., Mellado R.P. Dynamic metabolic modelling of overproduced protein secretion in Streptomyces lividans using adaptive DFBA. BMC Microbiol. 2019;19:233. doi: 10.1186/s12866-019-1591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres P., Saa P.A., Albiol J., Ferrer P., Agosin E. Contextualized genome-scale model unveils high-order metabolic effects of the specific growth rate and oxygenation level in recombinant Pichia pastoris. Metab Eng Commun. 2019;9 doi: 10.1016/j.mec.2019.e00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomàs-Gamisans M., Ferrer P., Albiol J. Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb Biotechnol. 2018;11:224–237. doi: 10.1111/1751-7915.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varoquaux G. Cross-validation failure: Small sample sizes lead to large error bars. NeuroImage. 2018;180:68–77. doi: 10.1016/j.neuroimage.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 62.Vabalas A., Gowen E., Poliakoff E., Casson A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim J.-H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput Stat Data Anal. 2009;53:3735–3745. [Google Scholar]
- 64.Xu Y., Goodacre R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. J Analysis Testing. 2018;2:249–262. doi: 10.1007/s41664-018-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodacre R. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics. 2007;3:231–241. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Worley B., Powers R. Multivariate Analysis in Metabolomics. Curr Metabolomics. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Berg R.A., Hoefsloot H.C.J., Westerhuis J.A., Smilde A.K., van der Werf M.J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Dijk D. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 2018;174:716–729.e727. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arisdakessian C., Poirion O., Yunits B., Zhu X., Garmire L.X. DeepImpute: an accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biol. 2019;20:211. doi: 10.1186/s13059-019-1837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong W., Kwak I.-Y., Pota P., Koyano-Nakagawa N., Garry D.J. DrImpute: imputing dropout events in single cell RNA sequencing data. BMC Bioinf. 2018;19:220. doi: 10.1186/s12859-018-2226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W.V., Li J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun. 2018;9:997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang M. SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods. 2018;15:539–542. doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolón-Canedo V., Sánchez-Maroño N., Alonso-Betanzos A. Feature selection for high-dimensional data. Progress Artificial Intelligence. 2016;5:65–75. [Google Scholar]
- 74.Bolón-Canedo V., Alonso-Betanzos A. Ensembles for feature selection: A review and future trends. Information Fusion. 2019;52:1–12. [Google Scholar]
- 75.Chandrashekar G., Sahin F. A survey on feature selection methods. Comput Electr Eng. 2014;40:16–28. [Google Scholar]
- 76.Guyon I., Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 77.Sánchez-Maroño N, Alonso-Betanzos A, Tombilla-Sanromán M, in Intelligent Data Engineering and Automated Learning - IDEAL 2007, H. Yin, P. Tino, E. Corchado, W. Byrne, X. Yao, Eds., Springer Berlin Heidelberg, Berlin, Heidelberg; 2007, p. 178–87.
- 78.Bommert A., Sun X., Bischl B., Rahnenführer J., Lang M. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput Stat Data Anal. 2020;143 [Google Scholar]
- 79.Kohavi R., John G.H. Wrappers for feature subset selection. Artif Intell. 1997;97:273–324. [Google Scholar]
- 80.Tibshirani R. Regression Shrinkage and Selection Via the Lasso. J Roy Stat Soc: Ser B (Methodol) 1996;58:267–288. [Google Scholar]
- 81.Clemmensen L., Hastie T., Witten D., Ersbøll B. Sparse Discriminant Analysis. Technometrics. 2011;53:406–413. [Google Scholar]
- 82.Guyon I., Weston J., Barnhill S., Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- 83.Lu M. Embedded feature selection accounting for unknown data heterogeneity. Expert Syst Appl. 2019;119:350–361. [Google Scholar]
- 84.Yamada M., Jitkrittum W., Sigal L., Xing E.P., Sugiyama M. High-Dimensional Feature Selection by Feature-Wise Kernelized Lasso. Neural Comput. 2013;26:185–207. doi: 10.1162/NECO_a_00537. [DOI] [PubMed] [Google Scholar]
- 85.Barrett C.L., Herrgard M.J., Palsson B. Decomposing complex reaction networks using random sampling, principal component analysis and basis rotation. BMC Syst Biol. 2009;3:30. doi: 10.1186/1752-0509-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.González-Martínez J.M. Metabolic flux understanding of Pichia pastoris grown on heterogenous culture media. Chemometrics Intelligent Lab Syst. 2014;134:89–99. [Google Scholar]
- 87.Folch-Fortuny A., Marques R., Isidro I.A., Oliveira R., Ferrer A. Principal elementary mode analysis (PEMA) Mol Biosyst. 2016;12:737–746. doi: 10.1039/c5mb00828j. [DOI] [PubMed] [Google Scholar]
- 88.Folch-Fortuny A., Teusink B., Hoefsloot H.C.J., Smilde A.K., Ferrer A. Dynamic elementary mode modelling of non-steady state flux data. BMC Syst Biol. 2018;12:71. doi: 10.1186/s12918-018-0589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.von Stosch M., Rodrigues de Azevedo C., Luis M., Feyo de Azevedo S., Oliveira R. A principal components method constrained by elementary flux modes: analysis of flux data sets. BMC Bioinf. 2016;17:200. doi: 10.1186/s12859-016-1063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhadra S., Blomberg P., Castillo S., Rousu J. Principal metabolic flux mode analysis. Bioinformatics. 2018;34:2409–2417. doi: 10.1093/bioinformatics/bty049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brunk E. Characterizing Strain Variation in Engineered E. coli Using a Multi-Omics-Based Workflow. Cell Syst. 2016;2:335–346. doi: 10.1016/j.cels.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bordbar A. Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics. Sci Rep. 2017;7:46249. doi: 10.1038/srep46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zelezniak A. Machine Learning Predicts the Yeast Metabolome from the Quantitative Proteome of Kinase Knockouts. Cell Syst. 2018;7:269–283.e266. doi: 10.1016/j.cels.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oyetunde T., Liu D., Martin H.G., Tang Y.J. Machine learning framework for assessment of microbial factory performance. PLoS ONE. 2019;14:e0210558. doi: 10.1371/journal.pone.0210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Occhipinti S. Lung Cancer Stigma across the Social Network: Patient and Caregiver Perspectives. J Thoracic Oncol. 2018;13:1443–1453. doi: 10.1016/j.jtho.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 96.Angione C., Lió P. Predictive analytics of environmental adaptability in multi-omic network models. Sci Rep. 2015;5:15147. doi: 10.1038/srep15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Segrè D., DeLuna A., Church G.M., Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 98.Magnúsdóttir S. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 99.Cortassa S. Metabolic remodelling of glucose, fatty acid and redox pathways in the heart of type 2 diabetic mice. J Physiol. 2020;598:1393–1415. doi: 10.1113/JP276824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yaneske E., Angione C. The poly-omics of ageing through individual-based metabolic modelling. BMC Bioinf. 2018;19:415. doi: 10.1186/s12859-018-2383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Angione C. Integrating splice-isoform expression into genome-scale models characterizes breast cancer metabolism. Bioinformatics. 2017;34:494–501. doi: 10.1093/bioinformatics/btx562. [DOI] [PubMed] [Google Scholar]
- 102.Barsacchi M, Terre HA, Lió P, GEESE: Metabolically driven latent space learning for gene expression data. bioRxiv, 365643 (2018).
- 103.Way G.P., Greene C.S. Extracting a biologically relevant latent space from cancer transcriptomes with variational autoencoders. Pac Symp Biocomput. 2018;23:80–91. [PMC free article] [PubMed] [Google Scholar]
- 104.Guo W, Xu Y, Feng X, DeepMetabolism: A Deep Learning System to Predict Phenotype from Genome Sequencing. arXiv e-prints, arXiv:1705.03094; 2017.
- 105.Angione C., Pratanwanich N., Lió P. A Hybrid of Metabolic Flux Analysis and Bayesian Factor Modeling for Multiomic Temporal Pathway Activation. ACS Synth Biol. 2015;4:880–889. doi: 10.1021/sb5003407. [DOI] [PubMed] [Google Scholar]
- 106.Jaumot J., Gargallo R., de Juan A., Tauler R. A graphical user-friendly interface for MCR-ALS: a new tool for multivariate curve resolution in MATLAB. Chemometrics Intelligent Lab Syst. 2005;76:101–110. [Google Scholar]
- 107.Folch-Fortuny A. MCR-ALS on metabolic networks: Obtaining more meaningful pathways. Chemometrics Intelligent Lab Syst. 2015;142:293–303. [Google Scholar]
- 108.Zhang X., Acencio M.L., Lemke N. Predicting Essential Genes and Proteins Based on Machine Learning and Network Topological Features: A Comprehensive Review. Front Physiol. 2016;7 doi: 10.3389/fphys.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang J.H. A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell. 2019;177:1649–1661.e1649. doi: 10.1016/j.cell.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zampieri G., Vijayakumar S., Yaneske E., Angione C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Plaimas K. Machine learning based analyses on metabolic networks supports high-throughput knockout screens. BMC Syst Biol. 2008;2:67. doi: 10.1186/1752-0509-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Acencio M.L., Lemke N. Towards the prediction of essential genes by integration of network topology, cellular localization and biological process information. BMC Bioinf. 2009;10:290. doi: 10.1186/1471-2105-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dale J.M., Popescu L., Karp P.D. Machine learning methods for metabolic pathway prediction. BMC Bioinf. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Res. 2004;14:1310–1314. doi: 10.1101/gr.2300204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li M., Zhang H., Wang J.-X., Pan Y. A new essential protein discovery method based on the integration of protein-protein interaction and gene expression data. BMC Syst Biol. 2012;6:15. doi: 10.1186/1752-0509-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Szappanos B. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43:656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kogadeeva M., Zamboni N. SUMOFLUX: A Generalized Method for Targeted 13C Metabolic Flux Ratio Analysis. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vodopivec M., Lah L., Narat M., Curk T. Metabolomic profiling of CHO fed-batch growth phases at 10, 100, and 1,000 L. Biotechnol Bioeng. 2019;116:2720–2729. doi: 10.1002/bit.27087. [DOI] [PubMed] [Google Scholar]
- 119.Kavvas E.S., Yang L., Monk J.M., Heckmann D., Palsson B.O. A biochemically-interpretable machine learning classifier for microbial GWAS. Nat Commun. 2020;11:2580. doi: 10.1038/s41467-020-16310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sridhara V. Predicting Growth Conditions from Internal Metabolic Fluxes in an In-Silico Model of E. coli. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0114608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zampieri G, Coggins M, Valle G, Angione C, A poly-omics machine-learning method to predict metabolite production in CHO cells; 2017.
- 122.Wu S.G. Rapid Prediction of Bacterial Heterotrophic Fluxomics Using Machine Learning and Constraint Programming. PLoS Comput Biol. 2016;12:e1004838. doi: 10.1371/journal.pcbi.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nandi S., Subramanian A., Sarkar R.R. An integrative machine learning strategy for improved prediction of essential genes in Escherichia coli metabolism using flux-coupled features. Mol BioSyst. 2017;13:1584–1596. doi: 10.1039/c7mb00234c. [DOI] [PubMed] [Google Scholar]
- 124.Alper H., Stephanopoulos G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 125.Alam M.T. The metabolic background is a global player in Saccharomyces gene expression epistasis. Nat Microbiol. 2016;1:15030. doi: 10.1038/nmicrobiol.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Millard P., Smallbone K., Mendes P. Metabolic regulation is sufficient for global and robust coordination of glucose uptake, catabolism, energy production and growth in Escherichia coli. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oliveira A.P. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol. 2012;8:623. doi: 10.1038/msb.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keller M.A., Piedrafita G., Ralser M. The widespread role of non-enzymatic reactions in cellular metabolism. Curr Opin Biotechnol. 2015;34:153–161. doi: 10.1016/j.copbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heckmann D. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models. Nat Commun. 2018;9:5252. doi: 10.1038/s41467-018-07652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Amin S.A., Chavez E., Porokhin V., Nair N.U., Hassoun S. Towards creating an extended metabolic model (EMM) for E. coli using enzyme promiscuity prediction and metabolomics data. Microb Cell Fact. 2019 doi: 10.1186/s12934-019-1156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Varma A., Palsson B. Stoichiometric Flux Balance Models Quantitatively Predict Growth and Metabolic By-Product Secretion in Wild-Type Escherichia coli W3110. Am Soc Microbiol. 1994;60 doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mahadevan R., Edwards J.S., Doyle F.J. Dynamic Flux Balance Analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Provost A., Bastin G. Dynamic metabolic modelling under the balanced growth condition. J Process Control. 2004;14:717–728. [Google Scholar]
- 134.Leighty R.W., Antoniewicz M.R. Dynamic metabolic flux analysis (DMFA): a framework for determining fluxes at metabolic non-steady state. Metab Eng. 2011;13:745–755. doi: 10.1016/j.ymben.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 135.Gao Y., Zhao Z., Liu F. DMFA-based operation model for fermentation processes. Comput Chem Eng. 2018;109:138–150. [Google Scholar]
- 136.Gomez J.A., Hoffner K., Barton P.I. DFBAlab: a fast and reliable MATLAB code for dynamic flux balance analysis. BMC Bioinf. 2014;15:409. doi: 10.1186/s12859-014-0409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carbonell P. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals. Commun Biol. 2018;1:66. doi: 10.1038/s42003-018-0076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang J.-Y., Lee H.-M., Ahmad S. Prediction and evolutionary information analysis of protein solvent accessibility using multiple linear regression. Proteins Struct Funct Bioinf. 2005;61:481–491. doi: 10.1002/prot.20620. [DOI] [PubMed] [Google Scholar]
- 139.Chen Y., Li Y., Narayan R., Subramanian A., Xie X. Gene expression inference with deep learning. Bioinformatics. 2016;32:1832–1839. doi: 10.1093/bioinformatics/btw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He J., Zelikovsky A. MLR-tagging: informative SNP selection for unphased genotypes based on multiple linear regression. Bioinformatics. 2006;22:2558–2561. doi: 10.1093/bioinformatics/btl420. [DOI] [PubMed] [Google Scholar]
- 141.Pan X.-M. Multiple linear regression for protein secondary structure prediction. Proteins Struct Funct Bioinf. 2001;43:256–259. doi: 10.1002/prot.1036. [DOI] [PubMed] [Google Scholar]
- 142.Müller F.-J. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shevade S.K., Keerthi S.S. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics. 2003;19:2246–2253. doi: 10.1093/bioinformatics/btg308. [DOI] [PubMed] [Google Scholar]
- 144.Liao J.G., Chin K.-V. Logistic regression for disease classification using microarray data: model selection in a large p and small n case. Bioinformatics. 2007;23:1945–1951. doi: 10.1093/bioinformatics/btm287. [DOI] [PubMed] [Google Scholar]
- 145.Golub T.R. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 146.Lewis D.P., Jebara T., Noble W.S. Support vector machine learning from heterogeneous data: an empirical analysis using protein sequence and structure. Bioinformatics. 2006;22:2753–2760. doi: 10.1093/bioinformatics/btl475. [DOI] [PubMed] [Google Scholar]
- 147.Liu Y. Active Learning with Support Vector Machine Applied to Gene Expression Data for Cancer Classification. J Chem Inf Comput Sci. 2004;44:1936–1941. doi: 10.1021/ci049810a. [DOI] [PubMed] [Google Scholar]
- 148.Hua S., Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001;17:721–728. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- 149.Cai Y.-D., Liu X.-J., Xu X.-B., Zhou G.-P. Support Vector Machines for predicting protein structural class. BMC Bioinf. 2001;2:3. doi: 10.1186/1471-2105-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yousef M., Jung S., Kossenkov A.V., Showe L.C., Showe M.K. Naïve Bayes for microRNA target predictions—machine learning for microRNA targets. Bioinformatics. 2007;23:2987–2992. doi: 10.1093/bioinformatics/btm484. [DOI] [PubMed] [Google Scholar]
- 151.Murakami Y., Mizuguchi K. Applying the Naïve Bayes classifier with kernel density estimation to the prediction of protein–protein interaction sites. Bioinformatics. 2010;26:1841–1848. doi: 10.1093/bioinformatics/btq302. [DOI] [PubMed] [Google Scholar]
- 152.Sgourakis N.G., Bagos P.G., Hamodrakas S.J. Prediction of the coupling specificity of GPCRs to four families of G-proteins using hidden Markov models and artificial neural networks. Bioinformatics. 2005;21:4101–4106. doi: 10.1093/bioinformatics/bti679. [DOI] [PubMed] [Google Scholar]
- 153.Li G, Rabe KS, Nielsen J, Engqvist MKM, Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. bioRxiv, 522342; 2019. [DOI] [PubMed]
- 154.Che D, Zhao J, Cai L, Xu Y, in 2007 IEEE Symposium on Computational Intelligence and Bioinformatics and Computational Biology, 2007, pp. 135–42.
- 155.Ge G., Wong G.W. Classification of premalignant pancreatic cancer mass-spectrometry data using decision tree ensembles. BMC Bioinf. 2008;9:275. doi: 10.1186/1471-2105-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen K.-H. Gene selection for cancer identification: a decision tree model empowered by particle swarm optimization algorithm. BMC Bioinf. 2014;15:49. doi: 10.1186/1471-2105-15-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh M, Singh P, Singh H, in 2006 International Conference on Advanced Computing and Communications. 2006, p. 564-8.
- 158.Lee M.S., Oh S. Alternating decision tree algorithm for assessing protein interaction reliability. Vietnam J Computer Sci. 2014;1:169–178. [Google Scholar]
- 159.Li L., Weinberg C.R., Darden T.A., Pedersen L.G. Gene selection for sample classification based on gene expression data: study of sensitivity to choice of parameters of the GA/KNN method. Bioinformatics. 2001;17:1131–1142. doi: 10.1093/bioinformatics/17.12.1131. [DOI] [PubMed] [Google Scholar]
- 160.Medjahed S.A., Saadi T., Benyettou A. Breast Cancer Diagnosis by using k-Nearest Neighbor with Different Distances and Classification Rules. Int J Computer Appl. 2013;62:1–5. [Google Scholar]
- 161.Costello Z., Martin H.G. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. npj Syst Biol Appl. 2018;4:19. doi: 10.1038/s41540-018-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shlomi T., Cabili M.N., Herrgård M.J., Palsson B., Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 163.Yang J.H., Wright S.N., Hamblin M., McCloskey D., Alcantar M.A., Schrubbers L. A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action. Cell. 2019;177:1649–1661.e1649. doi: 10.1016/j.cell.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang J, Petersen S, Radivojevic T, Ramirez A, Perez A, Abeliuk E, Sanchez BJ, Costello Z, Chen Y, Fero M, Martin HG, Nielsen J, Keasling JD, Jensen MK, Predictive engineering and optimization of tryptophan metabolism in yeast through a combination of mechanistic and machine learning models. bioRxiv, 858464; 2019.