Summary
Androgen receptor (AR) plays a fundamental role in most aspects of adult prostate homeostasis, and anti-androgen therapy represents the cornerstone of prostate cancer treatment. However, early prostate organogenesis takes place during pre-pubertal stages when androgen levels are low, raising the possibility that AR function is more limited during prostate development. Here, we use inducible AR deletion and lineage tracing in genetically engineered mice to show that basal and luminal epithelial progenitors do not require cell-autonomous AR activity during prostate development. We also demonstrate the existence of a transient bipotent luminal progenitor that can generate luminal and basal progeny, yet is also independent of AR function. Furthermore, molecular analyses of AR-deleted luminal cells isolated from developing prostates indicate their similarity to wild-type cells. Our findings suggest that low androgen levels correlate with luminal plasticity in prostate development and may have implications for understanding how AR inhibition promotes lineage plasticity in prostate cancer.
Keywords: prostate organogenesis, androgen receptor, luminal progenitor, lineage tracing, plasticity
Graphical Abstract

Highlights
-
•
Prostate epithelial cell types are specified when epithelial AR expression is low
-
•
Bipotent basal progenitors can generate luminal cells without cell-autonomous AR
-
•
Transient bipotent luminal progenitors can generate luminal and basal cells
-
•
AR-deleted luminal cells during development are molecularly similar to wild-type
Androgen receptor (AR) is fundamental for the adult prostate and is targeted in cancer treatment. Shibata and colleagues use lineage tracing to show that prostate progenitors do not require cell-autonomous AR during development. AR deletion does not affect bipotency of basal progenitors or activity of transient bipotent luminal progenitors. These findings suggest that luminal plasticity is correlated with low AR activity.
Introduction
Prostate tumors are initially dependent on the activity of androgen receptor (AR) and respond to androgen deprivation therapy. However, most prostate tumors will ultimately develop resistance, resulting in castration-resistant prostate cancer. In recent years, the development of potent anti-androgens has also led to emergence of tumors that are AR null or AR indifferent (Beltran et al., 2019). Consequently, understanding potential AR-indifferent functions in prostate biology would have significant translational impact.
In the mouse, organogenesis of the prostate begins before birth and continues through postnatal stages (Toivanen and Shen, 2017). In contrast to the adult prostate, postnatal prostate development is not strictly dependent on AR activity; following a transient burst of testosterone after birth (Motelica-Heino et al., 1988), levels of circulating androgens remain low until the onset of puberty (Jean-Faucher et al., 1978). Furthermore, tissue recombination and grafting studies using AR mutant (Tfm) mice have shown that AR is initially required in mesenchymal cells and not the epithelium during prostate induction, but is subsequently necessary in the prostate epithelium for production of secretory proteins (Cunha et al., 1987).
The adult prostate contains three major epithelial cell types, corresponding to basal cells, luminal cells, and rare neuroendocrine cells (Shen and Abate-Shen, 2010). Lineage-tracing studies have shown that the adult prostate epithelium is maintained by unipotent basal and luminal progenitors (Choi et al., 2012; Wang et al., 2013). In contrast, lineage tracing during postnatal prostate development has revealed multipotent basal progenitors that can differentiate into basal, luminal, and neuroendocrine cells, as well as unipotent basal and luminal progenitors (Ousset et al., 2012; Tika et al., 2019; Wuidart et al., 2016).
The requirement for AR in prostate luminal and basal progenitors has previously been studied during androgen-mediated prostate regeneration and homeostasis (Chua et al., 2018; Wu et al., 2007; Xie et al., 2017; Zhang et al., 2016). In the adult prostate, AR is required in luminal cells to suppress over-proliferation and to maintain epithelial tight junctions and cell polarity during homeostasis (Xie et al., 2017; Zhang et al., 2016). However, it has been unclear whether AR is necessary during prostate organogenesis for specification of luminal cells, either when derived from a luminal or a basal progenitor. Consequently, we have investigated the cell-autonomous epithelial requirements of AR during prostate organogenesis by lineage tracing combined with inducible deletion of AR.
Results
Specification of Luminal and Basal Cell Types Precedes High Prostate Epithelial AR Expression
Prostate epithelial buds arise from the urogenital sinus epithelium (UGE) at 17.5 days postcoitum and grow as solid epithelial cords which branch and develop into ducts with lumens surrounded by a pseudostratified epithelium (Toivanen and Shen, 2017). We examined AR expression in prostate cells from neonatal male mice from postnatal day 0 (P0) through 4 weeks of age by immunofluorescence (IF) staining (Figures 1A–1J and Figures S1A–S1D and S1J). We relied upon serial sectioning, histological landmarks, and Nkx3.1-lacZ reporter expression to distinguish prostate ducts (Figures S1G–S1I) (Kruithof-de Julio et al., 2013). Our analyses complement and extend previous analyses (He et al., 2018; Takeda and Chang, 1991).
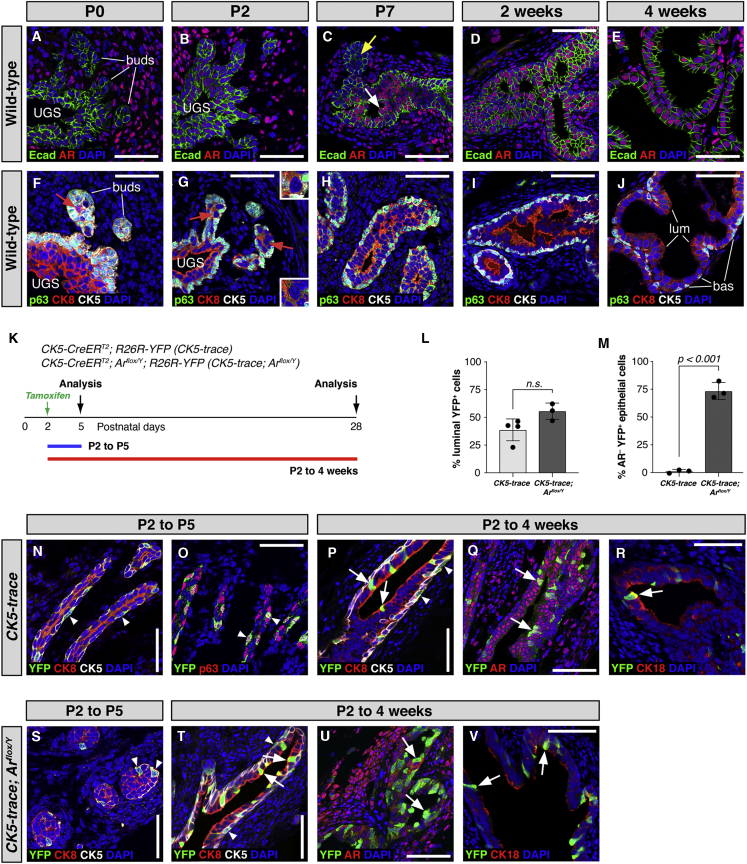
Figure 1.
AR Is Not Required in Bipotent Basal Progenitors in the Prostate Epithelium
(A–E) AR and E-cadherin (Ecad) expression in wild-type mouse prostates. Arrows in (C) indicate epithelial cells with low AR expression (yellow) and high AR expression (white) in prostate cells facing the lumen.
(F–J) Immunofluorescence (IF) staining for luminal (CK8) and basal (p63 and CK5) markers. Red arrow in (F) indicates co-expression of CK5 and CK8 at P0. Pre-basal cells (CK5highCK8lowp63+) and pre-luminal cells (CK5lowCK8highp63–; red arrows and insets in G) were observed at P2.
(K–V) AR deletion in basal cells during postnatal prostate development. (K) Analysis timeline. (L) Quantitation of YFP+ cells in the luminal layer that express CK8 at 4 weeks after lineage marking (n = 4 mice and n = 3 mice). (M) Quantitation of AR-negative YFP+ cells at 4 weeks after lineage marking (n = 3 mice each). (N–V) IF staining at 3 days (P2 to P5) or 4 weeks (P2 to 4 weeks) after lineage marking. Arrowheads indicate YFP+ basal cells, and arrows indicate YFP+ luminal cells. bas, basal; lum, luminal; P, postnatal day; UGS, urogenital sinus; n.s., not significant. Scale bars, 50 μm. Error bars represent standard deviation. The p values were calculated by t tests.
See also Figures S1–S3, and Tables S1A–S1E.
At P0 and P2 (n = 2 animals each), we observed strong nuclear AR expression in urogenital mesenchyme cells. We also detected nuclear AR staining in most UGE cells, with lower levels in prostate epithelial buds (Figures 1A, 1B, S1A, and S1B). At P7 (n = 2 animals), AR expression remained high in stromal cells, but was upregulated in prostate epithelial cells (Figure 1C). By the age of 2 and 4 weeks, we detected AR expression in almost all prostate epithelial cells, including basal cells (Figures 1D and 1E, n = 2 animals each; Figures S1C and S1D). In contrast, AR expression in stromal cells appeared lower compared with epithelial cells (Figures 1D, 1E, S1C, and S1D).
To correlate epithelial AR expression with specification of prostate basal and luminal cell types, we performed IF staining for basal (p63 and CK5) and luminal (CK8) markers (Figures 1F–1J). At P0, almost all prostate bud epithelial cells co-expressed p63, CK5, and CK8, although a few “inner” bud cells with reduced p63 expression and higher CK8 expression were observed (Figure 1F) (Ousset et al., 2012). At P2, we clearly distinguished two types of prostate epithelial cells, corresponding to “pre-basal” cells that expressed p63 and CK5 with low levels of CK8 (CK5highCK8lowp63+), and “pre-luminal” cells that lacked p63 expression and expressed low levels of CK5 together with higher levels of CK8 expression (CK5lowCK8highp63–) (Figure 1G; single channels in Figure S1E). p63-positive pre-basal cells were observed as a continuous layer of cells on the outer layer of the developing ducts, although occasional p63-negative cells with higher CK8 expression were found on the outer layer. By P7, the developing prostate ducts contained lumens with solid prostate bud tips, and were stratified into an outer cell layer expressing basal markers (p63 and CK5) with low levels of CK8 expression, and an inner cell layer expressing luminal markers (CK8) with low levels of CK5 and no p63 expression (Figures 1F–1J and S1F). At P7, the highest epithelial AR expression was observed in prostate epithelial cells facing the lumen (Figure 1C). These observations suggest that the bifurcation of luminal and basal cell types occurs after P0, before prostate lumen formation, and is essentially complete by P7.
AR Is Not Required in Bipotent Basal Progenitors
To determine whether androgen signaling is required in bipotent basal progenitors during prostate organogenesis, we used a tamoxifen-inducible CK5-CreERT2 transgenic driver together with an R26R-YFP reporter and a conditional Ar allele (Arflox/Y). Using the resulting CK5-trace (CK5-CreERT2; R26R-YFP) control mice and CK5-trace; Arflox/Y (CK5-CreERT2; Arflox/Y; R26R-YFP) mice, we treated P2 neonates with a low dose of tamoxifen to label p63+CK5highCK8low pre-basal cells. To confirm the specificity of basal lineage marking, we assessed YFP expression at P5, 3 days after tamoxifen treatment (Figures 1K, 1N, 1O, and S2A). We found that CK5-trace YFP+ cells were p63+CK5highCK8low, marking 27.0% of pre-basal cells (Figures 1N and 1O; single channels in Figure S2A; n = 102 YFP+ cells, 97.7% CK5+; quantitation in Table S1A). We also detected YFP expression specifically in p63+CK5highCK8low pre-basal cells in CK5-trace; Arflox/Y prostates (Figures 1S; single channels in S2C).
To investigate the consequences of AR deletion on basal progenitor activity, we examined YFP expression in CK5-trace and CK5-trace; Arflox/Y mice at 4 weeks after tamoxifen induction at P2. Tamoxifen treatment of P2 neonates resulted in slightly delayed prostate growth (Figures S4A–S4D). In CK5-trace prostates, we found that 39.1% ± 9.6% of YFP+ cells (n = 4 mice) were luminal, as determined by expression of CK8, whereas in CK5-trace; Arflox/Y prostates, 56.1% ± 6.9% of YFP+ cells (n = 3 mice) were luminal, as also shown by CK18 expression (Figures 1L, 1P, 1R, 1T, 1V, S2B, S2D, S3A, and S3B; Tables S1B and S1D). We confirmed that Ar, which is expressed in both basal and luminal cells (Figures 1D, 1E, S1C, and S1D) (Lee et al., 2012), was efficiently deleted, since AR staining was not observed in 73.8% ± 7.4% of YFP+ cells in CK5-trace; Arflox/Y prostates (n = 3); for comparison, AR staining was absent in only 1.5% ± 1.4% of YFP+ cells in control CK5-trace prostates (n = 3) (Figures 1M, 1Q, and 1U; Tables S1C and S1E). This finding suggests that cell-autonomous AR signaling is not required for basal progenitors to generate basal or luminal progeny during neonatal prostate development.
Bipotent Luminal Progenitors in Prostate Organogenesis
To investigate luminal progenitor activity during organogenesis, we marked luminal cells using a tamoxifen-inducible CK8-CreERT2 transgenic driver (Figure 2A). Administration of a low dose of tamoxifen to CK8-trace (CK8-CreERT2; R26R-YFP) neonates at P2 resulted in specific YFP labeling; 99.2% ± 1.8% (n = 5 mice) of YFP+ cells at P4 and P5 were CK5lowCK8high pre-luminal cells (Figures 2B and 2B'; Table S1F). In addition, CK8-trace marked cells did not express p63 at P4 (Figure 2C; n = 0/115 cells; Table S1G).
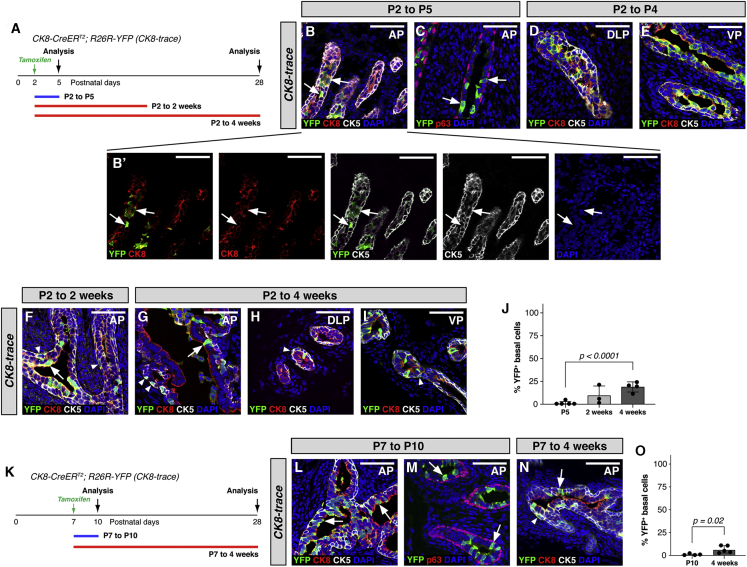
Figure 2.
Identification of Bipotent Luminal Cells during Postnatal Prostate Development
(A) Analysis timeline for P2 lineage marking.
(B–I) IF staining at 2 or 3 days (P2 to P4 or P5), 2 weeks (P2 to 2 weeks), or 4 weeks (P2 to 4 weeks) after lineage marking. (B′) Two-channel and single-channel images.
(J) Quantitation of YFP+ cells in the basal layer that express CK5 at P5, 2 weeks, and 4 weeks (n = 5, n = 3, and n = 4 mice).
(K) Analysis timeline for P7 lineage marking.
(L–N) IF staining at 3 days (P7 to P10) or 4 weeks (P7 to 4 weeks) after lineage marking.
(O) Quantitation of YFP+ cells in the basal layer that express CK5 at P10 and 4 weeks (n = 4 and n = 5 mice). Arrowheads point to YFP+ basal cells, and arrows point to YFP+ luminal cells. AP, anterior prostate, DLP, dorsal lateral prostate; VP, ventral prostate. Scale bars, 50 μm. Error bars represent standard deviation. The p values were calculated by t tests.
See also Figures S2 and S4, and Table S1F–S1L.
Unexpectedly, at 2 and 4 weeks after administration of tamoxifen to neonates at P2, both luminal and basal YFP-marked cells were observed in CK8-trace prostates; at 4 weeks, 19.6% ± 4.9% of YFP+ cells (n = 4 mice) were basal cells (Figures 2F, 2G, and 2J; single channels in Figures S2E and S2F; quantitation in Table S1H and S1I). Similar results were observed in both the dorsal lateral prostate and ventral prostate (Figures 2D, 2E, 2H, and 2I), indicating that p63–CK5lowCK8high pre-luminal cells in all three lobes can generate basal progeny.
Interestingly, lineage marking in CK8-trace neonates at P7, which was specific for luminal cells (Figures 2L, 2M, 2O, and S2G) (n = 575/578, 99.5%; Tables S1J and S1K), resulted in 6.0% ± 3.6% of YFP+ cells being basal at 4 weeks (n = 5 mice) (Figures 2K, 2N, and 2O; Table S1L; Figures S2H and S2I), suggesting that bipotent luminal progenitors remain at P7. These results indicate that luminal progenitors can generate basal cells during early postnatal development, and differ with the conclusions of an earlier study, which reported that luminal progenitors are unipotent during prostate organogenesis (Ousset et al., 2012).
AR Is Not Required in Luminal Progenitors during Prostate Organogenesis
To determine whether epithelial AR is required for luminal progenitor cells, we performed lineage tracing using CK8-trace and CK8-trace; Arflox/Y (CK8-CreERT2; Arflox/Y; R26R-YFP) mice (Figure 3A), inducing animals with tamoxifen at P2. By IF staining, we observed YFP+CK8+ luminal cells as well as YFP+CK5+ basal cells in CK8-trace; Arflox/Y prostates at 4 weeks after induction (Figures 3B, 3D, 3F, S2I, S2J, S3C, and S3D; Table S1N). The percentage of YFP+ cells that were CK5+ basal cells was 19.6% ± 4.9% in CK8-trace prostates and 22.6% ± 14.6% in CK8-trace; Arflox/Y prostates (Figure 3B). AR staining was not observed in 78.3% ± 14.4% of YFP-labeled cells in CK8-trace; Arflox/Y prostates (n = 4), whereas AR staining was absent in only 1.9% ± 2.6% of YFP+ cells in control CK8-trace prostates (n = 2), indicating that Ar was efficiently deleted (Figures 3C, 3E, and 3G; Tables S1M and S1O). At 4 weeks, AR-deleted epithelial cells in CK8-trace; Arflox/Y prostates were morphologically similar to AR-expressing epithelial cells (Figures 3D–3G). Furthermore, we found that AR-deleted luminal cells could contribute to the 8-week-old adult prostate, although these AR-deleted cells were less frequent (36.4% ± 27.4% of YFP-labeled cells, n = 4 mice) and were abnormal in appearance (Figures S4E–S4K; Tables S1P and S1Q), as described previously for AR deletion in the adult prostate (Xie et al., 2017; Zhang et al., 2016). These findings suggest that luminal cells do not have a cell-autonomous requirement for AR during prostate organogenesis.
Figure 3.
Cell Autonomous AR Is Not Essential for Luminal Prostate Progenitor Cells during Postnatal Prostate Development
(A) Timeline for lineage marking and analysis.
(B) Quantitation of YFP+ cells in the basal layer that express CK5 at 4 weeks after lineage marking (n = 4 and n = 3 mice).
(C) Quantitation of AR-negative YFP+ cells at 4 weeks after lineage marking (n = 2 and n = 4 mice). Error bars in (B and C) represent standard deviation. The p values were calculated by t tests. n.s., not significant.
(D–G) Immunofluorescence staining at 4 weeks after lineage marking. Arrowheads point to YFP+ basal cells, and arrows point to YFP+ luminal cells. Scale bars, 50 μm.
(H–N) RNA sequencing (RNA-seq) analysis of AR-deleted luminal cells. (H) Principal component analysis of RNA-seq samples from YFP+ luminal cells. (I–N) GSEA of an AR-deleted luminal signature comparing AR-deleted YFP+CD49flow/– luminal cells and wild-type YFP+CD49flow/– luminal cells with a signature of (I) AR-deleted adult luminal cells (Zhang et al., 2016), (J) AR-regulated genes (Carver et al., 2011), (K) neuroendocrine prostate cancer (Beltran et al., 2016), or (L) AR-negative non-neuroendocrine prostate cancer (Bluemn et al., 2017). (M) A signature defined between profiles of AR-deleted YFP+CD49flow/– luminal cells and bulk wild-type prostate shows significant enrichment with a signature defined between wild-type YFP+CD49flow/– luminal cells and bulk wild-type prostate. (N) Significant enrichment between basal-origin (CK5-trace versus bulk wild-type prostate) and luminal-origin (CK8-trace versus bulk wild-type prostate) signatures. NES, normalized enrichment score; p value was calculated using 1,000 gene permutations.
See also Figures S2–S4, and Tables S1M–S1O.
Molecular Similarity of Wild-Type and AR-Deleted Luminal Cells
Because we did not detect overt phenotypic differences between wild-type and AR-deleted luminal cells during organogenesis, we next examined whether differences might exist at the molecular level by expression profiling of isolated luminal cells. We dissociated prostate tissues and sorted YFP+CD49flow/– luminal-enriched cells from 4-week CK5-trace, CK5-trace; Arflox/Y, CK8-trace, and CK8-trace; Arflox/Y prostates induced with tamoxifen at P2 (Figures S3E–S3N). We performed RNA sequencing of the isolated wild-type and AR-deleted YFP+CD49flow/– luminal cells to compare their expression profiles. Notably, principal component analysis revealed that the wild-type (n = 3 samples) and AR-deleted luminal samples (n = 5 samples) did not cluster by genotype (Figure 3H). We also observed that basal-derived and luminal-derived luminal cells did not display strong clustering (Figure S4L).
To assess whether AR deletion affected luminal differentiation, we performed gene set enrichment analysis (GSEA) using an expression signature defined between profiles of AR-deleted YFP+CD49flow/– luminal cells and wild-type YFP+CD49flow/– luminal cells. We compared this AR-deleted luminal signature to an AR-deleted luminal signature from adult mouse prostates (Zhang et al., 2016), and found significant enrichment of AR downregulated genes (Figure 3I). Leading edge genes included Cldn3, which encodes a tight junction protein that is regulated by AR (Zhang et al., 2016), and Tgm4 and Spink3, which are prostate secretory proteins (Table S2). When we compared the AR-deleted luminal signature with an AR-regulated signature derived from adult mouse prostate (Carver et al., 2011), we observed negative enrichment of AR-upregulated genes (Figure 3J), consistent with the loss of AR function in the AR-deleted luminal cells. However, we did not observe any enrichment in comparison with a signature of neuroendocrine prostate cancer or with a signature of AR-negative non-neuroendocrine (“double-negative”) prostate cancer (Figures 3K and 3L) (Beltran et al., 2016; Bluemn et al., 2017).
To assess the potential similarity of AR-deleted and wild-type luminal cells, we generated another expression signature defined between profiles of wild-type YFP+CD49flow/– luminal cells and bulk wild-type 3-week prostate tissue. GSEA comparison of AR-deleted luminal and wild-type luminal signatures revealed significant enrichment, indicating strong similarities between the AR-deleted luminal cells and wild-type luminal cells (Figure 3M). We also generated signatures defined between expression profiles of YFP+ luminal cells from CK5-trace prostates with bulk wild-type prostates, and between profiles of YFP+ luminal cells from CK8-trace prostates with bulk wild-type prostates. GSEA revealed significant enrichment between these expression signatures, indicating that luminal cells derived from basal progenitors are similar to luminal cells derived from luminal progenitors (Figure 3N). These findings are consistent with our lineage-tracing results, and indicate that AR is not required cell autonomously in luminal cells during neonatal prostate organogenesis.
Discussion
In our study, we have defined key steps in the bifurcation of basal and luminal epithelial lineages during mouse prostate organogenesis, and have demonstrated the existence of a transient bipotent luminal progenitor. Based on these and previous findings (Ousset et al., 2012; Pignon et al., 2013; Tika et al., 2019; Wuidart et al., 2016), we propose a lineage hierarchy in which a prostate bud epithelial progenitor with mixed basal and luminal characteristics gives rise to distinct populations of pre-basal and pre-luminal cells, which in turn generate both bipotent and unipotent basal and luminal progenitors (Figure 4). We suggest that CK5+CK8+ cells during prostate organogenesis can be subdivided into basal and luminal progenitors by their relative levels of CK5 and CK8 expression, and by the presence or absence of p63 expression; such “double-positive” cells differ from “intermediate” cells in the adult mouse prostate that reside in the basal layer and co-express basal and luminal markers (Wang et al., 2013).
Figure 4.
Schematic Time Course of Basal and Luminal Differentiation during Prostate Organogenesis
Urogenital epithelial progenitors with mixed basal and luminal characteristics (CK5+, CK8+, and p63+, depicted in purple) give rise to distinct populations of “outer” pre-basal (CK5high, CK8low, and p63+) and “inner” pre-luminal cells (CK5low, CK8high, and p63−). Pre-basal cells generate both bipotent and unipotent basal progenitors. Pre-luminal cells generate both bipotent and unipotent luminal progenitors. However, bipotent luminal progenitors are more transient than bipotent basal progenitors. P, postnatal day.
Notably, generation of basal cells from luminal progenitors does not seem to be a rare event during early neonatal prostate development. In particular, CK5lowCK8highp63– pre-luminal cells, as well as subsequently arising CK5low/–CK8+p63– luminal cells, are specified but not fully lineage restricted, as bipotent luminal progenitors are present at P2 as well as P7. However, whereas bipotent basal progenitors can be identified at ductal tips until the onset of puberty (Tika et al., 2019; Wuidart et al., 2016), bipotent luminal progenitors appear to be more transient. Intriguingly, the initial expression of both basal and luminal markers in epithelial progenitors is similar in both the developing prostate and mammary glands, although the timing of their subsequent lineage restriction may differ (Rios et al., 2014; Wuidart et al., 2016, 2018).
Our findings of a transient bipotent progenitor in prostate organogenesis are consistent with previous evidence that luminal cells can display bipotent properties under non-homeostatic conditions. Luminal cells from adult mouse prostates can generate basal cells when cultured as prostate organoids (Chua et al., 2014; Karthaus et al., 2014; Zhang et al., 2018), and luminal cells can generate basal cells in renal grafts (Crowley et al., 2020; Kwon et al., 2016; Zhang et al., 2018); in addition, rare luminal castration-resistant progenitors in adult prostates can generate basal cells during androgen-mediated prostate regeneration (Wang et al., 2009). In the mammary gland, recent studies have revealed unexpected bipotency of luminal cells during pregnancy or upon hormone stimulation (Song et al., 2019).
Our results also indicate that AR is not essential in prostate epithelial progenitors for luminal specification during neonatal organogenesis. Consequently, the requirement for AR for production of secretory proteins (Cunha et al., 1987) is likely to reflect incomplete luminal differentiation rather than a significant defect in luminal specification. In contrast, AR is required for rare basal-to-luminal differentiation that occurs during adult homeostasis (Xie et al., 2017), and for basal-to-luminal differentiation in CK5-Cre; Ar f/Y mice analyzed at 4 weeks onward (Lee et al., 2012).
Interestingly, the plasticity of early postnatal luminal cells is associated with low levels of testicular androgen biosynthesis (Jean-Faucher et al., 1978) and nuclear AR expression (Figures 1A–1E and S1A–S1D). Since the ability of luminal progenitors to generate basal cells is lost as AR expression increases in epithelial cells (Figures 1A–1E and 4), AR activity could conceivably suppress luminal plasticity. However, deletion of epithelial AR is not sufficient to change cell fate, since the ratio of marked luminal/basal cells is not significantly altered in CK5-trace; Arflox/Y or CK8-trace; Arflox/Y prostates relative to controls. An alternative but not mutually exclusive possibility is that the progressive loss of bipotent luminal progenitors may be associated with a temporal transition in prostate outgrowth from proximal peri-urethral luminal cells to more distal luminal cells. Thus, the transient bipotency of the earliest luminal cells during organogenesis may resemble castration-resistant luminal cells in the proximal region of the adult mouse prostate that are bipotent in organoid and graft assays and have lower AR expression (Crowley et al., 2020; Karthaus et al., 2020; Kwon et al., 2016).
Overall, our findings suggest that the requirement for AR function in prostate luminal cells is more limited during organogenesis than in adulthood. Notably, the role of AR in inhibiting luminal plasticity in the normal and transformed adult prostate may be acquired during puberty, whereas pre-pubertal luminal progenitors can display lineage plasticity. Thus, we speculate that further studies on early prostate epithelial progenitors may provide translational insights into luminal plasticity in AR-indifferent prostate tumors.
Experimental Procedures
Mouse Strains and Genotyping
CK5–CreERT2 (Tg(KRT5-cre/ERT2)2Ipc [Indra et al., 1999]), CK8–CreERT2 (Tg(Krt8–cre/ERT2)17Blpn/J [Van Keymeulen et al., 2011]), and R26R–CAG–YFP (B6.Cg–Gt(ROSA)26Sortm3(CAG−EYFP)Hze/J [Madisen et al., 2010]) mice were obtained from the Jackson Laboratory Induced Mutant Resource. The Arflox (Artm1Verh/Ibcm [De Gendt et al., 2004]) allele was obtained from the EMMA mouse repository. TCF/Lef:H2B-GFP and Nkx3.1lacZ animals were obtained as described previously (Kruithof-de Julio et al., 2013). Animals were housed in specific pathogen-free conditions and maintained on a congenic C57BL/6N background; wild-type animals used were C57BL/6N. Primers used for genotyping are listed in Table S3. Animal studies were approved by and conducted according to standards set by the Columbia University and the George Washington University Institutional Animal Care and Use Committees.
Lineage Tracing
For lineage tracing, neonates were given a subcutaneous injection of tamoxifen (Sigma) in the skin over the neck, at a dose of 0.1 mg (20 μL of a 5-mg/mL solution) at P2 and 0.25 mg (50 μL of a 5-mg/mL solution) at P7, where the day of birth was considered P0. We limited our studies to doses of 0.1 mg at P2, as a dosage of 0.5 mg of tamoxifen at P2 resulted in frequent runting of pups and occasional loss of litters. Doses of tamoxifen were optimized to reduce toxicity and to label cells expressing higher levels of the CreERT2 driver to mark luminal versus basal populations.
Histological Analysis
Tissues were dissected in 1× PBS, fixed in 4% paraformaldehyde, immersed in 30% sucrose in PBS, and embedded with OCT (Tissue-Tek). P0, P2, and P5 prostates were embedded transversely sections and serially sectioned at 6 μm. We distinguished prostate buds from ductus deferens and seminal vesicle based on their morphology as well as the location of prostate buds in relation to the urogenital sinus epithelium, guided by parallel analyses of marker expression. In particular, at early postnatal stages, the seminal vesicle expresses high levels of TCF/Lef:H2B-GFP, but does not express Nkx3.1lacZ, whereas prostate expresses Nkx3.1lacZ but only patchy TCF/Lef:H2B-GFP (Kruithof-de Julio et al., 2013) (Figures S1G–S1I). Moreover, the seminal vesicle epithelium is more pseudostratified in appearance compared with the developing prostate. Since there are differences in the timing of prostate development between the lobes, we focused our analyses and quantification on the anterior prostate.
Immunofluorescence Analysis
Images were quantified using the multi-point tool in ImageJ. Basal and luminal cells were quantified based on expression of CK5 and CK8 or p63. We calculated the percentage of luminal or basal cells over the total number of YFP+ cells counted from each prostate, since we observed variability in the efficiency of YFP labeling between animals. For IF analyses, representative staining patterns were confirmed in at least three samples from at least two independent experiments.
Flow Cytometry and RNA Sequencing
For RNA sequencing of lineage-marked luminal cells, prostates from 4-week-old males that were treated with 0.1 mg tamoxifen at P2 as described above were dissected in cold 1× PBS and dissociated. Using conservative gating parameters, YFP+CD49flow/– luminal-enriched cells were sorted and RNA was extracted.
Statistical Analysis
Data were collected and statistical analysis was performed using Microsoft Excel v.16 and GraphPad Prism 7, using a two-tailed unpaired Student's t test; Gaussian distribution was assumed.
Data and Code Availability
Expression data have been deposited in GEO under accession number GEO: GSE136982.
Author Contributions
Conceptualization, M.S. and M.M.S.; Methodology, M.S. and M.M.S.; Investigation, M.S. and S.X.; Investigation (Bioinformatic Analyses), M.S., N.J.E., and A.M.; Writing – Original Draft, M.S. and M.M.S.; Writing – Review & Editing, M.S. and M.M.S.; Funding Acquisition, M.S. and M.M.S.
Acknowledgments
We thank Sarah Bergren for assistance with mouse breeding and genotyping, and Cory Abate-Shen, Francesco Cambuli, and Eva Leung for thoughtful comments on the manuscript. This work was supported by NCI K99/R00 CA194287 (to M.S.) and NCI R01 CA238005 (to M.M.S.). Research performed in the CCTI Flow Cytometry Core was supported in part by NIH S10 RR027050 and NIH P30 CA013696.
Published: November 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.10.004.
Contributor Information
Maho Shibata, Email: mshibata@gwu.edu.
Michael M. Shen, Email: mshen@columbia.edu.
Supplemental Information
References
- Beltran H., Hruszkewycz A., Scher H.I., Hildesheim J., Isaacs J., Yu E.Y., Kelly K., Lin D., Dicker A., Arnold J. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 2019;25:6916–6924. doi: 10.1158/1078-0432.CCR-19-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J., Marotz C., Giannopoulou E., Chakravarthi B.V., Varambally S. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemn E.G., Coleman I.M., Lucas J.M., Coleman R.T., Hernandez-Lopez S., Tharakan R., Bianchi-Frias D., Dumpit R.F., Kaipainen A., Corella A.N. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver B.S., Chapinski C., Wongvipat J., Hieronymus H., Chen Y., Chandarlapaty S., Arora V.K., Le C., Koutcher J., Scher H. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua C.W., Epsi N.J., Leung E.Y., Xuan S., Lei M., Li B.I., Bergren S.K., Hibshoosh H., Mitrofanova A., Shen M.M. Differential requirements of androgen receptor in luminal progenitors during prostate regeneration and tumor initiation. eLife. 2018;7:e28768. doi: 10.7554/eLife.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua C.W., Shibata M., Lei M., Toivanen R., Barlow L.J., Bergren S.K., Badani K.K., McKiernan J.M., Benson M.C., Hibshoosh H. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014;16:951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley L., Cambuli F., Aparicio L., Shibata M., Robinson B.D., Xuan S., Li W., Hibshoosh H., Loda M., Rabadan R. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife. 2020;9:59465. doi: 10.7554/eLife.59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha G.R., Donjacour A.A., Cooke P.S., Mee S., Bigsby R.M., Higgins S.J., Sugimura Y. The endocrinology and developmental biology of the prostate. Endocr. Rev. 1987;8:338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- De Gendt K., Swinnen J.V., Saunders P.T., Schoonjans L., Dewerchin M., Devos A., Tan K., Atanassova N., Claessens F., Lecureuil C. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl. Acad. Sci. U S A. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Hooker E., Yu E.J., Wu H., Cunha G.R., Sun Z. An indispensable role of androgen receptor in Wnt responsive cells during prostate development, maturation, and regeneration. Stem Cells. 2018;36:891–902. doi: 10.1002/stem.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Faucher C., Berger M., de Turckheim M., Veyssiere G., Jean C. Developmental patterns of plasma and testicular testosterone in mice from birth to adulthood. Acta Endocrinol. (Copenh) 1978;89:780–788. doi: 10.1530/acta.0.0890780. [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Hofree M., Choi D., Linton E.L., Turkekul M., Bejnood A., Carver B., Gopalan A., Abida W., Laudone V. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368:497–505. doi: 10.1126/science.aay0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof-de Julio M., Shibata M., Desai N., Reynon M., Halili M.V., Hu Y.P., Price S.M., Abate-Shen C., Shen M.M. Canonical Wnt signaling regulates Nkx3.1 expression and luminal epithelial differentiation during prostate organogenesis. Dev. Dyn. 2013;242:1160–1171. doi: 10.1002/dvdy.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O.J., Zhang L., Xin L. Stem cell antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential. Stem Cells. 2016;34:191–202. doi: 10.1002/stem.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.O., Tian J., Huang C.K., Ma Z., Lai K.P., Hsiao H., Jiang M., Yeh S., Chang C. Suppressor role of androgen receptor in proliferation of prostate basal epithelial and progenitor cells. J. Endocrinol. 2012;213:173–182. doi: 10.1530/JOE-11-0474. [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motelica-Heino I., Castanier M., Corbier P., Edwards D.A., Roffi J. Testosterone levels in plasma and testes of neonatal mice. J. Steroid Biochem. 1988;31:283–286. doi: 10.1016/0022-4731(88)90351-2. [DOI] [PubMed] [Google Scholar]
- Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B.D., Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Pignon J.C., Grisanzio C., Geng Y., Song J., Shivdasani R.A., Signoretti S. p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. U S A. 2013;110:8105–8110. doi: 10.1073/pnas.1221216110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang R., Jiang W., Yin Q., Peng G., Yang R., Yu Q.C., Chen J., Li J., Cheung T.H. Hormones induce the formation of luminal-derived basal cells in the mammary gland. Cell Res. 2019;29:206–220. doi: 10.1038/s41422-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., Chang C. Immunohistochemical and in-situ hybridization analysis of androgen receptor expression during the development of the mouse prostate gland. J. Endocrinol. 1991;129:83–89. doi: 10.1677/joe.0.1290083. [DOI] [PubMed] [Google Scholar]
- Tika E., Ousset M., Dannau A., Blanpain C. Spatiotemporal regulation of multipotency during prostate development. Development. 2019;146 doi: 10.1242/dev.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivanen R., Shen M.M. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development. 2017;144:1382–1398. doi: 10.1242/dev.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H.L., Halili M.V., Hu Y.P., Price S.M., Abate-Shen C., Shen M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.T., Altuwaijri S., Ricke W.A., Huang S.P., Yeh S., Zhang C., Niu Y., Tsai M.Y., Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc. Natl. Acad. Sci. U S A. 2007;104:12679–12684. doi: 10.1073/pnas.0704940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A., Ousset M., Rulands S., Simons B.D., Van Keymeulen A., Blanpain C. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A., Sifrim A., Fioramonti M., Matsumura S., Brisebarre A., Brown D., Centonze A., Dannau A., Dubois C., Van Keymeulen A. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 2018;20:666–676. doi: 10.1038/s41556-018-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Liu Y., Cai T., Horton C., Stefanson J., Wang Z.A. Dissecting cell-type-specific roles of androgen receptor in prostate homeostasis and regeneration through lineage tracing. Nat. Commun. 2017;8:14284. doi: 10.1038/ncomms14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kwon O.J., Henry G., Malewska A., Wei X., Zhang L., Brinkley W., Zhang Y., Castro P.D., Titus M. Non-cell-autonomous regulation of prostate epithelial homeostasis by androgen receptor. Mol. Cell. 2016;63:976–989. doi: 10.1016/j.molcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jeter C., Gong S., Tracz A., Lu Y., Shen J., Tang D.G. Histone 2B-GFP label-retaining prostate luminal cells possess progenitor cell properties and are intrinsically resistant to castration. Stem Cell Reports. 2018;10:228–242. doi: 10.1016/j.stemcr.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression data have been deposited in GEO under accession number GEO: GSE136982.