Summary
To date, it remains unclear if there are specific cell-surface markers for purifying glucose-responsive pancreatic β-like cells derived from human pluripotent stem cells (hPSCs). In searching for this, we generated an efficient protocol for differentiating β-like cells from human embryonic stem cells. We performed single-cell RNA sequencing and found that CD9 is a negative cell-surface marker of β-like cells, as most INS+ cells are CD9−. We purified β-like cells for spontaneous formation of islet-like organoids against CD9, and found significantly more NKX6.1+MAFA+C-PEPTIDE+ β-like cells in the CD9− than in the CD9+ population. CD9− cells also demonstrate better glucose responsiveness than CD9+ cells. In humans, we observe more CD9+C-PEPTIDE+ β cells in the fetal than in the adult cadaveric islets and more Ki67+ proliferating cells among CD9+ fetal β cells. Taken together, our experiments show that CD9 is a cell-surface marker for negative enrichment of glucose-responsive β-like cells differentiated from hPSCs.
Keywords: CD9, pancreatic β cells, single-cell transcriptomics, cell-surface marker, human pluripotent stem cells, human embryonic stem cells
Graphical Abstract

Highlights
-
•
scRNA-seq reveals the heterogeneity of hPSC-derived β-like cells
-
•
CD9 is preferentially expressed by immature and proliferating human β cells
-
•
CD9 may not have a functional role in human β-like cell differentiation
-
•
CD9 is a negative cell-surface marker for enrichment of GSIS+ human β-like cells
In this study, Dr. Lui and colleagues demonstrate an efficient differentiation protocol to generate human β-like cells from pluripotent stem cells (hPSC-β). Through single-cell RNA sequencing, they further identified CD9 as a marker for immature β-like cells that are also present in the human fetal islets. Altogether, CD9 is a negative cell-surface marker for the enrichment of GSIS+ hPSC-β.
Introduction
Insulin deficiency is detected in all type 1 (T1D) and some type 2 diabetic patients. However, conventional insulin therapy does not provide a real-time glycemic control, while transplantation of glucose-sensing insulin-secreting (GSIS) cadaveric islets requires at least two donors. To address the issue of donor shortage, much progress has been made in the past decades to generate literally an unlimited amount of pancreatic β-like cells from human pluripotent stem cells (hPSCs) by recapitulating signaling pathways essential in pancreatic development. For instance, the combination of epidermal growth factor and nicotinamide signaling pathways induces the generation of NK6 homeobox 1 (NKX6.1+) pancreatic progenitor cells from hPSCs (Nostro et al., 2015). Moreover, inhibition of the bone morphogenetic protein signaling pathway at specific stages promotes the generation of insulin-expressing β-like cells (Nostro et al., 2011). Modifications of these stepwise pathways facilitate the generation of neurogenin 3 (NGN3+) and/or neurogenic differentiation factor 1 (NEUROD1+) endocrine progenitor cells (Rezania et al., 2014), as well as monohormonal endocrine cells, including insulin-expressing β-like cells that are glucose responsive in vitro (Pagliuca et al., 2014; Rezania et al., 2014). More recently, Rho-associated coiled-coil containing protein kinase 2 (ROCKII) inhibition has also been found to promote the maturation of hPSC-derived β-like cells (Ghazizadeh et al., 2017).
Using these protocols, hPSC-derived pancreatic progenitor (Schulz, 2015) or β-like (Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015) cells reverse diabetes after transplantation; and therefore, implantation of encapsulated human embryonic stem cell (hESC)-derived pancreatic progenitor cells into T1D patients forms the basis of a clinical trial being conducted by ViaCyte, Inc. (ClinicalTrials.gov identifier: NCT02239354). Although these advances have increased our understanding of human β cell development with potential clinical applications, differentiation efficiency of the reported protocols has not been validated in a large number of hPSC lines, including those that showed poor efficiency in β cell lineage specification. This might attenuate the therapeutic potential of patient-specific induced PSCs (iPSCs), as we and others have demonstrated that their efficiency in lineage commitment varies greatly among different hiPSC lines (Chetty et al., 2015; Sahara et al., 2014; Xu et al., 2012). Moreover, the generated β-like cells express fewer maturation markers, such as musculoaponeurotic fibrosarcoma oncogene homolog A (MAFA) and insulin, compared with human islets (Ghazizadeh et al., 2017; Johnson, 2016; Millman and Pagliuca, 2017). Nevertheless, identification of specific cell-surface markers that enables the purification of mature and functional human β-like cells for transplantation awaits urgent investigations.
Here, we develop an efficient differentiation protocol to generate islet-like organoids from hESCs, particularly the H9 line that has been previously shown to have poor β cell specification. We then searched for cell-surface markers that marked β-like cells by using genome-wide single-cell RNA sequencing (scRNA-seq) at various developmental stages. We demonstrate that cluster of differentiation 9 (CD9) is a cell-surface marker that negatively marks NKX6.1+MAFA+C-PEPTIDE+ β-like cells that are glucose responsive. We also validate our findings in human islets by immunostaining on both immature fetal and mature adult cadaveric islets. Furthermore, we show that CD9 might not be essential in human β cell specification and function. Our results reveal that CD9 could be used as a cell-surface marker for negative enrichment of glucose-responsive human β-like cells.
Results
Generation of Pancreatic Islet-like Organoids from Human Pluripotent Stem Cells with High Efficiency
We attempted to differentiate human β-like cells from hESCs by generating an efficient protocol (Figure 1A). We used the H9 hESC line that has been previously found less efficient in its differentiation toward the β cell lineage compared with other hESC lines such as H1 (Nostro et al., 2015). We first followed two protocols previously published by Pagliuca et al. (Pagliuca et al., 2014) and Rezania et al. (Rezania et al., 2014) to generate β-like cells. The reported differentiation efficiency of insulin-expressing β-like cells in HUES8 hESCs was about 33% using the protocol derived by Pagliuca et al., while that in H1 hESCs was about 50% using the one derived by Rezania et al. However, the differentiation efficiency using these protocols in H9 hESCs was only about 10% in our laboratory as demonstrated by immunostaining for NKX6.1 and C-PEPTIDE (Figure 1B). We observed that the amount of NKX6.1+ cells was low, but NKX6.1 is a specific marker for the emerging β cell lineage (Schaffer et al., 2013); and NKX6.1− cells are found to be polyhormonal and non-glucose responsive after differentiation from hPSCs (Nostro et al., 2015). We sought to increase the amount of NKX6.1+C-PEPTIDE+ β-like cells by following the protocol by Nostro et al. (Nostro et al., 2015) that focused on increasing the number of pancreatic and duodenal homeobox 1 (PDX1+) NKX6.1+ pancreatic progenitor cells during the first four stages of differentiation. So our protocol was modified from those derived by Nostro et al., Pagliuca et al., and Rezania et al. with combined factors (Table S1).
Figure 1.
Generation of Pancreatic Islet-like Organoids from hPSCs with High Efficiency
(A) Schematic diagram outlining the modified protocol used in this study. Uncommon abbreviations: RA, retinoic acid; VC, vitamin C: Nic, nicotinamide; N-cys, N-acetyl cysteine; and Comp E, compound E.
(B) Immunostaining on frozen sections for NKX6.1 (green) and C-PEPTIDE (red) in S6 cells after differentiation using the protocols reported by Pagliuca et al. (n = 3) and Rezania et al. (n = 3), our protocol (n = 10), or adult cadaveric islets (n = 6). Scale bars: 20 μm. The percentage of NKX6.1+C-PEPTIDE+ cells among total cells of the hESC-islet organoids was also quantified. We counted ~20 islet-like clusters per group and repeated three times with a total of ~60 islet-like clusters per group. Scale bars: 20 μm.
(C) Immunostaining on frozen sections for PDX1, NKX6.1, MAFA, and C-PEPTIDE in S6 cells after differentiation using our protocol. Scale bars: 20 μm.
(D) Quantification of human insulin secreted by human β-like cells derived from the three protocols by in vitro GSIS.
Data were obtained from three independent experiments. (B and D) Data are presented as the mean ± SD, ∗p < 0.05, ∗∗p < 0.01.
To confirm if our protocol generated more pancreatic progenitors, we traced the marker gene expression at the first four stages of differentiation by qRT-PCR (Figure S1). We observed that there was no significant difference in the expression levels of stage (S) 1 markers of the definitive endoderm, such as C-X-C motif chemokine receptor 4 (CXCR4), forkhead box protein A2 (FOXA2), and SRY-box (SOX) 17 (SOX17), using S1 cells derived from protocols of Pagliuca et al., Rezania et al., and our team (Figure S1A). In S2, there was a significantly higher expression level of the primitive gut tube marker hepatocyte nuclear factor 1 homeobox A (HNF1A) in cells derived from the protocol by Rezania et al. compared with those by Pagliuca et al. and our team (Figure S1B). In S3, there were significantly greater expression levels of the posterior foregut markers PDX1 and SOX9 in cells derived from the protocol by Pagliuca et al. compared with those by Rezania et al. and our team (Figure S1C). In S4, there was significantly more expression of the pancreatic progenitor marker NKX6.1 in cells derived from our protocol compared with those of Pagliuca et al. and Rezania et al. (Figure S1D). Our findings showed that the major difference in the first four stages of differentiation using these protocols relied on the better differentiation of NKX6.1+ cells, potentially leading to more efficient β-like cell differentiation.
To verify this assumption, we performed immunostaining for NKX6.1 and C-PEPTIDE at S6, and found significantly increased %NKX6.1+C-PEPTIDE+ cells, from 10% to 35%, compared with those generated by Pagliuca et al. and Rezania et al., respectively (p < 0.01, Figure 1B). There was no significant difference in the %NKX6.1+C-PEPTIDE+ cells of hESC-derived islet-like organoids (hESC-islets) compared with human cadaveric islets (Figure 1B). Following differentiation until S6, we observed that the generated human β-like cells were PDX1+NKX6.1+ (Figure 1C). Moreover, almost all C-PEPTIDE+ cells expressed the maturation marker MAFA (Figure 1C). Our protocol also allowed the generation of glucagon (GCG)-expressing α- and somatostatin (SST)-expressing δ-like cells, albeit at lower efficiency (Figure S2). Notably, we demonstrated a similar percentage, if not significantly reduced, of polyhormonal cells compared with previous reports: C-PEPTIDE+GCG+ (5.5% versus 7.7% by Pagliuca et al. and 11% by Rezania et al.) and C-PEPTIDE+SST+ (3.7% versus 4.7% by Pagliuca et al.). Therefore, we generated hESC-islets that, at least in part, recapitulated human cadaveric islets containing three types of endocrine cells, with β-like cells being the predominant ones. We also repeated with the H1 and H7 hESCs and confirmed that this protocol was efficient in generating NKX6.1+C-PEPTIDE+ β-like cells, with the H1 line being the most efficient hESC line compared with H7 and H9 (Figure S3).
One key functional characteristic of a pancreatic β cell is its ability to respond to glucose challenge in GSIS assays in vitro (Pagliuca et al., 2014; Veres et al., 2019). Quantification of three independent experiments demonstrated that our protocol facilitated the generation of glucose-responsive human β-like cells, in line with those generated by Pagliuca et al. and Rezania et al. (Figure 1D). Importantly, we also showed that the human β-like cells derived from our protocol released a significantly greater amount of insulin after depolarization with KCl compared with the other two reported protocols, suggesting a better differentiation efficiency with greater insulin production.
Single-Cell Transcriptomic Profiling Reveals Heterogeneity during Human Pancreas Development
Genome-wide scRNA-seq has been used to characterize the heterogeneity of adult human pancreatic islets (Baron et al., 2016; Muraro et al., 2016; Segerstolpe et al., 2016; Xin et al., 2016). However, we cannot track individual cell-fate decision, as we are unable to obtain islet samples from the same human volunteer at multiple developmental stages. hESC derivatives could partly recapitulate human islet development in vivo; we, therefore, differentiated hESCs into the pancreatic lineage for large-scale, droplet-based scRNA-seq using a 10X Genomics system as recently reported by us (Leung et al., 2018; Li et al., 2019, 2020; Liang et al., 2020). In this system, individual cells were encapsulated with unique nucleotide barcodes and molecule identifiers (UMI) for tagging RNAs inside the droplets. We established a transcriptomic map by analyzing a total of ~10,000 cells at progressive developmental stages, namely, S4, S5, and S6 cells (Figure 2A). After filtering, we had 2,388 S4, 3,525 S5, and 3,984 S6 cells for subsequent analysis (Table S2).
Figure 2.
scRNA-Seq Reveals Heterogeneity during Human Pancreas Development
(A) t-SNE analysis showing transcriptomic clustering of single cells derived from three differentiation stages. Cells were colored individually according to their stage: green, pancreatic progenitor-like cells (S4); blue, endocrine progenitor-like and INS+ immature β-like cells (S5); and purple, INS+ mature β-like cells (S6).
(B) Quantification of expression levels of the pancreatic lineage-specific genes. Expression levels were quantified as log2-scaled UMI.
(C) t-SNE analysis showing transcriptomic clustering of single cells by graph-based clustering analysis.
(D) Bar chart of the cells constituting the 17 clusters of (C).
(E) Heatmap analysis depicting the transcript levels of genes that were overexpressed in at least one of the clusters. The clusters are shown on the left border. Genes that co-expressed with INS are highlighted at the bottom.
To identify distinct cell populations, we first performed unsupervised t-stochastic neighbor embedding (t-SNE) analysis with all 9,897 transcriptomes that revealed three major cell clusters with gene expression patterns specific for each of the three developmental stages (Figure 2A). We demonstrated an overexpression of PDX1, NKX6.1, NEUROD1, and insulin (INS) with the emergence of the β cell lineage in S5 and S6, respectively, and a downregulation of GCG from S5 to S6 upon β cell maturation (Figure 2B). Specifically, we observed expression of PDX1 (Figure S4A) and NKX6.1 (Figure S4B) by each cell cluster. NGN3 (Figure S4C) and NEUROD1 (Figure S4D) were expressed by the same cells within the S5 cluster; and NEUROD1, a direct target of NGN3 required for β cell maturation (Gu et al., 2010), was expressed by some cells of the S5 and S6 clusters (Figure S4D). In addition to INS+ cells (Figure S4E), we detected GCG+ α-like (Figure S4F), SST+ δ-like (Figure S4G), and pancreatic polypeptide (PPY+)-like (Figure S4H) cells, albeit at low frequency, consistent with the protein data (Figure 1B). Altogether, our transcriptomic analysis suggested that S4 could comprise PDX1+NKX6.1+ pancreatic progenitor-like cells, S5 could contain both NGN3+NEUROD1+ endocrine progenitor- and INS+ immature β-like cells, and S6 could harbor INS+ mature β-like cells.
After that, a graph-based clustering algorithm was used for further delineating different cell subsets of each developmental stage. We identified 17 clusters (Figure 2C) and the constitution of most clusters demonstrated a developmental stage-specific pattern (Figure 2D). To annotate these clusters, we examined gene expression for each cluster by comparing its expression level with the mean value of the remaining clusters. We revealed that 339 genes were overexpressed in at least one of the clusters (Table S3). In S4 (Figure 2C and Table S3), we observed that midkine (MDK), which has also been found in embryonic day 15.5 mouse pancreas (Krentz et al., 2018), was upregulated in C2, C6, and C11. Moreover, DNA-binding protein inhibitor 3 (ID3) was upregulated in clusters C2, C6, C11, and C16, and ID1 was upregulated in C2 and C11. In fact, ID1 and ID3 regulate cell growth, suggesting that C2, C6, C11, and C16 cells might be more proliferative than clusters at later stages of lineage specification (O'Brien et al., 2012).
In S5 (Figure 2C and Table S3), NGN3 and regulator of G-protein signaling 4 (RGS4) (Serafimidis et al., 2011) were upregulated; and NEUROD1 also began to be expressed in C4, indicating that C4 could likely represent endocrine progenitor cells. Moreover, β-like (gastrin/GAST [Krentz et al., 2018], NEUROD1, and INS), α-like (GCG), and δ-like (SST) cell markers were upregulated in C5, suggesting that C5 could be immature polyhormonal endocrine cells. On the other hand, β cell markers such as tryptophan hydroxylase 1 (TPH1) (Almaca et al., 2016), stanniocalcin-1 (STC1) (Zaidi et al., 2012), and insulin-like growth factor-binding protein 3 (IGFBP5) (Gleason et al., 2010) were overexpressed in C12 without INS. In addition to TPH1, DOPA decarboxylase (DDC), which is also involved in serotonin biosynthesis, was overexpressed in C12. C8, C9, and C13 were more heterogeneous. For instance, acinar cell markers c-FOS (FOS) and c-JUN (JUN) (Turner et al., 2001), as well as the developing islet marker SOX11 (Lioubinski et al., 2003), were upregulated in C8.
In C1 and C14 of S6 cells (Figure 2C and Table S3), we detected upregulation of genes associated with fat metabolism, such as apolipoprotein E (APOE) and fatty acid-binding protein 1 (FABP1), as well as claudin 7 (CLDN7), which has been previously found in acinar and ductal cells of the postnatal pancreas (Westmoreland et al., 2012). CLDN3, previously reported to be restricted to human endocrine cells (Borka et al., 2007), was overexpressed in C17, indicating that C17 could likely be endocrine cells, albeit with low INS expression. Furthermore, we detected upregulation of β cell markers, including NEUROD1 in C7, C10, and C15; INS in C7, C10, and C15; TPH1 in C10 and C15; and DDC in C10 and C15. FOXA2, which regulates insulin secretion (Lantz et al., 2004), was upregulated in C7, C10, and C15; and insulin-like growth factor 2 (IGF2), which regulates β cell mass (Modi et al., 2015), was overexpressed in C10. These results indicate that C7, C10, and C15 were β-like cells.
A heatmap of the hierarchical clustering analysis showed that most clusters displayed a distinct gene expression profile (Figure 2E). We highlighted genes co-localized with INS in C7, C10, and C15 of S6 cells, including carboxypeptidase E (CPE) and neuroendocrine protein 7B2 (encoded by SCG5), which have been found to be involved in the cleavage of proinsulin (Liew et al., 2014; Yoshida et al., 1995). We also found other unreported genes, such as calbindin 2 (CALB2), protein phosphatase 1 regulatory subunit 1A (PPP1R1A), and cationic amino acid transporter 2 (SLC7A2).
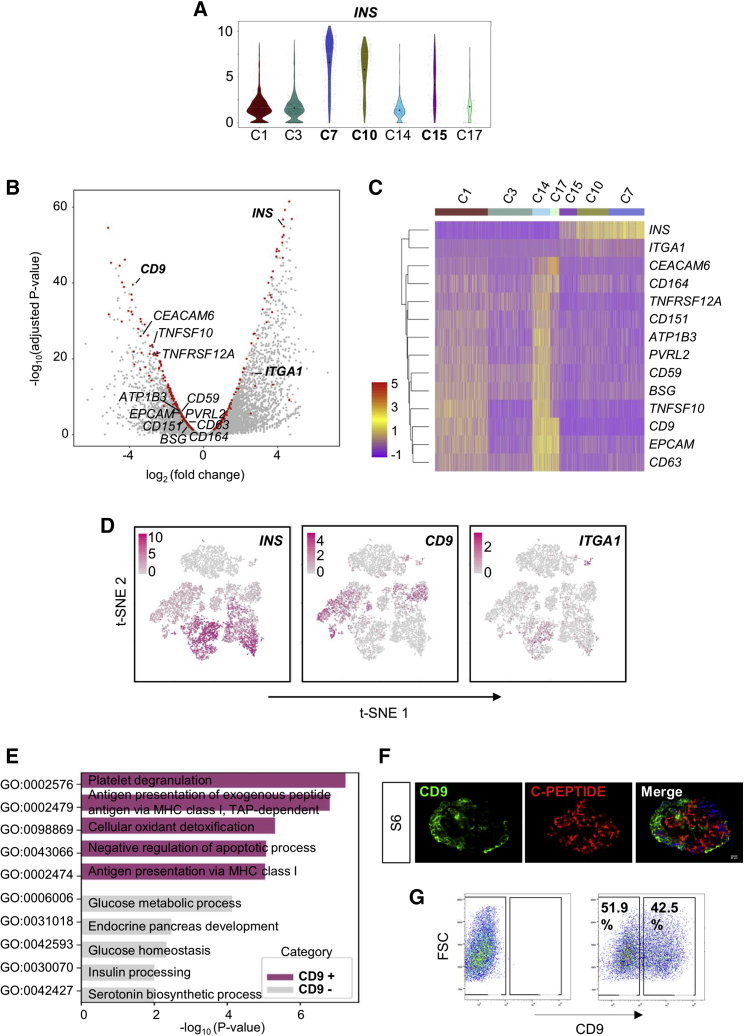
CD9 Negatively Marks INShi Human β-like Cells
We asked if there were specific cell-surface markers facilitating the purification and enrichment of INShi mature β cells at S6. We focused on C7, C10, and C15, which overexpressed INS (Figure 3A), and performed differential expression analysis by comparing them with clusters that showed low INS expression (C1, C3, C14, and C17). We found a total of 448 differentially expressed genes and focused on genes that code for cell-surface molecules (CD molecules, HUGO Gene Nomenclature Committee). We further identified 12 differentially expressed cell-surface markers (Figure 3B) and demonstrated their expression at the single-cell level by cluster-based heatmap (Figure 3C). In line with a recent report, we also observed that integrin subunit α1 (ITGA1) was a cell-surface marker co-localized with INS (Figure 3B) (Veres et al., 2019). However, our statistical analysis did not reveal that ITGA1 was a differentially expressed gene for INShi cells, at least for those derived from H9 hESCs, even though ITGA1 was clustered along with INS as demonstrated by heatmap analysis (Figure 3C). Despite being more specific for INS+ cells, ITGA1 did not label all INS+ cells efficiently, especially in C7, C10, and C15 (Figures 3C and 3D). On the other hand, CD9 was a more statistically significant marker, negatively correlating with INS expression (Figure 3B). CD9 also negatively marked cells with high INS expression in C7, C10, and C15 and positively marked cells with low INS expression in C1, C14, and C17 cells (Figure 3C). Moreover, t-SNE analyses showed that expression of CD9 was more restricted to INS− or INSlo cells at both S5 and S6 (Figure 3D).
Figure 3.
Identification of CD9 as a Negative Cell-Surface Marker for INS+ β-like Cells
(A) Violin plot showing expression levels of INS by S6 cell clusters. Expression levels were quantified as log2-scaled UMI.
(B) Volcano plot showing genes differentially expressed between INShi (C7, C10, and C15) and INSlo (C1, C3, C14, and C17) clusters. Highlighted in red are 448 differentially expressed genes, and cell-surface markers are further named.
(C) Heatmap analysis of cell-surface markers differentially expressed by INS+ S6 cell clusters.
(D) Expression patterns of INS, CD9, and ITGA1 in single cells of the three differentiation stages S4–S6 by t-SNE plot. Expression levels were quantified as log2-scaled UMI.
(E) Representation of the most significantly upregulated pathways in CD9+ and CD9− cells determined by GO functional annotation (see also Table S4).
(F) Immunostaining of frozen sections for CD9 (green) and C-PEPTIDE (red) in S6 cells after differentiation. Scale bars: 20 μm.
(G) Flow cytometric analysis showing the percentage of S6 cells expressing CD9 and the potential for enrichment of C-PEPTIDE+ cells by sorting against CD9.
We, therefore, hypothesized that CD9 could be a better cell-surface marker for negative selection of pancreatic β-like cells. Indeed, gene ontology (GO) functional annotations showed that the most significantly upregulated pathways in CD9+ S6 cells were not related to pancreatic development nor β cell function, while those of CD9− S6 cells were associated with glucose metabolism, endocrine pancreas development, glucose homeostasis, insulin processing, and serotonin biosynthesis (Figure 3E, Table S4). In addition, we validated protein expression by immunostaining for CD9 and C-PEPTIDE on frozen sections of S6 cells. Our data revealed that most C-PEPTIDE+ β cells did not express CD9 (Figure 3F). Furthermore, we consistently observed that efficient β cell differentiation would result in CD9 expression in less than half of the hESC-islet cell population by flow cytometry (Figure 3G). Altogether, our findings suggested that CD9 could be used as a cell-surface marker for negative enrichment of β-like cells derived from hESCs.
Negative Enrichment of Glucose-Responsive Human β-like Cells through CD9
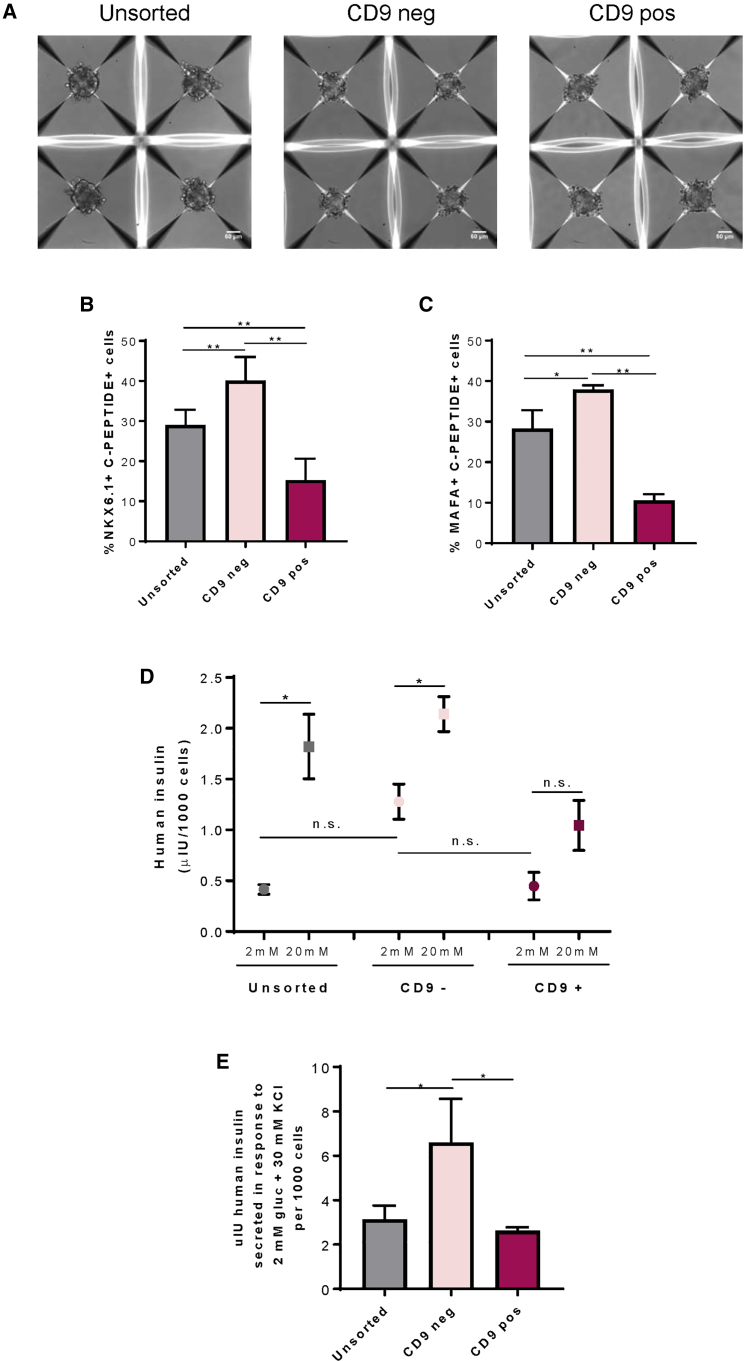
Next, we asked if negative selection against CD9 could purify glucose-responsive pancreatic β-like cells. We sorted CD9− and CD9+ cells by flow cytometry and dissociated hESC-islet cells containing both CD9− and CD9+ cells (unsorted) at S6. We then cultured 500 cells of each group for overnight (~16 h) reaggregation in AggreWell plates. The sorted or dissociated cells spontaneously self-organized into spherical 3D organoids (Figure 4A). We further quantified %NKX6.1+C-PEPTIDE+ (Figure S5A) and %MAFA+C-PEPTIDE+ (Figure S5B) cells by immunostaining. Our results demonstrated significantly more NKX6.1+C-PEPTIDE+ (Figure 4B) or MAFA+C-PEPTIDE+ (Figure 4C) cells in the CD9− compared with the CD9+ or unsorted population. Moreover, the percentage of these β-like cells was significantly reduced in the CD9+ compared with the unsorted population (Figures 4B and 4C).
Figure 4.
Negative Enrichment of Glucose-Responsive Human β-like Cells through CD9
(A) Representative images of four individual hESC-islet organoids reaggregated after overnight culture in AggreWell400. Each organoid contained 500 dissociated or sorted cells. Scale bars: 50 μm.
(B and C) Quantification of (B) %NKX6.1+C-PEPTIDE+ cells (see also Figure S5A) and (C) %MAFA+C-PEPTIDE+ cells (see also Figure S5B) among total cells of the hESC-islet organoids.
(D) Quantification of human insulin secreted by human β-like cells derived from dissociated unsorted, sorted CD9−, or sorted CD9+ cells by in vitro GSIS.
(E) Quantification of human insulin secreted from dissociated unsorted, sorted CD9−, or sorted CD9+ cells in response to 2 mM glucose and 30 mM KCl.
(B–E) Data were obtained from four independent experiments and presented as the mean ± SD, ∗p < 0.05, ∗∗p < 0.01, and n.s. denotes no statistically significant difference.
To determine whether overnight reaggregation would reduce the purity of NKX6.1+C-PEPTIDE+ cells among CD9− (Figure S6A) and CD9+ (Figure S6B) sorted cells, and whether a part of the purified CD9−C-PEPTIDE+ cells would become C-PEPTIDE− after reaggregation, we compared the efficiency using sorted S6 cells derived from the H1 hESCs immediately after sorting and after reaggregation overnight. We showed that there was no significant difference in the %NKX6.1+C-PEPTIDE+ cells before and after overnight reaggregation of either CD9− or CD9+ sorted cells. Similarly, we found the same results using sorted S6 cells derived from the H9 hESCs (Figures S6C and S6D). Therefore, our findings suggest that overnight reaggregation did not alter the purity of human β-like cells after negative selection against CD9 at S6.
We also performed GSIS assays in vitro, and quantification of four independent experiments demonstrated that CD9− cells responded better to glucose challenge compared with the CD9+ cells (Figure 4D). In contrast, CD9+ cells secreted very low amounts of insulin and the amounts did not differ in 2 and 20 mM glucose, suggesting that they failed to respond to a change in glucose concentration (Figure 4D). Furthermore, quantification of four independent experiments showed that CD9− cells had significantly increased levels of insulin after depolarization with KCl compared with CD9+ or unsorted controls (Figure 4E).
CD9 Is Preferentially Expressed by Human Immature and Proliferating β Cells
It is generally believed that fetal β cells are immature, as they acquire glucose responsiveness through functional maturation after birth. To ask if CD9 might be a marker for immature β cells, we compared CD9 expression in the immature fetal and the mature adult pancreata by collecting human pancreata from fetuses aborted at the second trimester and from adult patients. Immunostaining for CD9 and C-PEPTIDE showed significantly fewer %CD9+C-PEPTIDE+ cells among total C-PEPTIDE+ β cells of adult (~17%, n = 6) compared with fetal (~28.5%, n = 3) islets (Figures 5A and 5B). Moreover, we also performed immunostaining for the immature β cells that are capable of proliferation (Figure 5C) and showed that significantly more Ki67+C-PEPTIDE+ proliferating β cells (~75%) expressed CD9 compared with the Ki67−C-PEPTIDE+ non-proliferating β cells (~35%) of the fetal islets (Figure 5D, n = 3). Ki67+C-PEPTIDE+CD9+ proliferating β cells could be found only in the fetal and not in the adult islets (Figure 5E). Furthermore, we also observed that reaggregated CD9+ cells at S6 showed expression of SST (Figure S7A), and CD9 also co-localized with SST in the fetal (Figure S7B) and adult (Figure S7C) human islets, suggesting that CD9 could be a marker for SST+ δ cells. Altogether, our results showed that CD9 could be used as a negative marker to enrich human β-like cells, as CD9 marked immature β cells as well as other endocrine cells such as δ cells in the human islets of the fetal pancreas in particular.
Figure 5.
CD9 Is Preferentially Expressed by Human Immature β Cells
(A) Immunostaining of frozen sections for C-PEPTIDE (green) and CD9 (red) in fetal (n = 3, <100 μm) or adult (n = 6, 100–250 μm) cadaveric islets. Scale bars: 20 μm.
(B) Quantification of (A) showing %CD9+C-PEPTIDE+ among total C-PEPTIDE+ β-like cells in fetal and adult cadaveric islets.
(C) Immunostaining of frozen sections for C-PEPTIDE (blue), CD9 (green), and Ki67 (red) in fetal (n = 3) or adult (n = 6, data not shown) cadaveric islets. The white box indicates the amplified regions on the right. Scale bars: 20 μm.
(D) Quantification of (C) showing %CD9+ cells among Ki67+C-PEPTIDE+ or Ki67−C-PEPTIDE+ β-like cells in fetal cadaveric islets.
(E) Quantification showing %Ki67+CD9+C-PEPTIDE+ among total CD9+C-PEPTIDE+ β-like cells in fetal and adult cadaveric islets.
(B, D, and E) Data are presented as the mean ± SD, ∗p < 0.05.
CD9 May Not Have a Functional Role in the Differentiation of Human β-like Cells
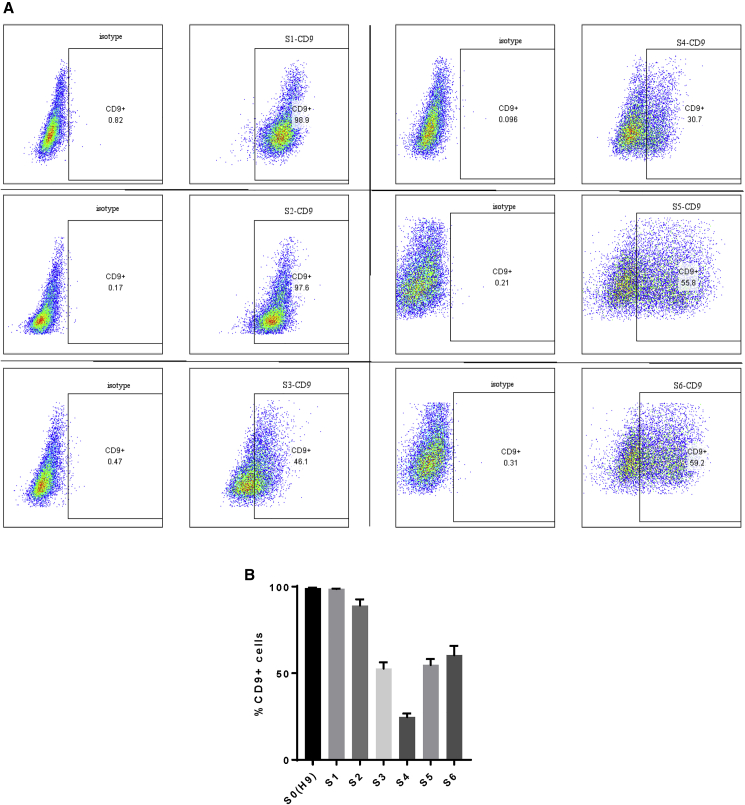
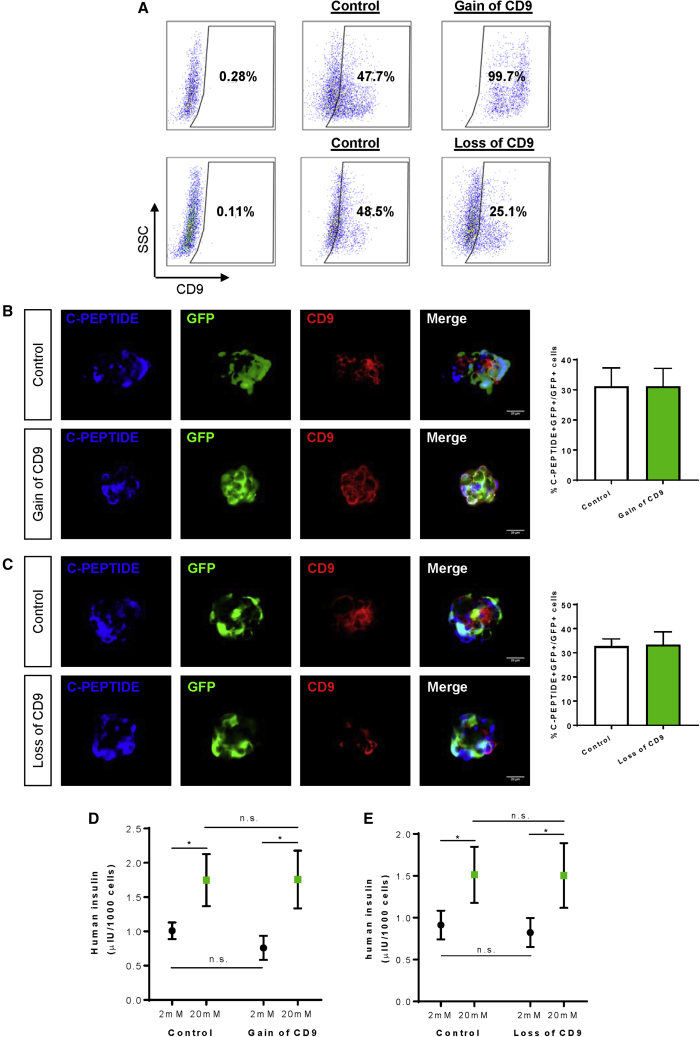
Last, we asked if CD9 plays any functional role in human β-like cell development. We performed flow cytometry to score for the temporal expression pattern of CD9 during human β-like cell differentiation from hESCs (Figure 6A). Our results revealed that the expression levels of CD9 were very high in the pluripotent and endodermal stages. About 100% of cells at S0–S2 expressed CD9, with a gradual reduction to ~30% at S4 and a slight increase to ~50% at S5–S6 (Figure 6B). Our data might suggest that the expression levels of CD9 started to decline upon pancreatic/endocrine lineage specification. We then overexpressed or knocked out CD9 during β cell specification at the end of S4 through ectopic expression of hCD9-GFP or CRISPR/Cas9-GFP targeting CD9, respectively. Flow cytometric analysis demonstrated that there were ~99% and ~25% cells, respectively, expressing CD9 in the overexpression and knockout groups, compared with ~50% cells of the control group (Figure 7A). We then performed immunostaining for C-PEPTIDE, GFP, and CD9 and found that neither overexpression (Figure 7B) nor knockout (Figure 7C) of CD9 altered the %C-PEPTIDE+GFP+ cells among total transduced GFP+ cells at S6. Furthermore, we performed GSIS in vitro at S6 to determine if altering the expression level of CD9 would influence the function of human β-like cells. Our results showed that neither overexpression (Figure 7D) nor knockout (Figure 7E) of CD9 at S5 influenced the ability of S6 human β-like cells to respond to a change in glucose concentration.
Figure 6.
Temporal Expression of CD9 during Human β-like Cell Differentiation
(A) Flow cytometric analysis showing the FACS plots and (B) quantification data showing the percentages of S1–S6 cells derived from the H9 hESCs expressing CD9. Data are presented as the mean ± SD.
Figure 7.
CD9 May Not Have a Functional Role in the Differentiation of Human β-like Cells
(A) Flow cytometric analysis showing the percentage of S5 cells expressing CD9 at day 7 (end of S5) after ectopic expression of CD9-GFP (gain of CD9) or CRISPR/Cas9-GFP targeting CD9 (loss of CD9).
(B and C) Immunostaining of frozen sections and quantification of S6 cells for C-PEPTIDE (blue), GFP (green), and CD9 (red) in S6 cells after (B) gain of CD9 or (C) loss of CD9. Scale bars: 20 μm.
(D and E) Quantification of human insulin secreted by S6 human β-like cells generated after (D) gain of CD9 or (E) loss of CD9 by in vitro GSIS.
(B–D) Data were obtained from three independent experiments and presented as the mean ± SD, ∗p < 0.05, and n.s. denotes no statistically significant difference.
Discussion
The use of hPSCs in the generation of β-like cells could provide an unlimited source of cells for potential treatment of diabetes through cell therapy, disease modeling, and drug discovery. Although current protocols produce human β-like cells with some functionality (Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015), the differentiation efficiency varies greatly among different hPSC lines. For instance, most studies utilize HUES8 (Pagliuca et al., 2014; Veres et al., 2019) or H1 (Ghazizadeh et al., 2017; Rezania et al., 2014) hESCs, which have been shown to be efficient in β cell lineage commitment. The same protocol, when applied to other cell lines, including patient-specific iPSCs, could result in poor β cell specification (Ghazizadeh et al., 2017). Moreover, the generated cells lack expression of maturation markers such as MAFA and show limited glucose responsiveness compared with human cadaveric islets (Ghazizadeh et al., 2017; Johnson, 2016).
In this study, we developed a protocol using reported differentiation factors (Nostro et al., 2015; Pagliuca et al., 2014; Rezania et al., 2014) for efficient generation of monohormonal NKX6.1+MAFA+C-PEPTIDE+ β-like cells from the H9 hESC line previously known to be less amenable to β cell differentiation. Unlike rodent islets, which are predominantly constituted of β cells (~80%), human islets have fewer (~50%) and more disperse β cells (Brissova et al., 2005). The H9 hESC-islets that we generated contained ~30%–40% β-like cells, ~5% α-like cells, and ~2% δ-like cells, recapitulating the cellular architecture of the endocrine cell populations of human islets. We performed high-resolution, genome-wide scRNA-seq to comprehensively characterize cellular heterogeneity during human islet development. During islet cell differentiation, S4 mainly harbored PDX1+NKX6.1+ pancreatic progenitor-like cells, S5 encompassed both NGN3+NEUROD1+ endocrine progenitor- and INS+ immature β-like cells, and S6 contained INS+ mature β-like cells. Moreover, we identified 17 cell clusters that could exhibit diverse roles in supporting endocrine cell function. It was also intriguing to observe that the expression level of INS differed among individual β-like cells. The heterogeneous β-like cell subsets might express genes not directly related to GSIS but associated with β cell maturation or maintaining the non-secretory roles that are important for the overall islet function (Liu and Hebrok, 2017). Our finding might provide a resource for identifying genes that marked and/or regulated the heterogeneous human β cell subsets.
We also sought to enrich β-like cells by identifying specific cell-surface markers. Our scRNA-seq data revealed that CD9 was the most statistically significant cell-surface marker negatively correlated with INS expression. We established a 3D culture system for spontaneous reaggregation of hESC-islet organoids after negative enrichment against CD9 at S6 by flow cytometry. CD9− β-like cells expressed more of the maturation marker MAFA and demonstrated better glucose responsiveness by in vitro GSIS compared with CD9+ cells. Importantly, we validated this by performing immunostaining for CD9 and C-PEPTIDE on frozen sections of fetal and adult human islets. Indeed, a previous report has shown that CD9− β cells form a distinct subtype of β cells within human islets, but its functional role in β cells has not been investigated (Dorrell et al., 2016). Here, we further demonstrated that CD9 could be used for negative enrichment of glucose-responsive human β-like cells, but CD9 may not have any functional role in β cell specification nor glucose responsiveness, as neither overexpression nor knockout of CD9 altered their differentiation, maturation, or function.
One might argue that we did not completely knock out CD9 expression in our experiments, but we showed that a 50% reduction in the expression level of CD9 did not change β cell function. Another possible role of CD9 may lie in its regulation of β cell proliferation, as we observed more proliferating β cells of human fetal islets expressing CD9 compared with the non-proliferating ones. However, we were unable to identify proliferating β cells in the hPSC differentiation system, as the differentiated β-like cells were rarely Ki67+ at both S5 and S6. Due to the intrinsic difference in human β cells derived from hPSCs and fetal tissues, we might not be able to delineate the physiological role of CD9 in human β cells through hPSC differentiation. Altogether, our results suggested that CD9 was a negative marker for enrichment of glucose-responsive human β-like cells derived from hPSCs, and CD9 did not have a functional role in β cell differentiation or function. Future studies are needed to explore the functional role of CD9 in β cells using human tissues. Essentially, our results give support to the application, particularly the purification, of hESC-derived β-like cells for potential cell replacement therapy in diabetic individuals.
Recently, ITGA1 has been identified as a specific cell-surface marker for human β-like cells derived from the HUES8 hESC line (Veres et al., 2019). Although ITGA1 was not shown to be co-localized with MAFA nor specifically expressed by β cells within the cadaveric islets (Veres et al., 2019), we confirmed that ITGA1 indeed labeled some of the β-like cells derived from the H9 hESC line using our protocol, albeit at very low efficiency. Our analyses may indicate that the efficiency of using cell-surface markers for the purification of human β-like cells could vary between sources of hPSCs. Moreover, it is worth noting that batch-to-batch variation in the differentiation of β-like cells from hPSCs is commonly seen even in the same laboratory (Pagliuca et al., 2014). Our protocol targeting negative enrichment against CD9 appeared to reduce the batch-to-batch variation. Nevertheless, future effort is needed to carefully examine the specificity and labeling efficiency of positive or negative cell-surface markers of β cells using more hPSC lines and human cadaveric islets. Taken together, our results demonstrate that CD9 is a negative enrichment marker for glucose-responsive human β-like cells derived from hPSCs that could support clinical research targeting the therapeutic use of human β-like cells in cell therapy, disease modeling, or drug screening.
Experimental Procedures
For details of this section, please also refer to the Supplemental Experimental Procedures.
Human Patients
All procedures were approved by The Chinese University of Hong Kong-Hospital Authority (NTEC) Joint Clinical Research Ethics Committee. Pregnant women who had decided on termination of pregnancy due to trisomy 13/21 in prenatal diagnosis provided consent. Those who took medications that are detrimental to fetal development were excluded from this study. Fetal pancreata were collected from aborted fetuses of the second trimester at gestational week 13–20. In addition, adult pancreatic biopsies were collected from patients with pancreatic cysts or gastrointestinal cancers (Table S5).
Human ESC Cultures and Pancreatic Islet Differentiation
The H9 hESC line (WA09, WiCell) was maintained in mTesR1 medium (Stemgent). To generate pancreatic islets, confluent cultures were dissociated into single-cell suspensions using Accutase (Thermo Fisher) and reseeded at a density of 1.5 × 105 cells/cm2 on Matrigel (Corning)-coated surfaces supplemented with 10 μM Y-27632 (Selleck Chemicals) in mTesR1 for 24 h (day −1). hESCs were then differentiated by stepwise administration of growth factors as follows. On day 0/S1, cells were cultured in RPMI medium (Thermo Fisher) containing 100 ng/mL Activin A (Peprotech) and 2 μM CHIR-99021 (Selleck Chemicals). On day 1–2, cells were refreshed with RPMI containing 100 ng/mL Activin A, 5 ng/mL hFGF-basic (Peprotech), and 50 μg/mL ascorbic acid (Sigma). On day 3–5/S2, cells were cultured in RPMI containing 1% B27 supplement without vitamin A (Thermo Fisher), 50 ng/mL KGF (Peprotech), and 50 μg/mL ascorbic acid. On day 6–7/S3, cells were refreshed with DMEM (Thermo Fisher) containing 1% B27 supplement without vitamin A, 0.25 μM SANT-1 (Sigma), 2 μM retinoic acid (RA, Sigma), 50 ng/mL Noggin (Peprotech), 50 ng/mL KGF, and 50 μg/mL ascorbic acid. On day 8–12/S4, cells were cultured in DMEM containing 1% B27 supplement without vitamin A, 50 μg/mL ascorbic acid, 50 ng/mL Noggin, 50 ng/mL EGF (Peprotech), and 10 mM nicotinamide (Sigma). Medium was refreshed every day. On day 13/S5, the cells were rinsed with 1× DPBS and then cultured with 1 U/mL dispase (Thermo Fisher) for 7 min at 37°C, followed by gentle pipetting to break cells into clumps of 100–200 μm. After that, the cell clumps were transferred into each well of a 6-well ultra-low attachment plate and cultured in MCDB131 (Corning) containing 1.5 g/L NaHCO3, 20 mM D-(+)-glucose, 2% fatty acid-free BSA (Proliant), 1% Glutamax, 1:200 ITS-X (Thermo Fisher), 10 μM ZnSO4, 10 μg/mL heparin, 50 μg/mL ascorbic acid, 0.25 μM SANT-1, 100 nM RA, 10 μM RepSox (Selleck Chemicals), 1 μM GC1 (R&D Systems), 100 nM LDN193189 (Sigma), 1 μM Compound E (StemCell Technologies), and 10 μM Y-27632 for 3 days. On day 16–19, cell clusters were refreshed with MCDB131 containing 1.5 g/L NaHCO3, 20 mM D-(+)-glucose, 2% fatty acid-free BSA, 1% Glutamax, 1:200 ITS-X, 10 μM ZnSO4, 10 μg/mL heparin, 50 μg/mL ascorbic acid, 10 μM RepSox, 1 μM GC1, 100 nM LDN193189, 1 μM Compound E, and 10 μM Y-27632. Medium was changed on alternate days. On day 20–28/S6, cell clusters were cultured in MCDB131 containing 1.5 g/L NaHCO3, 20 mM D-(+)-glucose, 2% fatty acid-free BSA, 1% Glutamax, 1:200 ITS-X, 10 μM ZnSO4, 10 μg/mL heparin, 50 μg/mL ascorbic acid, 10 μM RepSox, 1 μM GC1, 10 μM Trolox (Sigma), 2 μM R428 (Cayman), and 1 mM N-acetyl cysteine (Sigma). Medium was changed on alternate days. To reaggregate purified cells after fluorescence-activated cell sorting (FACS), 600,000 CD9+, CD9−, or dissociated cells were cultured in AggreWell400, 24 wells (StemCell Technologies), with S6 medium supplemented with 10 μM Y-27632 overnight to promote cell survival after sorting.
Identification of Cell-Surface Markers for INShi Mature β-like Cells
Cell clusters were defined as INShi or INSlo cells according to their expression level of INS. Differentially expressed genes were identified by pairwise comparison using Cell Ranger pipelines with the settings of minimum mean expression level of 0.5 and adjusted p value with a cutoff of 0.05. Cell-surface marker genes were obtained from the HUGO Gene Nomenclature Committee (Group 471, CD molecules) for further examination. GO enrichment analysis of upregulated genes in CD9+ or CD9− cells was performed by DAVID Bioinformatics Resources (v.6.8).
Statistical Analysis
The data were expressed as the arithmetic mean ± SD of biological replicates performed independently under the same conditions at least three times. Statistical analysis was performed using the unpaired Student t test with data from two groups, while data from more than two groups were analyzed using an ANOVA followed by Tukey's method for multiple comparisons. Significance was accepted when p < 0.05.
Data and Code Availability
scRNA-seq data are deposited under NCBI Bioproject (PRJNA594252).
Author Contributions
X.L. and K.O.L. designed the experiments; X.L. and V.C. performed the experiments; X.L. and K.Y.Y. analyzed the data; K.O.L. interpreted the data; K.T.L., X.Z., and A.S.W. provided reagents; C.C.N.C. and C.C.W. recruited human volunteers and provided clinical samples; M.K. provided optimized protocols for sequencing; K.O.L. supervised the research and wrote the manuscript.
Acknowledgments
We thank Professors Dennis Lo and Rossa Chiu (The Chinese University of Hong Kong [CUHK]) for their machinery support in the scRNA-seq experiments. This work was supported by the Research Grants Council of Hong Kong (14108420, C4024-16W, C4026-17WF); National Natural Science Foundation of China (81922077); Croucher Foundation (Innovation Award and Start-Up Allowance); University Grants Committee Research Matching Grant Scheme (2019, 2020); Faculty Innovation Award, Research Committee Funding, Direct Grant, and Postdoctoral Fellowship (K.Y.Y.); and Postgraduate Studentship (X.L.) from CUHK.
Published: October 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.09.009.
Supplemental Information
References
- Almaca J., Molina J., Menegaz D., Pronin A.N., Tamayo A., Slepak V., Berggren P.O., Caicedo A. Human beta cells produce and release serotonin to inhibit glucagon secretion from alpha cells. Cell Rep. 2016;17:3281–3291. doi: 10.1016/j.celrep.2016.11.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borka K., Kaliszky P., Szabo E., Lotz G., Kupcsulik P., Schaff Z., Kiss A. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 2007;450:549–557. doi: 10.1007/s00428-007-0406-7. [DOI] [PubMed] [Google Scholar]
- Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M., Powers A.C. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005;53:1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- Chetty S., Engquist E.N., Mehanna E., Lui K.O., Tsankov A.M., Melton D.A. A Src inhibitor regulates the cell cycle of human pluripotent stem cells and improves directed differentiation. J. Cel. Biol. 2015;210:1257–1268. doi: 10.1083/jcb.201502035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Schug J., Canaday P.S., Russ H.A., Tarlow B.D., Grompe M.T., Horton T., Hebrok M., Streeter P.R., Kaestner K.H. Human islets contain four distinct subtypes of beta cells. Nat. Commun. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh Z., Kao D.I., Amin S., Cook B., Rao S., Zhou T., Zhang T., Xiang Z., Kenyon R., Kaymakcalan O. ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nat. Commun. 2017;8:298. doi: 10.1038/s41467-017-00129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C.E., Ning Y., Cominski T.P., Gupta R., Kaestner K.H., Pintar J.E., Birnbaum M.J. Role of insulin-like growth factor-binding protein 5 (IGFBP5) in organismal and pancreatic beta-cell growth. Mol. Endocrinol. 2010;24:178–192. doi: 10.1210/me.2009-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Stein G.H., Pan N., Goebbels S., Hornberg H., Nave K.A., Herrera P., White P., Kaestner K.H., Sussel L. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11:298–310. doi: 10.1016/j.cmet.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.D. The quest to make fully functional human pancreatic beta cells from embryonic stem cells: climbing a mountain in the clouds. Diabetologia. 2016;59:2047–2057. doi: 10.1007/s00125-016-4059-4. [DOI] [PubMed] [Google Scholar]
- Krentz N.A.J., Lee M.Y.Y., Xu E.E., Sproul S.L.J., Maslova A., Sasaki S., Lynn F.C. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Rep. 2018;11:1551–1564. doi: 10.1016/j.stemcr.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz K.A., Vatamaniuk M.Z., Brestelli J.E., Friedman J.R., Matschinsky F.M., Kaestner K.H. Foxa2 regulates multiple pathways of insulin secretion. J. Clin. Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C.S., Yang K.Y., Li X., Chan V.W., Ku M., Waldmann H., Hori S., Tsang J.C.H., Lo Y.M.D., Lui K.O. Single-cell transcriptomics reveal that PD-1 mediates immune tolerance by regulating proliferation of regulatory T cells. Genome Med. 2018;10:71. doi: 10.1186/s13073-018-0581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li X., Liang C., Ling L., Chen Z., Wong C.K., Waldmann H., Lui K.O. Coreceptor blockade targeting CD4 and CD8 allows acceptance of allogeneic human pluripotent stem cell grafts in humanized mice. Biomaterials. 2020;248:120013. doi: 10.1016/j.biomaterials.2020.120013. [DOI] [PubMed] [Google Scholar]
- Li J., Yang K.Y., Tam R.C.Y., Chan V.W., Lan H.Y., Hori S., Zhou B., Lui K.O. Regulatory T-cells regulate neonatal heart regeneration by potentiating cardiomyocyte proliferation in a paracrine manner. Theranostics. 2019;9:4324–4341. doi: 10.7150/thno.32734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Yang K.Y., Chan V.W., Li X., Fung T.H.W., Wu Y., Tian X.Y., Huang Y., Qin L., Lau J.Y.W. CD8(+) T-cell plasticity regulates vascular regeneration in type-2 diabetes. Theranostics. 2020;10:4217–4232. doi: 10.7150/thno.40663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew C.W., Assmann A., Templin A.T., Raum J.C., Lipson K.L., Rajan S., Qiang G., Hu J., Kawamori D., Lindberg I. Insulin regulates carboxypeptidase E by modulating translation initiation scaffolding protein eIF4G1 in pancreatic beta cells. Proc. Natl. Acad. Sci. U S A. 2014;111:E2319–E2328. doi: 10.1073/pnas.1323066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioubinski O., Muller M., Wegner M., Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev. Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- Liu J.S., Hebrok M. All mixed up: defining roles for beta-cell subtypes in mature islets. Genes Dev. 2017;31:228–240. doi: 10.1101/gad.294389.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman J.R., Pagliuca F.W. Autologous pluripotent stem cell-derived beta-like cells for diabetes cellular therapy. Diabetes. 2017;66:1111–1120. doi: 10.2337/db16-1406. [DOI] [PubMed] [Google Scholar]
- Modi H., Jacovetti C., Tarussio D., Metref S., Madsen O.D., Zhang F.P., Rantakari P., Poutanen M., Nef S., Gorman T. Autocrine action of IGF2 regulates adult beta-cell mass and function. Diabetes. 2015;64:4148–4157. doi: 10.2337/db14-1735. [DOI] [PubMed] [Google Scholar]
- Muraro M.J., Dharmadhikari G., Grun D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Ogawa S., Holtzinger A., Corneo B., Li X., Micallef S.J., Park I.H., Basford C., Wheeler M.B. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Yang C., Holland A., Elefanty A.G., Stanley E.G., Greiner D.L., Keller G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien C.A., Kreso A., Ryan P., Hermans K.G., Gibson L., Wang Y., Tsatsanis A., Gallinger S., Dick J.E. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gurtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O'Dwyer S., Quiskamp N., Mojibian M., Albrecht T. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Russ H.A., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara M., Hansson E.M., Wernet O., Lui K.O., Spater D., Chien K.R. Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res. 2014;24:820–841. doi: 10.1038/cr.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A.E., Taylor B.L., Benthuysen J.R., Liu J., Thorel F., Yuan W., Jiao Y., Kaestner K.H., Herrera P.L., Magnuson M.A. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet. 2013;9:e1003274. doi: 10.1371/journal.pgen.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.C. Concise review: manufacturing of pancreatic endoderm cells for clinical trials in type 1 diabetes. Stem Cell translational Med. 2015;4:927–931. doi: 10.5966/sctm.2015-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe A., Palasantza A., Eliasson P., Andersson E.M., Andreasson A.C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafimidis I., Heximer S., Beis D., Gavalas A. G protein-coupled receptor signaling and sphingosine-1-phosphate play a phylogenetically conserved role in endocrine pancreas morphogenesis. Mol. Cell. Biol. 2011;31:4442–4453. doi: 10.1128/MCB.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.J., Cowles R.A., Segura B.J., Mulholland M.W. Cholinergic stimulation of rat acinar cells increases c-fos and c-jun expression via a mitogen-activated protein kinase-dependent pathway. J. Gastrointest. Surg. 2001;5:661–672. doi: 10.1016/s1091-255x(01)80110-4. [DOI] [PubMed] [Google Scholar]
- Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H., Harb G., Poh Y.C., Sintov E., Gurtler M., Pagliuca F.W. Charting cellular identity during human in vitro beta-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland J.J., Drosos Y., Kelly J., Ye J., Means A.L., Washington M.K., Sosa-Pineda B. Dynamic distribution of claudin proteins in pancreatic epithelia undergoing morphogenesis or neoplastic transformation. Dev. Dyn. 2012;241:583–594. doi: 10.1002/dvdy.23740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y., Kim J., Okamoto H., Ni M., Wei Y., Adler C., Murphy A.J., Yancopoulos G.D., Lin C., Gromada J. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Xu H., Yi B.A., Wu H., Bock C., Gu H., Lui K.O., Park J.H., Shao Y., Riley A.K., Domian I.J. Highly efficient derivation of ventricular cardiomyocytes from induced pluripotent stem cells with a distinct epigenetic signature. Cell Res. 2012;22:142–154. doi: 10.1038/cr.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Ohagi S., Sanke T., Furuta H., Furuta M., Nanjo K. Association of the prohormone convertase 2 gene (PCSK2) on chromosome 20 with NIDDM in Japanese subjects. Diabetes. 1995;44:389–393. doi: 10.2337/diab.44.4.389. [DOI] [PubMed] [Google Scholar]
- Zaidi D., Turner J.K., Durst M.A., Wagner G.F. Stanniocalcin-1 co-localizes with insulin in the pancreatic islets. ISRN Endocrinol. 2012;2012:834359. doi: 10.5402/2012/834359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data are deposited under NCBI Bioproject (PRJNA594252).