Single-cell proteomics capabilities have the potential to transform biomedical research and enable understanding of biological systems with a new level of granularity. Recent advances in sample processing, separations and MS instrumentation now make it possible to quantify >1000 proteins from individual mammalian cells.
Keywords: Cell biology, Cell sorting, Label-free quantification, Mass Spectrometry, Tandem Mass Spectrometry, Tissues*, FAIMS, nanoLC, nanoPOTS, single-cell, ultrasensitive
Graphical Abstract
Highlights
Label-free and isobaric labeling approaches for in-depth profiling of single cells.
Miniaturization and simplification of sample processing reduce surface losses.
Nanoflow separations enhance ionization efficiency and reduced chemical noise.
Ultrasensitive mass spectrometry and gas-phase separation add selectivity.
Abstract
MS-based proteome profiling has become increasingly comprehensive and quantitative, yet a persistent shortcoming has been the relatively large samples required to achieve an in-depth measurement. Such bulk samples, typically comprising thousands of cells or more, provide a population average and obscure important cellular heterogeneity. Single-cell proteomics capabilities have the potential to transform biomedical research and enable understanding of biological systems with a new level of granularity. Recent advances in sample processing, separations and MS instrumentation now make it possible to quantify >1000 proteins from individual mammalian cells, a level of coverage that required an input of thousands of cells just a few years ago. This review discusses important factors and parameters that should be optimized across the workflow for single-cell and other low-input measurements. It also highlights recent developments that have advanced the field and opportunities for further development.
Complex biological processes are rooted in dynamic interactions between individual cells, often spanning multiple cell types as well as different states, fates and susceptibilities within a given cell type. Global MS-based proteome measurements have historically been limited to bulk samples comprising thousands or millions of cells. Such measurements can provide a quantitative and nearly comprehensive profile of protein expression (1), yet they fail to account for heterogeneity within the sample. Single cell protein measurements can resolve this heterogeneity, yet the prevailing antibody-based approaches such as immunofluorescence, flow cytometry and mass cytometry have limited specificity and are confined to measuring a small number of preselected proteins per cell. Filling in this gap to achieve an in-depth and unbiased profile of protein expression within single cells is expected to have broad impact in biomedicine by, for example, elucidating microenvironmental factors that promote or inhibit tumor growth (2) and identifying novel cellular subpopulations (3) or developmental trajectories (4) that may be obscured in bulk measurements. Developing new technologies for single cell proteomics also renders accessible many other limited samples such as rare circulating tumor cells isolated from whole blood and fine needle aspiration biopsies, and enables protein expression to be mapped across tissues with high spatial resolution (5).
Although many of the analytical requirements for single cell and other low-input proteomics have been in place for some time, including efficient nanoflow separations (6) and sensitive MS instrumentation (7), innovations in sample processing have lagged. As such, thousands of mammalian cells or large single cells (e.g. oocytes or blastocysts) have been required to achieve an in-depth measurement (8–10). With recent innovations in sample preparation and experimental design, the first reports of profiling hundreds of proteins from single mammalian cells were published in 2018 (11, 12). The field has advanced rapidly since those initial reports. More than 1000 protein groups can now be reliably profiled from single cells using both label-free (13), and isobaric labeling workflows (14–16), and ∼6000 protein groups can be profiled from samples comprising just a few hundred cells (17). Thus, a tradeoff still exists between sample size and proteome coverage, but it has dramatically shifted in favor of much smaller samples. These advances have been applied to a wide variety of cell and tissue types, including dissociated cells and microdissected tissues as described below.
For such nanoscale analyses to be successful, every aspect of the workflow must be carefully optimized. This article takes a detailed look at sample isolation and preparation, liquid-phase separation and ionization, and gas-phase separation and MS. For each step, important factors affecting sample recovery and detection are discussed, examples are described, and opportunities for continued development are highlighted. The relative advantages of label-free and isobaric labeling workflows are also considered. Data analysis is of course a crucial component of any experiment, but as existing software packages using standard settings have thus far largely been employed for single-cell proteomics, data analysis is only touched on lightly here. It is hoped that the present work will emphasize the progress to date and the tremendous opportunities that lie ahead in advancing the nascent field of single-cell and low-input proteomics.
SAMPLE ISOLATION AND PREPARATION
Miniaturization
Sample preparation workflows for bottom-up proteomics aim to (1) quantitatively extract proteins from isolated cells or tissues, (2) chemically and enzymatically process the extracted proteins to generate peptides, and (3) deliver the peptides to a separation platform in ready-to-analyze form and in sufficient quantities to enable a robust measurement. Protein extraction, which encompasses cell lysis and protein solubilization, often requires harsh chemical or mechanical conditions such as SDS detergent, urea or bead beating to disrupt tissues and liberate proteins, which then necessitates extensive cleanup and buffer exchange steps to remove insoluble debris and reagents that may interfere with the downstream analysis (18). During these cleanup steps, and indeed throughout the entire sample preparation process, proteins and peptides are exposed to various surfaces that incur losses due to nonspecific adsorption (19). Assuming these surfaces contain a finite number of adsorptive sites, sample losses that may be negligible for a bulk analysis can become prohibitive or even total for a small sample subjected to the same workflow. As evidence of this, Wu et al. showed 15% sample losses following multiple sample transfer steps for 50 µg protein samples, which increased to 89% losses for 2 µg samples (20). These losses would have undoubtedly been far greater for single-cell-sized samples containing ∼4 orders of magnitude less protein. Minimizing adsorption is thus of crucial importance for trace sample analysis, and because no surface can eliminate adsorption of all proteins and peptides (19), this can most effectively be achieved by dramatically reducing sample processing volumes to avoid as much surface contact as possible.
Another key consideration for proteomic sample processing is the impact that sample and reagent concentrations have on reaction rates. For example, tryptic digestion follows Michaelis-Menten kinetics and under conditions of low substrate concentration, the digestion rate becomes directly proportional to both enzyme (trypsin) and substrate (protein sample) concentration (21). As such, digestion conditions that work well for bulk samples in standard processing volumes of tens of microliters may become ineffective when processing a single cell in the same volume because of the greatly reduced protein concentration. Dramatically increasing the concentration of trypsin can partially offset the effects of reduced substrate concentrations, but there is likely a limit beyond which autolysis and the resulting chymotryptic activity (22) from excess trypsin will interfere with the analysis. Minimizing sample processing volumes thus provides the dual benefits of reducing surface adsorption and increasing sample concentrations and thus should significantly enhance single-cell proteomics.
Simplification
Although miniaturizing sample preparation can be challenging, some aspects of the workflow fortunately become more favorable when preparing small samples. Buffer exchange and sample cleanup become increasingly unnecessary, as salts and insoluble cellular debris can be of insufficient quantities to interfere with the downstream analysis. Similarly, consider that a single cell has a surface-area-to-volume ratio (SA:V) that is hundreds of times larger than that of, for example, a tissue sample obtained by core needle biopsy. Sample processing reagents can therefore more readily access the single cell and its contents without requiring aggressive approaches for tissue disruption and protein extraction. Avoiding such steps can eliminate the need to expose a trace sample to surfaces of beads or filters during preparation, further reducing adsorptive losses. Given these factors, sample preparation workflows that are both smaller and simpler than their bulk-scale counterparts appear to be a winning combination for single-cell proteomics.
Open Format
An otherwise perfect sample processing platform would be of no value without a way to transfer samples to the platform in the first place. Although enclosed microchannel-based microfluidic devices (23) appear at first glance to be ideal for manipulating nanoliter volumes that benefit single cell analyses, the so-called “world-to-chip” interface problem (24) can be very difficult to overcome. In contrast, open microfluidic platforms (25) that reduce dimensions but maintain the general form factor of the microwell plate can be directly interfaced with all common single-cell sample isolation techniques including limiting dilution (11, 17, 26), micromanipulation (13, 27), FACS (4, 14, 28) and laser capture microdissection (LCM) (11, 13, 17, 26, 29–32), providing broad compatibility with cells in suspension as well as spatially resolved regions of tissue sections. In addition, open reactors minimize surface exposure, and sample retrieval can be accomplished through nanopipetting. This is not to say that there are no prospects for enclosed channel-based microfluidics for preparing and analyzing trace proteomic samples, but rather that there appear to be substantial headwinds for such developments.
Progress
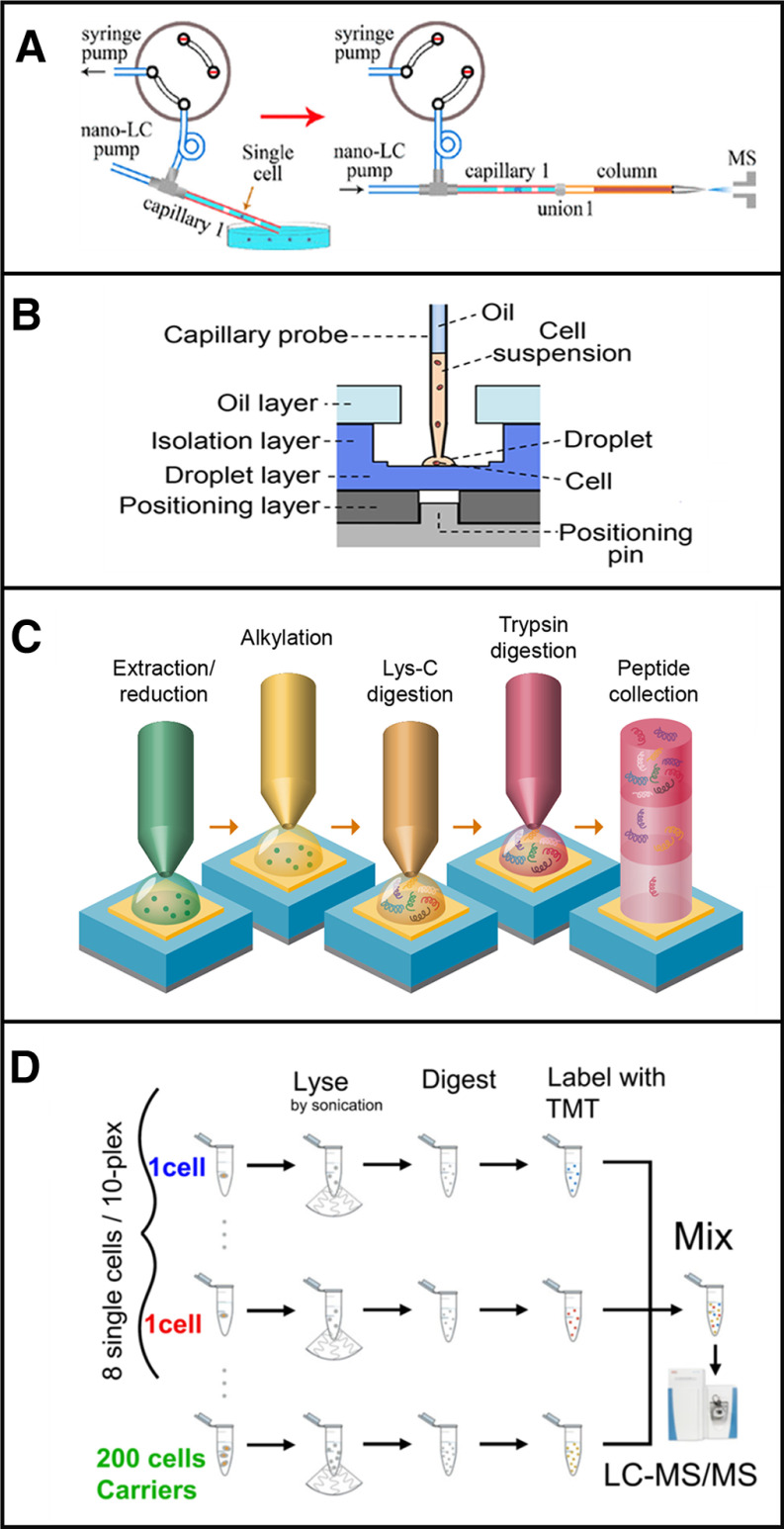
A variety of approaches have been employed to prepare trace samples and single cells for proteome profiling (33), many of which combine the benefits of miniaturization and simplification as discussed above. A few recent examples are described here for illustrative purposes and are depicted in Fig. 1. Shao et al. reported on an integrated proteome analysis device (iPAD) (34) in which a mixture of reagents including trypsin and the chaotrope guanidine hydrochloride were added directly to a suspension of cells. Single cells and the added reagents were then rapidly aspirated into a capillary in a volume of just 2 nL surrounded by plugs of air to create a nanoliter reactor (Fig. 1A). Cell lysis and digestion took place at elevated temperature and with sonication, after which the capillary was connected to a short separation column for rapid LC–MS analysis with a 30-min gradient. The entire preparation workflow consisted of just cell lysis and trypsin digestion, and any sample cleanup took place on the reversed-phase separation column. Using this approach, the authors identified an average of 128 protein groups by MS/MS and 271 including identifications from MS1-level feature matching using MaxQuant's Match Between Runs (MBR) algorithm.
Fig. 1.
Strategies for sample preparation of low input samples based on (A) iPAD, (B) OAD, (C) nanoPOTS and (d) SCoPE-MS. Adapted with permission from References 31, 32, 7, and 8, respectively.
Li et al. developed a disposable oil-air-droplet (OAD) device (35) (Fig. 1B) in which nanoliter volumes of cells and reagents were sequentially added and incubated in a reactor made of low binding polypropylene or hydrophobized glass for sample confinement. A suspended disc of oil over the sample could be penetrated with a capillary nanopipette tip, which then automatically resealed upon removal of the tip to minimize droplet evaporation. Following digestion, the device was inserted into a high-pressure chamber for direct transfer of the sample to the head of an LC column for analysis. An average of 35 and 108 proteins were identified from 1 and 10 HeLa cells, respectively.
Our group developed the nanoPOTS (Nanodroplet Processing in One pot for Trace Samples) platform (11) (Fig. 1C) in which a nanopipettor sequentially dispenses nanoliter volumes of cells and reagents into nanowells. The nanowells are ∼1-mm-diameter glass pedestals that are patterned on a standard glass microscope slide; each nanowell is surrounded by a hydrophobic barrier. Evaporation is minimized by dispensing reagents inside a humidified chamber and then applying a removable cover over the array of nanodroplets for extended incubations. Prepared samples can then be loaded into a capillary and transferred to a solid-phase extraction column that is subsequently interfaced with a nanoLC column. The minimal sample losses and efficient preparation, combined with highly sensitive nanoLC-MS, enabled 211 and 1517 protein groups to be identified from 1 and ∼10 HeLa cells, respectively, which increased to an average of 669 and 3092 identifications for the same samples with MBR identifications included (11, 28).
Budnik et al. prepared single cells for analysis using a Tandem Mass Tag (TMT) workflow termed SCoPE-MS. In the first-generation platform (12), single cells were lysed by sonication in 10 µL volumes and finally processed in a total volume of ∼12 µL before combining single cell samples with a larger carrier sample for analysis. In the second iteration (SCoPE 2) (16), lysis was accomplished through a combination of freeze/thaw and heat in a 1 µL volume. Although this preparation workflow was not miniaturized to the nanoliter range, it was nonetheless greatly simplified relative to a standard bulk preparation and thus avoided cleanup-related losses.
Prospects
Although there are undoubtedly as-yet-unimagined improvements to be made in nanoscale proteomic sample processing, a few points are becoming clear:
Minimizing sample preparation volumes is beneficial for reducing adsorptive losses and increasing sample concentrations for more efficient reaction with trypsin and other reagents.
Open microfluidic processing platforms have a substantial advantage over closed systems in terms of minimizing SA:V, and they facilitate direct coupling with widely used sample isolation strategies including micromanipulation, FACS and LCM.
Solution-based sample processing reduces surface exposure relative to immobilized enzymatic reactors (36) and filter-based protocols (e.g. FASP (37) and micro-FASP (38)) and have thus far proven more effective for trace samples. However, bead, column or functionalized surface-based methods may still be required for enrichment of post-translationally modified proteins, etc., despite some expected sample losses.
Sample cleanup and aggressive protein extraction steps that are sometimes required for bulk workflows may be unnecessary for low input samples, thus providing opportunities to simplify sample preparation and further reduce losses.
As the picture becomes clearer, there are additional areas relating to low-input sample preparation waiting to be explored. First, while reducing sample processing volumes to a few hundred nanoliters has greatly benefitted single-cell proteomics, the contents of a single cell (with a volume of a few picoliters) are still being diluted by a factor of ∼100,000. It remains to be seen whether further miniaturization will provide substantial additional gains or if a point of diminishing returns has been reached. Other aspects of sample preparation, including reagent compositions, incubation times and temperatures may be further optimized to provide increased sample recovery, time savings and compatibility with more challenging samples such as formalin-fixed, paraffin-embedded tissues. The stability of prepared samples during extended storage under both wet and dry conditions may differ from that of bulk samples and should be carefully studied. Finally, autosamplers are not typically compatible with nanoliter volumes, and interfacing single cell preparation with separations has largely required manual handling by a highly skilled operator. Automating the entire workflow will facilitate the dissemination of single-cell proteomics to many additional laboratories. To this end, Williams et al. have developed a custom autosampler for nanoliter samples (39), but opportunities for further automation and simplification remain.
SEPARATIONS AND IONIZATION
Efficient chemical separations reduce the complexity of the sample composition entering the mass spectrometer at a given time, expanding dynamic range and reducing ionization suppression. The characteristic elution times in combination with accurate mass measurements can also provide information for identification using, for example, accurate mass and time tag (40) or MBR (41) approaches. Reducing the flow rate of the separation and its corresponding electrospray ionization (ESI) source to the low-nanoliter-per-minute or even picoliter-per-minute range can dramatically increase ionization efficiencies and further reduce ionization suppression (42, 43). In addition, operating at low flow rates increases the proportion of ions resulting from peptides rather than solvent contaminants. Various separation strategies that have proven successful for low-input samples are discussed below.
Progress
Narrow-Bore Packed Columns
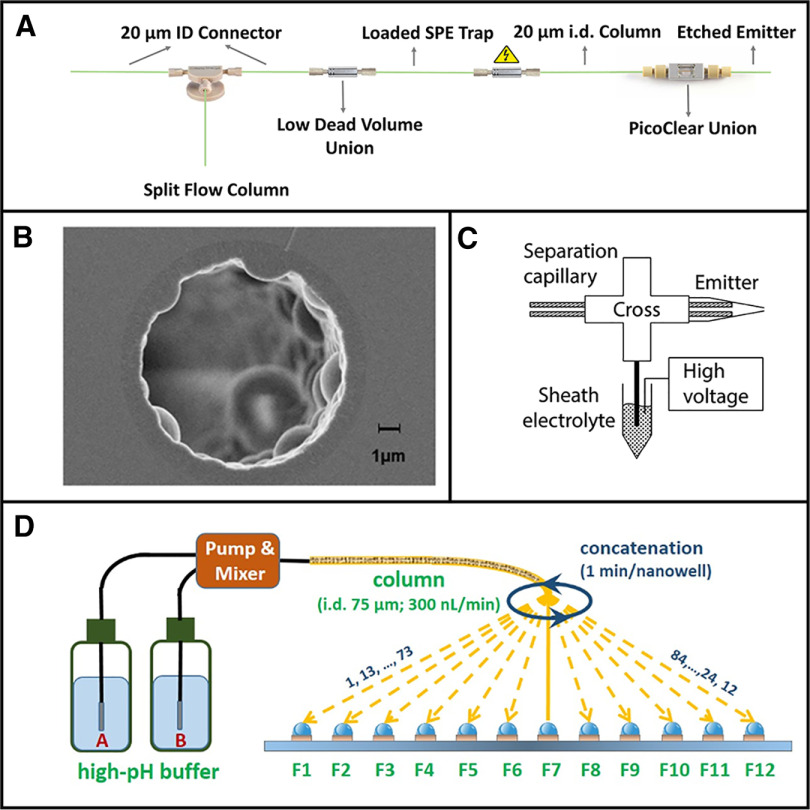
Most proteomics separations use particle-packed reversed-phase liquid chromatography (LC) columns having a 75-μm bore and operating at ∼300 nL/min (44). Not all such columns are created equal however, and high-efficiency, low-dead-volume columns such as those produced by Ion Opticks can produce very narrow peaks containing peptides at higher eluting concentrations for improved detection (45). Yet much lower flow rates are required to fully reap the sensitivity benefits provided by nanoESI (46). High-efficiency custom-packed nanoLC columns having inner diameters as small as 15 μm and operating optimally at ∼20 nL/min were developed and evaluated by Shen et al. in the early 2000s (6, 47, 48), thus predating single-cell proteome profiling by nearly two decades. These separations required highly custom apparatus as they predated nanovolume fittings, nanoLC pumps capable of delivering reliable programed linear gradients, and commercial nanoESI emitters. Of course, they also substantially predated effective sample processing methods for single cells and modern mass spectrometers that could rapidly perform tandem MS measurements on such small samples. Despite these challenges, the authors were impressively able to identify standard peptides present in 75 zmol quantities and identify hundreds of proteins from low-nanogram samples (6).
More recently, we have revisited similar nanoLC columns in the context of single-cell analysis and modern MS instrumentation. We showed a substantial increase in proteome coverage for 30-μm-i.d. columns operating at ∼50 nL/min relative to standard 75-μm-i.d. columns (49), and these 30-μm columns have been used for most of our work since. In analyzing single nanoPOTS-prepared HeLa cells with these columns, we identified at average of 211 protein groups/cell using an Orbitrap Fusion Lumos mass spectrometer, which increased to 669 including MBR identifications (28). We have since further miniaturized our LC separations for single-cell proteomics to 20-μm-i.d. columns operating at ∼20 nL/min (Fig. 2A), and the average MS/MS-derived identifications increased by >40% to nearly 300 protein groups using the same cell type, preparation method and mass spectrometer (27). This illustrates the benefits of further miniaturizing separations, although challenges with reproducibly preparing and operating these 20-μm-i.d. (or smaller) packed columns may preclude their routine use.
Fig. 2.
Separation methods for single cells and other trace samples. A, Narrow-bore packed LC columns. B, Porous layer open tubular columns. C, Capillary electrophoresis with electrokinetically driven sheath flow interface. D, Nanowell-mediated 2D LC. Adapted with permission from References 23, 48, 52 and 13, respectively.
PLOT Columns
Given the difficulties with extreme miniaturization of packed nanoLC columns, a variety of alternative approaches have been explored. One highly promising option is to instead use monolithic or narrow-bore porous layer open tubular (PLOT) columns (Fig. 2B). Yue et al. originally developed ∼4-m-long, 10-μm-i.d. PLOT LC columns that utilized in situ-polymerized poly(styrene-divinylbenzene) as stationary phase (50). These operated at ∼20 nL/min, had a low-fmol loading capacity and achieved impressive peak capacities of ∼400. More recently, the same group applied similar PLOT columns to low-input proteomics (51). They achieved detection limits ranging from 10–100 zmol for spiked peptides and identified >2500 protein groups from aliquots of spiked MCF-7 cells corresponding to ∼100 cells that were captured from whole blood.
µPAC
An exciting recent development has been the commercialization of microfluidic pillar array columns (µPAC) (52) for nanoLC by PharmaFluidics. Rather than randomly packing separation media into capillaries, the µPAC columns have an ordered stationary phase micropatterned onto a silicon wafer. These ordered separations reduce dispersion and provide highly efficient separations at much lower operating pressures than a typical nanoLC column. They also promise improved robustness with no clog-prone frits and have demonstrated outstanding retention time reproducibility. However, the cost of the columns is several times that of a typical packed column. At present, µPAC columns are designed to operate in a flow regime similar to that of a 75-μm-i.d. column (>100 nL/min) but creating narrower pillar beds that operate optimally at much lower flow rates should be possible. Using a commercial 50-cm-long µPAC column, Stadlmann et al. (53) reproducibly identified >2400 protein groups from 10-ng cell lysate digests, indicating substantial promise for this separation technology with further miniaturization.
Capillary Electrophoresis
Capillary electrophoresis (CE) separations have also proven highly capable for nanoscale bottom-up proteomics (54). CE separates liquid-phase ionic species according to their differential migration rates in an electric field. Because CE lacks a stationary phase, it should in principle reduce any adsorptive losses that may take place in an LC separation. CE is also largely orthogonal to LC separations, with advantages for analyzing species that are either unretained or fully retained on a RPLC column. Recent improvements in ionization sources such as the electrokinetically pumped sheath-flow electrospray interface (55) (Fig. 2C) have enabled sensitive analyses with detection limits as low as 1 zmol (56). Zhang et al. identified >3500 protein groups from 48 ng of mammalian protein digest (57). CE has been used extensively for single-cell proteomics, but so far the cells analyzed have generally been oocytes and blastocysts, etc. (9, 10, 38, 58–60), all of which are much larger than typical mammalian cells. One current disadvantage for CE relative to LC is that a packed LC column naturally cleans up and preconcentrates samples into focused plugs at the inlet of the column prior to gradient elution, whereas effective preconcentration prior to separation for CE is less straightforward. Still, it is likely that CE-based separations will continue to make important contributions to nanoscale proteomics.
2D Separations
Achieving deep proteome coverage of >5000 protein groups generally requires a multidimensional separation that provides greater peak capacity than that of a one-dimensional separation. However, fractionation schemes typically lose significant amounts of sample to the collection vessels between the two separation dimensions, thus posing a significant challenge for low-input proteomics (61). To address this, we recently developed nanowell-mediated 2D LC in which the same glass chips used for nanoPOTS sample preparation served as fraction collectors between two dimensions of nanoLC (17) (Fig. 2D). The reduced surface losses enabled identification of nearly 6000 proteins from 50 ng lysates as well as from low-input nanoPOTS-prepared cell and tissue samples comprising a few hundred cells. A similar fractionation scheme was implemented for LC coupled with ion mobility spectrometry (62), and with some modest upscaling, the platform was rendered compatible with a commercial autosampler and microwell plates (63). The latter implementation identified 6700 protein groups from 100 ng lysate samples and 20,000 phosphopeptides from 100 µg samples. Although these 2D separations have not been evaluated at the single-cell level, further optimization should make possible single-cell analyses with greater depth of coverage, albeit with a significant tradeoff in measurement throughput.
Prospects
Whether using LC or CE, cutting-edge separations have tended to operate at minimum flow rates of no less than 20 nL/min. This is not because optimum conditions are achieved at these flow rates, but rather that it becomes increasingly difficult to maintain a stable electrospray with further flow reduction. To explore the sub-nanoflow regime, we developed chemically etched fused silica emitters having 2-μm inner diameters that provided stable operation at flow rates as low as 400 pL/min (43). In direct infusion experiments, we found that detection limits decreased substantially at flows close to 1 nL/min. More recently, Xiang et al. employed 2-μm-i.d. open tubular “picoLC” columns for trace proteomic analyses (64). The separations operated at just 790 pL/min and achieved a respective proteome coverage of 78 and 949 protein groups for 7.5 and 75 pg of bacterial lysate tryptic digest, which is far less protein than is found in a typical mammalian cell. This platform is not yet ready for single cell applications as it currently lacks a mechanism for lossless sample introduction, but these studies strongly suggest that further miniaturization of separations to the ∼1 nL/min range will greatly benefit single-cell and subcellular proteome profiling.
MASS SPECTROMETRY AND GAS-PHASE SEPARATIONS
To determine protein expression from a sample as small as a single mammalian cell, analyte losses through every step of sample preparation and analysis must be carefully minimized. Before such trace analyses could be considered, MS instrumentation had to become far more efficient in transmitting electrosprayed ions, which are typically generated at atmospheric pressure, to the high vacuum region of the mass analyzer. Conventional gas conductance-limiting orifices such as the heated capillary inlet and skimmer are necessarily narrow to enable efficient differential pumping with modestly sized rough pumps. These were originally able to sample and transmit ≪1% of ions to high vacuum where more efficient ion optics could convey ions through the mass analyzer and to the detector (65). Fortunately, more efficient ion transmission, made possible by the electrodynamic ion funnel (66) and related advances such as the SPIN source (67), has made it possible to ionize and transmit ∼50% of solution-phase molecules to the high-vacuum region of the mass spectrometer (68). Also fortunate for the researcher interested in single-cell proteomics, many of these advances in ion transmission efficiency and MS sensitivity have been incorporated into commercial instrumentation such that one need not be an expert in MS instrumentation to benefit. Indeed, low-input proteome profiling efforts have generally utilized unmodified commercial MS instrumentation as described below.
Progress
Orbitrap MS
The vast majority of low-input proteome analyses have used Orbitrap-based instruments and have benefitted from ongoing improvements provided by successive generations of these mass spectrometers with respect to ion transmission efficiency, detector sensitivity, duty cycle, and resolution (69, 70). We have evaluated these cumulative benefits across different Orbitrap platforms. In analyzing 2 ng aliquots of bacterial lysate tryptic digest, we identified nearly 3 times more unique peptides using an Orbitrap Fusion Lumos compared with an LTQ Orbitrap XL mass spectrometer (49). More recently, we observed an increase in peptide and protein coverage of 36 and 20%, respectively, when analyzing single HeLa cells on the latest-generation Orbitrap Eclipse mass spectrometer compared with the Orbitrap Fusion Lumos (27). In all cases, MS acquisition settings tend to differ substantially from those used for bulk studies. Most importantly for label-free analyses, the decreased ion fluxes associated with trace samples require much longer MS2 ion injection times to reach the AGC target. As such, using a typical maximum injection time of, e.g. 50 ms will result in few usable spectra. On the other hand, an overly long maximum injection time will excessively reduce duty cycle, again reducing the number of productive MS2 spectra. For single-cell studies using an Orbitrap mass analyzer for MS2, we typically set the maximum injection time to ∼500 ms. For TMT-based studies employing a carrier channel, it is beneficial to increase the AGC target beyond a standard (instrument-dependent) setting to ensure a sufficient number of peptides from the sample channels (e.g. single-cells) are trapped along with the carrier peptides (15, 71). The higher AGC target reduces the duty cycle of the instrument but ensures a sufficient reporter ion signal for accurate quantification.
Ion Mobility
Drift tube-based ion mobility spectrometry (IMS) and high-field asymmetric ion mobility (FAIMS) can increase selectivity and separate or filter singly charged species from the multiply charged peptides that are generally used for identification and quantification in bottom-up proteomics (72, 73). Removing singly charged species is especially beneficial for trace sample analyses, as solvent clusters and contaminants that may be negligible in bulk studies can become predominant for low-input studies. These contaminants increase spectral complexity and occupy the finite charge capacity of ion trapping instruments, effectively crowding out the multiply charged peptides. We analyzed single HeLa cells and microdissected neurons from human spinal tissue by incorporating the FAIMS Pro interface into our workflow and identified ∼1100 protein groups per cell, with ready differentiation of closely related neuronal subtypes based on label-free quantification (13). This unprecedented coverage indicates that the added selectivity more than compensates for the signal attenuation that takes place during transmission through the FAIMS interface. Although we scanned between two compensation voltages (CVs) in our study, the added selectivity from additional CVs may provide additional coverage. Other ion mobility-based strategies are also being brought to bear on low-input proteomics. For example, researchers at Brüker Corp. reported identifying 3300 unique peptides and nearly 700 protein groups from 0.15 ng of K562 tryptic digest samples using their trapped ion mobility (TIMS) TOF platform despite employing an LC separation that operated at conventional flow rates (74). These results highlight the promise of ion mobility approaches for dramatically enhancing single cell proteomic analyses.
Prospects
Single cell proteomics has only become possible with the development of suitable MS instrumentation, and new generations of instruments will only improve in terms of sensitivity and speed. However, even without further improvements, the full parameter space available to researchers with available instrumentation remains to be fully explored in terms of instrument settings, acquisition modes (including data independent acquisition (75), BoxCar MS (76), etc.) and data analysis strategies. In addition, given recent results with FAIMS and other ion mobility platforms, gas-phase separation and filtering will lead to enhanced measurements at and near the single cell level.
To TMT or Not to TMT
Single cell proteome profiling efforts have employed both label-free and isobaric labeling workflows. For the latter approach, samples are labeled with distinct tandem mass tag (TMT) reagents and pooled and analyzed together in a single LC–MS experiment. Of particular interest for low-input proteomics, samples can be analyzed in the presence of a much larger carrier sample, which provides for strong MS1 and MS2 spectra while still enabling quantification of each sample based on reporter ion intensity. This general approach was first used by Russell et al. to identify low-abundance proteins in body fluids using solid tissue samples in carrier channels (77), and its application to proteome profiling of single mammalian cells was introduced by Budnik et al. in the SCoPE-MS workflow (12).
As with bulk proteome profiling, label-free and isobaric labeling experiments each have advantages and disadvantages that must be carefully considered (78). The ability to measure multiple samples in a single LC–MS analysis gives TMT experiments a clear throughput advantage, particularly with the 16-plex reagents that are now available. The combined signal from carrier and sample channels also enables detection of peptides that might otherwise fall below detection limits if analyzed individually, and the multiplexed analysis may reduce surface losses during separation (12). However, potential pitfalls also exist for TMT experiments. Although ratio compression resulting from precursor co-isolation is common to many TMT studies, this is likely more significant for single-cell experiments where offline fractionation and other strategies such as MS3 (79) are not generally employed. The added steps during sample preparation, and the need to combine samples in a larger well or vial prior to analysis also provide additional surface exposure with potential for losses. Finally, although the use of a carrier channel can improve detection, it can simultaneously impair quantification as carrier peptides limit the sample ion population in the mass analyzer. Thus, multiple groups (14, 15, 71) have shown that limiting the size of the carrier proteome is critical for maintaining accurate quantification. Importantly, Cheung et al. have developed a program called Single Cell Proteomics Companion that analyzes isobaric labeling datasets to evaluate data quality and recommend modifications to MS acquisition parameters for improved performance (71). Given the tradeoffs between label-free and isobaric labeling workflows, it seems likely that both approaches will continue to be developed and applied to single-cell studies. Regardless of the selected method, experiments will greatly benefit from optimizing each step of the workflow, from sample preparation to separation and MS analysis as described above to reduce sample losses and increase sensitivity.
SUMMARY
The field of single-cell proteomics is still in its infancy but is advancing very rapidly. For example, we have seen label-free proteome coverage increase from ∼200 protein groups for single HeLa cells (without MS1-level feature matching) to >1000 protein groups through improved separations, MS instrumentation and the incorporation of FAIMS filtering, etc. Progress has been similarly rapid for isobaric labeling workflows (15, 16). Although the technology has improved, the application space for low-input and single cell proteomics has also broadened from cultured cells to spatially resolved mammalian and plant tissues (29, 30, 32), with fundamental insights being provided by these studies. As mentioned throughout this perspective, there are still many ways to improve measurement throughput, proteome coverage and quantitative accuracy for global studies, as well as opportunities to extend functional measurements of post-translational modifications and the determination of proteoform activities to the nanoscale. We must also continue to invest in efforts to automate and simplify the techniques currently required for low-input proteomics without sacrificing performance, which will significantly broaden accessibility. Finally, data analysis strategies that maximize the information extracted from sparse and noisy datasets will be tremendously beneficial. With continued refinement and maturation, single cell proteomics will follow other techniques such as mass cytometry and scRNA-seq to become a powerful and widely used tool in biomedical research.
Footnotes
Funding and additional information—This work was supported by the National Cancer Institute of the National Institutes of Health under award number R33 CA225248. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—Author declares no competing interests.
Abbreviations—The abbreviations used are:
- SA:V
- surface-area-to-volume ratio
- LCM
- laser capture microdissection
- iPAD
- integrated proteome analysis device
- MBR
- Match Between Runs
- TMT
- Tandem Mass Tag.
REFERENCES
- 1. Richards A. L., Merrill A. E., and Coon J. J. (2015) Proteome sequencing goes deep. Curr. Opin. Chem. Biol. 24, 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binnewies M., Roberts E. W., Kersten K., Chan V., Fearon D. F., Merad M., Coussens L. M., Gabrilovich D. I., Ostrand-Rosenberg S., Hedrick C. C., Vonderheide R. H., Pittet M. J., Jain R. K., Zou W. P., Howcroft T. K., Woodhouse E. C., Weinberg R. A., and Krummel M. F. (2018) Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crow M., Paul A., Ballouz S., Huang Z. J., and Gillis J. (2018) Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun. 9, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu Y., Scheibinger M., Ellwanger D. C., Krey J. F., Choi D., Kelly R. T., Heller S., and Barr-Gillespie P. G. (2019) Single-cell proteomics reveals changes in expression during hair-cell development. Elife 8, e50777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snyder M. P., et al. (2019) The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen Y., Tolic N., Masselon C., Pasa-Tolic L., Camp D. G., Hixson K. K., Zhao R., Anderson G. A., and Smith R. D. (2004) Ultrasensitive proteomics using high-efficiency on-line micro-SPE-NanoLC-NanoESI MS and MS/MS. Anal. Chem. 76, 144–154 [DOI] [PubMed] [Google Scholar]
- 7. Belov M. E., Gorshkov M. V., Udseth H. R., Anderson G. A., and Smith R. D. (2000) Zeptomole-sensitivity electrospray ionization−Fourier transform ion cyclotron resonance mass spectrometry of proteins. Anal. Chem. 72, 2271–2279 [DOI] [PubMed] [Google Scholar]
- 8. Chen W. D., Wang S., Adhikari S., Deng Z. H., Wang L. J., Chen L., Ke M., Yang P. Y., and Tian R. J. (2016) Simple and integrated spintip-based technology applied for deep proteome profiling. Anal. Chem. 88, 4864–4871 [DOI] [PubMed] [Google Scholar]
- 9. Sun L. L., Dubiak K. M., Peuchen E. H., Zhang Z. B., Zhu G. J., Huber P. W., and Dovichi N. J. (2016) Single cell proteomics using frog (Xenopus laevis) blastomeres isolated from early stage embryos, which form a geometric progression in protein content. Anal. Chem. 88, 6653–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lombard-Banek C., Moody S. A., and Nemes P. (2016) Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew. Chem. Int. Ed. 55, 2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu Y., Piehowski P. D., Zhao R., Chen J., Shen Y. F., Moore R. J., Shukla A. K., Petyuk V. A., Campbell-Thompson M., Mathews C. E., Smith R. D., Qian W. J., and Kelly R. T. (2018) Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells. Nat. Commun. 9, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Budnik B., Levy E., Harmange G., and Slavov N. (2018) SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cong Y., Motamedchaboki K., Misal S. A., Liang Y., Guise A. J., Truong T., Huguet R., Plowey E. D., Zhu Y., Lopez-Ferrer D., and Kelly R. T. (2020) Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell. bioRxiv 2020.2006.2003.132449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dou M. W., Clair G., Tsai C. F., Xu K. R., Chrisler W. B., Sontag R. L., Zhao R., Moore R. J., Liu T., Pasa-Tolic L., Smith R. D., Shi T. J., Adkins J. N., Qian W. J., Kelly R. T., Ansong C., and Zhu Y. (2019) High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform. Anal. Chem. 91, 13119–13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai C.-F., Zhao R., Williams S. M., Moore R. J., Schultz K., Chrisler W., Pasa-Tolic L., Rodland K., Smith R. D., Shi T., Zhu Y., and Liu T. (2020) An improved boosting to amplify signal with isobaric labeling (iBASIL) strategy for precise quantitative single-cell proteomics. Mol. Cell. Proteomics 19, 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Specht H., Emmott E., Petelski A. A., Gray Huffman R., Perlman D. H., Serra M., Kharchenko P., Koller A., and Slavov N. (2019) Single-cell mass-spectrometry quantifies the emergence of macrophage heterogeneity. bioRxiv 665307 [Google Scholar]
- 17. Dou M. W., Zhu Y., Liyu A., Liang Y. R., Chen J., Piehowski P. D., Xu K. R., Zhao R., Moore R. J., Atkinson M. A., Mathews C. E., Qian W. J., and Kelly R. T. (2018) Nanowell-mediated two-dimensional liquid chromatography enables deep proteome profiling of < 1000 mammalian cells. Chem. Sci. 9, 6944–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. (2008) Proteomics Sample Preparation, Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 19. Goebel-Stengel M., Stengel A., Taché Y., and Reeve J. R. Jr (2011) The importance of using the optimal plasticware and glassware in studies involving peptides. Anal. Biochem. 414, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu R., Xing S., Badv M., Didar T. F., and Lu Y. (2019) Step-wise assessment and optimization of sample handling recovery yield for nanoproteomic analysis of 1000 mammalian cells. Anal. Chem. 91, 10395–10400 [DOI] [PubMed] [Google Scholar]
- 21. Nelson D. L., and Cox M. M. (2017) Principles of Biochemistry, 7th Ed, W. H. Freeman and Company, New York [Google Scholar]
- 22. Keil-Dlouhá V. V., Zylber N., Imhoff J., Tong N., and Keil B. (1971) Proteolytic activity of pseudotrypsin. FEBS Lett. 16, 291–295 [DOI] [PubMed] [Google Scholar]
- 23. Nielsen J. B., Hanson R. L., Almughamsi H. M., Pang C., Fish T. R., and Woolley A. T. (2020) Microfluidics: innovations in materials and their fabrication and functionalization. Anal. Chem. 92, 150–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J., Hansen C., and Quake S. R. (2003) Solving the “World-to-Chip” interface problem with a microfluidic matrix. Anal. Chem. 75, 4718–4723 [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Cordero J. L., and Fan Z. H. (2017) Sessile droplets for chemical and biological assays. Lab Chip. 17, 2150–2166 [DOI] [PubMed] [Google Scholar]
- 26. Xu K. R., Liang Y. R., Piehowski P. D., Dou M. W., Schwarz K. C., Zhao R., Sontag R. L., Moore R. J., Zhu Y., and Kelly R. T. (2019) Benchtop-compatible sample processing workflow for proteome profiling of < 100 mammalian cells. Anal. Bioanal. Chem. 411, 4587–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cong Y., Liang Y., Motamedchaboki K., Huguet R., Truong T., Zhao R., Shen Y., Lopez-Ferrer D., Zhu Y., and Kelly R. T. (2020) Improved single-cell proteome coverage using narrow-bore packed NanoLC columns and ultrasensitive mass spectrometry. Anal. Chem. 92, 2665–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu Y., Clair G., Chrisler W. B., Shen Y. F., Zhao R., Shukla A. K., Moore R. J., Misra R. S., Pryhuber G. S., Smith R. D., Ansong C., and Kelly R. T. (2018) Proteomic analysis of single mammalian cells enabled by microfluidic nanodroplet sample preparation and ultrasensitive NanoLC-MS. Angew. Chem. Int. Ed. Engl. 57, 12370–12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y., Dou M. W., Piehowski P. D., Liang Y. R., Wang F. J., Chu R. K., Chrisler W. B., Smith J. N., Schwarz K. C., Shen Y. F., Shukla A. K., Moore R. J., Smith R. D., Qian W. J., and Kelly R. T. (2018) Spatially resolved proteome mapping of laser capture microdissected tissue with automated sample transfer to nanodroplets. Mol. Cell. Proteomics 17, 1864–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liang Y. R., Zhu Y., Dou M. W., Xu K. R., Chu R. K., Chrisler W. B., Zhao R., Hixson K. K., and Kelly R. T. (2018) Spatially resolved proteome profiling of <200 cells from tomato fruit pericarp by integrating laser-capture microdissection with nanodroplet sample preparation. Anal. Chem. 90, 11106–11114 [DOI] [PubMed] [Google Scholar]
- 31. Zhu Y., Podolak J., Zhao R., Shukla A. K., Moore R. J., Thomas G. V., and Kelly R. T. (2018) Proteome profiling of 1 to 5 spiked circulating tumor cells isolated from whole blood using immunodensity enrichment, laser capture microdissection, nanodroplet sample processing, and ultrasensitive nanoLC-MS. Anal. Chem. 90, 11756–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piehowski P. D., Zhu Y., Bramer L. M., Stratton K. G., Zhao R., Orton D. J., Moore R. J., Yuan J., Mitchell H. D., Gao Y., Webb-Robertson B.-J. M., Dey S. K., Kelly R. T., and Burnum-Johnson K. E. (2020) Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-μm spatial resolution. Nat. Commun. 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu Y., Piehowski P. D., Kelly R. T., and Qian W. J. (2018) Nanoproteomics comes of age. Exp. Rev. Proteomics 15, 865–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao X., Wang X., Guan S., Lin H., Yan G., Gao M., Deng C., and Zhang X. (2018) Integrated proteome analysis device for fast single-cell protein profiling. Anal. Chem. 90, 14003–14010 [DOI] [PubMed] [Google Scholar]
- 35. Li Z. Y., Huang M., Wang X. K., Zhu Y., Li J. S., Wong C. C. L., and Fang Q. (2018) Nanoliter-scale oil-air-droplet chip-based single cell proteomic analysis. Anal. Chem. 90, 5430–5438 [DOI] [PubMed] [Google Scholar]
- 36. Huang E. L., Piehowski P. D., Orton D. J., Moore R. J., Qian W. J., Casey C. P., Sun X. F., Dey S. K., Burnum-Johnson K. E., and Smith R. D. (2016) SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology 157, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, U359–U360 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Z. B., Dubiak K. M., Huber P. W., and Dovichi N. J. (2020) Miniaturized filter-aided sample preparation (MICRO-FASP) method for high throughput, ultrasensitive proteomics sample preparation reveals proteome asymmetry in Xenopus laevis embryos. Anal. Chem. 92, 5554–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams S. M., Liyu A. V., Tsai C.-F., Moore R. J., Orton D. J., Chrisler W. B., Gaffrey M. J., Liu T., Smith R. D., Kelly R. T., Pasa-Tolic L., and Zhu Y. (2020) Automated coupling of nanodroplet sample preparation with liquid chromatography-mass spectrometry for high-throughput single-cell proteomics. Anal. Chem. 92, 10588–10596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pasa-Tolić L., Masselon C., Barry R. C., Shen Y., and Smith R. D. (2004) Proteomic analyses using an accurate mass and time tag strategy. BioTechniques 37, 621–639 [DOI] [PubMed] [Google Scholar]
- 41. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 42. Wilm M., and Mann M. (1996) Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 [DOI] [PubMed] [Google Scholar]
- 43. Marginean I., Tang K. Q., Smith R. D., and Kelly R. T. (2014) Picoelectrospray ionization mass spectrometry using narrow-bore chemically etched emitters. J. Am. Soc. Mass Spectrom. 25, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson S. R., Olsen C., and Lundanes E. (2019) Nano liquid chromatography columns. Analyst 144, 7090–7104 [DOI] [PubMed] [Google Scholar]
- 45. Sandow J. J., Infusini G., Dagley L. F., Larsen R., and Webb A. I. (2019) Simplified high-throughput methods for deep proteome analysis on the timsTOF Pro. bioRxiv 657908 [Google Scholar]
- 46. Schmidt A., Karas M., and Dülcks T. (2003) Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI?. J. Am. Soc. Mass Spectrom. 14, 492–500 [DOI] [PubMed] [Google Scholar]
- 47. Shen Y., Zhao R., Berger S. J., Anderson G. A., Rodriguez N., and Smith R. D. (2002) High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal. Chem. 74, 4235–4249 [DOI] [PubMed] [Google Scholar]
- 48. Shen Y., Tolić N., Masselon C., Paša-Tolić L., Camp Ii D. G., Lipton M. S., Anderson G. A., and Smith R. D. (2004) Nanoscale proteomics. Anal. Bioanal. Chem. 378, 1037–1045 [DOI] [PubMed] [Google Scholar]
- 49. Zhu Y., Zhao R., Piehowski P. D., Moore R. J., Lim S., Orphan V. J., Pasa-Tolic L., Qian W. J., Smith R. D., and Kelly R. T. (2018) Subnanogram proteomics: Impact of LC column selection, MS instrumentation and data analysis strategy on proteome coverage for trace samples. Int. J. Mass Spectrom. 427, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yue G., Luo Q., Zhang J., Wu S.-L., and Karger B. L. (2007) Ultratrace LC/MS proteomic analysis using 10-μm-i.d. porous layer open tubular poly(styrene−divinylbenzene) capillary columns. Anal. Chem. 79, 938–946 [DOI] [PubMed] [Google Scholar]
- 51. Li S. Y., Plouffe B. D., Belov A. M., Ray S., Wang X. Z., Murthy S. K., Karger B. L., and Ivanov A. R. (2015) An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Mol. Cell. Proteomics 14, 1672–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Malsche W., Eghbali H., Clicq D., Vangelooven J., Gardeniers H., and Desmet G. (2007) Pressure-driven reverse-phase liquid chromatography separations in ordered nonporous pillar array columns. Anal. Chem. 79, 5915–5926 [DOI] [PubMed] [Google Scholar]
- 53. Stadlmann J., Hudecz O., Krššáková G., Ctortecka C., Van Raemdonck G., Op De Beeck J., Desmet G., Penninger J. M., Jacobs P., and Mechtler K. (2019) Improved sensitivity in low-input proteomics using micropillar array-based chromatography. Anal. Chem. 91, 14203–14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Z., Qu Y., and Dovichi N. J. (2018) Capillary zone electrophoresis-mass spectrometry for bottom-up proteomics. TrAC Trends Anal. Chem. 108, 23–37 [Google Scholar]
- 55. Sun L. L., Zhu G. J., Zhang Z. B., Mou S., and Dovichi N. J. (2015) Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J. Proteome Res. 14, 2312–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Amenson-Lamar E. A., Sun L., Zhang Z., Bohn P. W., and Dovichi N. J. (2019) Detection of 1 zmol injection of angiotensin using capillary zone electrophoresis coupled to a Q-Exactive HF mass spectrometer with an electrokinetically pumped sheath-flow electrospray interface. Talanta 204, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Z., Hebert A. S., Westphall M. S., Qu Y., Coon J. J., and Dovichi N. J. (2018) Production of over 27 000 peptide and nearly 4400 protein identifications by single-shot capillary-zone electrophoresis–mass spectrometry via combination of a very-low-electroosmosis coated capillary, a third-generation electrokinetically-pumped sheath-flow nanospray interface, an orbitrap fusion lumos tribrid mass spectrometer, and an advanced-peak-determination algorithm. Anal. Chem. 90, 12090–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yan X. J., Sun L. L., Zhu G. J., Cox O. F., and Dovichi N. J. (2016) Over 4100 protein identifications from a Xenopus laevis fertilized egg digest using reversed-phase chromatographic prefractionation followed by capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry analysis. Proteomics 16, 2945–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baxi A. B., Lombard-Banek C., Moody S. A., and Nemes P. (2018) Proteomic characterization of the neural ectoderm fated cell clones in the xenopus laevis embryo by high-resolution mass spectrometry. ACS Chem. Neurosci. 9, 2064–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lombard-Banek C., Moody S. A., Manzin M. C., and Nemes P. (2019) Microsampling capillary electrophoresis mass spectrometry enables single-cell proteomics in complex tissues: developing cell clones in live Xenopus laevis and Zebrafish embryos. Anal. Chem. 91, 4797–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan H., Jiang B., Zhao B., Zhang L., and Zhang Y. (2019) Recent advances in multidimensional separation for proteome analysis. Anal. Chem. 91, 264–276 [DOI] [PubMed] [Google Scholar]
- 62. Dou M. W., Chouinard C. D., Zhu Y., Nagy G., Liyu A. V., Ibrahim Y. M., Smith R. D., and Kelly R. T. (2019) Nanowell-mediated multidimensional separations combining nanoLC with SLIM IM-MS for rapid, high-peak-capacity proteomic analyses. Anal. Bioanal. Chem. 411, 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dou M. W., Tsai C. F., Piehowski P. D., Wang Y., Fillmore T. L., Zhao R., Moore R. J., Zhang P. F., Qian W. J., Smith R. D., Liu T., Kelly R. T., Shi T. J., and Zhu Y. (2019) Automated nanoflow two-dimensional reversed-phase liquid chromatography system enables in-depth proteome and phosphoproteome profiling of nanoscale samples. Anal. Chem. 91, 9707–9715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiang P. L., Zhu Y., Yang Y., Zhao Z. T., Williams S. M., Moore R. J., Kelly R. T., Smith R. D., and Liu S. R. (2020) Picoflow liquid chromatography-mass spectrometry for ultrasensitive bottom-up proteomics using 2-mu m-i.d. open tubular columns. Anal. Chem. 92, 4711–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith R. D., Loo J. A., Edmonds C. G., Barinaga C. J., and Udseth H. R. (1990) New developments in biochemical mass spectrometry: electrospray ionization. Anal. Chem. 62, 882–899 [DOI] [PubMed] [Google Scholar]
- 66. Kelly R. T., Tolmachev A. V., Page J. S., Tang K., and Smith R. D. (2010) The ion funnel: theory, implementations, and applications. Mass Spectrom. Rev. 29, 294–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Page J. S., Tang K., Kelly R. T., and Smith R. D. (2008) Subambient pressure ionization with nanoelectrospray source and interface for improved sensitivity in mass spectrometry. Anal. Chem. 80, 1800–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Marginean I., Page J. S., Tolmachev A. V., Tang K., and Smith R. D. (2010) Achieving 50% ionization efficiency in subambient pressure ionization with nanoelectrospray. Anal. Chem. 82, 9344–9349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eliuk S., and Makarov A. (2015) Evolution of orbitrap mass spectrometry instrumentation. Annu Rev Anal. Chem. 8, 61–80 [DOI] [PubMed] [Google Scholar]
- 70. Nolting D., Malek R., and Makarov A. (2019) Ion traps in modern mass spectrometry. Mass Spectrom. Rev. 38, 150–168 [DOI] [PubMed] [Google Scholar]
- 71. Cheung T. K., Lee C.-Y., Bayer F., McCoy A., Kuster B., and Rose C. M. (2020) Defining the carrier proteome limit for single cell proteomics. Nat. Methods in press [DOI] [PubMed] [Google Scholar]
- 72. Baker E. S., Burnum-Johnson K. E., Ibrahim Y. M., Orton D. J., Monroe M. E., Kelly R. T., Moore R. J., Zhang X., Theberge R., Costello C. E., and Smith R. D. (2015) Enhancing bottom-up and top-down proteomic measurements with ion mobility separations. Proteomics 15, 2766–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bekker-Jensen D. B., Martinez-Val A., Steigerwald S., Ruther P., Fort K. L., Arrey T. N., Harder A., Makarov A., and Olsen J. V. (2020) A compact quadrupole-orbitrap mass spectrometer with FAIMS interface improves proteome coverage in short LC gradients. Mol. Cell. Proteomics 19, 716–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kosinski T., Chen N., Mahu E., and Subeck M. (2020) PASEF for sensitive shotgun proteomics: toward single cell analysis. 68th ASMS Conference on Mass Spectrometry and Allied topics MP 547 [Google Scholar]
- 75. Saha-Shah A., Esmaeili M., Sidoli S., Hwang H., Yang J., Klein P. S., and Garcia B. A. (2019) Single cell proteomics by data-independent acquisition to study embryonic asymmetry in Xenopus laevis. Anal. Chem. 91, 8891–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meier F., Geyer P. E., Virreira Winter S., Cox J., and Mann M. (2018) BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods 15, 440–448 [DOI] [PubMed] [Google Scholar]
- 77. Russell C. L., Heslegrave A., Mitra V., Zetterberg H., Pocock J. M., Ward M. A., and Pike I. (2017) Combined tissue and fluid proteomics with Tandem Mass Tags to identify low-abundance protein biomarkers of disease in peripheral body fluid: An Alzheimer's Disease case study. Rapid Commun. Mass Spectrom. 31, 153–159 [DOI] [PubMed] [Google Scholar]
- 78. Muntel J., Kirkpatrick J., Bruderer R., Huang T., Vitek O., Ori A., and Reiter L. (2019) Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res. 18, 1340–1351 [DOI] [PubMed] [Google Scholar]
- 79. Ting L., Rad R., Gygi S. P., and Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]