Summary
Direct reprogramming has been widely explored to generate various types of neurons for neurobiological research and translational medicine applications, but there is still no efficient reprogramming method to generate retinal ganglion cell (RGC)-like neurons, which are the sole projection neurons in the retina. Here, we show that three transcription factors, Ascl1, Brn3b, and Isl1, efficiently convert fibroblasts into RGC-like neurons (iRGCs). Furthermore, we show that the competence of cells to enter iRGC reprogramming route is determined by the cell-cycle status at a very early stage of the process. The iRGC reprogramming route involves intermediate states that are characterized by a transient inflammatory-like response followed by active epigenomic and transcriptional modifications. Our study provides an efficient method to generate iRGCs, which would be a valuable cell source for potential glaucoma cell replacement therapy and drug screening studies, and reveals the key cellular events that govern successful neuronal fate reprogramming.
Keywords: direct somatic cell reprogramming, induced neuron, retinal ganglion cell, Ascl1, Brn3b, Isl1, iRGC
Highlights
-
•
Ascl1+Brn3b+Isl1 efficiently reprograms fibroblasts into iRGCs
-
•
iRGC reprogramming route is committed early, which is determined by cell-cycle status
-
•
iRGC reprogramming route involves intermediate states
In this article, Chen and colleagues show that Ascl1+Brn3b+Isl1 transcription factor combination efficiently reprograms fibroblasts into iRGCs. The authors further show that the iRGC reprogramming route is committed early, which is determined by the cell-cycle status at the time, and that the iRGC reprogramming route involves intermediate states that are characterized by transient inflammatory-like response and active epigenomic and transcriptional modifications.
Introduction
Experiments using technologies, such as somatic nuclear transfer, induced pluripotent stem cells, and direct somatic cell reprogramming have revealed the plasticity of cell fates (Vierbuchen and Wernig, 2011; Xu et al., 2015). Due to the great interest in obtaining sufficient numbers of neurons for neural biological and translational research, great efforts have been made to explore protocols to directly induce neurons from easily accessible somatic cells, such as fibroblasts. Brn2/Ascl1/Myt1l (BAM) is the first transcription factor (TF) combination demonstrated to be able to efficiently reprogram mouse fibroblasts into functional induced neurons (iNs) with generic properties (Vierbuchen et al., 2010). Later, TF combinations that could directly convert fibroblasts into specific types of neurons were established (Blanchard et al., 2015; Caiazzo et al., 2011; Colasante et al., 2015; Pfisterer et al., 2011; Sheng et al., 2012; Son et al., 2011; Wainger et al., 2015). Investigations of iN reprogramming are starting to reveal the molecular and cellular events during the processes, which show that the process involves active epigenetic modifications (Luo et al., 2019; Wapinski et al., 2013) and needs to pass a metabolic checkpoint to avoid cell death (Gascon et al., 2016). Recent studies using single-cell RNA sequencing (scRNA-seq) techniques on small-scale iN reprogramming cells suggest that the reprogramming path is continuous and may involve a neural stem cell-like intermediate state (Karow et al., 2012; Treutlein et al., 2016). However, the detailed iN reprogramming route remains elusive.
RGCs are the projection neurons at the inner-most layer of the neural retina and are responsible for transmitting visual information from the eye to the brain. RGCs are vulnerable to various insults, such as increased intraocular pressure, genetic mutations, and aging, leading to the development of glaucoma. Glaucoma is the most prevalent retinal diseases that cause blindness and it affects approximately 1 out of every 40 adults over the age of 40 years worldwide (Quigley, 2011). None of the current treatments can reverse the progression of vision loss in glaucoma patients (Varma et al., 2011). RGCs, as with all other retinal neurons, are generated during development by multipotent retinal progenitor cells (RPCs) (Bassett and Wallace, 2012; Cepko, 2014). Math5 (Atoh7), a basic-helix-loop-helix (bHLH) family TF, determines the RGC-competent status of RPCs (Brzezinski et al., 2012; Wang et al., 2001; Yang et al., 2003). Downstream of Math5, a POU homeodomain TF, Brn3b (Pou4f2), and an LIM homeodomain TF, Isl1, work synergistically to control the cell fate specification, final differentiation, and refinement of neurite growth of RGCs (Gan et al., 1996, 1999; Li et al., 2014; Liu et al., 2001; Mu et al., 2005, 2008; Pan et al., 2008; Wu et al., 2015; Xiang, 1998). Between Math5 and Brn3b/Isl1, the SoxC family TFs, Sox4 and Sox11, have recently been demonstrated to work synergistically to control RGC development (Chang et al., 2017; Jiang et al., 2013).
In this study, we show that, by combining Ascl1 with two RGC-genic TFs, Brn3b and Isl1, mouse fibroblasts can be quickly and highly efficiently converted into RGC-like neurons (iRGCs). Supplementing these three TFs with Sox4, human fibroblasts can also be efficiently reprogrammed into iRGCs. By tracing the reprogramming process, we further show that the successful iRGC reprogramming route is committed at a very early stage, which is determined by the cell-cycle status of the reprogramming cell at the time, and that the reprogramming route includes intermediate states that involve transient inflammatory-like activity followed by active adjustments of the transcriptomic and epigenomic environments of the reprogramming cells.
Results
Ascl1, Brn3b, and Isl1 TF Combination Reprograms Fibroblasts into BRN3A+-iNs
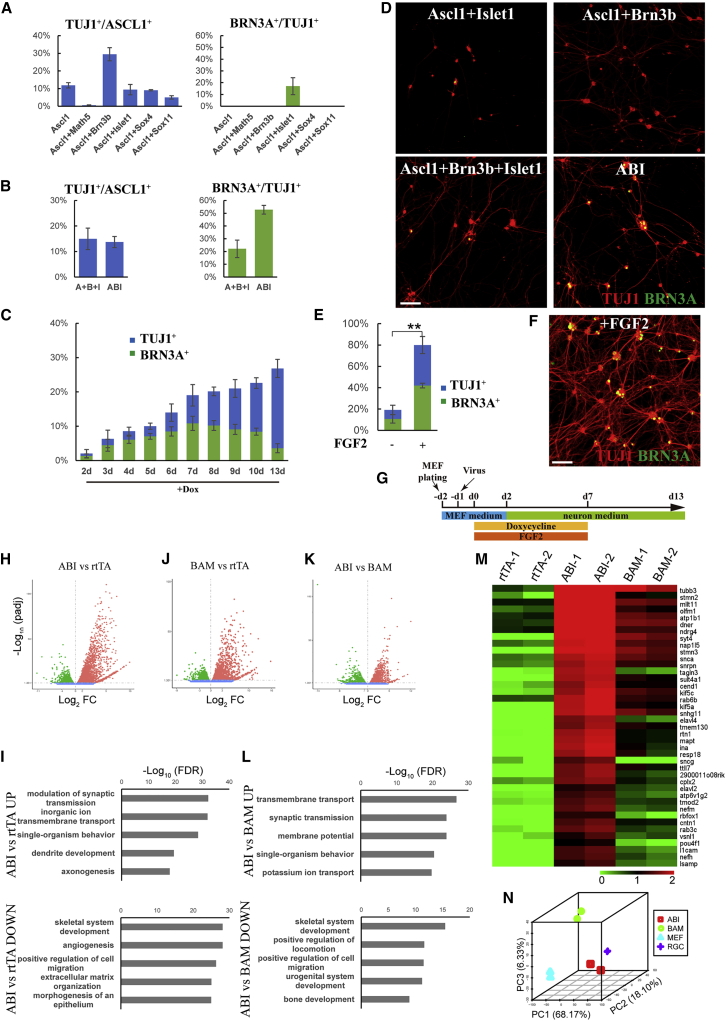
BRN3A is a widely used RGC marker that is expressed in most RGCs soon after they are generated (Xiang, 1998). We first tested whether BAM could reprogram mouse embryonic fibroblasts (MEFs) into BRN3A+ putative iRGCs. However, there was no BRN3A expression in BAM-induced iNs (Figure S1A). We then tested five RGC-genic TFs: Math5, Brn3b, Isl1, Sox4, and Sox11. In addition, considering the “pioneering factor” role of Ascl1 in inducing neuron properties (Wapinski et al., 2013), we included Ascl1 even though it is not expressed in most RGC-generating RPC lineages in vivo (Brzezinski et al., 2011). Ascl1 alone could not induce BRN3A+-iNs (Figure S1A). Ascl1+Isl1 induced BRN3A+-iNs (BRN3A+; TUJ1+), but the number was very low, and the induced neurons appeared morphologically immature (Figures 1A and 1D). Ascl1+Brn3b significantly improved the TUJ1+ iN induction efficiency of Ascl1, but there were no BRN3A+ cells among them (Figures 1A and 1D). Math5, Sox4, and Sox11 showed no improvement and sometimes even detrimental effects (Figure 1A). We next combined Ascl1, Brn3b, and Isl1. Encouragingly, Ascl1+Brn3b+Isl1 together could convert approximately 15% ± 4.2% of fibroblasts into iNs; among them, 22.1% ± 6.8% expressed BRN3A (Figures 1B and 1D). In the above experiment, Ascl1, Brn3b, and Isl1 were transduced by separate viruses; thus, only a portion of the plated cells received all three TFs with each at variable levels (Figure S1B). We speculated that efficient iRGC induction might require balanced expression levels between the three TFs. We thus constructed a polycistronic plasmid that expresses Ascl1, Brn3b, and Isl1 simultaneously and called the construct ABI. Although TUJ1+ generic iN fate induction was similar between the Ascl1+Brn3b+Isl1 group and the ABI group, the proportion of BRN3A+ cells among iNs increased dramatically in the ABI group (Figures 1B, 1D, and S1C), we thus used this ABI construct in all subsequent experiments. We next examined how long ABI is needed for efficient reprogramming and found that 7 days of induction was optimal (Figure 1C). Supplementing ABI with Math5, Sox4, or Sox11 did not further improve the induction efficiency and even showed detrimental effects, especially Math5 (Figure S1D, two experiments). Our previous work showed that fibroblast growth factor (FGF) signaling is required for the initiation of RGC development in vivo (Chen et al., 2013). Thus, we tested whether FGF2 could promote BRN3A+-iN induction. Excitingly, the addition of FGF2 significantly increased the TUJ1+ iN induction efficiency by approximately four times to 80.0% ± 8.0%, while the percentage of BRN3A+ cells among TUJ1+ iNs remained unchanged (Figures 1E, 1F, and S1C). It should be noted that FGF2 also improved the iN induction efficiency of Ascl1 and BAM, although not as dramatically as that of ABI (Figure S1E). Finally, we tested when FGF2 was needed to promote iN induction. The results showed that FGF2 was needed from the first day of reprogramming to efficiently induce TUJ1+ iNs (Figure S1F, two experiments). Taken together, we determined the optimal BRN3A+-iN induction scheme: ABI was induced for 7 days with FGF2 supplementation during the same period, after which both doxycycline (Dox) and FGF2 were withdrawn, and the cells were allowed to further mature in neuronal culture medium for an additional 6 days (Figure 1G).
Figure 1.
Ascl1/Brn3b/Isl1 Efficiently Reprogram MEFs into iRGCs
(A) Quantification of generic iN (TUJ1+) and BRN3A+ iN reprogramming efficiencies induced by combining Ascl1 with various RGC-genic TFs.
(B) Quantification of iN and BRN3A+ iN reprogramming efficiencies by viruses that carry Ascl1, Brn3b, and Isl1 separately (A+B+I) or by viruses that carry a polycistronic construct (ABI).
(C) Quantification of iN and BRN3A+ iN reprogramming efficiencies induced by ABI for different lengths of time.
(D) TUJ1 (red) and BRN3A (green) immunofluorescent images showing the reprogramming outcomes of different TF combinations.
(E) Quantification showing that FGF2 dramatically increased the iN reprogramming efficiency of ABI. FGF2 was supplemented from day 1 to 7 of reprogramming. ∗∗p < 0.01.
(F) TUJ1 (red) and BRN3A (green) immunofluorescent images showing efficient induction of BRN3A+ iNs by ABI supplemented with FGF2.
(G) The final reprogramming scheme.
(H) Volcano plot showing gene expression differences between ABI-induced neurons versus rtTA-transduced control cells.
(I) GO terms enriched in the groups of genes upregulated (top) or downregulated (bottom) in ABI-induced neurons compared with rtTA-transduced control cells.
(J) Volcano plot showing gene expression differences between BAM-induced neurons versus rtTA-transduced control cells.
(K) Volcano plot showing gene expression differences between ABI-induced neurons versus BAM-induced generic neurons.
(L) GO terms enriched in the groups of genes upregulated (top) or downregulated (bottom) in ABI-induced neurons compared with BAM-induced generic neurons.
(M) The heatmap shows that RGC marker genes were strongly expressed in ABI-induced neurons, which were much stronger than the levels in BAM-induced neurons.
(N) Principal component analysis (PCA) showing differences between the transcriptomes of ABI-induced neurons, BAM-induced neurons, MEFs, and native RGCs.
Data in (A)–(C) and (E) represent means ± standard deviation, corresponding to three independent experiments. Scale bars, 100 μm.
ABI-Induced iNs Resemble RGCs at the Transcriptome Level
To characterize the molecular signatures of ABI-induced iNs, we purified ABI-iNs on day 13 of reprogramming and performed RNA-seq on the transcriptome. Compared with that of control cells, the transcriptome of ABI-iNs was significantly enriched for gene ontology (GO) terms, such as “modulation of synaptic transmission,” “ion transportation,” “dendrite development,” and “axonogenesis,” consistent with the neuron identity of the reprogrammed cells; while downregulated genes involved in “regulation of cell migration” and “ECM organization,” consistent with the shutdown of fibroblast properties in the reprogrammed cells (Figures 1H and 1I). We also purify BAM-induced iNs and sequenced the transcriptome for comparison. Compared with that of control cells, the transcriptome of BAM-iNs was enriched for biological processes, such as “nervous system development,” and downregulated genes involved in “angiogenesis” and “wound healing” (Figures 1J and S2A). Compared with BAM-iNs, ABI-iNs had 1,834 significantly upregulated and 1,171 significantly downregulated genes (Figure 1K). The upregulated genes were important for neuronal functions, such as “synaptic transmission,” “membrane potential,” and “potassium ion transportation,” while many downregulated genes were involved in mesoderm fates development, such as “skeletal system development,” “urogenital system development,” and “bone development” (Figure 1L). We next examined the expression patterns of established RGC marker genes (Macosko et al., 2015). We found that all of these RGC marker genes were expressed at much higher levels in ABI-iNs than in BAM-iNs (Figure 1M). We then downloaded RNA-seq data of native mouse RGCs from the Gene Expression Omnibus for comparison (Park et al., 2019). Principal component analysis (PCA) showed that the transcriptomes of ABI-iNs were more closely related to the transcriptomes of native RGCs than to those of BAM-iNs (Figure 1N). Furthermore, our subsequent single-cell MCA (scMCA) analysis of the single-cell transcriptomes of ABI-iNs showed that they matched closely with those of native RGCs (please see Figure S4 below). Thus, we referred to ABI-induced iNs as iRGCs.
iRGCs Are Functional Neurons
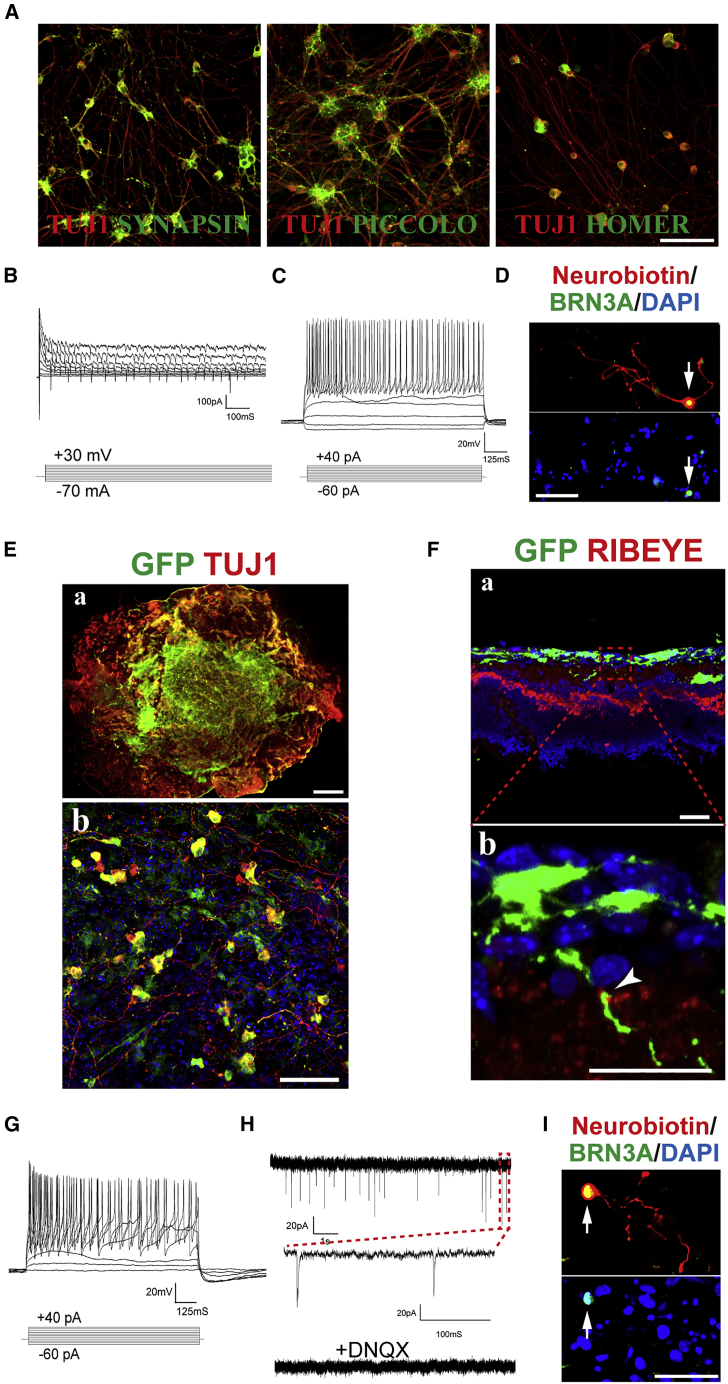
At day 13 of reprogramming, iRGCs strongly expressed proteins necessary for neuronal synapse formation, including SYNAPSIN, PICCOLO, and HOMER (Figure 2A). Patch-clamp examination showed that all iNs tested exhibited a negative resting membrane potential, averaging −70.66 ± 4.75 mV (n = 15); under the voltage-clamp mode, iNs exhibited fast inward currents and subsequent outward currents, corresponding to the activities of voltage-gated Na+ channels and K+ channels, respectively (Figure 2B); under the current-clamp mode, all the iRGCs tested fired repetitive action potentials (Figure 2C, n = 15), demonstrating the functionally mature neuron status of the iNs. BRN3A immunostaining after electrophysiological examination showed that most of the clamped cells were BRN3A+ iRGCs (seven out of nine neurobiotin-labeled clamped cells, Figure 2D). Next, we labeled ABI-transduced cells with GFP-expressing viruses and plated them on top of neonatal rat retinal explants on day 7 of reprogramming. After 7 days of coculture, many GFP-expressing cells expressed TUJ1 and extended thin long cellular processes (Figure 2E). Many of the iRGCs extended neurites into the inner plexiform layer of retinal explants, which closely opposed RIBEYE+ dots, suggesting the formation of ribbon synapses between transplanted iRGCs and explant retinal bipolar cells (Figure 2F). Under the current-clamp mode in the patch-clamp, 10 out of 11 cells fired repetitive action potentials (Figure 2G). More interestingly, in 3 out of 11 cells tested, we detected postsynaptic currents (PSCs), which could be blocked by DNQX (Figure 2H), suggesting the excitatory property of the PSCs. BRN3A immunostaining after electrophysiological examination showed that over half of the clamped cells were BRN3A+ iRGCs (five out of seven neurobiotin-labeled clamped cells, Figure 2I). The coculture experiment suggested that iRGCs could form synaptic connections with retinal interneurons.
Figure 2.
iRGCs Are Functional Neurons
(A) iRGCs expressed various synaptic proteins.
(B) Under voltage-clamp mode, iRGCs exhibited fast inward Na+ currents and outward K+ currents.
(C) Under current-clamp mode, iRGCs fired repetitive action potentials.
(D) A cell (arrow) labeled by neurobiotin during electrophysiological recording was a BRN3A+ iRGC.
(E) (a) Whole-mount immunofluorescence image showing ex-vivo-transplanted GFP-labeled iRGCs adhering to the top of the RGC layer of the rat retinal explants. (b) Higher-magnification image of the center of “a” showing transplanted iRGCs extended long neurites.
(F) (a) Tissue section immunofluorescence image showing ex-vivo-transplanted GFP-labeled iRGCs extended neurites into the inner plexiform layer of retinal explants. (b) Close-up image of the red-squared area in “a.” The arrowhead points to the RIBEYE+ dots, indicating ribbon synapses between the GFP+ iRGC and explant retinal bipolar cells.
(G) Ex-vivo-transplanted iRGCs fired repetitive action potentials.
(H) Ex-vivo-transplanted iRGCs exhibited spontaneous excitatory postsynaptic currents, which were blocked by DNQX.
(I) A transplanted cell (arrow) labeled by neurobiotin during electrophysiological recording was a BRN3A+ iRGC.
Scale bars: 100 μm (A, D, and I), 1 mm (E, a), 50 μm (E, b and F, a), and 25 μm (F, b).
Cellular Events during iRGC Reprogramming
To track the iRGC reprogramming process, we collected cells every day and stained them for TUJ1 and one of the ABI TFs or BRN3A. Twenty-four hours after Dox induction, approximately 62% of cells had started expressing ABI; the percentage of ABI-expressing cells slightly increased to approximately 73% on day 3 of reprogramming, then gradually decreased to 39% by day 7, and abruptly decreased to approximately 24% on day 8 when Dox was withdrawn from the medium; then, ABI was maintained only in TUJ1+ cells (Figure 3A). As early as day 3 of reprogramming, some ABI+ cells started to express strong TUJ1 signals with signs of elongated cellular processes. The proportion of TUJ1+ cells continued to increase between day 3 and 5, and the elongating neurites became more obvious. The induced cells exhibited typical neuronal morphology by day 9 of reprogramming (Figure 3A). BRN3A induction seemed to lag behind TUJ1 induction, but some presumptive TUJ1+ iN cells already showed BRN3A expression as early as day 5 of reprogramming, and the population continued to increase until it reached a plateau around day 9 of reprogramming (Figure 3A). These gene expression tracing experiments suggest that the iRGC reprogramming route was committed early.
Figure 3.
Cellular Events during iRGC Reprogramming
(A) TUJ1 (red) and ASCL1/BRN3b (green) or BRN3A (green) immunofluorescence images showing the changes in the expression patterns of the genes during iRGC reprogramming.
(B) Reprogramming cells treated without (top row) or with (bottom row) FGF2 were fed BrdU for 2 h, and stained for BrdU (red) and ASCL1 (green) during the first 8 days of reprogramming.
(C) Quantification of the percentage of BrdU+ cells during the first 8 days of reprogramming. “All” represents all the cells in the well; “ABI+” represents cells overexpressing ABI (judged by ASCL1 immunostaining).
(D) Quantification of the percentage of BrdU+ cells in all cells in the well after continuous BrdU feeding from day 2 to 13 of reprogramming.
(E) All TUJ1+ (red) iNs at day 13 of reprogramming in both FGF2-untreated (left) and FGF2-treated (right) groups did not incorporate BrdU (green) after continuous BrdU feeding from day 2 to 13 of reprogramming, while many other cells that failed the reprogramming process (TUJ1−) had proliferated (BrdU+).
(F) Immunofluorescence images of the TUNEL assay of FGF2-untreated (top row) and -treated (bottom row) cells on day 2 (left) and day 4 (right) of reprogramming.
(G) Quantification of the percentage of TUNEL+ cells in the ABI+ and ABI− groups of cells treated with or without FGF2 on day 2 and 4 of reprogramming. The percentages were calculated as follow: TUNEL+; ASCL1+/ASCL1+ for ABI+ groups, and TUNEL+; ASCL1−/ASCL1− for ABI− groups.
Scale bars, 100 μm. Data in (C), (D), and (G) represent means ± standard deviation, corresponding to three independent experiments.
We next examined the cell proliferation dynamics during iRGC reprogramming. We pulse-labeled cells with 2-h bromodeoxyuridine (BrdU) feeding. Initially, approximately 15% of MEFs were positive for BrdU, and FGF2 increased the population to approximately 24% (Figures 3B and 3C). On day 3 of reprogramming, when the culture medium was changed from serum containing MEF medium to serum-free neuronal medium, there was a sharp reduction in the number of BrdU+ cells in both the FGF2-treated and FGF2-untreated groups. In the untreated groups, BrdU+ cells quickly decreased to a negligible level by day 5 of reprogramming, but in the FGF2-treated groups, FGF2 maintained a significant number of cells in the cell cycle until day 7 of reprogramming, when FGF2 was withdrawn from the medium; on day 8, most cells in all groups had stopped proliferating (Figures 3B and 3C). Interestingly, we found that exogeneous ABI overexpression had no effect on the cell-cycle status, nor did it affect the response of transduced cells to FGF2 treatment: ABI-overexpressing cells proliferated at speeds similar to those of the cells that escaped viral infection (Figures 3B and 3C, compare ABI+ groups with all cell groups). In summary, our BrdU pulse-labeling experiment demonstrated that, with the help of FGF2, a significant number of ABI-overexpressing cells were actively proliferating during the early stage of iRGC reprogramming.
Next, we continued feeding cells from day 2 of reprogramming until day 13. Without FGF2, approximately 24% of ABI-infected cells had gone through the cell cycle during the 13-day reprogramming period, and FGF2 treatment increased the population by approximately 1-fold (Figure 3D). Without FGF2, all TUJ1+ iNs were negative for BrdU, meaning that they had exited the cell cycle before day 2 of reprogramming (Figure 3E, left). In the FGF2-treated group, to our surprise, even though nearly half of the cells in the well showed strong BrdU labeling, none of the TUJ1+ iNs were positive for BrdU (Figure 3E, right). These results demonstrated that all iNs had exited the cell cycle before day 2 of reprogramming.
We then used the TUNEL assay to examine apoptotic activity. On day 2 of reprogramming, apoptotic cells were only occasionally found in both the FGF2-treated and FGF2-untreated groups (Figures 3F and 3G). On day 4 of reprogramming, without FGF2, many cells underwent apoptosis (Figures 3F and 3G). Interestingly, ABI-expressing cells seemed less sensitive to the culture condition change (Figure 3G), possibly because ABI-overexpressing cells were adopting a neuronal fate, enabling better survival in neuronal medium. Supplementing FGF2 significantly protected cells from apoptosis, and the effect was even more prominent for ABI-overexpressing cells (Figures 3F and 3G). Thus, our results showed that FGF2 protected ABI-overexpressing cells from apoptosis.
Ascl1, Brn3b, and Isl1 Function Differently in Controlling iRGC Reprogramming
To explore the roles Ascl1, Brn3b, and Isl1 each contributed to iRGC reprogramming, we isolated MEFs that were transduced with Ascl1, Brn3b, and Isl1 individually, as well as cells transduced with ABI on day 2 of reprogramming, and sequenced their transcriptomes. Each of the three genes alone can significantly affect gene expression in MEFs (Figure 4A). PCA showed that Ascl1, Brn3b, and Isl1 affected the transcriptome of MEFs differently (Figure 4B). Interestingly, the transcriptome status of ABI-overexpressing cells resided between those of Ascl1/Brn3b/Isl1-overexpressing cells, but was mostly in the same direction as that of Asc1-overexpressing cells (Figure 4B); these results suggest that Ascl1 played a dominant role in modifying the transcriptome of ABI-overexpressing cells and that Brn3b and Isl1 might counteract each other, as well as some functions of Ascl1, to ensure the iRGC reprogramming route. In line with this, overlapping the lists of differentially expressed genes (DEGs) in different groups of cells compared with that of rtTA-transduced control cells showed that there were more common DEGs between ABI-overexpressing cells with Ascl1-overexpressing cells than with Brn3b- or Isl1-overexpressing cells (Figure 4C). GO term enrichment analyses showed that individually overexpressing Ascl1 and Isl1 was sufficient to upregulate many genes involved in neuronal development and/or maturation, and Ascl1 appeared more potent than Isl1 in promoting neuronal gene expression. All three factors alone suppressed the expression of genes involved in the mesenchymal properties of MEFs, such as ECM organization. Brn3b and Isl1 appeared to further suppress the expression of other tissue fates, such as bone formation. Interestingly, Brn3b suppressed the expression of genes involved in “skeleton system development,” indicating that one function of Brn3b during iRGC reprogramming might be to suppress the unwanted muscle cell fate often induced by Ascl1 during neuronal reprogramming (Karow et al., 2012; Treutlein et al., 2016) (Figure 4D). Comparing the transcriptome of ABI-overexpressing cells with the transcriptome of individual factor-overexpressing cells further supports the notion that Ascl1 play more important roles in promoting neuronal fate (Figures S2B–S2D).
Figure 4.
Ascl1, Brn3b, and Isl1 Function Differently in Controlling iRGC Reprogramming
(A) Volcano plots showing gene expression changes in cells infected with different TF-expressing viruses on day 2 of reprogramming.
(B) PCA showing differences between the transcriptomes of cells infected with different TF-expressing viruses on day 2 of reprogramming.
(C) Venn diagram showing overlapping differentially expressed genes (DEGs) in different groups of cells infected with different TF-expressing viruses.
(D) GO terms enriched in each group of DEGs.
(A) Ascl1 only; (B) Brn3b only; I, Isl1-only.
We also sequenced and compared the transcriptomes of reprogramming cells treated with or without FGF2 at day 2 of reprogramming. The results showed that FGF2 treatment greatly promoted the expression of genes involved in cell proliferation and DNA damage response and downregulated the expression of genes involved in cell adhesion, cell differentiation, and neuronal development (Figures S3A–S3C). Comparing ABI+ cells and ABI− cells suggested that survival-promoting effects of FGF2 were more prominent in ABI+ cells than in ABI− cells, while FGF2 treatment further repressed biological processes involved in neuronal development or maturation, such as nervous system development and “neurotransmitter secretion” in ABI+ cells (Figure S3D).
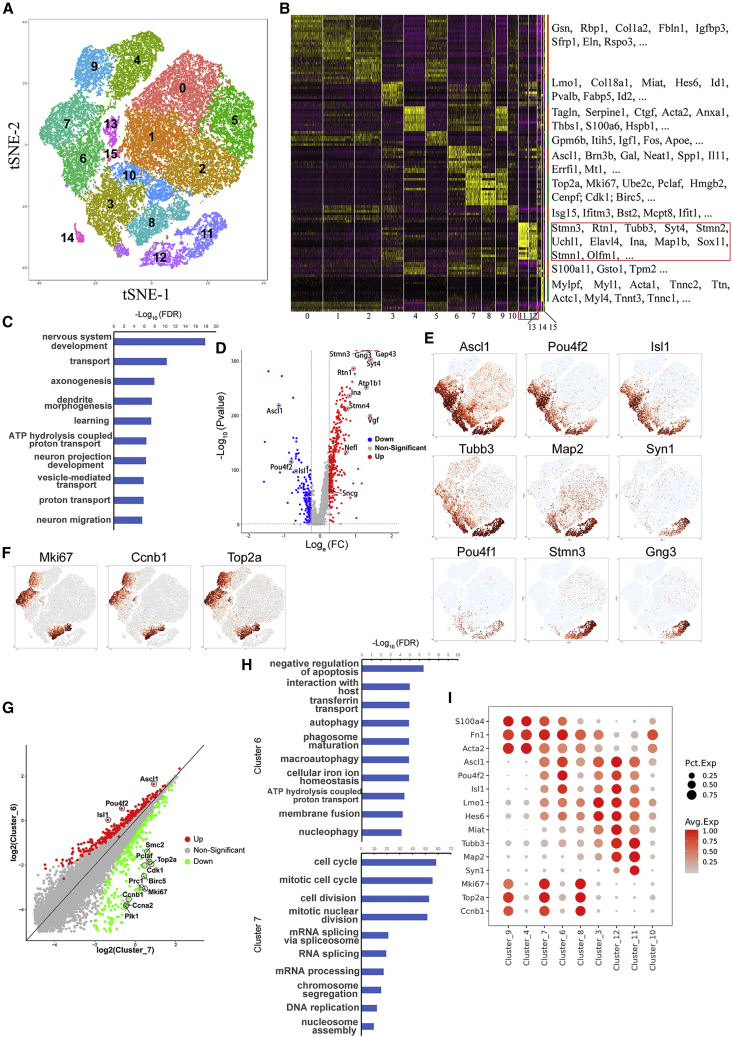
scRNA-Seq Analyses Reveal the iRGC Reprogramming Route that Is Committed Early and Is Determined by the Cell-Cycle Status of the Reprogramming Cell at the Time
To dissect the molecular events during iRGC reprogramming, we used scRNA-seq to sequence the transcriptomes of individual cells collected at six time points along the reprogramming path: the starting MEFs, cells at early reprogramming stages (day 2 and 4), the end of exogeneous gene overexpression (day 7), 2 days after exogenous gene withdrawal (day 9), and the end of reprogramming (day 13). Using the unsupervised dimensional reduction and visualization method of t-distributed stochastic neighbor embedding, we clustered cells from all stages into 16 cell clusters (Figure 5A). Based on the marker genes for each cluster (Figure 5B) and scMCA (Han et al., 2018) (Figure S4), we determined that cluster 11 and cluster 12 were the two clusters of cells that had been successfully reprogrammed into neurons (Figures 5B, 5C, 5E, and S5A). Comparison between cluster 11 and cluster 12 showed that cluster 11 represents the more mature neurons (Figures 5D, S5B, and S6). Furthermore, scMCA analysis showed that the transcriptomes of cells in cluster 11 closely matched the single-cell transcriptomes of retinal ganglion cells (Figure S4A, cluster 11 in the green circle), and a number of RGC-specific markers, including Pou4f1, Stmn3, and Gng3, were strongly expressed in cluster 11 (Figure 5E), further supporting our conclusion based on population sequencing that ABI-induced neurons resemble native RGCs.
Figure 5.
scRNA-Seq Analyses Reveal the iRGC Reprogramming Route
(A) Reprogramming cells collected on days 0 (MEF) 2, 4, 7, 9, and 13 were clustered into 16 clusters.
(B) The heatmap shows the expression patterns of calculated markers of each cluster across all cells.
(C) GO terms enriched in cluster 11-specific genes.
(D) Volcano plot showing gene expression differences between clusters 11 and 12.
(E) The expression patterns of reprogramming TFs, representative neuronal genes and RGC marker genes across all cells.
(F) The expression patterns of key cell-cycle regulators across all cells.
(G) Scatterplot showing gene expression differences between clusters 6 and 7.
(H) GO terms enriched in cluster 6- or cluster 7-specific genes.
(I) Bubble plot showing the expression dynamics of representative genes for mesenchymal cells, reprogramming TFs, intermediate reprogramming stage, neuronal cells, and proliferating cells across key clusters directly involved in iRGC reprogramming.
According to the marker genes for each cluster (Figure 5B), scMCA analysis (Figure S4), and the stages of the cells collected (Figure S6), we deduced cellular identities for the rest of the clusters: clusters 4 and 9 were starting MEF cells; clusters 6 and 7 were MEF cells at day 2 of reprogramming; cluster 3 contained cells that were starting to exhibit some neuronal properties; clusters 0, 1, 2, and 5 contained cells at the end stage that failed to be reprogrammed into neurons and adopted a stromal cell-like status; clusters 8 and 10 contained cells at the intermediate stage of the failed reprogramming route to the stromal fate; and cluster 14, although small in number, contained muscle-like cells, which also appeared in other iN reprogramming systems involving Ascl1 (Karow et al., 2012; Treutlein et al., 2016) (Figures 5B, and S4). Clusters 13 and 15 were two clusters of cells with low sequencing depth; otherwise, they were very similar to clusters 6 and 1, respectively. Three transgenes, Ascl1, Brn3b, and Isl1, were strongly expressed in clusters 7, 6, 3, 12, and 11 but mostly absent from clusters 4, 9, 0, 1, 2, and 5 (Figure 5E), which was consistent with the dynamics of transgene expression revealed by immunostaining tracking. Tuj1 (Tubb3) was induced as early as day 2 of reprogramming in clusters 6 and 7, grew stronger in cluster 3, and became the strongest in clusters 11 and 12, while Map2 lagged behind Tubb3, and other genes for more mature neurons were turned on only in cluster 12 and became stronger in cluster 11 (Figure 5E). Based on these expression dynamics of transgenes, neuronal genes, and cell-cycle genes (see below) (Figures 5E, 5F, and 5I), in combination with the molecular identities of each cluster and the stages at which the cells were collected, we speculate that the reprogramming route from MEF to iRGC was as follows: a portion of MEFs (clusters 4 and 9) infected with ABI gradually tuned down the mesenchymal properties of MEF during the period, represented by cluster 6/13, gained some neuronal properties in cluster 3, became neuron-like cells in cluster 12, and finally matured in cluster 11 (Figure S4A, red dashed arrows, and Figure S6D). A small portion of cells in the intermediate cluster 3 aberrantly adopted muscle cell fate (cluster 14), while the rest returned to a stromal-like status due to the loss of transgene expression (through cluster 10 to clusters 0, 1, 2, and 5). Actively proliferating MEFs infected with ABI in cluster 7 also turned on some neuronal properties, represented by cluster 8, but failed to go further and returned to a stromal-like status (through cluster 10 to clusters 0, 1, 2, and 5) (Figure S4A, black dashed arrows).
Interestingly, when we were inspecting starting MEFs and day 2 reprogramming cells, we noticed that cells at each stage were segregated into two clusters with dramatically different cell-cycle statuses (Figure 5F). To examine whether the two clusters at each stage represented two different reprogramming-competent statuses, we compared the gene expression patterns between the two clusters at each stage. The two MEF clusters, clusters 4 and 9, were very similar to each other except that genes involved in cell-cycle progression were dramatically downregulated in cluster 4 (Figures S5C and S5D). Our preliminary data indicated that some iRGCs came from cells that proliferated during days 1 and 2 of reprogramming. Thus, MEFs in clusters 4 and 9 were equally competent to become iRGCs, indicating that it is unlikely that there was a subset of MEFs predetermined to enter the iRGC reprogramming route upon induction. Regarding the two clusters of day 2 cells, cluster 7 was actively proliferating cells, and cluster 6 was cells that had dropped out of the cell cycle (Figure 5F). Our BrdU-tracing experiments showed that all iRGCs had exited the cell cycle before day 2 of reprogramming (Figure 3E), which means that they all came from cells in cluster 6. Taken together, these data suggested that the cell-cycle status had segregated the day 2 cells into two groups with different iRGC induction-competent statuses: proliferation-quiescent cluster 6 cells were competent to be reprogrammed into iRGCs, while actively proliferating cluster 7 cells were resistant to reprogramming. Comparing gene expression patterns between the two clusters, we found that the transcriptomes of cluster 6 cells dramatically upregulated genes involved in pathways, such as “negative regulation of apoptotic process,” “autophagy,” and “phagosome maturation” (Figures 5G and 5H), suggesting that cluster 6 cells were more resistant to cell death and had more active autophagy activity.
iRGC Reprogramming Route Involves Intermediate States
To explore whether the iRGC reprogramming route includes intermediate states, we used monocle2 to perform pseudotime ordering of reprogramming cells along the successful reprogramming route (cells in clusters 6, 3, 12, and 11). Pseudotime ordering showed that reprogramming was a continuous process that progressed from cluster 6 through clusters 3 and 12 to reach cluster 11 of mature neurons (Figures 6A and 6B). We identified genes that significantly changed their expression patterns during the reprogramming period and clustered them into five gene clusters based on their expression dynamics along the pseudotime sequence. GO term enrichment analysis of each gene cluster showed that infected MEFs quickly downregulated genes involved in the “angiogenesis,” “apoptotic process,” “positive regulation of cell proliferation,” and “cytoskeleton organization” pathways (Figure 6C, gene cluster 3) and gradually downregulated genes involved in “translation,” “cell-matrix adhesion,” and “positive regulation of cell migration” (Figure 6C, gene cluster 5). Neuronal function genes, such as genes involved in “synaptic transmission,” “neuron migration,” and “learning” (Figure 6C, gene cluster 1), were activated at the end stage of reprogramming. Interestingly, in the middle of the reprogramming route, there was a transient upregulation of genes involved in inflammatory response (Figure 6C, gene cluster 4), which was followed by a gradual upregulation of genes involved in the epigenetic and transcriptional regulation of gene expression (Figure 6C, gene cluster 2). Thus, the pseudotime analysis suggested that the iRGC reprogramming route involves intermediate states that are characterized by a transient inflammatory-like reaction followed by active epigenetic and transcriptional remodeling.
Figure 6.
iRGC Reprogramming Trajectory in Pseudotime
(A) iRGC reprogramming trajectory revealed by pseudotime ordering of cells in clusters 3, 6, 11, and 12.
(B) Distribution of cells collected at different time points along the reprogramming trajectory.
(C) Left: the heatmap shows the expression patterns of key dynamically expressed genes along the reprogramming pseudotime (2,000 genes). Middle: the 2,000 genes were categorized into 5 clusters based on their characteristic expression dynamics, and the enriched GO terms for each gene cluster were identified. Right: expression patterns of representative genes along the reprogramming trajectory.
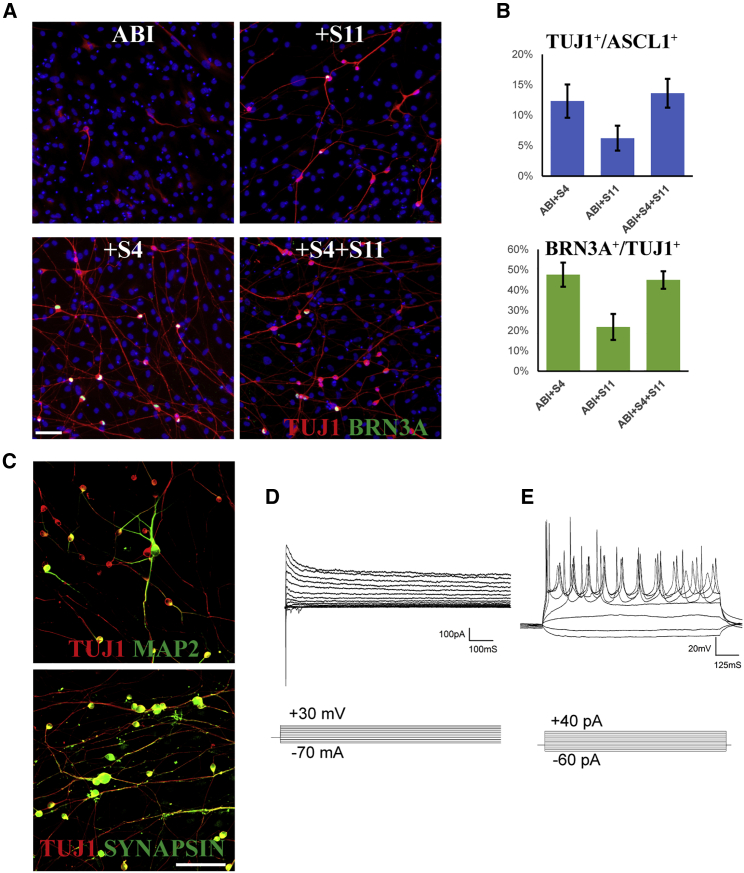
Human iRGC Induction
Finally, we investigated whether ABI could reprogram human fibroblasts into iRGCs. However, ABI failed to reprogram human neonatal primary dermal fibroblasts (HDFn) into iNs or iRGCs (Figure 7A). We then screened a set of neurogenic/RGC-genic TFs and found that ABI plus Sox4 or Sox11 induced BRN3A+; TUJ1+ RGC-like cells from HDFn, and Sox4 performed better than Sox11 (Figures 7A and 7B). Adding both Sox4 and Sox11 to ABI resulted in similar performance to ABI+Sox4 (Figure 7B), and we therefore focused on the ABI+Sox4 (ABIS4) factor combination for the rest of our experiment. A factor combination test between Ascl1, Brn3b, Isl1, and Sox4 showed that all four TFs were required for the induction of iRGCs from HDFn. The human iRGCs generated by ABIS4 expressed other mature neuron markers, such as MAP2 and SYNAPSIN (Figure 7C), exhibited a negative resting membrane potential, averaging −71.39 ± 3.99 mV (n = 20), fast inward currents, and subsequent outward currents under the voltage-clamp mode (Figure 7D), and produced repetitive action potentials under current-clamp mode (Figure 7E, n = 8), suggesting the functional maturation of induced human iRGCs.
Figure 7.
Ascl1/Brn3b/Isl1/Sox4 Can Efficiently Convert Human Fibroblasts into Functional iRGCs
(A) Immunofluorescence images showing that combining ABI with Sox4 could efficiently convert human fibroblasts into BRN3A+ neurons.
(B) Quantification of TUJ1+ generic iN and BRN3A+ iRGC reprogramming efficiency by combining ABI with Sox4 (S4) or Sox11 (S11).
(C) Human iRGCs express the mature neuron markers MAP2 and SYNAPSIN.
(D) Under voltage-clamp mode, human iRGCs exhibited fast inward Na+ currents and outward K+ currents.
(E) Under current-clamp mode, human iRGCs fired repetitive action potentials.
Scale bars, 100 μm. Data in (B) represent means ± standard deviation, corresponding to three independent experiments.
Discussion
Ascl1, Brn3b, and Isl1 Play Different Roles in iRGC Direct Induction
The mechanisms governing RGC development in vivo are relatively clear. It has long been known that bHLH TF-Math5 endows RPCs the competent state to generate RGCs and induces RGC fate when overexpressed in retinas (Liu et al., 2001; Mao et al., 2013). However, we found that Math5 failed to induce RGC fate from fibroblasts, either alone or together with other TFs, suggesting that the ability of Math5 to promote RGC fate requires a specific cellular environment found in RPCs. Downstream of Math5, Brn3b and Isl1 work synergistically to ensure RGC fate and to further regulate cell maturation (Gan et al., 1996; Li et al., 2014; Liu et al., 2001; Mu et al., 2008; Pan et al., 2008; Wu et al., 2015; Xiang, 1998). When we overexpressed Brn3b and Isl1 in MEFs, they each significantly changed the transcriptome of MEFs. However, overexpressing Brn3b and Isl1, either alone or together, failed to confer an RGC-like fate on fibroblasts. When we compared the genes that were regulated by Brn3b/Isl1 in fibroblasts with those in RPCs (Mu et al., 2008; Qiu et al., 2008), we found little overlap (Figure S7), suggesting that the gene regulatory effects of Brn3b/Isl1 are cell-type dependent. When Brn3b and Isl1 are combined with Ascl1, fibroblasts can be efficiently converted into iRGCs, suggesting that Ascl1 also functions as a “pioneering factor” in our iRGC reprogramming system, as in other iN reprogramming systems (Wapinski et al., 2013), to establish a suitable transcriptomic and epitranscriptomic environment for Brn3b and Isl1 to execute RGC fate-promoting functions. Consistently, when the three TFs are coexpressed, Ascl1 seems to play a dominant role in guiding fibroblast-to-neuron fate, while Brn3b and Isl1 counteract Ascl1 to repress unwanted cell fates and ensure the iRGC direction. This effect was especially associated with Brn3b, which seemed to play a more important role in suppressing the myocyte fate often induced during iN reprogramming involving Ascl1.
iRGC Reprogramming Route Is Committed at a Very Early Stage and Is Determined by the Cell-Cycle Status of the Reprogramming Cell at the Time
Our analyses on the iRGC reprogramming process showed that MEFs are generally homogeneous in terms of their competence to be reprogrammed into iRGCs, but ABI-transduced MEFs quickly segregate into two groups of cells with different iRGC induction-competent statuses at day 2 of reprogramming: proliferation-quiescent iRGC induction-competent cells and actively proliferating iRGC induction-incompetent cells. The BrdU-tracing experiments showed that all iRGCs had exited the cell cycle before day 2 of reprogramming (Figure 3E). The BrdU pulse-labeling experiment showed that, at this time point, approximately 20% of ABI-overexpressing cells were actively proliferating (Figure 3C), meaning that iRGCs came from the remaining 80% of ABI-overexpressing cells that had exited the cell cycle by this time point. The ABI-induced iN reprogramming efficiency calculated against ASCL1+ cells on day 2 of reprogramming was approximately 80% (Figure 1E), meaning that most ABI-overexpressing cells that had exited the cell cycle by day 2 of reprogramming were committed to an iN reprogramming route. These results suggest that the period around day 2 of reprogramming is the critical time window during which prospective iNs become committed to the successful reprogramming route, and cell-cycle status is the key factor that determines the iN induction competence of the reprogramming cells. Published reports suggested that Ascl1 overexpression causes reprogramming cells to exit the cell cycle during iN reprogramming (Treutlein et al., 2016); however, our BrdU-labeling experiments demonstrated that ABI-overexpressing cells proliferated at the same speed as ABI− cells (Figure 3C), suggesting that it is the nature of the culture environment that pushes cells out of the cell cycle and affects all the cells in culture equally. What makes proliferation-quiescent reprogramming cells competent to be converted into iNs? Comparing the differences in gene expression between proliferation-quiescent iN induction-competent cells and proliferation-active iN induction-incompetent cells showed that, in addition to cell-cycle-related genes, iN induction-competent cells upregulate genes involved in apoptosis and autophagy, suggesting that cell survival ability and autophagy activity might be involved in iN induction-competent state establishment. Future investigation on whether and how the cell cycle, apoptosis, and autophagy interact with each other to participate in the iN induction process is of interest.
iRGC Reprogramming Route Involves Intermediate States
A previous scRNA-seq study on the direct pericyte-to-neuron reprogramming process suggested that the iN reprogramming process proceeds through neural stem cell-like intermediates (Karow et al., 2012). We examined the expression patterns of the representative “switch genes” identified by Karow et al. (Nog, Lefty2, Dkk1, Notch2, Hes5, Hey1, and Id1) in our scRNA-seq data but found that these genes are not induced during iRGC reprogramming, possibly because our study and Karow et al.’s study used different starting somatic cell types and different reprogramming factors. In addition, classical markers for neural stem cells/RPCs, including Nestin, Musashi, Six3, Rax, Vsx1, Sox2, and Sox9, are not induced during iRGC reprogramming, suggesting that iRGC reprogramming does not involve a neural stem cell/RPC-like intermediate state. Instead, our scRNA-seq data suggest that the iRGC reprogramming route involves intermediate states during which reprogramming cells transiently upregulate the inflammatory-like response and actively modify the transcriptome and epigenome. In the future, it would be interesting to investigate whether and how these molecular events regulate the iRGC reprogramming process.
In summary, we show here that the Ascl1/Brn3b/Isl1 TF combination can quickly and efficiently reprogram fibroblasts into RGC-like functional neurons, which resemble native RGCs at the transcriptome level. Population transcriptome sequencing analyses indicate that Ascl1 plays a major role in inducing neuronal programs, while Brn3b and Isl1 counteract Ascl1 to suppress other tissue fates and promote RGC properties. By combining immunofluorescent imaging and scRNA-seq, we show that a successful iRGC reprogramming route is committed at a very early stage, and the competence of the reprogramming cells to enter the route is determined by the cell-cycle status of the cell at the time. Pseudotime ordering analyses show that the iRGC reprogramming process involves intermediate states that are characterized by a transient inflammatory-like response followed by active epigenomic and transcriptional modifications. The iRGC induction method established in this study provides a valuable cell source for glaucoma translational medicine applications, and the cellular and molecular mechanisms revealed in this study would facilitate future research on RGC and neuronal fate specification and reprogramming.
Experimental Procedures
iRGC Reprogramming
On the second day after plating, MEFs/HDFn were infected with lentiviruses. After 16–20 h, the cells were switched into fresh MEF medium containing Dox (Sigma) and FGF2 (PeproTech); 48 h later, the medium was switched to neuronal culture medium (DMEM/F-12, N2, B27) (all from Thermo Fisher Scientific), and forskolin (Sigma). Dox and FGF2 treatment was continued for 5 more days, unless otherwise specified. The medium was half-changed every other day. Reprogramming efficiency was calculated by dividing the number of TUJ1+ cells at the end of reprogramming with the number of ASCL1+ cells 48 h after Dox induction. See Supplemental Information for details.
Statistical Methods
All experiments subject to statistical analysis were performed with three biological independent replicates. Data are presented as mean ± standard deviation. Differences between experimental groups were tested using a two-tailed Student's t test. A p value of <0.05 was considered significant.
scRNA-Seq
Cells on reprogramming days 2, 4, 7, 9, and 13, as well as MEFs, were collected. The scRNA-seq libraries were generated using 10X Genomics Chromium Controller (10X Genomics, Pleasanton, CA) and sequenced using HiSeq X Ten (Illumina, San Diego, CA) by NovelBio. See Supplemental Information for details.
Data and Code Availability
The accession numbers for the RNA-seq and scRNA-seq data reported in this paper are NCBI Gene Expression Omnibus: GSE140689 and GSE140128.
Author Contributions
S.C. conceived, designed, supervised the project, and wrote the manuscript with inputs from all authors. J.W., Q.H., and K.Z. performed the experiments and data analyses with help from H.S., H.L., J.G., and L.H. G.Z. and J.K. helped with the electrophysiological study.
Acknowledgments
This work was supported by the Guangdong Provincial Department of Science and Technology (2015B020225003), and the National Natural Science Foundation of China (81870659).
Published: October 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.09.008.
Supplemental Information
References
- Bassett E.A., Wallace V.A. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Blanchard J.W., Eade K.T., Szucs A., Lo Sardo V., Tsunemoto R.K., Williams D., Sanna P.P., Baldwin K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.A.t., Kim E.J., Johnson J.E., Reh T.A. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.A.t., Prasov L., Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chang K.C., Hertz J., Zhang X., Jin X.L., Shaw P., Derosa B.A., Li J.Y., Venugopalan P., Valenzuela D.A., Patel R.D. Novel regulatory mechanisms for the SoxC transcriptional network required for visual pathway development. J. Neurosci. 2017;37:4967–4981. doi: 10.1523/JNEUROSCI.3430-13.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Li H., Gaudenz K., Paulson A., Guo F., Trimble R., Peak A., Seidel C., Deng C., Furuta Y. Defective FGF signaling causes coloboma formation and disrupts retinal neurogenesis. Cell Res. 2013;23:254–273. doi: 10.1038/cr.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G., Lignani G., Rubio A., Medrihan L., Yekhlef L., Sessa A., Massimino L., Giannelli S.G., Sacchetti S., Caiazzo M. Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell. 2015;17:719–734. doi: 10.1016/j.stem.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Gan L., Xiang M., Zhou L., Wagner D.S., Klein W.H., Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L., Wang S.W., Huang Z., Klein W.H. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev. Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Gascon S., Murenu E., Masserdotti G., Ortega F., Russo G.L., Petrik D., Deshpande A., Heinrich C., Karow M., Robertson S.P. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. Mapping the mouse cell atlas by Microwell-seq. Cell. 2018;172:1091–1107 e1017. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Ding Q., Xie X., Libby R.T., Lefebvre V., Gan L. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J. Biol. Chem. 2013;288:18429–18438. doi: 10.1074/jbc.M113.478503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M., Sanchez R., Schichor C., Masserdotti G., Ortega F., Heinrich C., Gascon S., Khan M.A., Lie D.C., Dellavalle A. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell. 2012;11:471–476. doi: 10.1016/j.stem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Li R., Wu F., Ruonala R., Sapkota D., Hu Z., Mu X. Isl1 and Pou4f2 form a complex to regulate target genes in developing retinal ganglion cells. PLoS One. 2014;9:e92105. doi: 10.1371/journal.pone.0092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Mo Z., Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl. Acad. Sci. U S A. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Lee Q.Y., Wapinski O., Castanon R., Nery J.R., Mall M., Kareta M.S., Cullen S.M., Goodell M.A., Chang H.Y. Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. eLife. 2019;8:e40197. doi: 10.7554/eLife.40197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C.A., Cho J.H., Wang J., Gao Z., Pan P., Tsai W.W., Frishman L.J., Klein W.H. Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development. 2013;140:541–551. doi: 10.1242/dev.085886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Fu X., Sun H., Beremand P.D., Thomas T.L., Klein W.H. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev. Biol. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Mu X., Fu X., Beremand P.D., Thomas T.L., Klein W.H. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Deng M., Xie X., Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.H., Snook J.D., Ostrin E.J., Kim S., Chen R., Frankfort B.J. Transcriptomic profiles of retinal ganglion cells are defined by the magnitude of intraocular pressure elevation in adult mice. Sci. Rep. 2019;9:2594. doi: 10.1038/s41598-019-39141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Bjorklund A., Lindvall O., Jakobsson J., Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F., Jiang H., Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J. Neurosci. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley H.A. Glaucoma. Lancet. 2011;377:1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- Sheng C., Zheng Q., Wu J., Xu Z., Sang L., Wang L., Guo C., Zhu W., Tong M., Liu L. Generation of dopaminergic neurons directly from mouse fibroblasts and fibroblast-derived neural progenitors. Cell Res. 2012;22:769–772. doi: 10.1038/cr.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A., Sim S., Neff N.F., Skotheim J.M., Wernig M. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R., Lee P.P., Goldberg I., Kotak S. An assessment of the health and economic burdens of glaucoma. Am. J. Ophthalmol. 2011;152:515–522. doi: 10.1016/j.ajo.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Wernig M. Direct lineage conversions: unnatural but useful? Nat. Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B.J., Buttermore E.D., Oliveira J.T., Mellin C., Lee S., Saber W.A., Wang A.J., Ichida J.K., Chiu I.M., Barrett L. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.W., Kim B.S., Ding K., Wang H., Sun D., Johnson R.L., Klein W.H., Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., Giresi P.G., Ng Y.H., Marro S., Neff N.F. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Kaczynski T.J., Sethuramanujam S., Li R., Jain V., Slaughter M., Mu X. Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. U S A. 2015;112:E1559–E1568. doi: 10.1073/pnas.1421535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev. Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Xu J., Du Y., Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Yang Z., Ding K., Pan L., Deng M., Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the RNA-seq and scRNA-seq data reported in this paper are NCBI Gene Expression Omnibus: GSE140689 and GSE140128.