Abstract
The coronavirus disease (COVID-19) was first identified in China, December 2019. Since then, it has spread the length and breadth of the world at an unprecedented, alarming rate. Severe acute respiratory syndrome coronavirus (SARS-CoV)-2, which causes COVID-19, has much in common with its closest homologs, SARS-CoV and Middle East respiratory syndrome-CoV. The virus–host interaction of SARS-CoV-2 uses the same receptor, ACE2, which is similar to that of SARS-CoV, which spreads through the respiratory tract. Patients with COVID-19 report symptoms including mild-to-severe fever, cough and fatigue; very few patients report gastrointestinal infections. There are no specific antiviral strategies. A few strong medications are under investigation, so we have to focus on proposals which ought to be taken to forestall this infection in a living host.
Keywords: : coronavirus, COVID-19, inhibitors, life cycle, RNA, SARS-CoV-2
Graphical abstract
Among the many illnesses that shook the world in recent history, coronavirus disease (COVID-19) has emerged as one of the most difficult pandemics to control as well as cure. In December 2019, cases of pneumonia of an unknown cause were found in patients, who had visited a seafood market in Wuhan, China [1]. This event has driven humanity into a trance of pandemic fear. COVID-19 is a serious medical concern in more than 195 countries. An alarming 9,703,330 cases have been confirmed, with over 490,989 dead as of 25 June 2020, and the world is still counting. COVID-19 is a respiratory disease caused by an infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It belongs to the β-Coronavirus subgenus of the Coronaviridae family, part of the order of viruses called the nidovirales (Figure 1) [2,3].
Figure 1. . Classification of severe acute respiratory syndrome coronavirus-2 starting from the family of nidovirales.
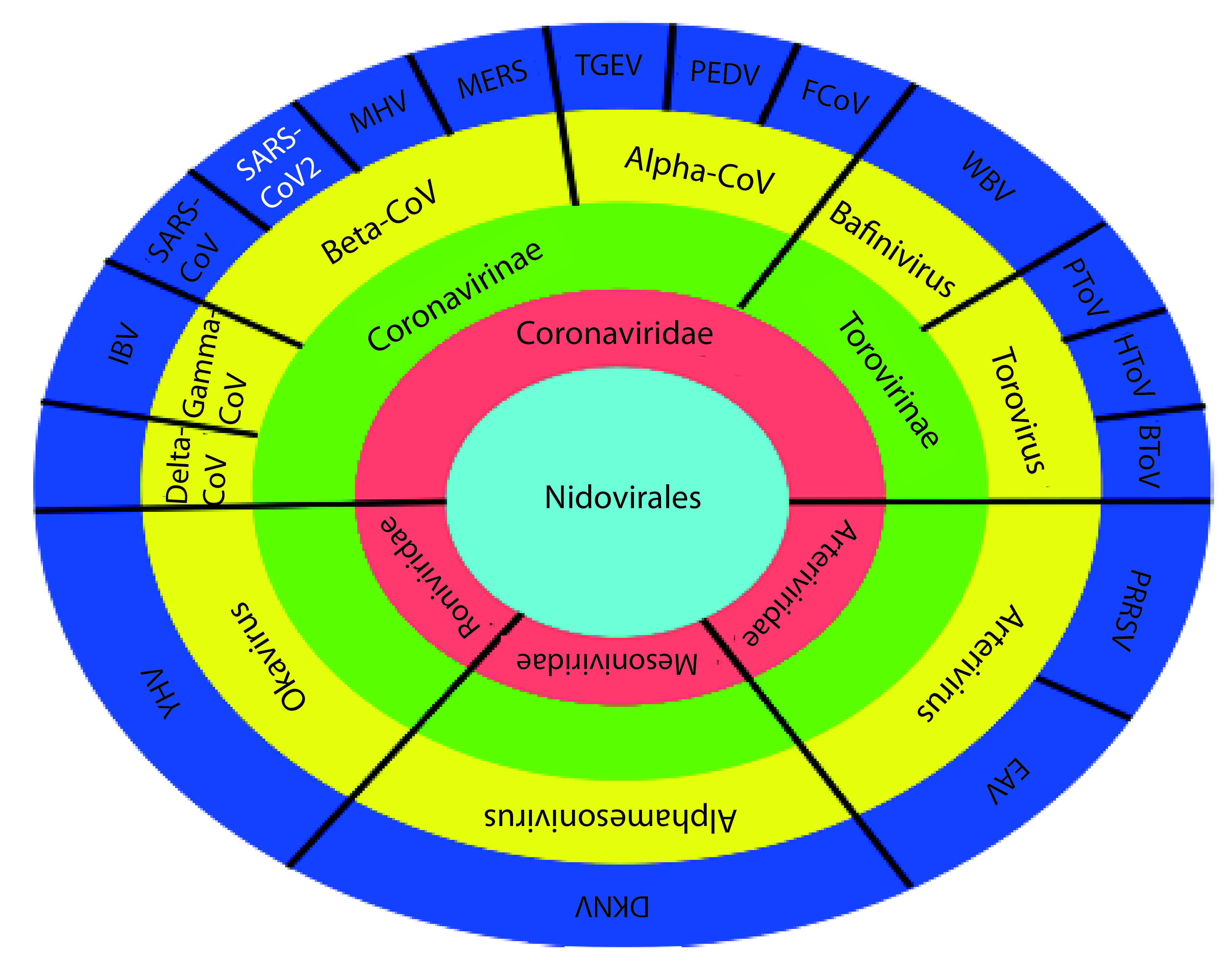
BToV: Bovine torovirus; DKNV: Dak nong virus; EAV: Equine arteritis virus; FCoV Feline coronavirus; HToV: Human torovirus; IBV: Infectious bronchitis virus; MERS-CoV: Middle East respiratory syndrome coronavirus; MHV: Mouse hepatitis virus; PRRSV: Porcine torovirus; TGEV: Transmissible gastroenteritis coronavirus; SARS-CoV: Severe acute respiratory syndrome coronavirus; YHV: Yellow head virus; WBV: White bream virus.
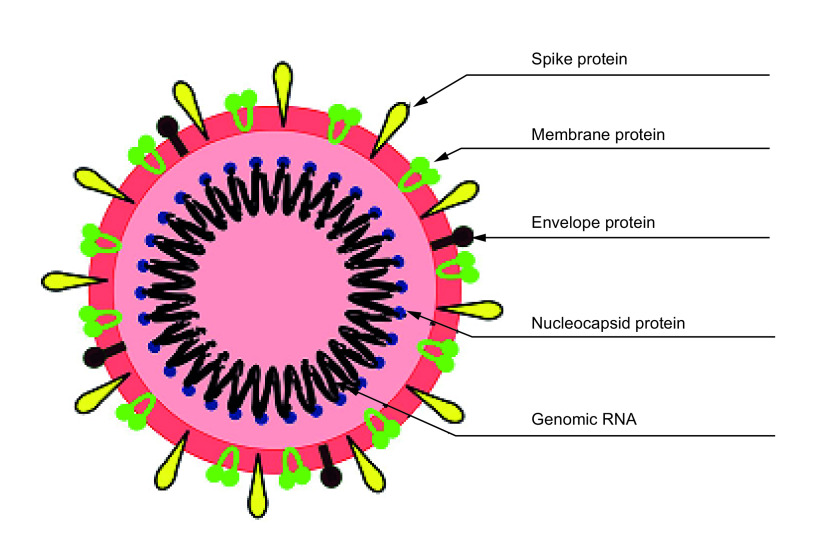
SARS-CoV-2 has been given this name because it is 85% similar to the SARS-CoV virus genome [1]. It is the seventh coronavirus (CoVs) that is known to have infected humans after HKU1, NL63, OC43 and 229E, associated with common cold symptoms, and bat-SL-CoVZC45, MG772933.11 [4,5], commonly known as SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) that cause severe respiratory illnesses. Electron micrographs of the virus revealed a spherical structure, like that of a solar corona or a sun-like morphology, giving its name Coronavirus, with a diameter of 60–140 nm, and presence of distinctive spikes of about 9–12 nm. The virus is made up of five types of protein, spike protein (S), envelope protein (E), membrane protein (M) and hemagglutinin-esterase dimer protein (HE) and nucleocapsid protein (N) [6–8], which are bound to the RNA (Figure 2). The S, E, M and HE proteins are on the surface, while the genetic material causing the replication is inside the virion, pegged together with N-protein [9], and the entire genome is 26,000–32,000 bases long [10].
Figure 2. . Structure of severe acute respiratory syndrome coronavirus-2 showing the genomic RNA and structural proteins.
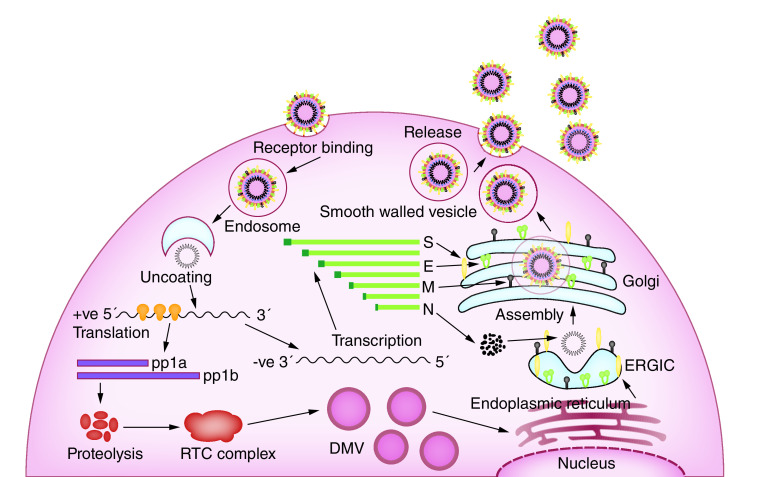
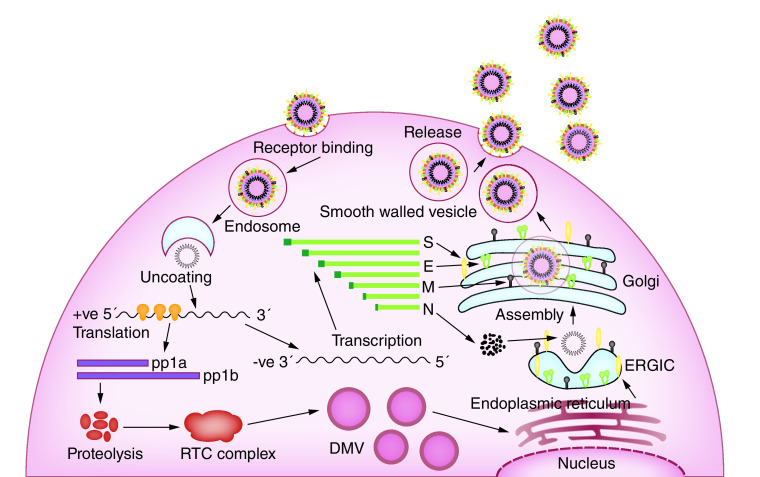
The coronavirus invades two types of cells in the lungs. They are the mucus producing goblet cells that keep the lungs from drying, as well as protecting them from pathogens, and the ciliated cells that are responsible for clearing debris from the lungs by beating the mucus to the body’s exterior. The cilia cells are the preferred viral host [11]. The spike protein, based on the virion surface, are responsible for binding with the host cell receptors, called ACE2 that are found in human cells in the lungs, heart, kidneys and intestines. The S-protein has two receptor binding domains (RBD’s), called the S1 and S2. These domains are used to bind with the ACE2 receptors, allowing the virion to penetrate eventually into the human cells by a process known as endocytosis [12]. The virus, once inside the cytoplasm, dissolves its own protein shells and releases the viral RNA payload inside the cell. The viral RNA hijacks the cell replication machinery and starts to replicate itself. It also manufactures the necessary proteins to assemble new viruses that Golgi bodies will transport out of the infected cell by a process called exocytosis (Figure 3). When the host cell loses its ability to maintain homeostasis, cell death or apoptosis occurs [13]. These dead cells fill the lung airways with debris and fluid, which causes clogging and eventually leads to pneumonia. Sometimes the immune system overreacts and damages healthy tissues in the lungs; then more cells die, further clogging the immune system and making the pneumonia worse. As damage to lungs keeps increasing, the patient experiences complete respiratory failure, eventually leading to death [14].
Figure 3. . Life cycle of severe acute respiratory syndrome coronavirus-2 corona virus in the human cell, based on the information acquired by studying coronavirus in general.
The investigation into the SARS-Cov-2 virus began when three patients with pneumonia from unknown cause were admitted to Wuhan Jinyintan Hospital, China, on 27 December 2019. The local health authority reported several other patients with similar symptoms, all of whom had visited the same seafood-animal market in Wuhan, China [15]. The China Center for Disease Control and Prevention (CCDC) began to explore the epidemiology of this novel virus. Their studies reported details of admitted patients. Patient 1 was a female shopkeeper (49 years old) with no chronic illness in her medical record but disclosed cough, fever (37–38°C) and chest pain on 23rd December 2019. After 4 days, cough and chest pain increased, but her fever reduced. The tomographic results proved pneumonia. An old man (61 years), who was a frequent visitor to the market, was patient 2. He reported a high fever with cough on 20 December 2019. After 7 days of illness, he developed respiratory distress, which worsened after 2 days. He was later transferred to the intensive care unit (ICU) for mechanical ventilation. Patient 3 was a young man (32 years) with an unknown clinical profile. Patient 1 and patient 3 were cured under continuous treatment and discharged on 16 January 2020. Patient 2 died in the hospital on 9 January 2020. Thus, cases were reported with minimum criteria, such as pneumonia with fever (≥38°C), dry cough, chest radiograph of pneumonia, lower lymphocyte or lower white blood cell count, etc. in the preliminary stage cases. The next stage was to observe the patient for 3 days under antimicrobial treatment with standard clinical practice guidelines. The target population were people who had the same symptoms linked with the seafood market, and also people with any travel history to or from, or directly or indirectly in social contact with patients from Wuhan [16].
After extensive literature review related to COVID-19, most of the researchers discussed only mild-to-severe symptoms, translucent mechanism and general inhibition process [17–19]. But the current review paper integrates, not only symptoms but also stepwise virus life cycle mechanism and inhibition methodology. At the same time, the graphical representation of different parameters was illustrated. This updated information will definitely guide present researchers belonging to different fields to develop a safe and effective vaccine.
Symptoms & behaviors
The main pathogens for respiratory disease were CoVs, found in many animal species and possible to isolate. All CoVs belong to +ssRNA stranded large family (RNA virus) [20]. The reason is unclear, but the virus can travel through species barriers and can cause moderate-to-severe disease in humans, like other MERS and SARS. It is assumed that these two deadly viruses originated from bats and later moved through mammalian hosts, like the Himalayan palm civet (SARS-CoV) and the Arabian camel (MERS-CoV) before making the jump into humans. Interestingly, COVID-19 is also thought to have originated from animals; genome analysis suggests it was also from bats [21] but additional analysis is required to confirm this. Pangolin may play an important role as an intermediate host for transmission of virus to human. The transfer process and virus pattern can be compared with the SARS virus, where an infected person was diagnosed in Guangdong, China, in 2002. Initially, the virus generated from bats, passing through host civet cats and finally targeting humans [22]. However, few researchers explained similarities and divergences between genomic sequences of SARS-CoV-2 and RaTG13, and clarified the effect of differences in calculation methodology due to RNA modification, which can further assist in understanding the genesis as well as evolution of SARS-CoV-2 [23,24].
Even after parsing several literary articles on the origin and evolution of coronavirus, we found that there is still much ambiguity on how the virus has originated and evolved. It is still ambiguous whether the SARS-CoV-2 virus has originated from bat, pangolin or any other wild species, also there is uncertainty regarding its evolution from SARS-CoV or any other coronavirus. However, the study on mutations and polymorphisms has very much assisted in clearing all such doubts and has enhanced further understanding of coronavirus as well as helped to map its transmission [25]. The genome sequence of SARS-CoV-2 changes over time due to mutations occurring randomly at single nucleotide polymorphism (SNP) sites, and any changes that occur in a given virus will be inherited by all copies of the next generation. DNA sequencing technologies cannot directly decode the +ssRNA SARS-CoV-2 virus. Scientists first transcribe the RNA of the virus into complementary DNA which can later be sequenced [26]. Studies have shown that single nucleotide polymorphism DNA microarray has been developed for the detection and genotype sequencing of the SARS coronavirus [27]. Sequencing the SARS-CoV-2 virus at different times in history can tell us how it is mutating and can give an idea on the direction it is heading. The coronavirus that has continually been propagating through people by its infection acquires mutation and thus evolves into different virus strains. These strains help us in the tracing the mutation, by giving us an evidence as to whether the virus was picked up by the patient from the community spread or foreign travel. Scientists from all over the world have started to deposit their analysis on the sequence alignment of the patient’s virus infection on integrated international websites such as GISAID [28], nextstrain [29], this has immensely helped in following the infection, mapping the mutation as well as tracing the transmission of different patients everywhere. Researchers from all around the globe are working to create new algorithms with the help of developed software (MUSCLE, PhyML, Figtree, MAFFT, etc.,) and computer hardware to make a real-time analysis of the pandemic more feasible. In future, we can have an extensive collection of this data in the form of database, which can assist in the study of mutations and polymorphisms of the coronavirus on the origin and evolution can help in co-relating the clinical parameters with the severity of the infected strain in comparison with other and thus guide in the formation of antiviral drugs and vaccine for preventing the further spread of this deadly pathogen.
The virus life cycle predominantly depends on the involvement of nonstructural protein enzymes, and structural glycoproteins are essential during the virus’s entry into the cell [30]. The catalytic enzyme site was protected for antiviral drugs. A similar kind of action has been noted with SARS and MERS [31]. It is extensively sensitive to high temperatures, and ultraviolet rays, in which the virus’s performance can disintegrate by introducing at least 60% ethanol, ether, peracetic acid, trichloro methane and sodium hypochlorite [32]. The genome, collected from several patients, came from Wuhan, China. Its nucleotide is 89% identical to bat SARS-COVZXC21 and 82% to human SARS-CoV [33], which may be behind the reason COVID-19 resembles SARS-COV-2. Viral transmission through infected patients has been enhanced through human-to-human social contacts and is directly related with number of secondary cases. The preliminary response period to be infected within 4 days; the average is 1.61 days [34]. The initial investigation was based on the population collected from epidemiological studies, as well as community and contact characteristics records with the help of the susceptible–exposed–infectious–recovered transmission model [34]. According to Huang et al. [35], COVID-19 was consistent with beta coronavirus infections, including major symptoms such as fever, dry cough, dyspnea, etc. The extended infection was noted graphically on a chest CT scan. The characteristics of SARS-CoV and MERS-CoV infections were consistent with COVID-19. Some patients infected with COVID-19 had symptoms such as sore throat, sneezing, etc., which was related to respiratory tract infection, indicating that the infected cells were present in the lower respiratory system. More than 25% of people infected with SARS-CoV and MERS-CoV had diarrhea, which was the gastrointestinal infection, which is rare in patients infected with COVID-19. Testing of urine and faucal samples will explain the right transmission route, which remains unclear. Around 40 patients reported fever whereas with cough 31 and 18 were diagnosed with myalgia or fatigue. Headache, hemoptysis and diarrhea were the minor symptoms of this virus because they occurred in fewer than 10% of cases, which was clearly shown on the graph. However, around 50% (22) had difficulty breathing, resulting in 41 infected patients ( Supplementary Figure 1 & Supplementary Table 1).
The study, based on the gender and average age, conducted by Real-Time Quantitative Reverse Transcription PCR reported that 99 infected patients (67 males and 32 females) had visited the seafood market in Wuhan [36]. The analysis suggested that fever, cough and breathing issues were the major symptoms; other symptoms, occurring less frequently, included chest pain, diarrhea and vomiting. The survey results indicated about 83% (82) presented with fever, 82% (81) with cough and 30% with breathing issues (31). Symptoms such as sore throats, headaches, nausea and vomiting were lower than 8% ( Supplementary Figure 2 & Supplementary Table 2). The other report represented in the graph ( Supplementary Figure 2) explained only few symptoms were major but other symptoms were minor for this deadly virus. It was found that 99% patients having fever with other symptoms like dry cough (82%), myalgia (48%), dyspnea (43%) and fatigue (96%), accumulated with 136 infected patients [37].
The distribution periods for incubation were established by Backer et al. [38], with 88 confirmed cases, ranging in age from 2 to 72 years, having similar travel histories. The four types of viruses with the long duration of up to 14 days [38] and 24 days from onset to the invasive condition for COVID-19 [39], followed by MERS for 5 days [40] as reported by center for disease control and prevention, whereas 1–4 days [41] for swine flu H1N1 influenza. Estimating and understanding the incubation period plays a vital role in controlling viral transmission and reducing its reproductive rate since there is no vaccine yet. Several case studies imply that the incubation period varies in the range of 2–24 days from the beginning of symptoms to the cure and deaths of infected patients, as illustrated in the Supplementary Figure 3& Supplementary Table 3.
Life cycle of coronavirus in human host cell
The spike protein of the SAR-CoV-2 virus
The spike proteins (S glycoprotein) are spike-like protrusions present on the coronavirus’s transmembrane, which helps the virus enter host cells [42]. The S1 subunit helps bind the virion to the host cell receptor at the N terminus. The S2 subunit mediates the fusion of the viral and cellular membranes at the C terminus [43]. Furthermore, the S glycoprotein is cleaved between the boundary of the S1 and S2 subunits, forming a polybasic furin cleavage site S2′. S2’ acts as a viral fusion peptide that is uncoated by cleavage of the S2 subunit during virus endocytosis. The presence of such a furin cleavage site distinguishes the SARS-CoV-2 from other coronaviruses such as the SAR-CoV, in that it has a monobasic cleavage site between the said S1/S2 subunits that is processed further by the target entry host cells. The non covalently bound S1/S2 prefusion conformation is further cleaved by host proteases, located at the top of the fusion peptide. This cleavage site activates the proteins for the fusion of the viral and cellular membranes by many irreversible conformations, making the virus entry into susceptible cells a complex process of receptor binding and proteolytic processing of S proteins [44–48]. The striking similarity between the SAR-CoV and the SAR-CoV-2 viruses is that the structure and sequence conservation of the spike protein emphasizes a similar receptor-binding mechanism. However, in the case of the SAR-CoV-2 virus, the presence of a furin cleavage site and its expression of furin-like proteases may create a SAR-CoV like cell and tissue tropism, but with increased pathogen transmissibility [49].
Receptor binding
The spike proteins of the SARS-CoV-2 behave like RBDs, specifically targeting the host cell receptor ACE2 of the human cells and forming the initial viral attachment step between the virus and the cell receptor. Studies conducted on the crystal structures of SARS-CoV-2 RBD, complexed with ACE2, revealed that the RBDs contain a core structure and a receptor-binding motif. It binds itself to the claw-like structure of the ACE2 host cell receptor [50–54]. Two virus-binding hotspots, hot spot 31 and 353 of the host cell, showed compatibility with specific amino acids, such as glutamine493, asparagine501, leucine455, phenylalanine486 and serine494 of the SARS-CoV-2. These hotspots enhanced viral binding with the host cell’s ACE2 receptor [55]. The super affinity of the spike protein RBDs with the ACE2 receptor mediates a superefficient viral entry into human cells [55–57]. The SARS-CoV-2 virus does not use previous coronavirus receptors, such as APN and DPP4 [58]. The spike protein and ACE2 receptor’s interaction and distribution further determines the tissue tropism and host range [51,59]. Once the S1 subunit binds itself with the host cell receptor, the S2 subunit assists with the fusion mechanism by bringing the viral and cellular membrane in close proximity, such that fusion occurs [60–62]. The fusion process between the viral and host membrane is a crucial step in the viral infection process. Conformational changes in the s protein, and interaction of the fusion peptides, complete the deformation and membrane binding step [30,63]. CoVs enter into the host cell through either the non endosomal pathway, the endosomal pathway or both. Cysteine protease cathepsin help the membrane fusion and endosomal entry of CoV due to low pH in the cellular environment [64–67]. TMPRSS2 and TMPTESS11D help the S1/S2 cleave to activate the S protein for non endosomal virus entry. The IFITM3 blocks the virus entry with a plasma membrane or endosomal membrane through modeling the host membrane’s fluidity [68,69].
Endocytosis, endosomal pathway
Endocytosis is the process of bringing a macromolecule into the human cell, in this case the macromolecule is the SARS-CoV-2 virus. It is a structurally complex virus, encoding five structural proteins [6]. These proteins encounter the cellular membrane at its entry and perform various ranges of functions to assist complete entry. The structural proteins comprise a third of the coding capacity of the gRNA, S, M and HE protein that encounter the cellular membrane upon infection. This combination of multiple interaction factors at multiple membrane sites makes the coronavirus-virus membrane interaction study a challenging process. The endosomal pathway of the spike protein containing virus is considered a pH-sensitive process, in which the endocytosed virions undergo an activation step mediated by acidic endosomal pH, leading to the fusion of viral and endosomal membranes, thus releasing the viral genome into the cell cytoplasm [62]. Studies have shown that the SARS-CoV virus enters the cellular membrane by pH-dependent endocytosis employing endosomal protease cathepsin L [60,70]. The virus cargo fuses with the early endosome, becoming late endosome. The receptors are then disassociated from the viral cargo and recycled back to the cell membrane [71]. In the clatherin-dependent endocytic pathway, after the receptors bind with the spike protein present in the virus, the AP2 complex is present in the cell surface’s cytoplasmic tail. The ACE2 receptor attracts the clatherin proteins present in the cell [72,73]. When clatherin triskelions start to bind with the receptor, it forms a honeycomb like structure, pulling the cell membrane inside to construct a clatherin-coated, spherical, cage-like pit. The pit engulfs the virus molecule and eventually pinches it off from the cell membrane by the action of helical dynamin forming a vesicle, also known as an early endosome, with a pH of 6–6.5. Once the vesicle gets into the cytoplasm, the uncoating starts off with the disassociation of clatherin protein. As the clatherin breaks off, the pH reduces, and the endosomes mature to a late endosome with a pH of 5.5–6 inside of the naked vesicle. The receptors present in the vesicle disassociate and attach back to the cell membrane. Lysosomal fusion occurs causing the late endosome to fuse with the lysosomes present in the cell [74–76].
Uncoating of genomic RNA from nucleocapsid protein
The genomic RNA of CoV is one of the largest, with a size between 26,000 and 32,000 bases [10]. It is now inside the lysosome, packed with nucleocapsid protein and covered by an envelope. The cell’s lysosome immunoresponse tries to digest the endosome by enzymatic reaction, thereby uncoating the gRNA present inside the endosome and releasing the viral genome. The enzymes present in the lysosome fail to stop the gRNA from entering the cell cytoplasm. After this process, the life cycle of the gRNA begins [77,78].
Translation
After the entry and uncoating of the viral endosome in the host cell cytoplasm, the coronavirus replicative cycle begins with the translation of mRNA. In eukaryotic cells, translation happens in four stages: initiation, elongation, termination and recycling [79]. The virus heavily depends on the host cell’s translation system to form its proteins [80]. The sgmRNA here has 5′ methylated cap and a 3′ polyadenylated tail [81]. The capped 5′ terminal makes up for two thirds of the genome that contains the two open reading frames ORF1a and ORF1b. These ORF’s allows the RNA to attach to the host cell’s ribosome for translation of the viral replicase gene (ORF1a and ORF1b) [80]. Encoding the two large ORF’s results in the synthesis of two large polyproteins (pp1a and pp1ab), which takes place by ribosomal frameshifting of ORF1a and ORF1b. These polyproteins are processed further by viral proteases into 16 nonstructural proteins, which will be used in synthesizing viral RNA [80]. Ribosomal frameshifting translates ORF1a between the start and stop codons to yield pp1a, and ORF1b to yield pp1a polyproteins [82,83]. These polyproteins, pp1a and pp1ab, contain the NSP1–11 and NSP11–16, respectively [84].
Replication–transcription complex
The NSP intrudes with the function of the host defenses and sets off the formation of double membrane vesicles (DMVs) as well as convoluted membranes. These virus induced vesicles together form the replication–transcription complexes (RTCs) [3]. These complexes intermediate genomic RNA synthesis. The coronavirus’s RNA possesses a large genome and a fairly unique replication strategy. The polyproteins formed during mRNA translation are then processed into 16 NSPs, which drive the viral genome replication and the transcription of subgenomic mRNA (sgmRNA) [85]. An RdRp is the central catalytic subunit and the RNA synthesis machinery responsible for synthesizing all positive-strand RNA viruses. The RdRp consists of two processes: genome replication and transcription of a collection of sgmRNAs that encode the viral RNA and accessory proteins [86,87]. The intermediate negative strand RNA serves as a template to produce progeny positive-strand RNA [6,88]. In coronavirus transcription, there is a discontinuous step which is unique among the other known RNA viruses, and an ultimate nested set of sgmRNAs with 5′and 3′terminal is produced [89,90]. Studies indicate that the positive-stranded RNA viruses are then encapsidated as RTC complexes and could act as a starting point replication machinery after another round of genome amplification, leading to improved virus infection efficiency [91].
Role of nonstructural proteins in the replication–transcription complex formation
The genomic RNA encodes 28 proteins, comprised of four structural proteins and 16 nonstructural proteins and the rest are a collection of accessory proteins (Figure 4) [92].
Figure 4. . Genomic representation of severe acute respiratory syndrome coronavirus-2 consisting of open reading frames that encode structural and nonstructural proteins, along with their structurally characterized 3D images.
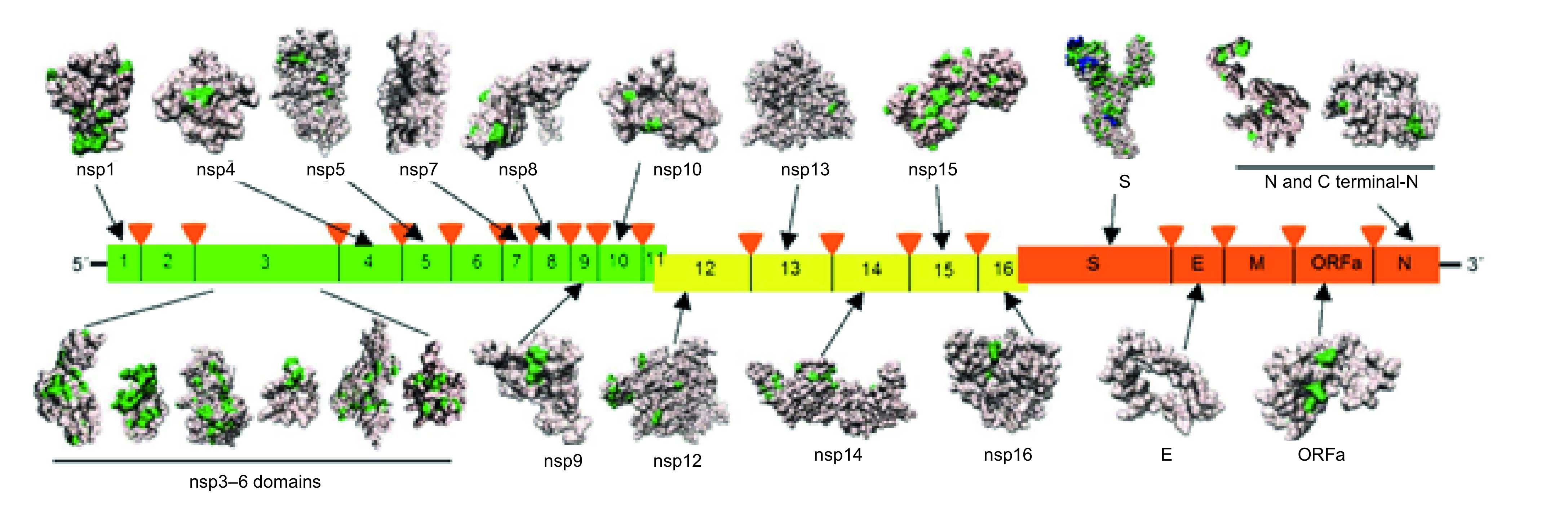
Mutations found when aligning the proteins against their closest homologs such as human SARS-CoV, bat coronavirus BtCoV and bat betacoronavirus BtRf-BetaCoV coronaviruses are highlighted in green and novel protein inserts found in Spike protein are highlighted in blue.
SARS-CoV: Severe acute respiratory syndrome coronavirus.
NSP1
NSP1, a small protein made up of 179 amino acids, is the first to be produced by the virus [93,94]. Although NSP1 is not used in viral replication, it plays an important role in suppressing the host cell’s immune response, thereby allowing the SARS coronavirus to infect and replicate freely [95–98]. Furthermore, NSP1 blocks host cell translation by binding to the ribosome’s 40S subunit, thereby inactivating their translation functions and promoting cellular mRNA degradation [99–101].
NSP2
Not many known NSP2 functions have been recorded across coronaviruses, but NSP2 does interact with two host proteins, called prohibitin1 and 2 (PHB1 & PHB2), which interact with various transcription factors modulating transcriptional activity. This alters the host cell environment and disrupts the intracellular host that signals the virus infection [102].
NSP3
NSP3 is multi domain and is the biggest encoded protein with the SAR-CoV genome. It plays a major role RTC complex formation. It contains two transmembrane regions, TM1 and TM2, along with eight domains: Ubl1 and Ubl2, Glu-rich, PL2pro, nsp3 ectodomain (3Ecto), macrodomain (X), CoV-Y and Y1 domains [103–106]. Along with PLpro, NSP3 is responsible for cleaving NSP1, NSP2 and itself from polyproteins. It interacts with other NSPs and the RNA during RTC complex formation [107] and modifies host proteins, such as RCHY1, to support viral survival and blocks the host’s innate immune response by de-ubiquitination [108].
NSP4
NSP4 is known as a membrane-spanning protein. Along with NSP3 and NSP6, it is important for the proper structure of DMV formation, and is thought to bind the viral RTC complexes to the modified ER [109–111].
NSP5
NSP5 is located between two hydrophobic membranes, spanning regions of NSP4 and NSP6. It is the viral main protease, referred to as 3CLpro or Mpro, which cleaves viral polyproteins pp1a and pp1b at 11 sites and acts as a catalyst in the maturation processing of NSP4–NSP16 [112–114].
NSP6
NSP6 is a multiple-spanning transmembrane protein near the ER. It induces autophagosome formation via an omegasome intermediate. Autophagosomes activation removes host proteins involved with inhibiting replication. The autophagosomes are small in diameter; therefore, limiting its expansion favors coronavirus infection by reducing the ability to deliver the viral components to lysosomes required for degradation [115–117].
NSP7 & NSP8
NSP7 forms a hexadecameric complex, composed of eight identical copies of NSP7 with NSP8, which may act as a processivity clamp for RNA polymerase. In SARS-CoV, a sequence-specific, oligonucleotide-synthesizing activity was found for NSP8. Oligonucleotides synthesized in this manner may be used as primers by the primer-dependent RNA-dependent RNA polymerase RdRp SNP12, thus making NSP8 the primase and SNP12 its main RdRp of SARS-CoV [118,119].
NSP9
NSP9 is a ssDNA/RNA-binding protein. Studies show that NSP9 dimerization favors efficient viral growth and replication. It acts as a key ingredient that intimately engages other proteins in the RTC complex, mediating efficient transcription and replication of the virus [120,121].
NSP10
NSP10 was proposed to play important roles in the synthesis of viral RNA’s and polyprotein processing, interacting with NSP5 protease [122]. Further studies indicated that NSP10 interacts with NSP14 and NSP16 and forms NSP10-NSP14 and NSP10-NSP16 complexes, thereby stimulating their ExoN and RNA Cap 2′-O-methyltransferase activities [123–125]. NSP10 acts as a platform that recruits NSP14 or NSP16 for the RTC complex to enhance NSP14 ExoN activity or NSP16 2′-O-MTase activity [126].
NSP12
NSP12 is a large protein containing 932 amino-acid residues. It is primer-dependent RdRp on both homo- and hetero-polymer templates, similar to close enzymatic collaboration with the coronavirus NSP8’s RNA primase activity. The replication and transcription of the large SARS-CoV RNA genome is mainly catalyzed by NSP12’s RdRp activity [127–129].
NSP13
NSP13, an RNA helicase, is reported to unwind double-stranded RNA (dsRNA) or DNA (dsDNA) up to several hundred base pairs using the energy of nucleotide hydrolysis. Studies show that NSP13’s helicase activity is stimulated by the presence of NSP12, and this protein is involved in forming an RTC complex and improving the viral replication efficiency [130]. NSP13 is also involved in the mediation of RNA 5′-triphosphatase activity, thereby suggesting its involvement in capping viral RNAs [131,132].
NSP14
NSP14 is a bifunctional protein, ExoN activity in the N terminal domain, and N7-MTase activity in the C-terminal domain. The N7-MTase domain adds 5′ cap to viral RNA [121]. NSP14's ExoN domain can hydrolyze ssRNA and dsRNA. It is proposed to be involved in proofreading, repair and recombination of the coronavirus genome [123].
NSP15
NSP15 (NendoU) is a Mn2+-dependent viral endoribonuclease, which undergoes cleavage to form 2′-3′-cyclic phosphates [133]. Although coronavirus NSP15’s endoribonuclease activity seems dispensable for viral RNA synthesis and replication, its EndoU activity infects primary macrophages with EndoU-deficient coronaviruses and mediates the evasion of host cell recognition of viral dsRNA [134–136].
NSP16
NSP16 or 2′O-MTase forms NSP10/NSP16 complex and is responsible for executing 2′-O-MTase activity that is active only in the presence of NSP10. These complexes shield viral RNA from being recognized by MDA5 receptor and ensure formation of a protective cap that prevents recognition by either MDA5 or IFIT proteins. This capping process permits viral infection with reduced host recognition and, consequently, robust viral replication in the absence of the host cell’s innate immune response [137–139].
ER insertion
After the replication and subgenomic RNA synthesis, the N-protein's mRNA undergoes translation by the cytosolic ribosomes to form the viral nucleocapsid protein. The mRNAs of S, E and M proteins are inserted into the ER, where they undergo translation by the ribosomes present in the ER and are then anchored to the ER [81,140,141].
Budding & endoplasmic reticulum–Golgi intermediate compartment trafficking
Budding of coronaviruses happens in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC), which is a structural, as well as a functional, continuation of the ER. The proteins formed during the ER insertion and translation travel in a secretory path that leads into the ERGIC compartment [142,143]. Here the viral genomes are encapsidated by nucleocapsid protein, and then budding happens with the ERGIC membranes containing necessary viral structural proteins to form mature virions [144].
Virus assembly
The M protein directs almost all the protein–protein interactions needed to assemble coronaviruses. The M protein, along with the E protein, helps to form virus-like particles because the M protein alone is not enough for virion formation. These two proteins function together to produce coronavirus envelopes [145]. Furthermore, the virus-like particle formation process is catalyzed by the N protein, which explains the fusion of encapsidated genomes into the ERGIC enhancing viral envelopment [146]. The S protein enters virions at this point, which is not needed for assembly since the M protein is more available than the E protein in the virion. The M protein union gives the force for envelope maturation. Still, it is unclear how this combination takes place. Different explanations were provided. Some work states the E protein induces membrane curvature [147–149]; another investigation suggested the E protein prevents the aggregation of M protein [150]. The E protein catalyzes the viral release by changing the secretory pathways [151]. The M protein, which binds to the nucleocapsid, increases the virion assembly completion. These interactions have been mapped to the C terminus of the endodomain of M with CTD 3 of the N-protein [152].
Exocytosis
The virions are then transported to the cell surface in vesicles and released by exocytosis process. It is unclear whether the virions use the same pathway as the transport of large cargo from the Golgi or if the virus uses another unique pathway [20,81].
Apoptosis
When protein synthesis exceeds the ER’s folding capacity, unfolded proteins accumulate, leading to an ER stress. Studies show that ER stress can also be activated by increased lipids or pro-inflammatory cytokines [153,154]. To maintain homeostasis, cells send signaling pathways that are known as the unfolded protein response[155]. The unfolded protein response signaling activates ER stress transducers, such as PERK, which ATF6 or IRE1. After activation, these transducers transmit the signal from the ER to the cytosol and then the nucleus; consequently, the cell responds by lowering protein synthesis and increasing the ER’s folding capacity. During this process, if homeostasis cannot be maintained, apoptosis occurs for the benefit of the entire organism [141,156].
Inhibition
The life-threatening illness human coronavirus (COVID-19), similar to SARS-CoV, spread from Wuhan to other parts of the world. It is a public health concern according to both the WHO and CDC. The glycoprotein, which interacts with the receptor at its surface region, is the major difference between SARS-CoV and SARS-CoV-2. The different inhibitors, discussed below, will clarify the coronavirus replication mechanisms, which will lead to the discovery of antivirals since there is no approved treatment of this virus.
Furin inhibitors
Furin can activate other proteins, and low pH enhances its capability [157]. The compounds having lower binding energies have more affinity toward the target protein. For example, hydroxystilbamidine’s calculated binding energy is lower and estimated to bind perfectly. Later reports indicate excellent competency toward the substrate’s binding site. A similar conformation corresponds with the inhibitor-developed docking model [158]. The PDB model suggested that hydroxystilbamidine contain two arginine and present imine group in the compounds formed hydrogen bonds with Asp159, Asp259, Asp306 but conformation stabilized between His194, Leu227, Trp254 and Asn295 due to a weak hydrophobic interaction. A similar effect also can be predicted with imatinib, an anticancer agent. Glu236, Gly255 forms two hydrogen bonds with the compound, upheld due to the interaction between Gly294 and Val231, Trp254, and Pro256. Ten hydrogen bonds formed with Suramin and are stabilized by an interaction between the compound and A532, Trp531, Tyr308, Leu227 and His194 [159–161]. These compounds may be useful for treating COVID-19 but require in vitro and in vivo studies to prove the drug’s efficacy.
Clathrin-mediated endocytosis inhibitors
The NAK family member baricitinib may be responsible for reducing the effect of the virus during CME. It has a high affinity with AAK1. It was selected as a possible treatment for SARS-CoV-2 but required clinical testing to prove its ability to work as an inhibitor. The most important pathway for GPCRs is CME. All inhibitors block the CME. Anti-arthritic drugs, such as ruxolitinib and fedratinib, are also suitable because of their lower plasma binding ability, interacting with enzymes and working as drug transporters [162–165].
Cathepsin inhibitors
The cathepsins are well-established enzymes involved in the cytosolic pathway. Inhibitors inbred the immune response and non lysosomal cysteine proteases calpain; they also affect auto lysosomal digestion. The inhibitors (E64D) restrict COVID-19's entrance into the Caco-2 (intestine) and Vero cells (kidney) [166,167] through the endosomal pathway.
TMPRSS2 inhibitors
A serine protease inhibitor was reportedly capable of blocking COVID-19’s entry into Caco-2 and Vero cells. The combined effect of E-64d and camostat mesylate only work to stop entire inhibition. A single inhibitor was unable to move through the non endosomal pathway. Therefore, camostat mesylate can protect lung cells [168].
Endosomal acidification inhibitors
Chloroquine, an endosomal inhibitor, influences the virus cycle process. Viral protein maturation was conventionally reduced by pH modulation. Cytosol, present in dendritic cells, increased with soluble antigen, enhanced cytotoxic CD8+T-cell and decreased cytokine production, which indirectly inhibits the immune response [169].
PLpro inhibitors
Rivavirin, an antiviral drug, is expected to work as an inhibitor. It maintained low binding energy and had a higher affinity with PLpro. According to docking model, it is confirmed that the PLpro inhibitors are fully fitted in active site of enzyme and connected by hydrogen bonds with Gly164, Gln270, Tyr274, Asp303. A weak hydrophobic interaction between the present triazole ring in the compound and Tyr265 stabilized the conformation. Therefore, ribavirin is assumed to be a potent drug against the enzyme because of its stronger hydrogen bonding as well as weak hydrophobic interactions. Another anti bacterial drug, chloramphenicol, may also be useful for treating COVID-19 viruses [170].
3CLpro inhibitors
3CLpro is the main proteinase for the deadly coronavirus. It originated from poly proteins through automatic cleavage as NSP5 and split again to NSP4–NSP16. The active site present within domain I and domain II between residues 8–184. Anti bacterial drugs and anti hypertensive drugs can be useful against its action [171,172]. Montelukast has a lower binding energy with 3CLpro, indicating a possible higher affinity. It is nicely fitted in the active pocket by forming a hydrogen bond between the carbonyl group, Asn14 and hydrophobic interaction involved with Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168 and His172 to stabilize the conformation. A similar effect may be expected with a combination of lopinavir and ritonavir [173].
Helicase inhibitors
Helicase, a multiple catalytic active protein, exhibits helicase domain C-terminus and metal binding N-terminal, having 26 cysteine residues. SARS-NSP13 is mainly responsible for replicating viruses necessary to prevent its action. Saquinavir, an HIV drug, showed affinity toward the target protein and restrained the formation of double-stranded RNA. It also blocked COVID-19 virus replication [174–176].
RNA-dependent RNA polymerase inhibitors
NSP12 is an important protein for the COVID-19 virus for its replication and transcription. The NSP8 protein plays a vital role as a primer for synthesizing NSP12-RdRp RNA. The binding capacity increased between NSP12 and RNA by the NSP7–NSP8 complex, consequently enhancing its enzyme activity [177]. Remdesivir, a novel antiviral nucleotide analog, was successfully used in the USA with a patient infected with the SARS-CoV-2 virus [178,179]. The compound adapted stable conformation in the active site through hydrogen bonds with Asn497, Arg569 and Asp684. The weak hydrophobic interactions between Leu576, Ala685 and Tyr687 and the compound play an important role for its stability [170,180,181].
CYP inhibitors
Cobicistat and darunavir drugs were potent inhibitors against the CYP enzyme, increasing the drug concentration in plasma and minimizing the adverse effect during metabolism [182]. It inhibits CYP function, including all parameters, and enhances cellular penetration and bioavailability [183,184].
Membrane bound RNA synthesis inhibitors
K22, (Z)-N-(3-(4-(4-bromophenyl)-4-hydroxypiperidin-1-yl)-3-oxo-1-phenylprop-1-en-2-yl) benzamide is an effective inhibitor of membrane bound RNA synthesis. Initially, its antiviral activity prevents the DMVs responsible for replication from forming at the early entry stage of coronavirus, decreasing the virus’s infection capability. Therefore, it is recommended for treating SARS-CoV-2 [185,186].
Statistical analysis
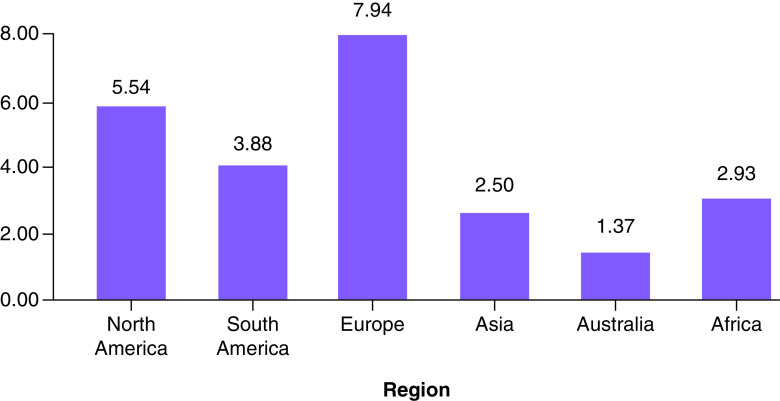
One of this section’s major purposes is to analyze, accumulate and investigate the data. This information is based on the results of extensive research which has been carried out since December 2019. On 27 December 2019, three patients diagnosed with COVID-19 were examined to determine, with no ambiguity due to sex or age factors, confirmed COVID-19 cases, recovered cases, critical cases and fatality rates in order to raise public awareness and take necessary precautions in an effective manner. The goal is to contribute to eradication of this global pandemic. The data analytics from various case studies, literature and governmental organizations will help with forecasting during the crossover and forthcoming period since a solution needs to be identified. The daily death rate was relatively low until 7 February 2020, then increased rapidly between the end of February 2020 and the end of June 2020 (Figure 5 & Supplementary Table 4). A similar rapid growth trend for the total case of COVID-19 patients has been noted between 29 February 2020 and 25 June 2020 (Figure 6 & Supplementary Table 5), which clearly shows this virus’s long incubation period, along with a lack of awareness among people and poor hygienic practices. On the other hand, COVID-19-infected patients were recovering at lower rates than the affected people on daily basis, as shown (Figure 7 & Supplementary Table 6) [187–189]. According to the data obtained from the CCDC report published on 17 February 2020, the confirmed death rate of males is higher than females, as shown in Supplementary Figure 4 & Supplementary Table 7, since men were more mobile and had smoking habits [190]. The summary of the report, taken from 72,314 cases from the CCDC published on 24 February 2020, clearly shows a higher fatality rate of COVID-19-affected patients who were diagnosed with co-morbidities, mostly cardiovascular problems (10.5%), followed by the other illness such as diabetes (7.3%), chronic respiratory disease (6.3%), hypertension (6%) and cancer (5%). The death rate among patients with no co-morbidities was low (0.9%) ( Supplementary Figure 5 & Supplementary Table 8) [190]. From the available data, the growth factor was calculated and found to be 1.11 ( Supplementary Figure 6 & Supplementary Table 4) between January 2020 and June 2020 [191,192]. On the other hand, COVID-19-infected patients were recovering at a lower rate compared with the daily addition of new cases ( Supplementary Figure 7 & Supplementary Table 6). Regional data for the case fatality rate have been collected as represented (Figure 8 & Supplementary Table 9), which undoubtedly concludes greater no for Europe and lesser with Australia [189,190,193,194].
Figure 5. . Death rate of novel coronavirus disease.
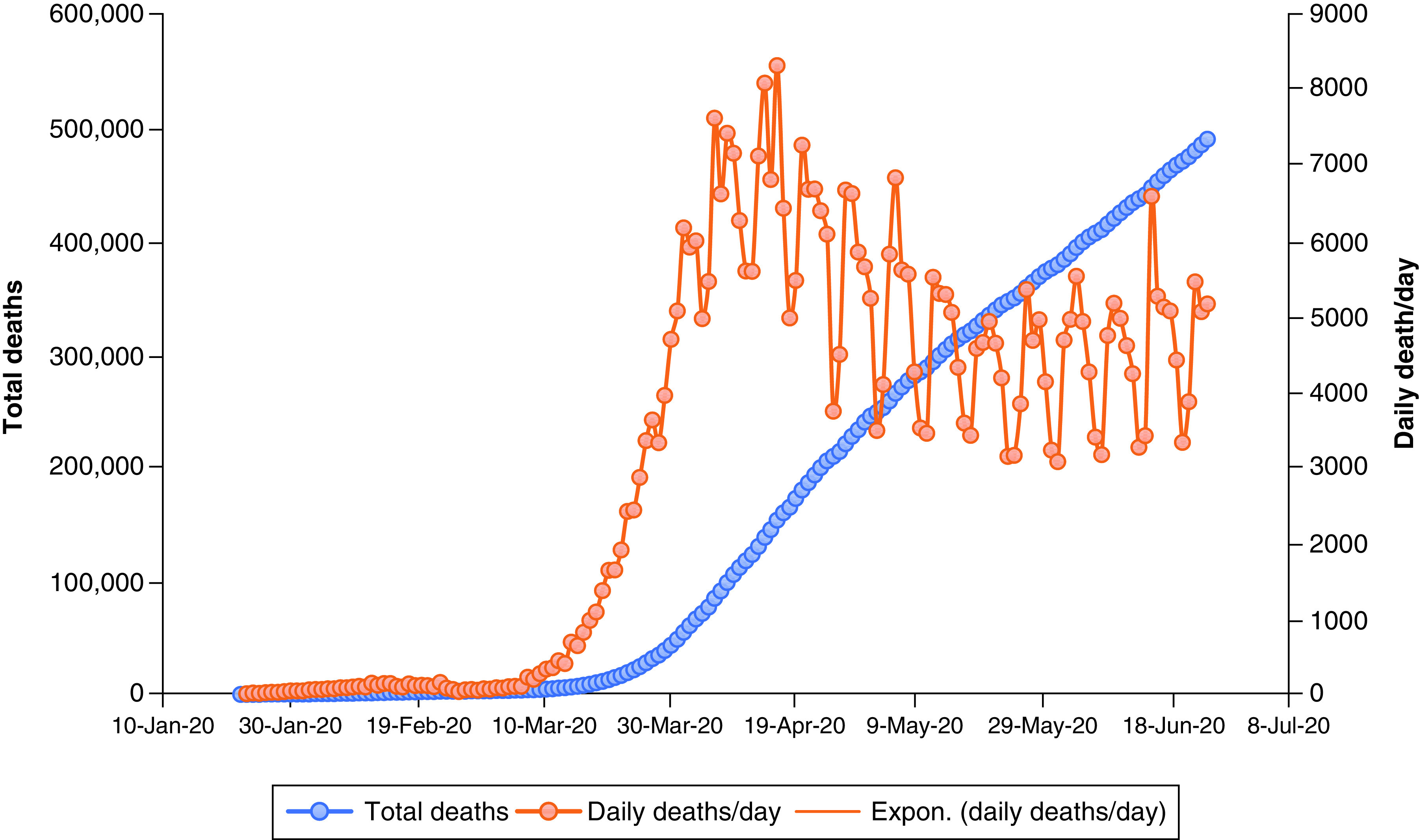
Expon.: Exponential.
Figure 6. . Total cases of novel coronavirus disease.
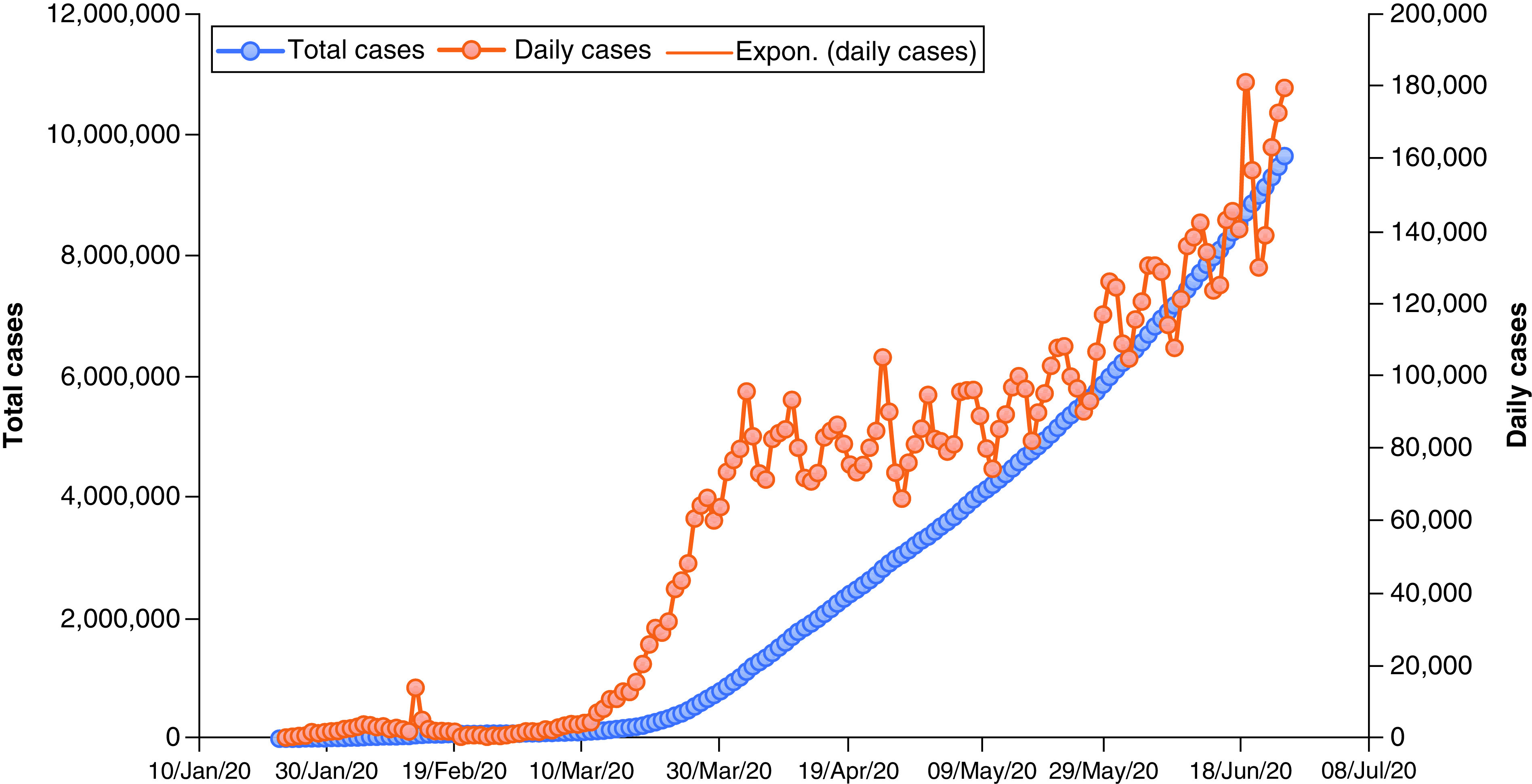
Expon.: Exponential.
Figure 7. . Total recovered cases of novel coronavirus disease.
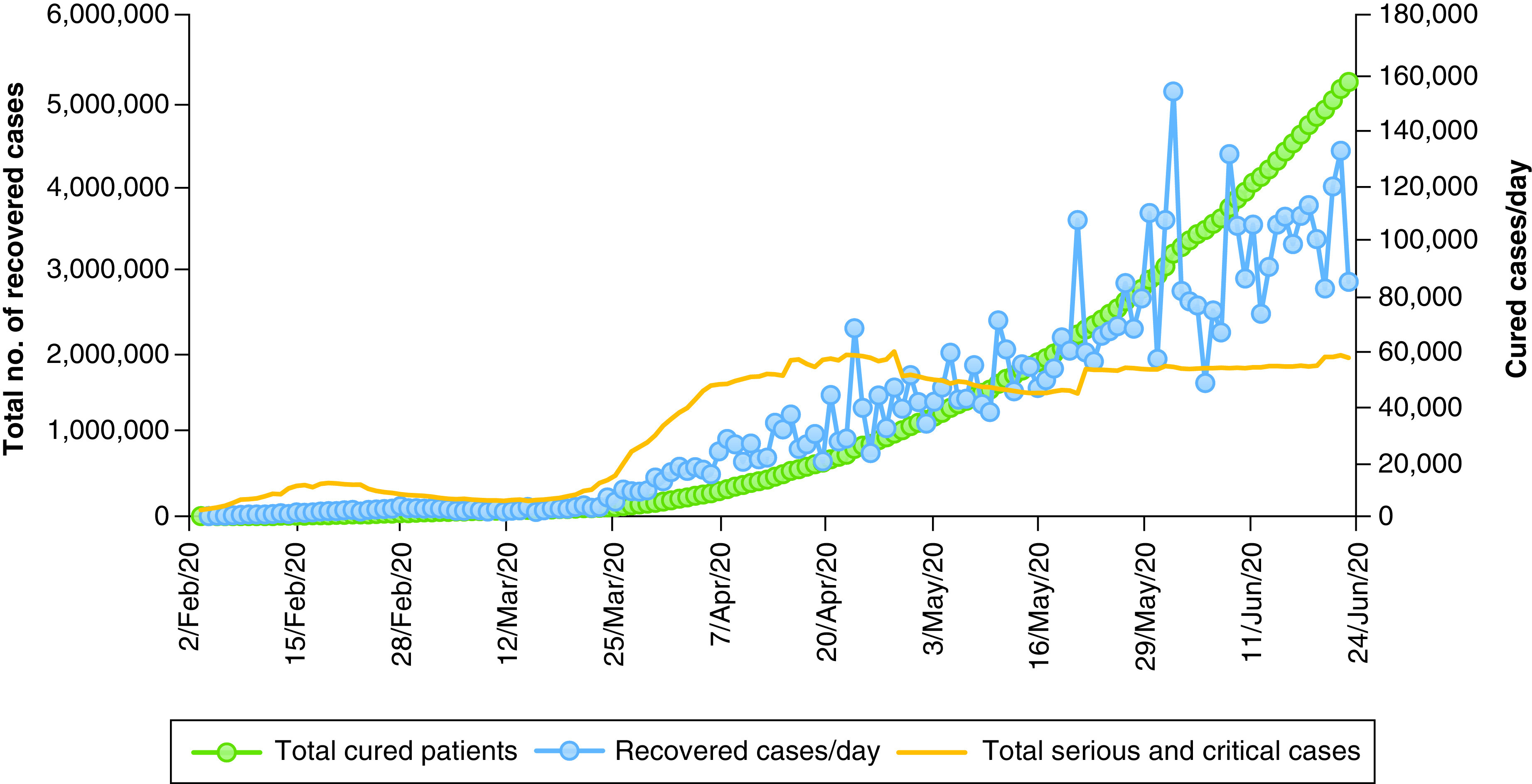
Figure 8. . Case fatality rate of different regions due to coronavirus disease.
Recommendations & advice
On 30 January 2020, the WHO declared COVID-19 a global public health emergency. This new virus is spreading at a disturbing rate throughout the globe. Currently, there is no vaccine; therefore, prevention is the only option, which is possible if every one of us works together. Each country has their own recommendations to reduce COVID-19’s spread. Together we can and will make a difference. Some proposals from different countries were included here. Home management is highly recommended for patients with mild infections, who should be isolated in an outpatient environment [193–195]. Managing these types of patients should focus on preventing transmission to others and monitoring their clinical conditions in case hospitalization is necessary. The WHO recommends that health personnel and others with respiratory symptoms must wear surgical masks to avoid contaminating others. A special mask (FFP2) is required for healthcare personnel and laboratory technicians who handle samples for analysis. At the same time, they should raise awareness by providing COVID-19 information to the general public and rulemaking bodies. Follow preliminary safety measures such as frequently cleansing hands with soap and water for at least 20 s, refrain from touching eyes, nose and mouth before and after washing hands, and use an alcohol-based sanitizer whenever necessary [195]. People should cough or sneeze into a tissue or the elbow crease. Tissues should be thrown into a bin with a lid. Further instructions include avoiding shaking hands, kissing and other activities involving physical contact. Avoid coming in contact with sick individuals maintain a safe distance of at least 2 meters. Stay home as much as possible and if you need to leave home for domestic necessities, observe the recommendations posed by health authorities outside of peak hours, and avoid places where it is impossible to maintain safe distances [195]. Extra care is needed for individuals who are over 65 years of age, have diabetes/cardiovascular diseases/chronic diseases of the respiratory tract/cancer/an immune deficiency due to a condition or therapy because they are more susceptible to this virus. Patients with COVID-19 with acute symptoms should be cared for at home while following the recommended isolation measures. Isolation is essential to those who are positive with SARS-COV-2 because they may infect healthy people (R.O. No. of SARS-COV-2 = 2.5) [196]. Patients exhibiting symptoms must be quarantined for a minimum of 14 days, close contact with other people must be avoided during this period, and a surgical mask must be worn by the patient at all times [195]. Every one of us who is treating a hospitalized person must disinfect our hands and maintain strict hygiene. Outside visitors to the hospital should be prohibited if possible. To minimize any chances of transmission of the coronavirus from a mother to her newborn child, healthcare professionals must consider distancing the new born from the infected mother temporarily [195]. Healthcare providers must comply with all applicable laws, policies and rules. Regular exercise and a balanced diet will help boost immunity to the virus [190].
Bioinformatics a study uniting important aspects of biology, information technology, mathematics and statistics is foremost in analyzing the blueprint of the novel coronavirus and provides an innovative approach in visualizing host-pathogen data as well as in the manufacture of antiviral drugs that are required to boost the fight against the deadly pandemic. As we know, the protease inhibitor needed to inhibit this virus is obtained from terpenoids. The terpenoids successfully inhibit virus protease compounds, effectively restricting key amino acids. These compounds can be introduced as effective antidrug due to fewer side effects. It is one of the best drugs to treat new coronavirus infections. Remdesivir is a novel nucleotide which has strong activity against severe acute respiratory syndrome [197,198]. Its efficiency on COVID-19 is being studied [199,200]. Chloroquine and hydroxychloroquine blocked SARS-CoV-2 in vitro [179] but remain under investigation [201–205]. The resulting data conclude that chloroquine could decrease the number days spent in the hospital and the evolution of coronavirus 19 pneumonia. As per the recommendation’s, mild-to-severe forms of COVID-19, pneumonia can be treated with 500 mg of chloroquine twice a day. The mechanisms of both chloroquine and hydroxychloroquine are the same, but hydroxychloroquine may be a better option for treating SARS-CoV-2; the required amount is under clinical trial. IL-6 inhibitor tocilizumab provides better results for patients with severe COVID-19, but its properties are under clinical trial [206]. Combined protease inhibitor lopinavir-ritonavir was used for HIV infection [207] and has some activity against MERS-CoV in animal studies [208] and is under clinical trial for COVID-19 [209,210]. As per the WHO and CDC recommendations, glucocorticoids cannot be used in patients with COVID-19 pneumonia unless there are other indications [193,195], since it is associated with a high mortality rate [211]. The European Medicines Agency (EMA) and the WHO do not suggest using non steroidal anti-inflammatory drugs [212,213], since there are a number of evidence of a negative impact on the disease outcome [214]. Men are more affected by novel coronavirus pneumonia than women because they have greater quantities of hemoglobin than women. Middle and old age diabetics are more affected by this virus because deoxyhemoglobin, a combination of glucose and hemoglobin, is higher for diabetic patients. The clinical effect of chloroquine phosphate on novel coronavirus pneumonia shows that the virus is closely related to abnormal hemoglobin metabolism. It is concluded that open reading frames (orf1ab, orf3a and orf10) attack the heme of hemoglobin’s beta chain. Hemoglobin, with or without oxygen (oxyhemoglobin or deoxyhemoglobin), were both attacked. During this process, the positions of orf1ab, orf3a and orf10 were different, indicating that greater quantities of hemoglobin increase the risk of disease [215]. Behavior of contacting residues present in ACE2, of the new deadly virus are, to a greater extent, favorable to nonhuman primates as well cats, ferrets and pigs. Earlier, RBD of SARS-CoV was adapted efficiently with the palm civet in their ACE2 [53]. Therefore, it is particularly important to screen bats and said animals in Wuhan for COVID-19. The WHO should continue to engage with international organizations, its other groups, and industries to implement travel-related health measures that are commensurate with public health risks and other issues. Epidemic SARS-CoV-2 (COVID-19) crosses all international boundaries [195]. The initial conformation of this virus in the human body will help us quarantine those affected and reduce further spread. At the same time, a drug or vaccine is the only option that will eradicate it completely. As we know, the clinical trial process for a drug or vaccine takes time; exactly how long will be recorded in history.
Future perspective
With the seemingly never-ending trend of the continually evolving coronavirus situation, it occurs to us that its treatment, as well as containment, will be a long weary battle. However, a ray of hope is seen in the enormous efforts put by researchers in the study of inhibitors and vaccination. A variety of inhibitors could soon be available to help us contain the situation to a great extent, and with the advent of an effective vaccine, the current situation woul slowly return to normal. However, the post-COVID-19 world will be a seriously changed world of new and unique opportunities. What looks obvious is that the transformation of physical spaces that could soon wind up being internationally connected internet spaces.
However, the treatment and containment of this pandemic over the next 5–10 years will largely depend upon clearing all uncertainties regarding the virus–host cell interaction and life cycle mechanism of the virus. What is imminent is the possibility of the existing virus configuration to mutate into several forms posing a constant threat for human existence. Vigorous research and investigation of SARS-CoV-2 virus about its possible polymorphisms, mutations and angle of evolution is the matter of immediate concern. Moreover, several alternative pathways need to be established to inhibit their proliferation.
Executive summary.
A thorough and comprehensive introduction gives a jumpstart to any researcher who wishes to indulge in knowing the current situation as well as the background of this ongoing pandemic.
Early and quick detection of the infected person will help control the spread of the virus.
A neat summary of findings made in the understanding of life cycle mechanism, based on virus and host cell interaction of the unique RNA replication process of SAR-CoV-2 virus.
The stepwise inhibition mechanism will help the researchers to develop effective inhibitors that can break the life cycle at its different stages.
The statistical records indicate that the pandemic has made its mark in every region of the world, the way ahead would be to network between all health centers of the world effectively and to create a global movement in finding a permanent solution against this deadly virus.
Until an effective and safe vaccine is developed, the rate of infection should be controlled as much as possible by complying to the recommendations furnished by health organizations.
More investment in the research of virus–host interaction and life cycle mechanism should be of topmost priority in the future to effectively curtail the spread of the virus.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/fvl-2020-0124
Author contributions
SKM Haque formulated the study and contributed with abstract, background, symptoms and behaviors, mechanism of viruses, inhibition, recommendation, advice, data interpretation, future perspective, executive summary and references. O Ashwaq participated with abstract, background, mechanism of viruses, future perspective, executive summary and references. A Sarief contributed with symptoms and behaviors, mechanism of viruses, recommendation, advice, future perspective and executive summary. AKAJ Mohamed participated with collection of data and created graphs, future perspective and references.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Zhu N, Zhang D, Wang W. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382(8), 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Reports the first novel CoV (2019-nCoV) infected hospitalized patient and discussed its cytopathic effects with morphology in the host cell.
- 2.Gorbalenya AE, Baker SC, Baric RS. et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5(4), 536–544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Categorical classification of 2019-nCoV based on virus taxonomy.
- 3.Cong Y, Verlhac P, Reggiori F. The interaction between nidovirales and autophagy components. Viruses 9(7), 182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S, Wong G, Shi W. et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24(6), 490–502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17(3), 181–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters PS. The molecular biology of coronaviruses. Adv. Virus Res. 65(06), 193–292 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortola E, Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 576(1–2), 174–178 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Zheng X, Gai W. et al. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget 8(8), 12686–12694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol. J. 16(1), 1–22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the importance of envelope protein and its unique structure in the virus life cycle.
- 10.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117(1), 17–37 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahy JV, Dickey BF. Airway mucus function and dysfunction. N. Engl. J. Med. 363(23), 2233–2247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y-R, Cao Q-D, Hong Z-S. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 7(1), 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrbod P, Ande SR, Alizadeh J. et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 10(1), 376–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moldoveanu B, Otmishi P, Jani P. et al. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2, 1–11 (2009). [PMC free article] [PubMed] [Google Scholar]
- 15.European Centre for Disease Prevention and Control. ‘Risk assessment: outbreak of acute respiratory syndrome associated with a novel coronavirus, China; First cases imported in the EU/EEA; second update’. www.ecdc.europa.eu/en/publications-data/risk-assessment-outbreak-acute-respiratory-syndrome-associated-novel-0
- 16.Deng S-Q, Peng H-J. Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J. Clin. Med. 9(2), 575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Analysis of symptoms and possible treatments based on public health responses of target population.
- 17.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 215, 108427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unhale SS, Ansar QB, Sanap S, Thakhre S, Wadatkar S. A review on corona virus (COVID-19). World J. Pharm. Life Sci. 6(4), 109–115 (2020). [Google Scholar]
- 19.Xie P, Ma W, Tang H, Liu D. Severe COVID-19: a review of recent progress with a look toward the future. Front. Public Heal. 8, 189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7(6), 439–450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Yang X Lou, Wang XG. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798), 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Zeng LP, Yang X Lou. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13(11), 1–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Yang X, Wang N. et al. The divergence between SARS-CoV-2 and RaTG13 might be overestimated due to the extensive RNA modification. Future Virol. (2020) (Epub ahead of print). [Google Scholar]
- 24.Tang X, Wu C, Li X. et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci. Rev. 7(6), 1012–1023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Zai J, Zhao Q. et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 92(6), 602–611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying SY. Complementary DNA libraries: an overview. Appl. Biochem. Biotechnol. – Part B Mol. Biotechnol. 27(3), 245–252 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Guo X, Geng P, Wang Q, Cao B, Liu B. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J. Microbiol. Biotechnol. 24(10), 1145–1454 (2014). [DOI] [PubMed] [Google Scholar]
- 28.‘GISAID – Enabling rapid and open access to epidemic and pandemic virus data’. www.gisaid.org/
- 29.‘Nextstrain – Real-time tracking of pathogen evolution’. https://nextstrain.org/ [DOI] [PMC free article] [PubMed]
- 30.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen KY. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 15(5), 327–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem 21(5), 730–738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). StatPearls 1–12 (2020). www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed] [Google Scholar]
- 33.Chan JFW, Kok KH, Zhu Z. et al. Genomic characterization of the 2019 novel human–pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9(1), 221–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read JM, Bridgen JRRE, Cummings DATA, Ho A, Jewell CP. Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. medRxiv 53(9), 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C, Wang Y, Li X. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223), 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen N, Zhou M, Dong X. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223), 507–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Hu B, Hu C. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 323(11), 1061–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Euro Surveill. 25(5), 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung GM, Hedley AJ, Ho L-M. et al. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann. Intern. Med. 141(9), 662–673 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Centers for Disease Control and Prevention. ‘MERS symptoms & complications’. www.cdc.gov/coronavirus/mers/about/symptoms.html
- 41.Jilani TN, Jamil RT, Siddiqui H Abdul. H1N1 Influenza (Swine Flu). U.S. National Library of Medicine, FL, USA: (2019). www.ncbi.nlm.nih.gov/books/NBK513241/ [Google Scholar]
- 42.Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 105, 93–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4(6), 1011–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl Acad. Sci. USA 106(14), 5871–5876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchdoerfer RN, Cottrell CA, Wang N. et al. Pre-fusion structure of a human coronavirus spike protein. Nature 531(7592), 118–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkard C, Verheije MH, Wicht O. et al. Coronavirus cell entry occurs through the Endo-/Lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 10(11), e1004502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walls AC, Tortorici MA, Bosch BJ. et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531(7592), 114–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 202, 120–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2), 281–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Moore MJ, Vasllieva N. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 89(4), 1954–1964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, Greenough TC, Moore MJ. et al. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 78(20), 11429–11433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Zhang C, Sui J. et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24(8), 1634–1643 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCray PB, Pewe L, Wohlford-Lenane C. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81(2), 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 94(7), 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Investigation reveals the structural similarities of spike protein of 2019-nCoV with SARS-CoV confirming the use of ACE2 receptors of the host cell for virus entry.
- 56.Moore MJ, Dorfman T, Li W. et al. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78(19), 10628–10635 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu XX, Hao P, Song XJ. et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 280(33), 29588–29595 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raj VS, Mou H, Smits SL. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495(7440), 251–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3(1), 237–261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets 21(2), 131–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Alsaadi EA J, Jones IM. Membrane binding proteins of coronaviruses. Future Virol. 14(4), 275–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes an extensive mechanism about the binding of virus with the host membrane.
- 63.Xia S, Liu M, Wang C. et al. Inhibition of SARS-CoV-2 infection (previously 2019-nCoV) by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30(4), 343–355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosch BJ, Bartelink W, Rottier PJM. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class i fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82(17), 8887–8890 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human middle east respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 8(10), 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl Acad. Sci. USA 102(33), 11876–11881 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mille JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl Acad. Sci. USA 111(42), 15214–15219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li K, Markosyan RM, Zheng YM. et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9(1), e1003124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim Y, Ng Y, Tam J, Liu D. Human coronaviruses: a review of virus–host interactions. Diseases 4(4), 26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang IC, Bosch BJ, Li F. et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281(6), 3198–3203 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H, Yang P, Liu K. et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18(2), 290–301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pearse BM, Smith CJ, Owen DJ. Clathrin coat construction in endocytosis. Curr. Opin. Struct. Biol. 10(2), 220–228 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Sorkin A. Cargo recognition during clathrin-mediated endocytosis: a team effort. Curr. Opin. Cell Biol. 16(4), 392–399 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Marsh M, Helenius A. Virus entry: open sesame. Cell 124(4), 729–740 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pelkmans L, Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 15(4), 414–422 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Inoue Y, Tanaka N, Tanaka Y. et al. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81(16), 8722–8729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tok TT, Tatar G. Structures and functions of coronavirus proteins: molecular modeling of viral nucleoprotein. Int. J. Virol. Infect. Dis. 2(1), 001–007 (2017). [Google Scholar]
- 78.Greber UF, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 2(2), 52–56 (1994). [DOI] [PubMed] [Google Scholar]
- 79.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73(1), 657–704 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Nakagawa K, Lokugamage KG, Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. : Advances in Virus Research Academic Press Inc., MA, USA: Volume 96, Chap. 5 165–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. : Coronaviruses: Methods and Protocols. Maier HJ, Bickerton E, Britton P (Eds). Springer, NY, USA, Volume 1282, Chap. 1, 1–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332(2), 498–510 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57(4), 537–547 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Araki K, Gangappa S, Dillehay DL, Rouse BT, Larsen CP, Ahmed R. Pathogenic virus-specific T cells cause disease during treatment with the calcineurin inhibitor FK506: implications for transplantation. J. Exp. Med. 207(11), 2355–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.King AMQ, Lefkowitz E, Adams MJ, Carstens EB. Virus taxonomy. : Virus Taxonomy (1st Edition). Elsevier academic press, MA, USA, 193–210 (2012). [Google Scholar]
- 86.Enjuanes L, Almazán F, Sola I, Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60(1), 211–230 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 48, 1–100 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sola I, Mateos-Gomez PA, Almazan F, Zuñiga S, Enjuanes L. RNA–RNA and RNA–protein interactions in coronavirus replication and transcription. RNA Biol. 8(2), 237–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mahy BWJ, van Regenmortel MHV. Assembly of viruses: enveloped particles. Encyclopedia of Virology. (3rd Edition). Brian WJ, Mahy Marc HV (). Elsevier academic press, MA, USA: (2008). [Google Scholar]
- 90.Sawicki SG, Sawicki DL. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440, 215–219 (1998). [DOI] [PubMed] [Google Scholar]
- 91.Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2(1), 265–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Srinivasan S, Cui H, Gao Z. et al. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 12(4), 360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes the structural and functional behaviors of proteins present in the genomic RNA of SARS-CoV-2 and comparison with other coronaviruses.
- 93.Almeida MS, Johnson MA, Herrmann T, Geralt M, Wuthrich K. Novel-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 81(7), 3151–3161 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bartlam M, Xu Y, Rao Z. Structural proteomics of the SARS coronavirus: a model response to emerging infectious diseases. J. Struct. Funct. Genomics 8(2–3), 85–97 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Züst R, Cervantes-Barragán L, Kuri T. et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3(8), 1062–1072 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81(21), 11620–11633 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Narayanan K, Huang C, Lokugamage K. et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of Type I interferon, in infected cells. J. Virol. 82(9), 4471–4479 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pfefferle S, Schöpf J, Kögl M. et al. The SARS-Coronavirus-host interactome: identification of cyclophilins as target for pan-Coronavirus inhibitors. PLoS Pathog. 7(10), e1002331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16(11), 1134–1140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 7(12), e1002433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 86(24), 13598–13608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cornillez-Ty CT, Liao L, Yates JR, Kuhn P, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 83(19), 10314–10318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neuman BW. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 135, 97–107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Neuman BW, Joseph JS, Saikatendu KS. et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82(11), 5279–5294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 149(January), 58–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harcourt BH, Jukneliene D, Kanjanahaluethai A. et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78(24), 13600–13612 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ziebuhr J, Thiel V, Gorbalenya AE. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276(35), 33220–33232 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Serrano P, Johnson MA, Chatterjee A. et al. Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J. Virol. 83(24), 12998–13008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Manolaridis I, Wojdyla JA, Panjikar S. et al. Structure of the C-terminal domain of nsp4 from feline coronavirus. Acta Crystallogr. Sect. D Biol. Crystallogr. 65(8), 839–846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clementz MA, Kanjanahaluethai A, O'Brien TE, Baker SC. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375(1), 118–129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gadlage MJ, Sparks JS, Beachboard DC. et al. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 84(1), 280–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) Structure: basis for design of anti-SARS drugs. Science 300(5626), 1763–1767 (2003). [DOI] [PubMed] [Google Scholar]
- 113.Yang H, Yang M, Ding Y. et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl Acad. Sci. USA 100(23), 13190–13195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu Y, Lu X, Denison MR. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69(6), 3554–3559 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cottam EM, Maier HJ, Manifava M. et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7(11), 1335–1347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10(8), 1426–1441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oostra M, Hagemeijer MC, van Gent M. et al. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82(24), 12392–12405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Imbert I, Guillemot JC, Bourhis JM. et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25(20), 4933–4942 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhai Y, Sun F, Li X. et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12(11), 980–986 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 83(7), 3007–3018 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y, Cai H, Pan J. et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. USA 106(9), 3484–3489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Donaldson EF, Graham RL, Sims AC, Denison MR, Baric RS. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 81(13), 7086–7098 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl Acad. Sci. USA 109(24), 9372–9377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Decroly E, Debarnot C, Ferron F. et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7(5), e1002059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen Y, Su C, Ke M. et al. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 7(10), e1002294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bouvet M, Lugari A, Posthuma CC. et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 289(37), 25783–25796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.te Velthuis AJW, Arnold JJ, Cameron CE, van den Worm SHE, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38(1), 203–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.te Velthuis AJW, van den Worml SHE, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 6(11), 1–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu X, Liu Y, Weiss S, Arnold E, Sarafianos SG, Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 31(24), 7117–7130 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Adedeji AO, Marchand B, Te Velthuis AJW. et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE 7(5), e36521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78(11), 5619–5632 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ivanov KA, Ziebuhr J. Human coronavirus 229e nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-Triphosphatase activities. J. Virol. 78(14), 7833–7838 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ricagno S, Egloff MP, Ulferts R. et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl Acad. Sci. USA 103(32), 11892–11897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deng X, Baker SC. An “Old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology 517, 157–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng X, Hackbart M, Mettelman RC. et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl Acad. Sci. USA 114(21), E4251–E4260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kindler E, Gil-Cruz C, Spanier J. et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13(2), 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Menachery VD, Debbink K, Baric RS. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 194, 191–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Decroly E, Imbert I, Coutard B. et al. Coronavirus nonstructural protein 16 is a Cap-0 binding enzyme possessing (Nucleoside-2′O)-methyltransferase activity. J. Virol. 82(16), 8071–8084 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Züst R, Cervantes-Barragan L, Habjan M. et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12(2), 137–143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ma HC, Fang CP, Hsieh YC, Chen SC, Li HC, Lo SY. Expression and membrane integration of SARS-CoV M protein. J. Biomed. Sci. 15(3), 301–310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 5(JUN), 296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Krijnse-Locker J, Ericsson M, Rottier PJM, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124(1–2), 55–70 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33(2), 281–293 (1984). [PubMed] [Google Scholar]
- 144.de Haan CAM, Rottier PJM. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64(05), 165–230 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bos ECW, Luytjes W, van der Meulen H, Koerten HK, Spaan WJM. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218(1), 52–60 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Siu YL, Teoh KT, Lo J. et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 82(22), 11318–11330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Raamsman MJB, Locker JK, de Hooge A. et al. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74(5), 2333–2342 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Corse E, Machamer CE. Infectious bronchitis virus e protein is targeted to the golgi complex and directs release of virus-like particles. J. Virol. 74(9), 4319–4326 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fischer F, Stegen CF, Masters PS, Samsonoff WA. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72(10), 7885–7894 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Boscarino JA, Logan HL, Lacny JJ, Gallagher TM. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 82(6), 2989–2999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye Y, Hogue BG. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 81(7), 3597–3607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hurst KR, Kuo L, Koetzner CA, Ye R, Hsue B, Masters PS. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79(21), 13285–13297 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kharroubi I, Ladrière L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Free fatty acids and cytokines induce pancreatic β-cell apoptosis by different mechanisms: role of nuclear factor-κB and endoplasmic reticulum stress. Endocrinology 145(11), 5087–5096 (2004). [DOI] [PubMed] [Google Scholar]
- 154.Pineau L, Colas J, Dupont S. et al. Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic 10(6), 673–690 (2009). [DOI] [PubMed] [Google Scholar]
- 155.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8(7), 519–529 (2007). [DOI] [PubMed] [Google Scholar]
- 156.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Feliciangeli SF, Thomas L, Scott GK. et al. Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J. Biol. Chem. 281(23), 16108–16116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dahms SO, Arciniega M, Steinmetzer T, Huber R, Than ME. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Natl Acad. Sci. USA 113(40), 11196–11201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dahms SO, Hardes K, Steinmetzer T, Than ME. X-ray structures of the proprotein convertase furin bound with substrate analogue inhibitors reveal substrate specificity determinants beyond the S4 pocket. Biochemistry 57(6), 925–934 (2018). [DOI] [PubMed] [Google Scholar]
- 160.Dahms SO, Jiao GS, Than ME. Structural studies revealed active site distortions of human furin by a small molecule inhibitor. ACS Chem. Biol. 12(5), 1211–1216 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Li H, Wu C, Yang Y. et al. Furin, a potential therapeutic target for COVID-19. ChinaXiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu TY, Frieman M, Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 15, 247–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bekerman E, Neveu G, Shulla A. et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 127(4), 1338–1352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pu SY, Xiao F, Schor S. et al. Feasibility and biological rationale of repurposing sunitinib and erlotinib for dengue treatment. Antiviral Res. 155, 67–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stebbing J, Phelan A, Griffin I. et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 20(4), 400–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang Y, Islam S, Wang J, Li Y, Chen X. Traditional Chinese Medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int. J. Biol. Sci. 2020(10), 1708–1717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bossowska-Nowicka M, Mielcarska MB, Romaniewicz M. et al. Ectromelia virus suppresses expression of cathepsins and cystatins in conventional dendritic cells to efficiently execute the replication process. BMC Microbiol. 19(1), 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hoffmann M, Kleine-Weber H, Schroeder S. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2), 271–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Devaux CA, Rolain J-M, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents 55(5), 105938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu C, Liu Y, Yang Y. et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 10(5), 766–788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yang H, Xie W, Xue X. et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3(10), 1742–1752 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 59(14), 6595–6628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen YW, Yiu C-PB, Wong K-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 9, 129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu C, Zhou Q, Li Y. et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 6(3), 315–331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shum KT, Tanner JA. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem 9(18), 3037–3045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jang KJ, Lee NR, Yeo WS, Jeong YJ, Kim DE. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 366(3), 738–744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Subissi L, Imbert I, Ferron F. et al. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 101(1), 122–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295(15), 4773–4779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Holshue ML, DeBolt C, Lindquist S. et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382(10), 929–936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reports an antiviral drug, remedesivir which was successfully used on the first patient in USA infected with 2019-nCoV.
- 180.Sheahan TP, Sims AC, Leist SR. et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 11, 222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang M, Cao R, Zhang L. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30(3), 269–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tseng A, Hughes CA, Wu J, Seet J, Phillips EJ. Cobicistat versus ritonavir: similar pharmacokinetic enhancers but some important differences. Ann. Pharmacother. 51(11), 1008–1022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Antimicrob. Chemother. 53(1), 4–9 (2004). [DOI] [PubMed] [Google Scholar]
- 184.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 38(4), 379–381 (2020). [DOI] [PubMed] [Google Scholar]
- 185.Lundin A, Dijkman R, Bergström T. et al. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog. 10(5), e1004166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Khan S, Siddique R, Shereen MA. et al. The emergence of a novel coronavirus (SARS-CoV-2), their biology and therapeutic options. J. Clin. Microbiol. 58(5), e00187–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.National Health Commission (NHC) of the People's Republic of China. www.nhc.gov.cn/
- 188.Health and family planning Commission of Hubei Province. www.grid.ac/institutes/grid.468046.8
- 189.Worldometer: coronavirus cases. www.worldometers.info/coronavirus/
- 190.Epidemiology working group for NCIP epidemic response. C center for disease control and prevention. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua liu xing bing xue za zhi 41(2), 145–151 (2020). [DOI] [PubMed] [Google Scholar]
- 191.Khafaie MA, Rahim F. Osong Public Health and Research Perspectives cross-country comparison of case fatality rates of COVID-19/SARS-COV-2. Osong Public Health Res. Perspect. 11(2), 74–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kucharski AJ, Russell TW, Diamond C. et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis. 20(5), 553–558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Centers for Disease Control and Prevention. Interim clinical guidance for management of patients with confirmed coronavirus disease (COVID-19). www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html
- 194.Wu X, Cai Y, Huang X. et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with Pneumonia, China. Emerg. Infect. Dis. 26(6), 1324–1326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.World Health Organization. Novel coronavirus (2019-nCoV) technical guidance: patient management. www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/patient-management
- 196.Zhang J, Litvinova M, Liang Y. et al. Age profile of susceptibility, mixing, and social distancing shape the dynamics of the novel coronavirus disease 2019 outbreak in China. medRxiv (2020). www.medrxiv.org/content/10.1101/2020.03.19.20039107v1 [Google Scholar]
- 197.Gilead Sciences Statement on the Company's Ongoing Response to the 2019 Novel Coronavirus (2019-nCoV). www.gilead.com/news-and-press/company-statements/gilead-sciences-statement-on-the-company-ongoing-response-to-the-2019-new-coronavirus
- 198.Sheahan TP, Sims AC, Graham RL. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9(396), eaal3653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.European Medicines Agency. EMA gives advice on the use of non-steroidal anti-inflammatories for COVID-19. www.ema.europa.eu/en/news/ema-gives-advice-use-non-steroidal-anti-inflammatories-covid-19
- 200.Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Agents 55(5), 105955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Colson P, Rolain JM, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents 55(3), 105923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yao X, Ye F, Zhang M. et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome main point: hydroxychloroquine was found to be more potent than chloroquine at inhibiting SARS-CoV-2 in vitro. Clin. Infect. Dis. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 14(1), 72–73 (2020). [DOI] [PubMed] [Google Scholar]
- 204.Colson P, Rolain J-M, Lagier J-C, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 55(4), 105932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care 57(June), 279–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Centers for Disease Control and Prevention. ‘Therapeutic options for patients with COVID-19. Centers for disease control and prevention (CDC)’. www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html
- 207.Reuters. China approves use of Roche drug in battle against coronavirus complications. www.reuters.com/article/us-health-coronavirus-china-roche-hldg/china-approves-use-of-roche-arthritis-drug-for-coronavirus-patients-idUSKBN20R0LF
- 208.Groneberg DA, Poutanen SM, Low DE, Lode H, Welte T, Zabel P. Treatment and vaccines for severe acute respiratory syndrome. Lancet Infect. Dis. 5(3), 147–155 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chan JFW, Yao Y, Yeung ML. et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERSCoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 212(12), 1904–1913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kenneth McIntosh M. Coronavirus disease 2019 (COVID-19). (2020) www.uptodate.com/contents/coronavirus-disease-2019-covid-19?topicRef=8298&source=see_link
- 211.Centers for Disease Control and Prevention. Interim guidance for persons who may have 2019 Novel Coronavirus (2019-nCoV) to prevent spread in homes and residential communities. www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-prevent-spread.html#First_heading
- 212.Day M. COVID-19: ibuprofen should not be used for managing symptoms, say doctors and scientists. BMJ 368(March), m1086 (2020). [DOI] [PubMed] [Google Scholar]
- 213.Sciencealert. ‘Updated: WHO now doesn't recommend avoiding ibuprofen for COVID-19 symptoms’. www.sciencealert.com/who-recommends-to-avoid-taking-ibuprofen-for-covid-19-symptoms
- 214.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 395(10223), 473–475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu W, Li H. COVID-19: attacks the 1-beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism 1 introduction. ChemRxiv 11938173 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.