Abstract
SARS CoV-2 is the causative agent of the pandemic disease COVID-19. There is an urgent need for effective drugs or vaccines which can effectively combat this outbreak. The main protease (Mpro), a key component for the SARS CoV-2 replication, is considered to be one of the important drug targets for developing anti-COVID-19 drugs. This SARS CoV-2 Mpro/cysteine protease has high sequence similarity with the same protease from SARS CoV-1. Previously, it has been shown experimentally that eight diterpenoids and four biflavonoids derived from the leaf of Torreya nucifera show inhibitory effect on the cleavage/catalytic activity of the SARS CoV-1 Mpro. But whether these phytochemicals exhibit any inhibitory effect on SARS CoV-2 Mpro is unclear. To understand this fact, here, we have adopted various in-silico approaches. Diterpenoids and biflavonoids those qualified pharmacological test (hinokiol, amentoflavone, bilobetin and ginkgetin) and two well-known Mpro inhibitors (N3 and lopinavir) were subjected for molecular docking studies. Only three biflavonoids (amentoflavone, bilobetin and ginkgetin) were selected by comparing their binding affinities with N3 and lopinavir. They interacted with two most important catalytic residues of Mpro (His41 and Cys145). Molecular dynamics studies further revealed that these three Mpro-biflavonoid complexes are highly stable and share a similar degree of compactness. Besides, these complexes experience less conformational fluctuations and more expansion than Mpro-N3 and/or Mpro-lopinavir complex. MM-GBSA and H-bond analysis further corroborated these findings. Altogether, our study suggested that these three biflavonoids could possibly inhibit the proteolytic/catalytic activity of SARS CoV-2 Mpro and might be useful for COVID-19 treatment.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS CoV-2 main protease, docking, molecular dynamics simulation, Torreya nucifera diterpenoids and biflavonoids
Graphical Abstract
1. Introduction
In December 2019, a novel disease namely COVID-19 has emerged from China. Since then, this disease is rapidly spread worldwide, infecting nearly 215 countries. As of September 5, 2020, the total number of COVID-19 infected cases has reached 26 million and over 875,000 people died worldwide due to this infectious disease (https://www.worldometers.info/coronavirus/). It has been declared as a global pandemic by the World Health Organization (WHO) on the 11th of March 2020 (Cucinotta & Vanelli, 2020). The United States remains the most affected country in terms of the total number of cases, followed by Brazil, India and Russia. Initial symptoms of COVID-19 infection include fever, myalgia, cough, and headache (Chen et al., 2020; Ren et al., 2020; Zhu et al., 2020). Infection usually resolves without active medical intervention, but for a subset of cases, the infection can progress to viral pneumonia and a variety of complications including acute lung damage leading to death (Zhao et al., 2020). However, the complications are atypical in most cases and the mortality rates increase dramatically with the age.
Severe acute respiratory syndrome coronavirus 2 (SARS CoV-2), a member of the Coronaviridae family and β-coronavirus genus, is the causative agent of the disease COVID-19 (Zhou et al., 2020). Since 2003, three coronaviruses have been associated with pneumonia; the first was severe acute respiratory syndrome coronavirus (SARS CoV-1) which affected 8,098 people causing 774 deaths between 2002 and 2003 (Drosten et al., 2003; Walls et al., 2020). The second was Middle-East respiratory syndrome coronavirus (MERS-CoV) which affected 27 countries and infected a total of 2,494 individuals. 858 people died due to MERS CoV (Zaki et al., 2012). SARS CoV-1 and SARS CoV-2 are genetically closely related to nearly 80% genome identity (Zhou et al., 2020). Additionally, both the coronaviruses are thought to have the same origin (bats) which most likely serve as a reservoir host for these two viruses (Zhou et al., 2020).
To date, much of our knowledge of COVID-19 virology has been inferred from the study of similar SARS CoV-1 and related coronaviruses including Middle East Respiratory Syndrome. Like other two coronaviruses, SARS CoV-2 is an enveloped virus and is made up of large single-stranded ∼30 kb long positive-sense RNA. The viral gene essential for the replication of the virus is expressed as a polyprotein that must be broken into functional subunits for replication and transcription activity (Zhang et al., 2020). The proteolytic cleavage of viral polyproteins is primarily accomplished by the main protease (Mpro), also known as 3CLpro (the 3C-like protease). Mpro cleaves the viral polyprotein at eleven sites. The main protease (Mpro or 3CL protease) is one of the best-characterized drug targets for coronaviruses (Anand et al., 2003; Hilgenfeld, 2014; Xue et al., 2008). The sequence of the main protease (Mpro) is highly conserved throughout coronavirus species (Mirza & Froeyen, 2020). The Mpro from SARS CoV-2 is reported to share more than 96% sequence similarity with the same protease from SARS CoV-1 and MERS which makes it an ideal target for broad-spectrum anti-CoV therapy (Ghosh et al., 2020b). Even more sequence similarity is observed between the main protease from SARS CoV-1 and SARS CoV-2 with a difference in only twelve amino acid residues (Supplemental Figure 1; Sequence homology performed using CLUSTALW (1.83) multiple sequence alignment program (Chakraborty et al., 2018; Nandi et al., 2016)) (Macchiagodena et al., 2020).
The SARS CoV-2 Mpro is a member of homologous cysteine proteases that are needed for viral replication (Jin, Du, et al., 2020; Zhang et al., 2020). This viral protease cleaves the initially translated viral polyprotein into its component proteins (Fan et al., 2004; Rota et al., 2003). Cleavage generally occurs immediately after a Gln residue, and the Gln residue is typically preceded by a hydrophobic residue, most often Leu. The residue that follows the Gln is often a small amino acid such as Ser, Ala, or Asn. Mpro autolytically cleaves itself from the polyprotein (Hegyi & Ziebuhr, 2002). Inhibiting Mpro activity slows or halts viral replication, offering the promise of improved clinical outcomes for COVID-19 and other coronavirus diseases. Furthermore, there are no known human proteases with similar cleavage specificity to Mpro, suggesting that it should be possible to develop inhibitors that target Mpro without off-target toxicity (Kim et al., 2016).
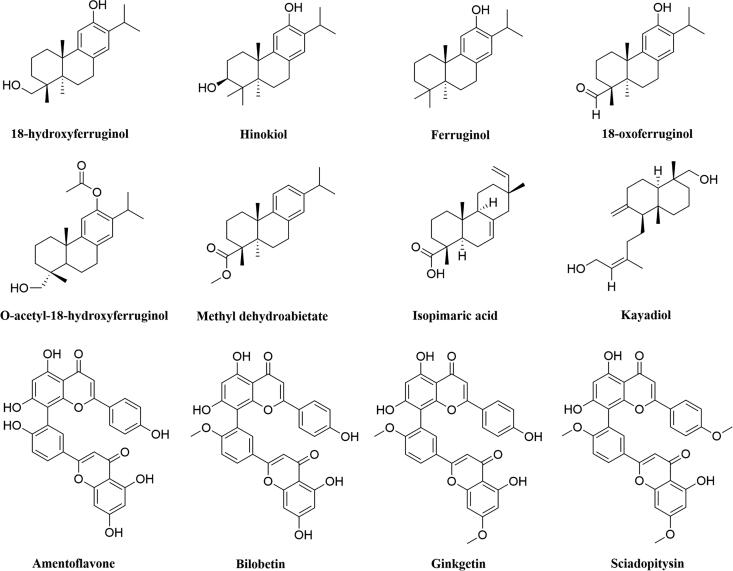
SARS CoV-2 Mpro is a two protomer homodimer in its active form with one active site per the homodimer chain (Jin, Du, et al., 2020; Muramatsu et al., 2016). Each of the protomers of the Mpro consists of three domains: domain I (amino acid residues 8-101), domain II (amino acid residues 102-184) and domain III (amino acid residues 201-303) (Jin, Du, et al., 2020). Domain I and II are mainly β-barrels, while, domain III mainly consists of α-helices (Jin, Du, et al., 2020). The catalytic site/active site/substrate binding site comprising of cysteine (Cys145) and histidine (His41) amino acid moiety and is located at the cleft of domain I and domain II (Jin, Du, et al., 2020). Cysteine145 serves as a common nucleophile and plays a vital role in the proteolytic functioning of Mpro (Anand et al., 2003; Chou et al., 2003; Hsu et al., 2005). The major research effort to date has been focused to inhibit the catalytic activity of SARS CoV-2 Mpro, i.e. by directly targeting its (Mpro) active site with molecules that competitively bind to the active site “pockets” (Dai et al., 2020; Jin, Du, et al., 2020; Jin, Zhao, et al., 2020; Zhang et al., 2020). Studies have been carried out to find suitable inhibitors of Mpro using drug repurposing strategy (Baby et al., 2020; Beck et al., 2020; Bharadwaj et al., 2020; Hage-Melim et al., 2020; Hakmi et al., 2020; Jimenez-Alberto et al., 2020; Kandeel & Al-Nazawi, 2020). Furthermore, various phytochemicals are also proposed as the SARS CoV-2 main protease inhibitors through screening and structure-based design approach (Gorla et al., 2020; Gurung et al., 2020; Joshi, Joshi, et al., 2020; Joshi, Sharma, et al., 2020; Mazzini et al., 2020). In recent times, it has been found that plant-derived polyphenols can serve as potent SARS CoV-2 Mpro inhibitors (Bhardwaj et al., 2020; Ghosh et al., 2020a, 2020b; Tripathi et al., 2020). Our group has identified three polyphenols from green tea [epigallocatechin gallate (EGCG), epicatechingallate (ECG) and gallocatechin-3-gallate (GCG)] can inhibit the catalytic activity of SARS CoV-2 Mpro (Ghosh et al., 2020a). In another independent study, we have identified six polyphenols from Broussonetia papyrifera which have the potency to inhibit the proteolytic activity SARS CoV-2 Mpro (Ghosh et al., 2020b). Purohit and coworkers have found that polyphenols such as oolonghomobisflavan-A and theaflavin-3-O-gallate from the tea plant (Camellia sinensis L.) can act as effective SARS CoV-2 Mpro inhibitors (Bhardwaj et al., 2020). Apart from polyphenolic compounds, plant-derived terpenoid compounds are also found to inhibit the activity of Mpro (Orhan & Senol Deniz, 2020; Sayed et al., 2020; Wen et al., 2007). The leaves of the traditional medicinal plant Torreya nucifera contains eight well-known diterpenoids (18-hydroxyferruginol, hinokiol, ferruginol, 18-oxoferruginol, O-acetyl-18-hydroxyferruginol, methyl dehydroabietate, isopimaric acid, kayadiol) and four biflavonoids (amentoflavone, bilobetin, ginkgetin, sciadopitysin) (Figure 1) (Ryu et al., 2010). Some of these diterpenoid and biflavonoid compounds from the leaf extract of T. nucifera exhibited antiviral property against a broad spectrum of viruses. Among the diterpenoids, ferruginol and its derivatives were reported to show anti-DENV effect against Dengue virus serotype 2 (Islam & Mubarak, 2020; Roa-Linares et al., 2016). Another ferruginol derivative 18-oxoferruginol had shown antiviral activity against the Dengue virus as well as human herpesvirus type 2 (Roa-Linares et al., 2016). Methyl dehydroabietate had also exhibited antiherpetic activity against herpes simplex virus type 1 (HHV-1) (Agudelo-Gómez et al., 2012).
Figure 1.
Chemical structure of T. nucifera diterpenoids and biflavonoids. The two-dimensional structure of eight diterpenoids and four biflavonoids from T. nucifera with their respective names are shown.
Among the biflavonoids of T. nucifera, amentoflavone exhibited antiviral potential against the Dengue virus (Coulerie et al., 2013). Henke and coworkers also demonstrated that this biflavonoid can decrease the replication of Coxsackievirus B3 (CVB3) (Wilsky et al., 2012). Amentoflavone, another biflavonoid of T. nucifera, showed potent antiviral activity against the respiratory syncytial virus (RSV), with an IC50 value of 5.5 μg/ml (Ma et al., 2001). This biflavonoid also showed inhibitory effects against the hepatitis-C virus (Lee et al., 2018). It has also been demonstrated that amentoflavone exhibits significant antiviral activity against influenza-A and influenza-B viruses whereas it exhibits moderate anti-herpes virus activity against HSV-1 and HSV-2 (Fritz et al., 2007; Lin et al., 1999). Moreover, amentoflavone also displayed evidence of anti-HIV activity with an IC50 value of 119 µM (Lin et al., 1997). A computational study has also predicted the inhibitory activity of amentoflavone against the Zika virus (Bhargava et al., 2019). Two other T. nucifera biflavonoids (bilobetin and ginkgetin) can serve as effective inhibitors of the influenza virus (Zhang & Wang, 2020). Bilobetin is reported to inhibit the expression of the Epstein-Barr virus (EBV) genes (Lin et al., 1997). Different computational studies further revealed that bilobetin is a potent candidate for the treatment of herpes virus infection and hepatitis-B virus infection (Pathak et al., 2014; Ramaiah & Suresh, 2013). Another T. nucifera biflavonoid (ginkgetin) showed anti-herpesvirus activity against herpes simplex virus type 1 and type 2 (Adnan et al., 2020; Hayashi et al., 1992). Ginkgetin also exhibited inhibitory effects against human cytomegalovirus (Adnan et al., 2020; Hayashi et al., 1992). Anti-influenza virus activity of ginkgetin has also been reported by many investigators (Adnan et al., 2020; Miki et al., 2007).
Attempt has also been taken to assess the antiviral activity of these T. nucifera phytochemicals against SARS CoV-1. Lee and coworkers first isolated eight diterpenoids and four biflavonoids (mentioned in Figure 1) from the ethanol extract of T. nucifera leaves and evaluated the SARS CoV-1 Mpro activity experimentally (Ryu et al., 2010). With the aid of fluorescence resonance energy transfer (FRET) analysis, they found that all the twelve (12) tested phytochemicals inhibited SARS CoV-1 Mpro in a dose-dependent manner. The IC50 values for the SARS CoV-1 Mpro inhibition of the eight diterpenoids range between 49.6 µM to 283.5 µM with ferruginol showing the lowest IC50 value. On the other hand, biflavonoids inhibited SARS CoV-1 Mpro activity with IC50 value ranging from 8.3 µM to 72.3 µM. Among the four biflavonoids of T. nucifera, amentoflavone exhibited the most potent SARS CoV-1 Mpro inhibitory effect with the lowest IC50 value (Ryu et al., 2010). But whether these eight diterpenoids and four biflavonoids which possess SARS CoV-1 Mpro inhibitory activity, can also exhibit any antiviral activity against SARS CoV-2 by inhibiting the catalytic/proteolytic activity of Mpro is not clear till now. Therefore, in this study, we have examined the inhibitory efficacy of these eight diterpenoids (18-hydroxyferruginol, hinokiol, ferruginol, 18-oxoferruginol, O-acetyl-18-hydroxyferruginol, methyl dehydroabietate, isopimaric acid, kayadiol) and four biflavonoids (amentoflavone, bilobetin, ginkgetin, sciadopitysin) from T. nucifera against SARS CoV-2 Mpro with the aid of in-silico docking studies, molecular dynamics simulations and MM-GBSA analysis. This study has revealed that three biflavonoids of T. nucifera (amentoflavone, bilobetin and ginkgetin) have a stronger binding affinity towards Mpro and may possibly act as inhibitors for the SARS CoV-2 Mpro.
2. Materials and methods
2.1. Preparation of Mpro
The SARS CoV-2 Mpro crystal structure was retrieved from the RCSB Protein Data Bank (http://www.rcsb.org) (PDB ID: 6LU7) (Jin, Du, et al., 2020). Then the presence of improper bonds, missing hydrogens, side-chain anomalies were checked and corrected accordingly. After correcting all those aspects, the structure file was inserted into AutoDock Tools and regular procedures were followed to get pdbqt file format of Mpro (Morris et al., 2008, 2009).
2.2. Preparation of the ligands
Each of the structures of T. nucifera diterpenoids and biflavonoids was retrieved from the PubChem database server in MDL/SDF format (https://pubchem.ncbi.nlm.nih.gov). Then B3LYP/6-31G* basis set in Gaussian09 software was used for the optimization of each of the T. nucifera diterpenoids and biflavonoids (Frisch et al., 2016). These ligands were then prepared using the AutoDock Tools involving the addition of hydrogen followed by assignment of the appropriate ionization state of each ligand and regular processes were used in AutoDock Tools to obtain the pdbqt files for T. nucifera diterpenoids and biflavonoids.
2.3. Molecular docking
We have used AutoDock Vina for the entire docking calculations of Mpro with N3, lopinavir and T. nucifera diterpenoids and biflavonoids where the grid box with a 10.0 Å radius throughout the active site region was chosen (Morris et al., 2008, 2009). The lowest root mean square deviation (RMSD), and the highest Vina score conformations of each complex were selected. The output from AutoDock Vina was rendered with DS visualizer software (Biovia, 2017).
2.4. Molecular dynamics simulation
We have performed molecular dynamics (MD) simulations of unligated Mpro, Mpro-N3, Mpro-lopinavir, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes using the GROMACS 2019 (Abraham et al., 2015). We have used the GROMOS9653a6 force field and SPC water model for all the simulations, where the PRODRG server was used for the ligand topologies (Oostenbrink et al., 2004; Schuttelkopf & van Aalten, 2004). The LINCS algorithm was used to constrained all bond the lengths of protein and N3/lopinavir/T. nucifera biflavonoids, while to restrain water molecules we have used the SETTLE algorithm (Hess et al., 1997; Miyamoto & Kollman, 1992). A total number of 30226, 30205, 30208, 30198, 30196 water molecules containing the unligated Mpro, Mpro-N3, Mpro-lopinavir, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes, respectively, were placed in a cubic box system. Each system was energy-minimized using the steepest descent algorithm and equilibrated to achieve the appropriate volume. The leapfrog algorithm with time step 2 fs was used and at every 5 steps, the neighbour list was updated. The Long-range electrostatics was calculated using the Particle Mesh Ewald method with cut off 1.2 nm and a Fourier grid spacing of 1.2 nm (Essmann et al., 1995). The periodic boundary conditions were applied and equilibration of the systems was carried out mainly in two stages. First, the system was equilibrated at 300 K in the NVT ensemble using the v-rescale algorithm for 10 ns. Then the system was further equilibrated in the NPT ensemble for 10 ns by positional restraining of the complexes (unligated Mpro, Mpro-N3, Mpro-lopinavir and Mpro-T. nucifera biflavonoid complexes). The Parrinello-Rahman method and Berendsen barostat were employed to maintain the pressure and temperature, respectively (Berendsen et al., 1984; Parrinello & Rahman, 1981). For each system, the average temperature and pressure values remained close to the desired values. The equilibrated systems were then subjected to unrestrained production MD simulations for 100 ns each, maintaining target pressure (1 bar) and temperature (300 K). In order to check the reproducibility of the results, repeating simulations for each of the system for five times with independent equilibration times and initial velocities were carried out. The overall trends in the several properties after MD simulation remained the same. Finally, we have used the MD trajectories for each system to calculate some important parameters in order to check protein-ligand interactions, namely, root mean square deviation (RMSD), root mean square fluctuation (RMSF), the radius of gyration (Rg), and solvent accessible surface area (SASA) (Ghosh et al., 2020a, 2020b). We also analyzed the secondary structure of Mpro in all these systems as well as analyzed the number of hydrogen bonds formed between Mpro and different compounds (N3, lopinavir, amentoflavone, bilobetin and ginkgetin) throughout the entire MD run (Bharadwaj et al., 2020).
2.5. MM-GBSA analysis
Recently to calculate the binding free energies of ligands to the receptor, several implicit solvent models have been used such as a) the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) b) Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) and c) Free energy perturbation, etc (Chen, 2016; Chen et al., 2015; Das et al., 2019; Hou et al., 2011). Here we have used the MM-GBSA method to calculate the relative binding free energies of N3, lopinavir and T. nucifera biflavonoids to Mpro. We have calculated the MM-GBSA from the entire trajectory of each MD simulation run for different systems (Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoids complexes). For that, we have taken the entire 100 ns trajectory of each MD simulation and extracted the coordinate file at every 5 ns interval. From these 20 points, we have calculated the MM-GBSA value for each system. The resultant/average binding free energy value was computed from these five independent analysis. The free energy of binding can be calculated as ΔGbind = Ecomplex − (Eligand + Ereceptor). Actually, ΔGbind is the energy difference between the complex and sum of the energies of the receptor and ligand. The energy for complex (Ecomplex), receptor (Ereceptor) and ligand (Eligand) can be further divided into molecular mechanics (electrostatic and van der Waals) and solvation (polar and non-polar) components, ETotal = EMM + ESol.
Prime MM-GBSA uses the VSGB 2.0 implicit solvation model to estimate the binding energy of the receptor-ligand complex. The generalized Born model with an external dielectric constant of 80 and an internal dielectric constant of 1, for the estimation of the polar contribution of the free energy, while the non-polar energy contribution is calculated from the solvent-accessible surface area (SASA). The prime module of the Schrodinger suite (Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY, 2020) was used for all MM-GBSA calculations.
2.6. Pharmacokinetic properties analysis
SwissADME and pkCSM-pharmacokinetics online softwares were used for the prediction of different pharmacokinetic properties of diterpenoids and biflavonoids from T. nucifera (Daina et al., 2017; Pires et al., 2015). Levels of toxicity along with the drug-likeness properties of these polyphenols such as absorption, distribution, metabolism and excretion parameters were mainly scrutinized.
3. Result and discussion
In 2005, Rao and coworkers designed a Michael inhibitor named N3 which efficiently inhibited the proteolytic activity of SARS CoV-1 Mpro as well as MERS CoV Mpro (Yang et al., 2005). Yang and coworkers took this inhibitor and docked into the constructed homology modeled SARS CoV-2 Mpro (Jin, Du, et al., 2020). They observed that N3 binds to the substrate-binding site of SARS CoV-2 Mpro. With the aid of experimental studies, the authors have shown that N3 inhibits the enzymatic activity of SARA CoV-2 Mpro via two-step irreversible inactivation process. The authors also determined the crystal structure of SARS CoV-2 Mpro in complex with N3 (Jin, Du, et al., 2020). The three-dimensional structure of Mpro in this co-crystal revealed that N3 binds to the Cys-His catalytic dyad of Mpro. Later, many investigators including us have chosen N3 as standard substrate and compared the binding affinity and/or binding modes between various small molecules including green tea polyphenols with that of “Mpro-N3 complex” (Ghosh et al., 2020a; Odhar et al., 2020). Besides N3, many anti-HIV drugs also have a good binding affinity towards the active site of Mpro (Beck et al., 2020). One such important anti-HIV drug is lopinavir. In the recent past, lopinavir has been also taken as a standard Mpro inhibitor by us and other investigator (Bhardwaj et al., 2020; Ghosh et al., 2020b). Thus, we have decided to take these two compounds (N3 and lopinavir) as standard inhibitors for this study. We also decided to screen the diterpenoids (18-hydroxyferruginol, hinokiol, ferruginol, 18-oxoferruginol, O-acetyl-18-hydroxyferruginol, methyl dehydroabietate, isopimaric acid, kayadiol) and biflavonoids (amentoflavone, bilobetin, ginkgetin, sciadopitysin) from T. nucifera with the aid of pharmacokinetic analysis before assessing their binding propensity towards Mpro.
3.1. Screening of diterpenoids and biflavonoids of Torreya nucifera using pharmacokinetic analysis
Understanding the pharmacokinetic behavior of a particular compound is extremely essential for assessing its suitability towards human administration. Pharmacokinetics properties obtained from SwissADME and pkCSM-pharmacokinetics tools were listed in Table 1.
Table 1.
Pharmacokinetic properties of diterpenoids and biflavonoids from Torreya nucifera.
| Compound | MW | H-Ac | H-Do | Nrot | TPSA | LogP | IA | TC | LD50 | HT | AT | MTD | NLV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18-hydroxyferruginol | 302.45 | 2 | 2 | 2 | 40.46 | 4.5182 | 92.113 | 0.693 | 2.601 | No | No | −0.194 | 0 |
| Hinokiol | 302.45 | 2 | 2 | 1 | 40.46 | 4.5166 | 91.639 | 0.658 | 2.423 | No | No | 0.057 | 0 |
| Ferruginol | 286.45 | 1 | 1 | 1 | 20.23 | 5.5458 | 91.755 | 0.688 | 2.559 | No | No | −0.09 | 1 |
| 18-oxoferruginol | 300.44 | 2 | 1 | 2 | 37.30 | 4.7248 | 92.986 | 0.743 | 2.545 | No | No | −0.238 | 0 |
| O-acetyl-18-hydroxyferruginol | 344.49 | 3 | 1 | 4 | 46.53 | 4.7379 | 93.263 | 0.659 | 2.524 | No | No | −0.7 | 1 |
| Methyl dehydroabietate | 314.46 | 2 | 0 | 3 | 26.30 | 4.9933 | 95.12 | 1.055 | 2.377 | No | No | −0.188 | 1 |
| Isopimaric acid | 302.45 | 2 | 1 | 2 | 37.30 | 5.2062 | 97.652 | 0.717 | 1.881 | Yes | No | −0.246 | 1 |
| Kayadiol | 306.48 | 2 | 2 | 5 | 40.46 | 4.4764 | 93.48 | 1.244 | 3.051 | No | No | −1.511 | 0 |
| Amentoflavone | 538.46 | 10 | 6 | 3 | 181.80 | 5.134 | 84.356 | 0.484 | 2.527 | No | No | 0.438 | 2 |
| Bilobetin | 552.48 | 10 | 5 | 4 | 170.80 | 5.437 | 86.049 | 0.571 | 2.56 | No | No | 0.437 | 1 |
| Ginkgetin | 566.51 | 10 | 4 | 5 | 159.80 | 5.74 | 95.376 | 0.646 | 2.733 | No | No | 0.427 | 1 |
| Sciadopitysin | 580.54 | 10 | 3 | 6 | 148.80 | 6.043 | 98.322 | 0.833 | 3.06 | Yes | No | 0.419 | 1 |
MW = Molecular weight (g/mol); H-Ac = No. of hydrogen bond acceptor; H-Do = No. of hydrogen bond donors; Nrot = No. of rotatable bonds; TPSA = Topological polar surface area (Å2); LogP = Predicted octanol/water partition coefficient; IA = Intestinal absorption (% Absorbed); TC = Total clearance (log ml/min/kg); LD50 = Oral rat acute toxicity (mol/kg); HT = Hepatoxicity; AT = AMES toxicity; MTD = Maximum tolerated dose for human (log mg/kg/day); NLV = No. of Lipinski rule violation.
The molecular weight of all twelve phytochemicals was ranging from ∼286 to ∼581 g/mol. Thus, their transportation, diffusion and absorption inside the body could be easier. The TPSA value of all the phytochemicals was less than 200 Å2 (∼20–182 Å2) which indicating their good bioavailability. Thus analysis further revealed that isopimaric acid (diterpenoid) and sciadopitysin (biflavonoid) are hepatotoxic molecules. Furthermore, an unfavorable (negative) tolerance dose of seven diterpenoids (18-hydroxyferruginol, ferruginol, 18-oxoferruginol, O-acetyl-18-hydroxyferruginol, methyl dehydroabietate, isopimaric acid and kayadiol) was evidenced. Such unfavorable tolerance dose ruled out the possibility of their usage as a potential drug for humans. The remaining one diterpenoid (hinokiol) and three biflavonoids (amentoflavone, bilobetin and ginkgetin) were found non-toxic and satisfied all the drug-likeness characteristics. Therefore, we selected these four phytochemicals for probing their binding propensity towards SARS CoV-2 Mpro using molecular docking studies.
3.2. Assessment of binding affinity and binding modes of different diterpenoids and biflavonoids using molecular docking studies
N3, lopinavir and phytochemicals of T. nucifera those possessing favorable drug-likeness characteristics were docked to assess the phytochemical(s) exhibiting the higher or comparable binding energy to that of “Mpro-N3/lopinavir interaction”. The binding energy of N3 and lopinavir towards Mpro was −7.0 and −7.3 kcal/mol, respectively (Table 2) (Ghosh et al., 2020a, 2020b).
Table 2.
The binding energy of N3, lopinavir and different phytochemicals (diterpenoid & biflavonoids) of T. nucifera with the active site of SARS CoV-2 Mpro.
| Drug | Binding energy (kcal/mol) |
|---|---|
| N3 | −7.0 |
| Lopinavir | −7.3 |
| Hinokiol | −6.4 |
| Amentoflavone | −9.2 |
| Bilobetin | −9.1 |
| Ginkgetin | −9.0 |
We also determined the binding energy of four phytochemicals (hinokiol, amentoflavone, bilobetin and ginkgetin) towards Mpro using molecular docking studies. It was found that the binding energy of the only diterpenoid i.e. hinokiol (−6.4 kcal/mol) was lower as compared to the binding energy of selected standards (N3 and lopinavir) towards Mpro (Table 2). On the contrary, three biflavonoids (amentoflavone, bilobetin and ginkgetin) exhibited higher binding affinity (−9.0 to −9.2 kcal/mol) towards Mpro than that of N3 and lopinavir (Table 2). As amentoflavone, bilobetin and ginkgetin had higher binding affinity than N3 and lopinavir, we decided to proceed further with these three biflavonoids.
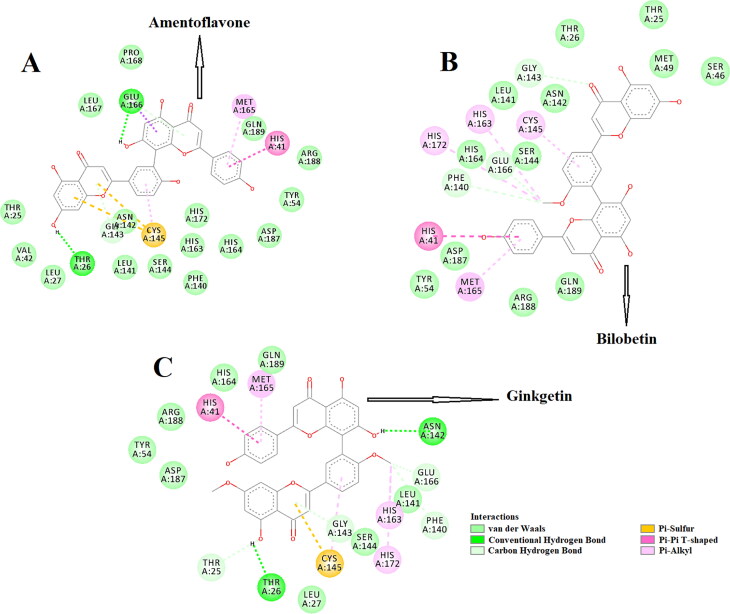
The amino acid residues within the active site of Mpro which were interacting with these three selected biflavonoids were carefully examined with the aid of discovery studio visualizer. It was evidenced that amentoflavone, bilobetin and ginkgetin efficiently interacted with different amino acid residues of domain I and II of Mpro (Figure 2).
Figure 2.
Molecular docking of three T. nucifera biflavonoids (amentoflavone, bilobetin and ginkgetin) with Mpro. The docked conformation of the Mpro-amentoflavone complex (panel A), Mpro-bilobetin complex (panel B) and Mpro-ginkgetin complex (panel C) depicting the possible interactions with various amino acids of Mpro. All of them interact with various amino acid residues including His41 and Cys145 of Mpro.
When amentoflavone was docked into the active site of Mpro, two hydrogen bond interactions [Thr26 (2.3 Å) and Glu166 (2.2 Å)], seventeen van der Waals interactions (Thr25, Leu27, Leu42, Tyr54, Phe140, Leu141, Asn142, Gly143, Ser144, His163, His164, Leu167, Pro168, His172, Asp187, Arg188 and Gln189) were evidenced (Figure 2(A)). Besides these, one π-sulfur (Cys145), one π-alkyl (Met165) and one π-π (His41) interactions were evidenced in the Mpro-amentoflavone complex (Figure 2(A)). Interestingly, no hydrogen bond interaction was observed when bilobetin interacted with Mpro (Figure 2(B)). But a total number of twelve van der Waals interactions (Thr25, Thr26, Ser46, Met49, Tyr54, Leu141, Asn142, Ser144, His164, Asp187, Arg188, Gln189), three C-H interactions (Phe140, Gly143 and Glu166), four π-alkyl interactions (Cys145, His163, Met165, His172) and a single π-π interaction (His41) was observed in the Mpro-bilobetin complex (Figure 2(B)). On the other hand, a couple of H-bond interactions [Thr26 (2.3 Å) and Asn142 (2.3 Å)] were observed when ginkgetin was docked to the active site of Mpro (Figure 2(C)). Mpro-ginkgetin complex was further stabilized by four C-H interactions (Thr25, Phe140, Gly143 and Glu166), eight van der Waals interactions (Leu27, Tyr54, Leu141, Ser144, His164, Asp187, Arg188 and Gln189), three π-alkyl interactions (His163, Met165 and His172), one π-sulfur (Cys145) interaction and a single π-π interaction (His41) (Figure 2(C)). Even, the interaction of two selected standards (N3 and lopinavir) with several critical residues within the active site of Mpro was well-evidenced in our previous studies (Ghosh et al., 2020a, 2020b). The Mpro-N3 complex was stabilized by multiple hydrogen bond interactions (especially with His41 and Cys145 of Mpro) and many other non-covalent interactions (Ghosh et al., 2020a). We also evidenced that Cys145, Met49 and Pro168 of Mpro interacted with lopinavir via hydrogen bond, π-sulfur bond and alkyl bond, respectively (Ghosh et al., 2020b). Lopinavir also formed numerous van der Waals interactions with various amino acid residues (Thr25, Thr26, Leu141, Tyr54, Phe140, Asn142, Gly143, His163, His164, Met165, Glu166, Leu167, Asp187, Arg188, Gln189, Thr190 and Gln192) of Mpro (Ghosh et al., 2020b). Overall, molecular docking studies clearly revealed that selected three biflavonoids (amentoflavone, bilobetin and ginkgetin) interacted with two key residues (His41 and Cys145) of Mpro via alkyl bond(s) interactions (Figure 2). In fact, there are many reports available in the literature where investigators noticed the formation of non-covalent bonds (other than H-bond) including alkyl bond(s) between compounds and residues of catalytic site (His41 and Cys145) of the Mpro (Bharadwaj et al., 2020; Bhardwaj et al., 2020; Ghosh et al., 2020b; Gurung et al., 2020; Joshi, Joshi, et al., 2020; Odhar et al., 2020). We believe that the availability and/or accessibility of Cys145 as well as His41 of Mpro may be ceased down due to the formation of alkyl bond(s) between compounds and these two residues of Mpro and such alterations possibly can inhibit its (Mpro) catalytic activity. Moreover, their binding affinity towards Mpro was more than the binding affinity of N3/lopinavir to Mpro (Table 2). Thus, it can be concluded that amentoflavone, bilobetin and ginkgetin may possibly inhibit the proteolytic activity of Mpro and may potentially be used to treat patients with COVID-19. We also computed the inhibition constant (ki) so as to get the idea about the inhibition potency of amentoflavone, bilobetin and ginkgetin towards Mpro (Gurung et al., 2020). Inhibition constant (ki) was estimated using the following equation
| (1) |
where ΔG is the binding energy in kcal/mol, R is the universal gas constant (1.987 calK−1 mol−1) and T is the temperature (298 K).
For the docked structure (having lowest energy) of Mpro-amentoflavone complex, Mpro-bilobetin complex and Mpro-ginkgetin complex, the in silico determined ki value at 298 K was 1.7 × 10−7 M, 2.1 × 10−7 M and 2.6 × 10−7 M, respectively. These computed ki values are far lower than their toxicity dose (LD50) range (mentioned in Table 1) which further validates the strong candidature of these three biflavonoids as target drugs to bind SARS CoV-2 Mpro.
These three Mpro-biflavonoid complexes were further subjected to molecular dynamics simulations as well as binding free energy computations to assess the stability of these complexes.
3.3. Molecular dynamics simulation studies
MD simulations were performed for 100 ns using the GROMOS9653a6 force field. Prior to assessing overall complex stability and various other important structural properties, we determined the binding modes/residues within the Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complex after the end of the MD run.
One π-π interaction (His41), one π-sigma interaction (Glu166), two π-alkyl interaction (Cys145, Met165), fifteen van der Waals interactions (Thr25, Thr26, Leu27, Ser46, Tyr54, Phe140, Leu141, Asn142, Ser144, His163, His164, His172, Asp187, Arg188, Gln189) and two hydrogen bond interactions [Met49 (2.4Å), Gly143 (2.4Å)] were evidenced in Mpro-amentoflavone system (Supplemental Figure 2A). Interestingly, two hydrogen bond interactions [Ser46 (2.4Å), Gly143 (2.4Å)] were observed in the Mpro-bilobetin system (Supplemental Figure 2B). The other interactions which stabilized the Mpro-bilobetin system were eleven van der Waals interactions (Thr25, Thr26, Leu27, Tyr54, Leu141, Asn142, Ser144, His164, Asp187, Arg188, Gln189), two C-H bond interactions (Phe140 and Glu166), two alkyl interactions (His163 and His172), three π-alkyl interactions (Met49, Cys145 and Met165) and one π-π interaction (His41) (Supplemental Figure 2B). On the other hand, two C-H bond interactions (Phe140, Leu141), two π-alkyl interactions (Cys145, Met165), one alkyl interaction (His163), one π-π interaction (His41), one π-sulfur interaction (Met49), ten van der Waals interactions (Thr25, Thr26, Tyr54, Ser144, His164, Glu166, His172, Asp187, Arg188, Gln189) and two hydrogen bond interactions [Asn142 (2.2Å), Gly143 (2.3Å)] were evidenced in Mpro-ginkgetin complex (Supplemental Figure 2C). This analysis clearly revealed that the interaction of these three biflavonoids with many important amino acid residues (including His41 and Cys145) of Mpro remained intact even after the MD run. These findings validated the stability of these complexes as well as strengthen their candidatures as SARS CoV-2 Mpro inhibitors.
3.3.1. Conformational stability and conformational fluctuation analysis
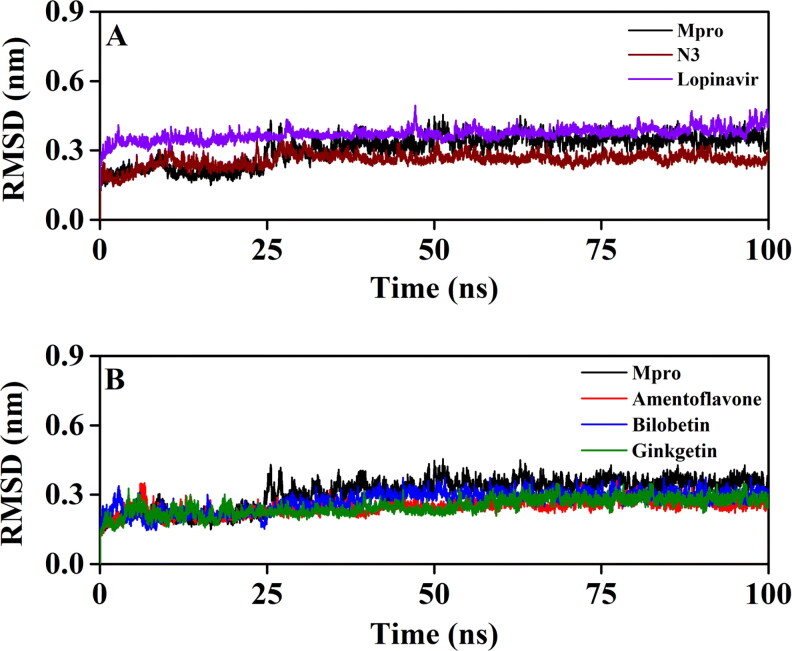
To corroborate the stability of Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes, we estimated the RMSD of backbone alpha carbon atoms of these three systems and compared the same with that of three other systems (unligated Mpro, Mpro-N3 and Mpro-lopinavir) (Figure 3). For unligated Mpro, the RMSD value from 2 ns to 17 ns maintained a constant value (∼0.21–0.22 nm). The value increased progressively and reached ∼0.31 nm (with some fluctuations) at 65 ns and remained almost the same till the end of the MD run (Figure 3(A, B)) (Ghosh et al., 2020b). The RMSD value of the Mpro-N3 complex was ∼0.18 nm at 2 ns, which rose to ∼0.28 nm at 10 ns. The RMSD magnitude remains the same (∼0.28 nm) until 25 ns. Then, within the next 6 ns, the value was slightly decreased (∼0.26 nm) and persisted at the same value till 100 ns (Figure 4(A)). The RMSD values for Mpro-lopinavir complexes were found to remain almost constant (∼0.36–0.37 nm) from 10 ns to 100 ns with some marginal fluctuations (Figure 4(A)) (Ghosh et al., 2020b). The RMSD value of the Mpro-amentoflavone complex increased from ∼0.18 nm (at 2 ns) up to ∼0.25 nm (at 30 ns) and remained almost constant till the end of the MD run with some marginal fluctuations (Figure 3(B)). For the Mpro-bilobetin complex, the RMSD value from 2 ns to 24 ns maintained a constant value (∼0.22 nm). Thereafter, the value increased gradually and reached to ∼0.4 nm at 40 ns. Then, the RMSD value slightly decreased and persisted at ∼0.38 nm from 53 ns till the end of the MD run (Figure 3(B)). For Mpro-ginkgetin complex, the RMSD value from 2 ns to 21 ns oscillated between ∼0.19–0.21 nm. Thereafter a gradual increase of RMSD value was observed upto 57 ns (∼0.26 nm) and maintained equilibrium upto 100 ns of MD run (Figure 3(B)). The average RMSD values (computed from 5 independent analysis) for unligated Mpro, Mpro-N3 and Mpro-lopinavir complex were found to be 0.297 nm, 0.249 nm and 0.364 nm, respectively (Table 3). Whereas, the average RMSD values of, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes (yielded from 5 independent analysis) were 0.248 nm, 0.279 nm and 0.246 nm, respectively (Table 3), suggesting that these three complexes are stable. However, the stability of Mpro-bilobetin is least among all of them. Also we can say that Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes are more stable than the Mpro-lopinavir system.
Figure 3.
The RMSD plots of unligated Mpro, Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoid complexes. The RMSD plots of unligated Mpro, Mpro-N3 and Mpro-lopinavir systems are plotted in panel A while panel B represents the RMSD plots of unligated Mpro, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes. The MD simulations for each system were performed for 100 ns.
Figure 4.
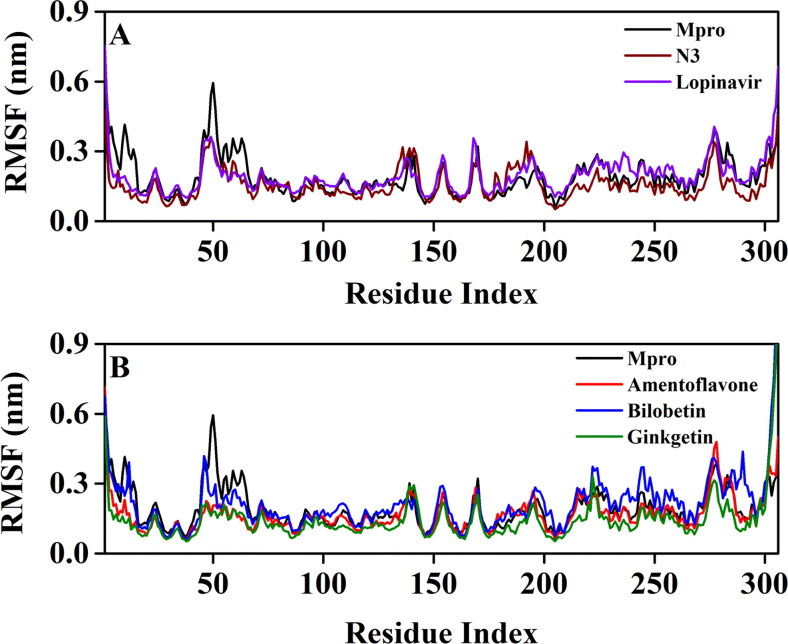
RMSF profiles of unligated Mpro and Mpro complexed with two selected standards and three T. nucifera biflavonoids. The RMSF values of unligated Mpro, Mpro-N3 and Mpro-lopinavir complexes are plotted in panel A against the amino acid residues of Mpro. Panel B represents the RMSF plots of unligated Mpro, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes against the amino acid residues of Mpro.
Table 3.
Average values of the RMSD and RMSF for the simulated systems.
| System | RMSD* (nm) |
RMSF* (nm) |
|---|---|---|
| Unligated Mpro | 0.297 ± 0.0531 | 0.1825 ± 0.0248 |
| Mpro-N3 | 0.249 ± 0.0438 | 0.1463 ± 0.0219 |
| Mpro-lopinavir | 0.364 ± 0.0572 | 0.1836 ± 0.0318 |
| Mpro-amentoflavone | 0.248 ± 0.0418 | 0.1372 ± 0.0219 |
| Mpro-bilobetin | 0.279 ± 0.0446 | 0.1413 ± 0.0312 |
| Mpro-ginkgetin | 0.246 ± 0.0414 | 0.1334 ± 0.0246 |
For obtaining the average value, 5 independent MD trajectories were analyzed with the aid of RMSD and RMSF.
The conformational flexibility of unligated Mpro, Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoid complexes was assessed by calculating the RMSF of alpha carbon atoms corresponding to these systems (Figure 4). It was quite evident from the RMSF profiles that the Mpro-lopinavir and Mpro-bilobetin complexes suffer more conformational fluctuations in domain III. In the case of unligated Mpro system, most of the amino acid residues within the domain I and II of this system had RMSF fluctuation below 0.3 nm (Figure 4(A, B)). Only residues 45-60 pertaining to this system experienced higher fluctuations (up to ∼0.6 nm). The average RMSF value (obtained from 5 independent analysis) for the unligated Mpro system was ∼0.183 nm (Table 3). The RMSF plot of the Mpro-N3 complex reflected that very few amino acid residues within domain I, II and III have an RMSF value of more than 0.25 nm (Figure 4(A)). Interestingly, the RMSF values of several stretches within these three domains of this system (residues 132-138 and 177-195) were more compared to that of the unligated system. The Mpro-lopinavir system experienced more or less similar conformational fluctuations to that of the unligated Mpro system (Figure 4(A)). The fluctuations for many amino acid residues of domain I were reduced upon the binding of lopinavir to Mpro. The average RMSF value for Mpro-N3 and Mpro-lopinavir system was ∼0.146 nm and ∼0.184 nm, respectively (Table 3). Upon analyzing all the RMSF profiles, it was clearly observed that Mpro-amentoflavone and Mpro-ginkgetin complexes showed lower fluctuations (especially in domain I and II; except residues 177-184 for Mpro-amentoflavone complex) as compared to the unligated Mpro and Mpro-N3/lopinavir complexes (Figure 4(B)). The average RMSF values of Mpro-amentoflavone and Mpro-ginkgetin complexes were ∼0.137 nm and ∼0.133 nm, respectively (Table 3). Even in Mpro-bilobetin complexes, the fluctuations of most of the amino acid residues (residues 98-111, 152-155 and 175-184 in Mpro-bilobetin complex) residing at the domain I and II were reduced (Figure 7). The average RMSF value of this complex is ∼0.141 nm (Table 3). In fact, it was clearly evident from the RMSF plots that many key amino acid residues in the catalytic/active site of Mpro (especially His41 and Cys145) were significantly reduced after binding to these three biflavonoids. These results further suggested that all three Mpro-biflavonoids complexes experience less conformational fluctuations than that of the Mpro-N3/Mpro-lopinavir complex.
3.3.2. Hydrogen bond analysis
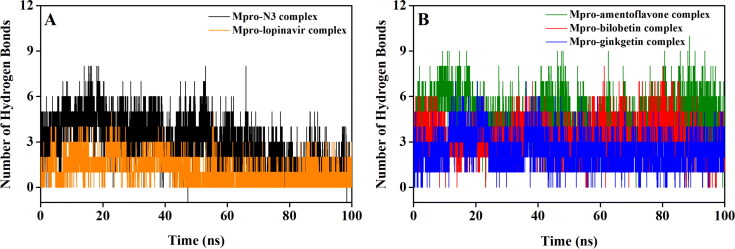
Hydrogen bonds are usually very specific bonds and are often responsible for receptor-ligand stability. In order to get the idea about the binding strength/stability of Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoid complexes, we estimated the number of hydrogen bonds formed throughout the MD run and presented in Figure 5. In the Mpro-N3 complex, the majority of conformations formed up to 3 hydrogen bonds during the MD simulation (Figure 5(A)). A small number of conformations exhibited less than 1 and greater than 5 hydrogen bonds. When lopinavir was complexed with Mpro, the existence of an average number of 1 hydrogen bond was evidenced (Figure 5(A)). Interestingly, in both Mpro-bilobetin and Mpro-ginkgetin complex, the average number of hydrogen bonds formed was 3 (Figure 5(B)). However, the biflavonoid amentoflavone had most of the intermolecular hydrogen bonding with Mpro between 2 to 7 throughout the whole simulation process with an average value of 5 (Figure 5(B)). These results clearly indicated that these three biflavonoids form a greater number of hydrogen bonds with Mpro during the simulation than lopinavir and/or N3. Also, it can be suggested that these Mpro-biflavonoid complexes are highly stable.
Figure 5.
Hydrogen bond profiles of Mpro complexed with selected standards and T. nucifera biflavonoids. The hydrogen bond profiles of Mpro-N3 complex (black) and Mpro-lopinavir complex (orange) are plotted in panel A while panel B represents the hydrogen bond profiles of Mpro-amentoflavone (green), Mpro-bilobetin (red) and Mpro-ginkgetin (blue) complexes throughout the 100 ns of MD run.
3.3.3. Secondary structural analysis during MD simulation
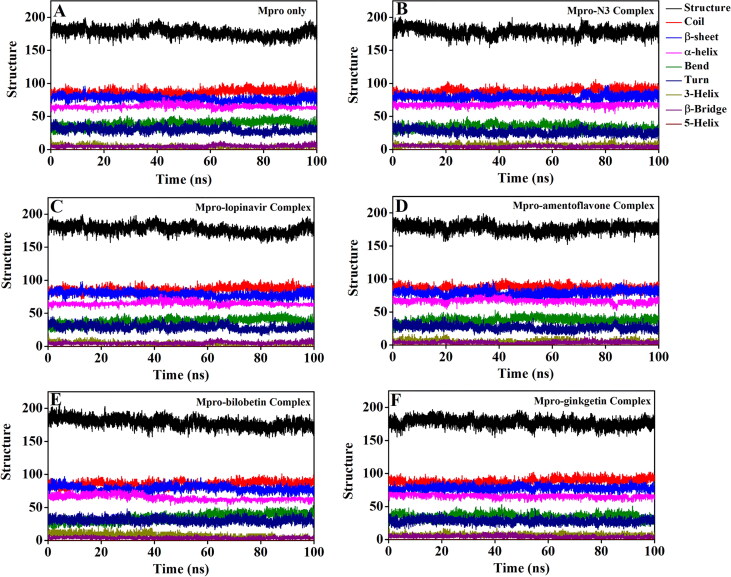
In order to assess the binding effect of N3, lopinavir and selected biflavonoids (amentoflavone, bilobetin and ginkgetin) to the structure/conformation (especially secondary structure) of Mpro during the entire MD run, we investigated the conformations/snapshots from trajectory corresponding to every 25 ns time interval. The snapshots presented in Supplemental Figures 3 and 4 indicated that the secondary structural elements of Mpro (α-helix and β-sheet) remain unaffected upon complexation with N3, lopinavir and three biflavonoids during 100 ns MD simulation. We also observed that the biflavonoids are persistently bound to the active/catalytic site without any structural modification, suggesting that the Mpro-biflavonoid complexes are highly stable.
The secondary structural analysis of unligated Mpro, as well as Mpro complexed with N3, lopinavir and three biflavonoids were estimated with the aid of using gmx_do_dssp option (Figure 6, Supplemental Figure 5 and Table 4).
Figure 6.
Analysis of Mpro secondary structure in different systems during 100 ns MD simulation. The secondary structure content of Mpro in (A) unligated Mpro system, (B) Mpro-N3 system, (C) Mpro-lopinavir system, (D) Mpro-amentoflavone system, (E) Mpro-bilobetin system and (F) Mpro-ginkgetin system.
Table 4.
The overall percentage of secondary structure elements of Mpro in unligated Mpro, Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoid complexes.
| Systems | Structure (α-helix + β-sheet + β-bridge + Turn) (%) | Coil (%) | β-sheet (%) | β-bridge (%) | Bend (%) | Turn (%) | α-helix (%) | 5-helix (%) | 3-helix (%) |
|---|---|---|---|---|---|---|---|---|---|
| Unligated Mpro | 58.0 | 28.0 | 25.0 | 2.0 | 13.0 | 10.0 | 21.0 | 0.0 | 1.0 |
| Mpro-N3 | 58.0 | 28.0 | 26.0 | 1.0 | 12.0 | 9.0 | 22.0 | 0.0 | 2.0 |
| Mpro-lopinavir | 59.0 | 27.0 | 27.0 | 2.0 | 13.0 | 9.0 | 21.0 | 0.0 | 1.0 |
| Mpro-amentoflavone | 59.0 | 28.0 | 26.0 | 2.0 | 12.0 | 9.0 | 22.0 | 0.0 | 1.0 |
| Mpro-bilobetin | 60.0 | 28.0 | 27.0 | 1.0 | 11.0 | 10.0 | 22.0 | 0.0 | 1.0 |
| Mpro-ginkgetin | 59.0 | 29.0 | 26.0 | 2.0 | 11.0 | 9.0 | 22.0 | 0.0 | 1.0 |
The content of α-helix, β-sheet, β-bridge and turn in Mpro only/unligated Mpro was 21%, 25%, 2% and 10%, respectively (Table 4). The determined secondary structural content for unligated Mpro/Mpro only is in close agreement with the previously published estimation (Bharadwaj et al., 2020). It was noticed that all these secondary structural components of Mpro undergo no significant alterations upon binding to N3, lopinavir and three biflavonoids (Table 4). Even, the coil content of Mpro was almost conserved when it formed a complex with these above-mentioned compounds (Table 4). Thus, we can say that the binding of these three biflavonoids to Mpro has no effect on the rigidity of Mpro structure. Altogether, these findings suggested that overall structural conformation including secondary conformation of Mpro is unaltered even when complexed with T. nucifera biflavonoids (amentoflavone, bilobetin and ginkgetin).
3.3.4. Structural compactness and solvent accessibility analysis
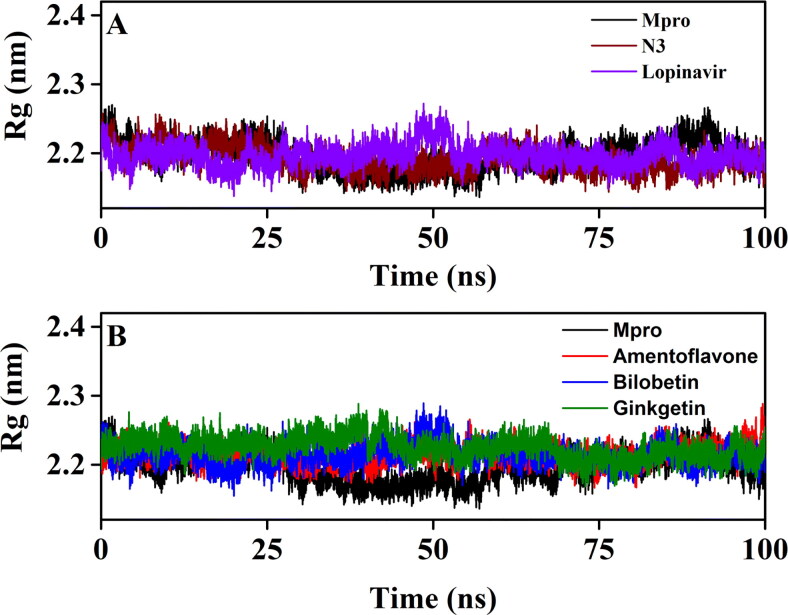
In order to assess the compactness of all the complexes, the Rg value has also been estimated (Figure 7 and Table 5).
Figure 7.
The Rg plots of unligated Mpro and Mpro complexed with two selected standards and three T. nucifera biflavonoids. The Rg plots of unligated Mpro, Mpro-N3 and Mpro-lopinavir complexes are plotted in panel A while panel B represents the Rg plots of unligated Mpro, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes. The MD simulations for each system were performed for 100 ns.
Table 5.
Average values of the Rg and SASA of the simulated systems.
| System | Rg* (nm) |
SASA* (nm2) |
|---|---|---|
| Unligated Mpro | 2.183 ± 0.0217 | 151.4341 ± 0.0236 |
| Mpro-N3 | 2.181 ± 0.0213 | 155.3633 ± 0.0328 |
| Mpro-lopinavir | 2.185 ± 0.0228 | 151.2614 ± 0.0225 |
| Mpro-amentoflavone | 2.181 ± 0.0215 | 152.0211 ± 0.0216 |
| Mpro-bilobetin | 2.182 ± 0.0319 | 152.7821 ± 0.0326 |
| Mpro-ginkgetin | 2.181 ± 0.0311 | 153.6813 ± 0.0218 |
For obtaining the average value, 5 independent MD trajectories were analyzed with the aid of Rg and SASA.
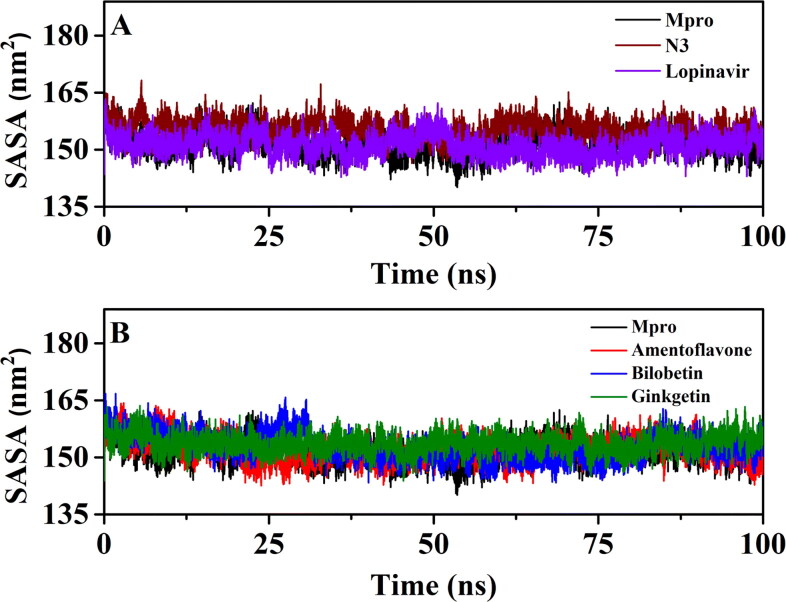
The average Rg values of Mpro-N3 complex (2.181 nm) and Mpro-lopinavir complex (20,185 nm) were found to be in a similar range with unligated Mpro (20,183 nm) (Table 5). Even, the average Rg value for Mpro-amentoflavone (2.181 nm), Mpro-bilobetin (20,182 nm) and Mpro-ginkgetin (2.181 nm) systems was almost identical to that of the other three systems (unligated Mpro, Mpro-N3 and Mpro-lopinavir) (Table 5). These results suggested that all these three Mpro-biflavonoid complexes as well as the Mpro-N3/lopinavir complex share a similar degree of compactness. SASA values were also calculated to assess the extent of expansion of protein volume in each system (Figure 8 and Table 5). The average SASA values of Mpro-N3 complex (∼155.363 nm2) was higher than all other studied systems suggesting an expansion of Mpro during the interaction with N3. The average SASA values of Mpro-lopinavir, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin were ∼151.261 nm2, ∼152.021 nm2, ∼152.782 nm2 and ∼153.681 nm2, respectively (Table 5). These values indicated that all three Mpro-biflavonoid complexes experience slightly more expansion than that of the Mpro-lopinavir complex.
Figure 8.
The SASA plots of unligated Mpro and Mpro complexed with selected standards and T. nucifera biflavonoids. The SASA plots of unligated Mpro, Mpro-N3 and Mpro-lopinavir complexes are plotted in panel A while panel B represents the SASA plots of unligated Mpro, Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes. The MD simulations for each system were performed for 100 ns.
3.4. Binding free energy estimation using MM-GBSA analysis
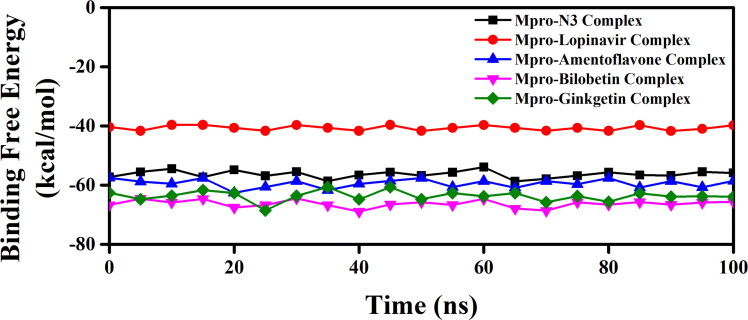
We have taken the configurations from 100 ns trajectory corresponding to every 5 ns time interval and determined the binding free energy (ΔGbind) of all these three Mpro-biflavonoids complexes as well as Mpro-N3 and Mpro-lopinavir complexes by using the MM-GBSA method. The determined binding free energy values throughout the trajectory for each system were presented in Figure 9.
Figure 9.
MM-GBSA binding free energy profiles of two Mpro-standard complexes and three Mpro-T. nucifera biflavonoids. The binding free energy values of Mpro-N3 complex, Mpro-lopinavir complex, Mpro-amentoflavone complex, Mpro-bilobetin complex and Mpro-ginkgetin complex were represented throughout the entire 100 ns simulation trajectory.
The resultant/average binding free energy value computed from five independent analysis was listed in Table 6. The average ΔGbind of Mpro-N3 and Mpro-lopinavir complexes were found to be −56.29 kcal/mol and −40.66 kcal/mol (Table 6). On the contrary, the binding free energy for Mpro-amentoflavone, Mpro-bilobetin and Mpro-ginkgetin complexes, the values were −59.57 kcal/mol, −66.31 kcal/mol and −63.62 kcal/mol, respectively (Table 6).
Table 6.
MM-GBSA values of Mpro-N3, Mpro-lopinavir and three Mpro-biflavonoid complexes.
| System | Binding Free Energy (kcal/mol) |
|---|---|
| Mpro-N3 | −56.29 ± 0.18 |
| Mpro-lopinavir | −40.66 ± 0.85 |
| Mpro-amentoflavone | −59.57 ± 0.35 |
| Mpro-bilobetin | −66.31 ± 0.16 |
| Mpro-ginkgetin | −63.62 ± 0.41 |
Among all the Mpro-biflavonoids complexes, the Mpro-bilobetin complex exhibited the highest binding free energies, while the Mpro-amentoflavone system showed the lowest binding free energies. The contribution of energy components towards binding free energy (ΔGbind) for N3, lopinavir and three biflavonoids at the binding site of SARS CoV-2 Mpro is presented in Supplemental Table 1. The van der Waals energy term (ΔGbind-vdW) and coulombic energy term (ΔGbind-Coulomb) for amentoflavone were found to be −48.67 and −19.22 kcal/mol, respectively. Similarly, the ΔGbind-vdW and ΔGbind-Coulomb energy terms were found to favor the strong binding of the other two biflavonoids towards Mpro (Supplemental Table 1). The non-polar salvation energy (ΔGbind-Sol SA) term was ranging from −4.11 kcal/mol to −6.08 kcal/mol which indicated some contribution of SASA towards these three biflavonoids binding at the binding pocket of Mpro. Even, the energy term due to H-bond also had some contribution towards the binding of biflavonoids to Mpro. Therefore, it can be suggested that van der Waals energy term (ΔGbind-vdW) and coulombic energy term (ΔGbind-Coulomb) are the key contributing factors for thermodynamically stable binding of these three biflavonoids at the Mpro binding pocket.
In order to rank different proposed SARS CoV-2 Mpro inhibitors, the binding free energy of these inhibitors was compared with the ΔGbind of “Mpro-N3 interaction” (Alamri et al., 2020; Ghosh et al., 2020a, 2020b; Kumar et al., 2020; Tripathi et al., 2020). Many proposed Mpro inhibitors including phytochemicals (such as withanone, caffeic acid, epigallocatechin gallate, epicatechingallate, gallocatechin-3-gallate etc.), antiviral drugs (such as ritonavir, paritaprevir, simeprevir, etc) were observed with lower binding free energy than N3 (Alamri et al., 2020; Ghosh et al., 2020a, 2020b; Kumar et al., 2020; Tripathi et al., 2020). Even, in this manuscript, we also observed that the lopinavir (the previously recommended antiviral drug for COVID-19 treatment) also exhibited a lesser ΔGbind value than N3 (Table 6). Very few reports are available in the literature where investigators identified potent SARS CoV-2 Mpro inhibitors having binding free energy more than N3 (Choudhury, 2020; Mittal et al., 2020). Asthama and coworkers have identified six potential molecules as SARS Mpro inhibitors (Leupeptin Hemisulphate, Pepstatin A, Nelfinavir, Birinapant, Lypression and Octreotide) which have shown the higher MM-GBSA score/binding free energy than N3 (Mittal et al., 2020). In another independent study, a researcher has proposed 17 potential binders (named as MP-In1 to MP-In17) of SARS CoV-2 Mpro, but only three of those (MP-In1 to MP-In3) have exhibited higher binding free energy values than N3 (Choudhury, 2020). Interestingly, our MM-GBSA analysis also revealed that the binding free energy of all the Mpro-biflavonoid interactions i.e. “Mpro-amentoflavone interaction”, “Mpro-bilobetin interaction” and “Mpro-ginkgetin interaction” are higher than both the “Mpro-N3 interaction” and “Mpro-lopinavir interaction”. Thus, it can be concluded from these findings that these three biflavonoids may be more effective SARS CoV-2 Mpro inhibitors than N3 (the well-known Mpro inhibitor). The relatively higher binding free energy and reasonably higher ki (inhibition constant) exhorted by these three Torreya nucifera biflavonoids made then promising SARS CoV-2 Mpro inhibitors.
4. Conclusion
In this study, we decided to work with eight diterpenoids and four biflavonoids of Torreya nucifera, which were potent SARS CoV-1 Mpro inhibitors. Among them, a single diterpenoid (hinokiol) and three biflavonoids (amentoflavone, bilobetin and ginkgetin) were found to be safe for human use based on their pharmacokinetic properties. The inhibition potency of these four phytochemicals against SARS CoV-2 Mpro was examined using a computational approach. Molecular docking studies were conducted so as to compare their binding affinities (towards Mpro) with two well-known Mpro inhibitors (N3 and lopinavir). Only three biflavonoids [amentoflavone (ki = 0.17 µM), bilobetin (ki = 0.21 µM) and ginkgetin (ki = 0.26 µM)] among the four phytochemicals had higher binding affinities than the N3 and lopinavir. Their interaction with both the key catalytic residues (His41 and Cys145) of Mpro was also evidenced. MD trajectories corresponding to these three Mpro-biflavonoid complexes and three other systems [unligated Mpro, Mpro-N3 and Mpro-lopinavir] were further analyzed with the aid of RMSD, RMSF, Rg and SASA. These analyses revealed that these three Mpro-biflavonoid complexes are highly stable and suffer less conformational fluctuations than that of the Mpro-N3 complex and/or Mpro-lopinavir complex. All these three Mpro-biflavonoid systems shared almost similar degree of compactness and these complexes were slightly more expanded compared to that of the Mpro-N3/lopinavir system. The binding of these three biflavonoids did not alter the conformation of Mpro. H-bond analysis as well as MM-GBSA analysis reconfirmed the high stability of these three Mpro-biflavonoid complexes. Overall, this study showed that three important biflavonoids of Torreya nucifera leaves (amentoflavone, bilobetin and ginkgetin) can act as SARS CoV-2 Mpro inhibitors. These proposed molecules might be useful in controlling COVID-19, but the experimental validation is highly essential.
Supplementary Material
Acknowledgements
RG acknowledges IIT Bhubaneswar for providing fellowship. The authors thank IIT Delhi HPC facility for computational resources.
Glossary
Abbreviations
- COVID-19
corona virus disease 2019
- SARS CoV-2
severe acute respiratory syndrome corona virus-2
- Mpro
main protease
- MD
molecular dynamics
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- Rg
radius of gyration
- SASA
solvent accessible surface area
Disclosure statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Adnan, M., Rasul, A., Hussain, G., Shah, M. A., Zahoor, M. K., Anwar, H., Sarfraz, I., Riaz, A., Manzoor, M., Adem, S., & Selamoglu, Z. (2020). Ginkgetin: A natural biflavone with versatile pharmacological activities. Food and Chemical Toxicology: An International Journal Published for the British Industrial Biological Research Association, 145, 111642. 10.1016/j.fct.2020.111642 [DOI] [PubMed] [Google Scholar]
- Agudelo-Gómez, L. S., Betancur-Galvis, L., & González, M. A. (2012). Anti HHV-1 and HHV-2 activity in vitro of abietic and dehydroabietic acid derivatives. Pharmacologyonline, 1(1), 36–42. [Google Scholar]
- Alamri, M. A., Tahir Ul Qamar, M., Mirza, M. U., Bhadane, R., Alqahtani, S. M., Muneer, I., Froeyen, M., & Salo-Ahen, O. M. H. (2020). Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CL(pro). Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1782768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Baby, K., Maity, S., Mehta, C. H., Suresh, A., Nayak, U. Y., & Nayak, Y. (2020). Targeting SARS-CoV-2 main protease: A computational drug repurposing study. Archives of Medical Research, 1–10. 10.1016/j.arcmed.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, B. R., Shin, B., Choi, Y., Park, S., & Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal, 18, 784–790. 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. v., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., & Kang, S. G. (2020). Computational insights into tetracyclines as inhibitors against SARS-CoV-2 Mpro via combinatorial molecular simulation calculations. Life Sciences, 257, 118080. 10.1016/j.lfs.2020.118080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, S., Patel, T., Gaikwad, R., Patil, U. K., & Gayen, S. (2019). Identification of structural requirements and prediction of inhibitory activity of natural flavonoids against Zika virus through molecular docking and Monte Carlo based QSAR Simulation. Natural Product Research, 33(6), 851–857. 10.1080/14786419.2017.1413574 [DOI] [PubMed] [Google Scholar]
- Biovia, D. S. (2017). Discovery studio modeling environment. Release.
- Chakraborty, A., Nandi, S. K., Panda, A. K., Mahapatra, P. P., Giri, S., & Biswas, A. (2018). Probing the structure-function relationship of Mycobacterium leprae HSP18 under different UV radiations. International Journal of Biological Macromolecules, 119, 604–616. 10.1016/j.ijbiomac.2018.07.151 [DOI] [PubMed] [Google Scholar]
- Chen, J. (2016). Drug resistance mechanisms of three mutations V32I, I47V and V82I in HIV-1 protease toward inhibitors probed by molecular dynamics simulations and binding free energy predictions. RSC Advances, 6(63), 58573–58585. 10.1039/C6RA09201B [DOI] [Google Scholar]
- Chen, J., Wang, X., Zhu, T., Zhang, Q., & Zhang, J. Z. (2015). A comparative insight into amprenavir resistance of mutations V32I, G48V, I50V, I54V, and I84V in HIV-1 protease based on thermodynamic integration and MM-PBSA methods. Journal of Chemical Information and Modeling, 55(9), 1903–1913. 10.1021/acs.jcim.5b00173 [DOI] [PubMed] [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Qiu, Y., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (London, England), 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, K. C., Wei, D. Q., & Zhong, W. Z. (2003). Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochemical and Biophysical Research Communications, 308(1), 148–151. 10.1016/s0006-291x(03)01342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, C. (2020). Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1771424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulerie, P., Nour, M., Maciuk, A., Eydoux, C., Guillemot, J. C., Lebouvier, N., Hnawia, E., Leblanc, K., Lewin, G., Canard, B., & Figadere, B. (2013). Structure-activity relationship study of biflavonoids on the Dengue virus polymerase DENV-NS5 RdRp. Planta Medica, 79(14), 1313–1318. 10.1055/s-0033-1350672 [DOI] [PubMed] [Google Scholar]
- Cucinotta, D., & Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Bio-Medica: Atenei Parmensis, 91(1), 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W., Zhang, B., Jiang, X. M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F., Li, C., Li, Y., Bai, F., Wang, H., Cheng, X., Cen, X., Hu, S., Yang, X., Wang, J., … Liu, H. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.), 368(6497), 1331–1335. 10.1126/science.abb4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, B. K., Pv, P., & Chakraborty, D. (2019). Computational insights into factor affecting the potency of diaryl sulfone analogs as Escherichia coli dihydropteroate synthase inhibitors. Computational Biology and Chemistry, 78, 37–52. 10.1016/j.compbiolchem.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H. R., Becker, S., Rabenau, H., Panning, M., Kolesnikova, L., Fouchier, R. A., Berger, A., Burguiere, A. M., Cinatl, J., Eickmann, M., Escriou, N., Grywna, K., Kramme, S., Manuguerra, J. C., Muller, S., … Doerr, H. W. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. The New England Journal of Medicine, 348(20), 1967–1976. 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fan, K., Wei, P., Feng, Q., Chen, S., Huang, C., Ma, L., Lai, B., Pei, J., Liu, Y., Chen, J., & Lai, L. (2004). Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. The Journal of Biological Chemistry, 279(3), 1637–1642. 10.1074/jbc.M310875200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M., Clemente, F., Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., & Zhe, G. (2016). Gaussian 09, Revision A. 01. Gaussian, Inc. [Google Scholar]
- Fritz, D., Venturi, C. R., Cargnin, S., Schripsema, J., Roehe, P. M., Montanha, J. A., & von Poser, G. L. (2007). Herpes virus inhibitory substances from Hypericum connatum Lam., a plant used in southern Brazil to treat oral lesions. Journal of Ethnopharmacology, 113(3), 517–520. 10.1016/j.jep.2007.07.013 [DOI] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020. a). Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1779818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020. b). Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1802347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorla, U. S., Rao, G. K., Kulandaivelu, U. S., Alavala, R. R., & Panda, S. P. (2020). Lead finding from selected flavonoids with antiviral (SARS-CoV-2) potentials against COVID-19: An in-silico evaluation. Combinatorial Chemistry & High Throughput Screening. 10.2174/1386207323999200818162706 [DOI] [PubMed] [Google Scholar]
- Gurung, A. B., Ali, M. A., Lee, J., Farah, M. A., & Al-Anazi, K. M. (2020). Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sciences, 255, 117831. 10.1016/j.lfs.2020.117831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Melim, L., Federico, L. B., de Oliveira, N. K. S., Francisco, V. C. C., Correia, L. C., de Lima, H. B., Gomes, S. Q., Barcelos, M. P., Francischini, I. A. G., & da Silva, C. (2020). Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sciences, 256, 117963. 10.1016/j.lfs.2020.117963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakmi, M., Bouricha, E. M., Kandoussi, I., Harti, J. E., & Ibrahimi, A. (2020). Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation, 16(4), 301–306. 10.6026/97320630016301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., Hayashi, T., & Morita, N. (1992). Mechanism of action of the antiherpesvirus biflavone ginkgetin. Antimicrobial Agents and Chemotherapy, 36(9), 1890–1893. 10.1128/aac.36.9.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi, A., & Ziebuhr, J. (2002). Conservation of substrate specificities among coronavirus main proteases. The Journal of General Virology, 83(Pt 3), 595–599. 10.1099/0022-1317-83-3-595 [DOI] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472 [Google Scholar]
- Hilgenfeld, R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, T., Wang, J., Li, Y., & Wang, W. (2011). Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of Chemical Information and Modeling, 51(1), 69–82. 10.1021/ci100275a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M. F., Kuo, C. J., Chang, K. T., Chang, H. C., Chou, C. C., Ko, T. P., Shr, H. L., Chang, G. G., Wang, A. H., & Liang, P. H. (2005). Mechanism of the maturation process of SARS-CoV 3CL protease. The Journal of Biological Chemistry, 280(35), 31257–31266. 10.1074/jbc.M502577200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. T., & Mubarak, M. S. (2020). Diterpenes and their derivatives as promising agents against dengue virus and dengue vectors: A literature-based review. Phytotherapy Research: PTR, 34(4), 674–684. 10.1002/ptr.6562 [DOI] [PubMed] [Google Scholar]
- Jimenez-Alberto, A., Ribas-Aparicio, R. M., Aparicio-Ozores, G., & Castelan-Vega, J. A. (2020). Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Computational Biology and Chemistry, 88, 107325. 10.1016/j.compbiolchem.2020.107325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jin, Z., Zhao, Y., Sun, Y., Zhang, B., Wang, H., Wu, Y., Zhu, Y., Zhu, C., Hu, T., Du, X., Duan, Y., Yu, J., Yang, X., Yang, X., Yang, K., Liu, X., Guddat, L. W., Xiao, G., Zhang, L., Yang, H., & Rao, Z. (2020). Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nature Structural & Molecular Biology, 27(6), 529–532. 10.1038/s41594-020-0440-6 [DOI] [PubMed] [Google Scholar]
- Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., & Chandra, S. (2020). In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. European Review for Medical and Pharmacological Sciences, 24(8), 4529–4536. 10.26355/eurrev_202004_21036 [DOI] [PubMed] [Google Scholar]
- Joshi, T., Sharma, P., Joshi, T., Pundir, H., Mathpal, S., & Chandra, S. (2020). Structure-based screening of novel lichen compounds against SARS Coronavirus main protease (Mpro) as potentials inhibitors of COVID-19. Molecular Diversity, 1–19. 10.1007/s11030-020-10118-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Liu, H., Galasiti Kankanamalage, A. C., Weerasekara, S., Hua, D. H., Groutas, W. C., Chang, K. O., & Pedersen, N. C. (2016). Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathogens, 12(3), e1005531. 10.1371/journal.ppat.1005531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., Dhanjal, J. K., Kaul, S. C., Wadhwa, R., & Sundar, D. (2020). Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (M(pro)) of SARS-CoV-2 and inhibit its activity. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. P., Lan, K. L., Liao, S. X., Huang, Y. H., Hou, M. C., & Lan, K. H. (2018). Inhibitory effects of amentoflavone and orobol on daclatasvir-induced resistance-associated variants of Hepatitis C Virus. The American Journal of Chinese Medicine, 46(4), 835–852. 10.1142/S0192415X18500441 [DOI] [PubMed] [Google Scholar]
- Lin, Y. M., Anderson, H., Flavin, M. T., Pai, Y. H., Mata-Greenwood, E., Pengsuparp, T., Pezzuto, J. M., Schinazi, R. F., Hughes, S. H., & Chen, F. C. (1997). In vitro anti-HIV activity of biflavonoids isolated from Rhus succedanea and Garcinia multiflora. Journal of Natural Products, 60(9), 884–888. 10.1021/np9700275 [DOI] [PubMed] [Google Scholar]
- Lin, Y. M., Flavin, M. T., Schure, R., Chen, F. C., Sidwell, R., Barnard, D. L., Huffman, J. H., & Kern, E. R. (1999). Antiviral activities of biflavonoids. Planta Medica, 65(2), 120–125. 10.1055/s-1999-13971 [DOI] [PubMed] [Google Scholar]
- Ma, S. C., But, P. P., Ooi, V. E., He, Y. H., Lee, S. H., Lee, S. F., & Lin, R. C. (2001). Antiviral amentoflavone from Selaginella sinensis. Biological & Pharmaceutical Bulletin, 24(3), 311–312. 10.1248/bpb.24.311 [DOI] [PubMed] [Google Scholar]
- Macchiagodena, M., Pagliai, M., & Procacci, P. (2020). Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling . Chemical Physics Letters, 750, 137489. 10.1016/j.cplett.2020.137489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzini, S., Musso, L., Dallavalle, S., & Artali, R. (2020). Putative SARS-CoV-2 M(pro) inhibitors from an in-house library of natural and nature-inspired products: A virtual screening and molecular docking study. Molecules, 25(16), 3745. 10.3390/molecules25163745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, K., Nagai, T., Suzuki, K., Tsujimura, R., Koyama, K., Kinoshita, K., Furuhata, K., Yamada, H., & Takahashi, K. (2007). Anti-influenza virus activity of biflavonoids. Bioorganic & Medicinal Chemistry, 17(3), 772–775. 10.1016/j.bmcl.2006.10.075 [DOI] [PubMed] [Google Scholar]
- Mirza, M. U., & Froeyen, M. (2020). Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis, 10(4), 320–328. 10.1016/j.jpha.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1768151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., & Kollman, P. A. (1992). Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry, 13(8), 952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., & Olson, A. J. (2008). Using AutoDock for ligand-receptor docking. Current Protocols in Bioinformatics, 24(1), 8–14. 10.1002/0471250953.bi0814s24 [DOI] [PubMed] [Google Scholar]
- Muramatsu, T., Takemoto, C., Kim, Y. T., Wang, H., Nishii, W., Terada, T., Shirouzu, M., & Yokoyama, S. (2016). SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proceedings of the National Academy of Sciences of the United States of America, 113(46), 12997–13002. 10.1073/pnas.1601327113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, S. K., Chakraborty, A., Panda, A. K., & Biswas, A. (2016). Conformational perturbation, hydrophobic interactions and oligomeric association are responsible for the enhanced chaperone function of Mycobacterium leprae HSP18 under pre-thermal condition. RSC Advances, 6(67), 62146–62156. 10.1039/C6RA00167J [DOI] [Google Scholar]
- Odhar, H. A., Ahjel, S. W., Albeer, A., Hashim, A. F., Rayshan, A. M., & Humadi, S. S. (2020). Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation, 16(3), 236–244. 10.6026/97320630016236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink, C., Villa, A., Mark, A. E., & van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry, 25(13), 1656–1676. 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- Orhan, I. E., & Senol Deniz, F. S. (2020). Natural products as potential leads against Coronaviruses: Could they be encouraging structural models against SARS-CoV-2? Natural Products and Bioprospecting, 10(4), 171–186. 10.1007/s13659-020-00250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pathak, R. K., Baunthiyal, M., Taj, G., & Kumar, A. (2014). Virtual screening of natural inhibitors to the predicted HBx protein structure of Hepatitis B Virus using molecular docking for identification of potential lead molecules for liver cancer. Bioinformation, 10(7), 428–435. 10.6026/97320630010428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah, R., & Suresh, P. C. (2013). Molecular docking studies of phytochemical compounds with viral proteases. International Journal of Pharmaceutical Sciences and Research, 4(1), 475. [Google Scholar]
- Ren, L. L., Wang, Y. M., Wu, Z. Q., Xiang, Z. C., Guo, L., Xu, T., Jiang, Y. Z., Xiong, Y., Li, Y. J., Li, X. W., Li, H., Fan, G. H., Gu, X. Y., Xiao, Y., Gao, H., Xu, J. Y., Yang, F., Wang, X. M., Wu, C., … Wang, J. W. (2020). Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chinese Medical Journal, 133(9), 1015–1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa-Linares, V. C., Brand, Y. M., Agudelo-Gomez, L. S., Tangarife-Castano, V., Betancur-Galvis, L. A., Gallego-Gomez, J. C., & Gonzalez, M. A. (2016). Anti-herpetic and anti-dengue activity of abietane ferruginol analogues synthesized from (+)-dehydroabietylamine. European Journal of Medicinal Chemistry, 108, 79–88. 10.1016/j.ejmech.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J. L., Chen, Q., Wang, D., Erdman, D. D., Peret, T. C., … Bellini, W. J. (2003). Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science (New York, N.Y.), 300(5624), 1394–1399. 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J. Y., Kim, D., Nguyen, T. T., Park, S. J., Chang, J. S., Park, K. H., Rho, M. C., & Lee, W. S. (2010). Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorganic & Medicinal Chemistry, 18(22), 7940–7947. 10.1016/j.bmc.2010.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed, A. M., Khattab, A. R., AboulMagd, A. M., Hassan, H. M., Rateb, M. E., Zaid, H., & Abdelmohsen, U. R. (2020). Nature as a treasure trove of potential anti-SARS-CoV drug leads: A structural/mechanistic rationale. RSC Advances, 10(34), 19790–19802. 10.1039/D0RA04199H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf, A. W., & van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Tripathi, M. K., Singh, P., Sharma, S., Singh, T. P., Ethayathulla, A. S., & Kaur, P. (2020). Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1790425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181(2), 281–292. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.-C., Kuo, Y.-H., Jan, J.-T., Liang, P.-H., Wang, S.-Y., Liu, H.-G., Lee, C.-K., Chang, S.-T., Kuo, C.-J., Lee, S.-S., Hou, C.-C., Hsiao, P.-W., Chien, S.-C., Shyur, L.-F., & Yang, N.-S. (2007). Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. Journal of Medicinal Chemistry, 50(17), 4087–4095. 10.1021/jm070295s [DOI] [PubMed] [Google Scholar]
- Wilsky, S., Sobotta, K., Wiesener, N., Pilas, J., Althof, N., Munder, T., Wutzler, P., & Henke, A. (2012). Inhibition of fatty acid synthase by amentoflavone reduces coxsackievirus B3 replication. Archives of Virology, 157(2), 259–269. 10.1007/s00705-011-1164-z [DOI] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Xie, W., Xue, X., Yang, K., Ma, J., Liang, W., Zhao, Q., Zhou, Z., Pei, D., Ziebuhr, J., Hilgenfeld, R., Yuen, K. Y., Wong, L., Gao, G., Chen, S., Chen, Z., Ma, D., Bartlam, M., & Rao, Z. (2005). Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biology, 3(10), e324. 10.1371/journal.pbio.0030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367(19), 1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- Zhang, J., & Wang, Y. (2020). Bilobetin, a novel small molecule inhibitor targeting influenza virus polymerase acidic (PA) endonuclease was screened from plant extracts. Natural Product Research, 1–4. 10.1080/14786419.2020.1808636 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W., Zhong, Z., Xie, X., Yu, Q., & Liu, J. (2020). Relation between chest CT findings and clinical conditions of Coronavirus Disease (COVID-19) pneumonia: A multicenter study. AJR. American Journal of Roentgenology, 214(5), 1072–1077. 10.2214/AJR.20.22976 [DOI] [PubMed] [Google Scholar]
- Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y., Shen, X. R., Wang, X., … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.