Abstract
Recently long non-coding RNAs (lncRNAs) were highlighted for their regulatory role in tumor biology. The novel human lncRNA cancer susceptibility candidate 2 (CASC2) has been characterized as a potential tumor suppressor in several tumor types. However, the roles of CASC2 and its interplay with miR-21 in different malignancy grade patient gliomas remain unexplored. Here we screened 99 different malignancy grade astrocytomas for CASC2, and miR-21 gene expression by real-time quantitative polymerase chain reaction (RT-qPCR) in isocitrate dehydrogenase 1 (IDH1) and O-6-methylguanine methyltransferase (MGMT) assessed gliomas. CASC2 expression was significantly downregulated in glioblastomas (p = 0.0003). Gliomas with low CASC2 expression exhibited a high level of miR-21, which was highly associated with the higher glioma grade (p = 0.0001), IDH1 wild type gliomas (p < 0.0001), and poor patient survival (p < 0.001). Taken together, these observations suggest that CASC2 acts as a tumor suppressor and potentially as a competing endogenous RNA (ceRNA) for miR-21, plays important role in IDH1 wild type glioma pathogenesis and patients’ outcomes.
Keywords: CASC2, miR-21, glioma, IDH1 status, patient survival
1. Introduction
Malignant gliomas, especially glioblastomas, are highly infiltrative, rapidly growing, aggressive, heterogeneous, chemo-resistant, and lethal neoplasms [1]. The accurate distinction between the different malignancy types has significant prognostic and therapeutic implications [2]. A thorough study of the molecular mechanisms of the formation and progression of glioma is essential for the screening of valuable diagnostic and prognostic molecular markers. Long non-coding RNAs (lncRNAs) were first recognized as being crucial regulators of gene expression in a wide range of biological context, including cancer [3]. Various lncRNAs, including homeobox (HOX) transcript antisense RNA (HOTAIR), metastasis associated lung adenocarcinoma transcript (MALAT), colorectal neoplasia differentially expressed (CRNDE), have been identified as novel players in glioma pathogenesis demonstrating associations with tumor subtype, histological stage, tumor isocitrate dehydrogenase (IDH) mutational status, chemosensitivity, and patient survival [4,5,6,7,8].
Steadily growing evidence on the ability of lncRNAs to interact with DNA, RNA, and proteins acting as tethers, guides, decoys, and scaffolds, includes them in the posttranscriptional regulatory network in cancer biology [9]. Moreover, the increasing evidence suggests an interplay between microRNAs and lncRNAs [10,11]. A large number of lncRNAs act as a competing endogenous RNAs (ceRNA) or sponges for microRNAs, for example, phosphatase and tensin homolog pseudogene 1 (PTENpg1), HOTAIR [11,12]. The lncRNA gene cancer susceptibility gene 2 (CASC2) has been characterized as tumor suppressor in various human malignancies [13,14,15,16,17,18,19]. Although the deregulated expression of CASC2 in cancer enhances its tumorigenic properties, however, the literature evidence limits current knowledge on the pathophysiological implications and the roles of CASC2, and its interplay with miR-21 in the pathology of gliomas [20].
In this study, we assessed levels of CASC2 and miR-21 and their interplay in different grades of glioma. Our findings indicate that CASC2 was proportionally downregulated in progressed gliomas, while miR-21 expression was inversely associated with CASC2 expression, malignancy grade, and patient survival. Here we demonstrate CASC2 acting as a tumor suppressor and likely interacting with miR-21 in IDH1 wild type gliomas.
2. Results
2.1. CASC2 and miR-21 Associations with Patient Clinical Parameters
To evaluate whether CASC2 and miR-21 were associated with glioma patient clinical parameters, we divided the samples into “low” and “high” (below and above the gene’s mean expression of all samples, respectively) gene expression groups. The threshold for “low” and “high” expression group of CASC2 and miR-21 was −5.865 and 3.448 (log2(2^−∆Ct)), respectively. As shown in Table 1, lower CASC2 and higher miR-21 expression were observed more frequently in patients with advanced tumor stage (IV grade gliomas/glioblastomas) (p < 0.0001). Furthermore, IDH1 wild-type gliomas more frequently had lower CASC2 and higher miR-21 expression (p = 0.037 and p < 0.0001, respectively).
Table 1.
The relationship between cancer susceptibility gene 2 (CASC2) and miR-21 gene expression in glioma tissue and patient clinical characteristics. Pearson’s χ2-test was used for comparison of categorical variables.
| Variable | Total No | CASC2 Expression | Total No | miR-21 Expression | ||||
|---|---|---|---|---|---|---|---|---|
| Low (%) | High (%) | p-Value | Low (%) | High (%) | p-Value | |||
| Gender | ||||||||
| Male | 45 | 25 (55.6) | 20 (44.4) | 0.422 | 37 | 13 (35.1) | 24 (64.9) | 0.182 |
| Female | 54 | 25 (46.3) | 29 (53.7) | 46 | 24 (52.2) | 22 (47.8) | ||
| Age, yr | ||||||||
| <56 | 47 | 23 (48.9) | 24 (51.1) | 0.841 | 42 | 24 (57.1) | 18 (42.9) | 0.027 |
| ≥56 | 52 | 27 (51.9) | 25 (48.1) | 41 | 13 (31.7) | 28 (68.3) | ||
| Grade | ||||||||
| II-III | 17 | 2 (11.8) | 15 (88.2) | <0.0001 | 17 | 16 (94.1) | 1 (5.9) | <0.0001 |
| IV | 82 | 48 (58.5) | 34 (41.5) | 66 | 21 (31.8) | 45 (68.2) | ||
| IDH1 | ||||||||
| Wt | 78 | 44 (56.4) | 34 (43.6) | 0.037 | 64 | 21 (32.8) | 43 (67.2) | <0.0001 |
| Mut | 18 | 5 (27.8) | 13 (72.2) | 16 | 14 (87.5) | 2 (12.5) | ||
| MGMT | ||||||||
| Unmeth | 48 | 25 (52.1) | 23 (47.9) | 1 | 40 | 16 (40) | 24 (60) | 0.812 |
| Meth | 41 | 22 (53.7) | 19 (46.3) | 33 | 15 (45.5) | 18 (54.5) | ||
| miR-21 | ||||||||
| low | 37 | 11 (29.7) | 26 (70.3) | 0.002 | ||||
| high | 46 | 30 (62.2) | 16 (34.8) | |||||
2.2. CASC2 and miR-21 Expression in High Grade and IDH1wt Gliomas
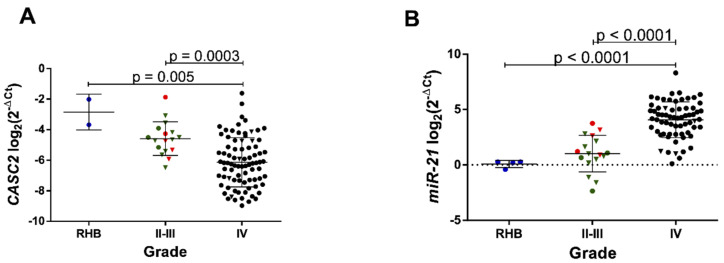
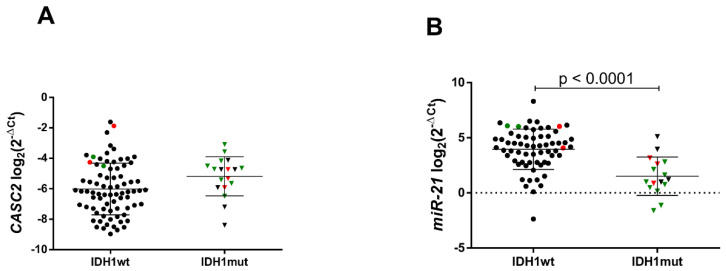
Whether the activity of CASC2 and miR-21 was linked to the clinical progression of gliomas, we examined gene expression in grade II–III and IV gliomas. Here we show significant CASC2 expression loss and drastic rise of miR-21 expression in glioblastomas as compared to the average expression levels in lower grade (II–III) gliomas (p = 0.0003 and p < 0.0001, respectively) and control non-cancerous brain tissues (p = 0.005 and p < 0.0001, respectively) (Figure 1A,B). When all samples were divided into IDH1 gene mutated (IDH1mut, n = 18) and IDH1 wild-type (IDH1wt, n = 78) gliomas, we observed a tendency of lower expression of CASC2 in IDH1wt (p = 0.053) and highly significant relationship between higher miR-21 expression and IDH1wt gliomas (p < 0.0001) (Figure 2A,B).
Figure 1.
Cancer susceptibility gene 2 (CASC2) and miR-21 gene expression are associated with glioma malignancy grade. (A) CASC2 expression measured by RT-qPCR in RHB (reference human brain, n = 2), II-III malignancy grade gliomas (n = 17) and IV grade gliomas, n = 82 (glioblastomas). (B) miR-21 expression measured in the same patient postoperative tumor tissue by RT-qPCR in RHB (n = 4), II-III (n = 17) and IV grade (n = 66) gliomas. The lines in the graphs indicate mean with the SD. Color corresponds to different glioma malignancy grade: green—grade II, red—grade III, black—grade IV gliomas. Triangle shape corresponds to isocitrate dehydrogenase 1 mutated C. 395G > A (IDH1mut) glioma, circle shape—isocitrate dehydrogenase 1 wild-type (IDH1wt).
Figure 2.
CASC2 (A) and miR-21 (B) gene expression in IDH1wt and IDH1mut gliomas. The lines in the graphs indicate mean with the SD. Color corresponds to different glioma malignancy grade: green reflects grade II, red—grade III, black—grade IV gliomas. Triangle shape corresponds to isocitrate dehydrogenase 1 mutated C. 395G > A (IDH1mut) glioma, circle shape—isocitrate dehydrogenase 1 wild-type (IDH1wt).
2.3. CASC2 and miR-21 Interplay in Gliomas
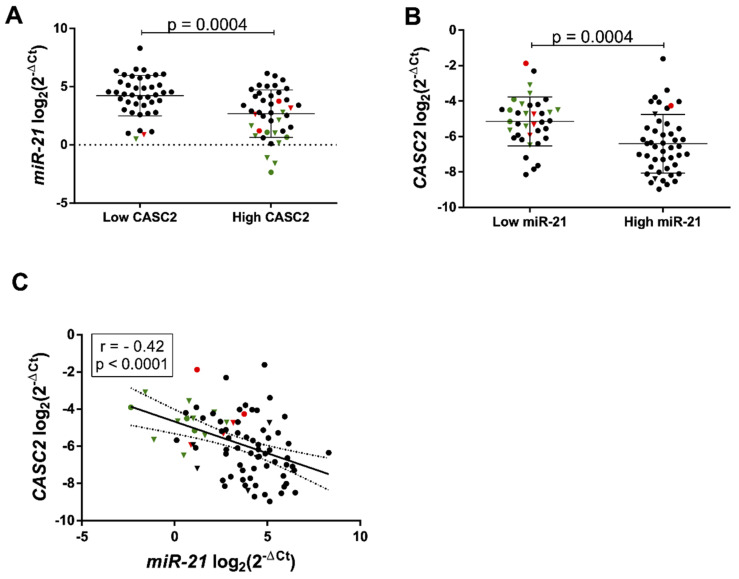
Several reports have suggested that lncRNAs may function as a molecular sponge or competing endogenous RNA in modulating miRNAs, suggesting that it could be an inverse correlation between lncRNA and miRNAs [21]. It was shown that miR-21 can bind to CASC2 directly by the putative miRNA response element (MRE) [20]. Here at the clinical level, we further confirm the recently reported interaction between CASC2 and miR-21 in glioma cell lines. We show that significantly higher miR-21 gene expression was observed in the “low” CASC2 group as compared to “high” and vice versa (p = 0.0004, Figure 3A,B). Correlation analysis revealed moderate negative association between CASC2 and miR-21 expression in gliomas (r2 = −0.42, n = 83, p < 0.0001, Figure 3C).
Figure 3.
CASC2 and miR-21 interplay in gliomas. (A) miR-21 gene expression in low (n = 41) and high (n = 42) CASC2 expression groups in all patient gliomas. (B) CASC2 gene expression in low (n = 37) and high (n = 46) miR-21 gene expression groups in all patient gliomas. (C) Expression correlation between CASC2 and miR-21 in gliomas (r = −0.42, p < 0.0001, n = 83) visualized as a scatter plot. The lines in the graphs indicate mean with the SD. Color corresponds to different glioma malignancy grade: green reflects grade II, red—grade III, black—grade IV gliomas. Triangle shape corresponds to IDH1mut glioma, circle shape—IDH1wt.
2.4. Survival Analysis
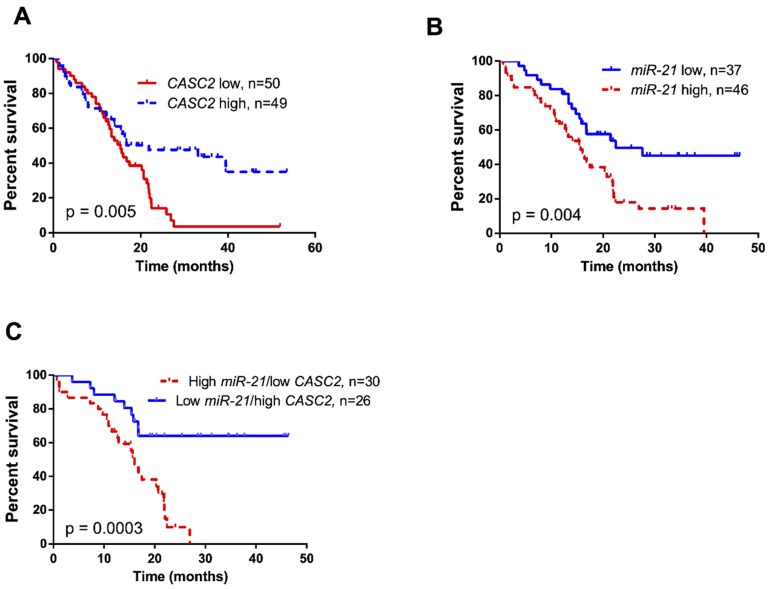
Kaplan–Meier survival analysis showed highly significant association between low CASC2 expression levels (n = 50) and worse patient outcome (Log-rank test, χ2 = 7.777, df = 1, p = 0.0053; Figure 4A), while patients with low miR-21 expression (n = 37) showed significantly increased overall survival, compared to patients with high miR-21 expression (n = 46) (Log-rank test, χ2 = 8.518, df = 1, p = 0.0035; Figure 4B). The combined effect of low CASC2 and high miR-21 expression (n = 30) in glioma was shown to be associated with significantly decreased overall survival compared to patients with the combination of high CASC2 and low miR-21 expression (n = 26) in tumor tissue (Log-rank test, χ2 = 12.91, df = 1, p = 0.0003; Figure 4C). Univariate Cox regression model revealed that patients’ clinical characteristics such as age and tumor stage, IDH1 status was associated with their survival as well as CASC2, miR-21, and combined CASC2/miR-21 expression. However, multivariate analysis showed that only patient age and tumor stage were covariates associated with the overall survival of glioma patients (Table 2).
Figure 4.
Kaplan–Meier curves for glioma patient survival correlation with (A) CASC2 expression (Log-rank test, χ2 = 7.777, df = 1, p = 0.005), (B) miR-21 expression (Log-rank test, χ2 = 8.518, df = 1, p = 0.004), and (C) combined miR-21 and CASC2 expression (Log-rank test, χ2 = 12.91, df = 1, p = 0.0003) in glioma tumor tissue.
Table 2.
Cox regression analysis of different clinicopathological variables, CASC2 and miR-21 expression.
| Characteristics | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age (<56 vs. ≥56) | 0.216 (0.123–0.381) | <0.0001 | 0.408 (0.215–0.775) | 0.006 |
| Gender (female vs. male) | 0.868 (0.537–1.404) | 0.564 | NA | |
| Tumor grade (II–III vs. IV) | 0.069 (0.017–0.284) | <0.0001 | 0.100 (0.019–0.526) | 0.007 |
| IDH1R132H | 0.160 (0.064–0.404) | <0.0001 | 0.809 (0.244–2.682) | 0.729 |
| MGMT (methylated vs. non methylated) | 0.722 (0.435–1.199) | 0.208 | NA | |
| CASC2 | 0.497 (0.301–0.821) | 0.006 | 0.751 (0.389–1.450) | 0.393 |
| miR-21 | 2.285 (1.290–4.045) | 0.005 | 1.173 (0.584–2.358) | 0.653 |
| CASC2 high/miR-21 low | 0.259 (0.118–0.570) | 0.001 | NA |
3. Discussion
The key finding of the current study is that downregulation of lncRNA CASC2 and upregulation of miR-21 expression is associated with glioma progression. Our results show that CASC2 downregulation is associated with highly expressed miR-21 and poor patient outcome. Moreover, here we show that highly active CASC2 significantly might have suppressed miR-21 levels in IDH1 wild-type gliomas.
Current knowledge on the involvement and function of lncRNA CASC2 in glioma evidences the availability of a small amount of data from clinical samples. Downregulation of CASC2 in glioma tissue was showed by Wang et al. [20] in a limited sample of 24 patients, while Liao et al. revealed CASC2 playing a role in modulating glioma temozolomide (TMZ) chemoresistance in 57 patient samples [22]. In agreement with our data, CASC2 expression in both studies was shown to correlate with glioma malignancy grade inversely. Recently, several studies including patient samples have been carried out on CASC2 expression in other malignancies. In particular, CASC2 acts as a tumor suppressor in endometrial, colorectal, lung, stomach, renal, gastric cancers, and osteosarcomas [13,14,15,16,17,18,19]. In NSCLC (non-small-cell lung carcinoma) patients (n = 76), CASC2 expression was downregulated proportionally to the pathological stage and associated with tumor size, and this gene was an independent predictor for overall survival [13]. In 76% CRC (colorectal cancer) patients (n = 68) CASC2 low expression was associated with tumor stage [14]. In RCC (renal cell carcinoma) (n = 32), CASC2 was significantly downregulated compared with the matched normal tissue [15]. In gastric cancer tissue (n = 67) and cell lines, CASC2 expression was downregulated [17] and low CASC2 level in tissue correlated with the vessel invasion, tumor stage, metastasis, and poor patient survival [19]. In osteosarcoma, CASC2 expression downregulation was observed in patient tissue samples and cell lines, and low expression in tissue was associated with poor tumor differentiation, higher malignancy grade, and shortened patient survival [18]. To sum up, recent scientific work and our research in astrocytic gliomas support evidence that CASC2 gene expression is downregulated proportionally to tumor stage, indicating the suppressive role of CASC2 in malignancy progression. Functional studies in vitro in various cancer cell lines confirmed CASC2 acting as a tumor suppressor as when overexpressed CASC2 was able to inhibit cell proliferation, cell growth, migration and invasion, and to induce apoptosis [20].
Emerging evidence revealed a new mechanistic role of lncRNAs as part of a posttranscriptional regulatory network in cancer biology. Recent data suggest that coding and non-coding RNAs can regulate one another through their ability to compete for miRNA binding through typical MREs (miRNA response elements). LncRNAs can act as competing endogenous RNAs (ceRNA) or miRNA “sponges”, which can sequester miRNAs, therefore preventing single or multiple miRNA from binding to their proper target RNAs and protecting them from suppression [23]. Importantly, micro RNAs also regulate lncRNAs [10]. Recently, it was demonstrated that CASC2 in colorectal cancer is functioning as ceRNA for miR-18a, thereby modulating the expression of target gene PIAS3, and subsequently inhibiting CRC cell proliferation and tumor growth [14]. Studies in hepatocellular carcinoma revealed that CASC2 prohibited mesenchymal–epithelial transition progression and exerted anti-metastatic effect via CASC2/mirR-396/FBXW7 axis [9]. Wang and colleagues demonstrated CASC2 and mir-21 reciprocal interaction in glioma cell lines U251 and U87 [20]. Similarly, Liao et al. [22] study showed CASC2 interaction with miR-181a and PTEN gene in regulating chemosensitivity in temozolomide resistant glioma cells. However, as to our knowledge, no studies are demonstrating CASC2 and miR-21 interaction in patient glioma samples and evaluating its clinical relevance.
Consistent with published reports on CASC2/mir-21 interaction in glioma and non-small cell lung cancer cells in vitro [20,24], we provide evidence in patient gliomas that CASC2 and miR-21 play antagonistic roles and potentially interact in glioma progression. In support of this, the RT-qPCR analysis showed that miR-21 expression is moderately upregulated in low-grade astrocytoma and even highly upregulated in malignant glioblastoma, while high expression of CASC2 in tumors might be responsible for the decrease of miR-21 expression. Agreeing with our findings, miR-21 has been well studied in gliomas with particularly high expression. miR-21 is consistently upregulated in astrocytic tumors (grade II–IV) and downmodulates an entire set of oncosuppressor genes, for example, PTEN [25]. High miR-21 expression in tumor tissue was highly associated with aggressive clinicopathological features and poor overall patient survival (n = 152) [26]. In the current study, we found a correlation between high mir-21 expression and IDH1wt gliomas. It is known that mutation in isocitrate dehydrogenase 1 (IDH1mut) is associated with distinct glioma cell metabolic profile, hypermethylated phenotype, and significantly longer overall survival as compared to patients with IDH1wt [27,28]. Our results further indicate miR-21 predictive value in IDH1wt associated gliomagenesis.
In summary, lncRNA CASC2 was found as a tumor suppressor and downregulated in low-grade astrocytomas and highly malignant glioblastomas as compared to healthy brain tissue. miR-21 was inversely expressed with CASC2 in gliomas and correlated with IDH1wt glioma and poor patient prognosis.
4. Material and Methods
4.1. Ethics
The research was reviewed and approved by the Kaunas Regional Bioethics Committee (protocol: L6.1-07/09, permission code: P2-9/2003, date: 10 October 2010) and performed following the Lithuanian regulations alongside with the principles of the Helsinki and Taipei Declarations [29,30].
4.2. Patient Sample
Due to rare occurrence of the disease, the maximum possible number of samples were included into the study. A total of 99 samples of different malignancy grade astrocytomas were analyzed for CASC2 and miR-21 expression: 17 grade II–III astrocytic gliomas and 82 grade IV astrocytic gliomas/glioblastomas. CASC2 expression was analyzed in 99 samples, while miR-21 in 83 samples. IDH1 status was obtained for 96 patients, MGMT promoter methylation status was determined for 89 samples. Tissue samples were prospectively collected at the Department of Neurosurgery of Lithuanian University of Health Sciences, during the period of 2015–2018. The pathological review was performed on each sample to confirm the diagnosis of astrocytic glioma. All tissue samples were stored in liquid. None of the patients had received preoperative chemotherapy or radiotherapy. All patients signed written consent forms. Overall survival was calculated from the day of surgery to the death or last follow-up.
4.3. RNA and DNA Extraction
Total and small RNAs (<200 nt) were extracted from 30–40 mg snap-frozen (−196 °C) post-surgical tumor samples applying cryogenic mechanical grinding, ultrasonic homogenization at 20% amplitude for 1 s on/off pulsation and using mirVana™ miRNA Isolation Kit (Thermo Fisher Scientific, Carlsbad, CA, USA). Procedures were done according to the manufacturer’s instructions. The RNA concentration was determined using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). Quality of extracted small RNAs was evaluated with a Small RNA analysis kit (Agilent, Santa Clara, CA, USA) on a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
DNA was extracted from ≈40 mg frozen tumor tissue using the desalting method with chloroform, and Proteinase K. DNA concentration was measured with a NanoDrop 2000 system.
4.4. CASC2 Gene Expression Analysis
cDNA synthesis was performed using 2 µg of RNA, hexamer primers, “Multiscribe™ Reverse Transcriptase” reverse transcriptase, and according to the manufacturer’s recommendations, using “High-Capacity cDNA Reverse Transcription Kit” (Applied Biosystems, Foster City, CA, USA). RT-qPCR was conducted using a “AB 7500 Fast Real-time PCR system” (Applied Biosystems, Foster City, CA, USA). CASC2 gene primer sequences were as follows: forward 5‘-GCACATTGGACGGTGTTTCC-3’; reverse 5’-CCCAGTCCTTCACAGGTCAC-3’ [31]. All amplification reactions were performed in 96-well plates and each sample was tested in three replicates. For normalization, the geometric average of five housekeeping genes’ expression glyceraldehyde 3-phosphate dehydrogenase (GAPDH), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), β-actin, 18S ribosomal RNA (18s rRNA), hypoxanthine phosphoribosyltransferase 1 (HPRT1) was used. As endogenous control “FirstChoice Human Brain Reference Total RNA” (RHB) (Ambion, Austin, TX, USA) was used. In order to quantify samples in 95% of the cases, samples with a standard deviation of more than 0.25 were eliminated from the analysis. Gene expression was calculated as 2−∆Ct values and in figures presented as log-transformed values.
4.5. miR-21 Gene Expression Analysis
In total, 10 ng of purified micro RNAs was synthesized to cDNA using “TaqMan Advanced miRNA cDNA Synthesis Kit” (Thermo Fisher Scientific, Pleasanton, CA, USA). Expression profile of mature micro RNA 21 was detected performing RT-qPCR on “7500 Fast Real-Time PCR system” (Applied Biosystems, Foster City, CA, USA) in three replicates using “TaqMan Fast Advanced Master Mix” (Thermo Fisher Scientific, Austin, TX, USA) and hsa-miR-21-5p probes (Assay ID: 477975_mir). In addition, hsa-miR-191-5p (Assay ID: 477952_mir) and hsa-miR-361-5p (Assay ID: 478056_mir) were measured in order to normalize the data. Relative quantitation of hsa-miR-21-5p expression for each sample was calculated according to Equation (1):
| (1) |
4.6. IDH1 Mutation Detection
The most common IDH1 gene mutation R132H in gliomas was analyzed in all the specimens applying custom TaqMan single nucleotide polymorphism (SNP) genotyping assays. PCR was carried out in a total volume of 12 µL consisting of “TaqMan™ Universal Master Mix II” (Thermofisher Scientific, Carlsbad, CA, USA), TaqMan probes and 20 ng of purified tumor DNA. All the procedures were accomplished according to the manufacturer’s recommendations. Fluorescence was measured with “7500 Fast Real-Time PCR System” (Applied Biosystems, Foster City, CA, USA). Amplification of DNA with Wild or Mutant allele labelled with VIC or FAM dyes indicated different gene variants, respectively.
4.7. MGMT Methylation Detection
MGMT promoter methylation status was determined using methylation-specific PCR (MSP). The reaction performed in 15 µL of total volume consisted of 7.5 µL “Hot Start PCR Master Mix” (Thermofisher Scientific, Carlsbad, CA, USA); 4.5 µL nuclease-free water (Thermofisher Scientific, Carlsbad, CA, USA); 1 µL (10 pmol/µL) of each primer, specific to methylated/unmethylated promoter; ≈20 ng of bisulfite-treated DNA as a template. Primer sequences for methylated MGMT sequence were 5’-GGACGTTAAGGGTTTAGAGC-3’ (sense), 5’-CAATACACGACCTCGTCAC-3’ (antisense), for unmethylated—5’-GGATGTTAAGGGTTTAGAGT-3’ (sense), 5’-CAATACACAACCTCATCAC-3’ (antisense). In addition, three controls were performed: positive—“Bisulfite converted Universal Methylated Human DNA Standard and Control primer” (ZymoResearch, Irvine, CA, USA); negative—bisulfite treated human blood lymphocytes DNA; water control. PCR products were visualized using agarose gel electrophoresis. Each sample methylation status was evaluated according to visible signals and documented using 0 (unmethylated) and 1 (methylated) system.
4.8. Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 6.0 (San Diego, CA, USA). Continuous variables were checked for normal distribution using a Shapiro–Wilk statistics and compared by a Student’s t-test when normally distributed or by a Mann–Whitney U test when data distributed non-normally. Pearson’s correlation coefficient was calculated to test the association between different gene expression. Pearson’s chi-squared test was used for comparison of categorical variables. Kaplan–Meier curves were compared using a Log-rank analysis in different gene expression groups. For regression analysis, gene expression values were categorized as “low” or “high” according to a log-transformed gene expression values which were above or below all sample expression means, respectively. The significance level was considered when p-value < 0.05 (*).
Acknowledgments
We kindly thank Jūrate Žeglienė for help in tissue sampling and acquiring patient data, also we thank Vilnius University Master’s degree student Aistė Stefanskytė for technical support in part of CASC2 gene expression experiments.
Author Contributions
Conceptualization, D.S. and P.V.; Data curation, P.V.; Formal analysis, D.S., R.S. and G.S.; Funding acquisition, P.V.; Investigation, D.S.; Methodology, R.S. and P.V.; Project administration, P.V.; Resources, A.T.; Supervision, D.S. and A.T.; Validation, D.S. and G.S.; Visualization, G.S.; Writing—original draft, D.S.; Writing—review and editing, R.S., G.S., A.T. and P.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Research Council of Lithuania, grant number S-MIP-17-108.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cuddapah V.A., Robel S., Watkins S., Sontheimer H. A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wesseling P., Kros J.M., Jeuken J.W.M. The pathological diagnosis of diffuse gliomas: Towards a smart synthesis of microscopic and molecular information in a multidisciplinary context. Diagn. Histopathol. 2011;17:486–494. doi: 10.1016/j.mpdhp.2011.08.005. [DOI] [Google Scholar]
- 3.Liz J., Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J.-X., Han L., Bao Z.-S., Wang Y.-Y., Chen L.-Y., Yan W., Yu S.-Z., Pu P.-Y., Liu N., You Y.-P., et al. HOTAIR, a cell cycle–associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-Oncology. 2013;15:1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma K., Wang H., Li X., Li T., Su G., Yang P., Wu J. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumor Biol. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X., Sun S., Pu J.K.S., Tsang A.C.O., Lee D., Man V.O.Y., Lui W.M., Wong S.T.S., Leung G.K.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X.-Q., Kiang K.M.-Y., Wang Y.-C., Pu J.K.-S., Ho A., Cheng S.Y., Lee D., Zhang P.-D., Chen J.-J., Lui W.-M., et al. IDH1 mutation-associated long non-coding RNA expression profile changes in glioma. J. Neuro-Oncol. 2015;125:253–263. doi: 10.1007/s11060-015-1916-9. [DOI] [PubMed] [Google Scholar]
- 8.Li W., Ma X., Zhan R., Jiang P., Wang P., Sun X., Yuan Z. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. OncoTargets Ther. 2016;9:3501. doi: 10.2147/OTT.S96278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Liu Z., Yao B., Li Q., Wang L., Wang C., Dou C., Xu M., Liu Q., Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnsson P., Ackley A., Vidarsdottir L., Lui W.-O., Corcoran M., Grandér D., Morris K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X.-H., Sun M., Nie F.-Q., Ge Y.-B., Zhang E.-B., Yin D.-D., Kong R., Xia R., Lu K.-H., Li J.-H., et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X., Liu Z., Su J., Yang J., Yin D., Han L., De W., Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumor Biol. 2016;37:9503–9510. doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 14.Huang G., Wu X., Li S., Xu X., Zhu H., Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci. Rep. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Xu R., Xu X., Zhou Y., Cui L., He X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Mol. Med. Rep. 2016;14:1019–1025. doi: 10.3892/mmr.2016.5337. [DOI] [PubMed] [Google Scholar]
- 16.Baldinu P., Cossu A., Manca A., Satta M.P., Sini M.C., Palomba G., Dessole S., Cherchi P., Mara L., Tanda F., et al. CASC2 Gene is Down-regulated in Endometrial Cancer. Anticancer Res. 2007;27:235–244. [PubMed] [Google Scholar]
- 17.Li P., Xue W.-J., Feng Y., Mao Q.-S. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am. J. Transl. Res. 2016;8:3522. [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L., Dai Z., Luo Q., Lv G. The long noncoding RNA cancer susceptibility candidate 2 inhibits tumor progression in osteosarcoma. Mol. Med. Rep. 2017;17:1947–1953. doi: 10.3892/mmr.2017.8080. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., Huang H., Tong S., Huo R. Overexpression of long non-coding RNA cancer susceptibility 2 inhibits cell invasion and angiogenesis in gastric cancer. Mol. Med. Rep. 2017;16:5235–5240. doi: 10.3892/mmr.2017.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P., Liu Y., Yao Y., Li Z., Li Z., Ma J., Xue Y. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y., Shen L., Zhao H., Liu Q., Fu J., Guo Y., Peng R., Cheng L. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J. Cell. Biochem. 2017;118:1889–1899. doi: 10.1002/jcb.25910. [DOI] [PubMed] [Google Scholar]
- 23.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Zhang H., Wang X., Wang J., Wei H. Long non-coding RNA CASC2 enhanced cisplatin-induced viability inhibition of non-small cell lung cancer cells by regulating the PTEN/PI3K/Akt pathway through down-regulation of miR-18a and miR-21. RSC Adv. 2018;8:15923–15932. doi: 10.1039/C8RA00549D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng F., Henson R., Wehbe–Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L., Li G., Feng D., Qin H., Gong L., Zhang J., Zhang Z. MicroRNA-21 expression is associated with overall survival in patients with glioma. Diagn. Pathol. 2013;8:200. doi: 10.1186/1746-1596-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen A.L., Holmen S.L., Colman H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013;13:345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo C., Pirozzi C.J., Lopez G.Y., Yan H. Isocitrate dehydrogenase mutations in gliomas. Curr. Opin. Neurol. 2011;24:648–652. doi: 10.1097/WCO.0b013e32834cd415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Medical Association World Medical Association declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA J. Am. Med. Assoc. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 30.Assembly W.M.A.G., Assembly W.M.A.G., Declaration T., Declaration T., Databases H., Declaration T., Database H. Annexe 2. WMA Declaration of Taipei on ethical considerations regarding health databases and biobanks. J. Int. Bioéthique D’éthique Sci. 2017;28:113. doi: 10.3917/jib.283.0113. [DOI] [PubMed] [Google Scholar]
- 31.Pei Z., Du X., Song Y., Fan L., Li F., Gao Y., Wu R., Chen Y., Li W., Zhou H., et al. Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:18145–18153. doi: 10.18632/oncotarget.15210. [DOI] [PMC free article] [PubMed] [Google Scholar]