Abstract
The algorithms of electroencephalography (EEG) decoding are mainly based on machine learning in current research. One of the main assumptions of machine learning is that training and test data belong to the same feature space and are subject to the same probability distribution. However, this may be violated in EEG processing. Variations across sessions/subjects result in a deviation of the feature distribution of EEG signals in the same task, which reduces the accuracy of the decoding model for mental tasks. Recently, transfer learning (TL) has shown great potential in processing EEG signals across sessions/subjects. In this work, we reviewed 80 related published studies from 2010 to 2020 about TL application for EEG decoding. Herein, we report what kind of TL methods have been used (e.g., instance knowledge, feature representation knowledge, and model parameter knowledge), describe which types of EEG paradigms have been analyzed, and summarize the datasets that have been used to evaluate performance. Moreover, we discuss the state-of-the-art and future development of TL for EEG decoding. The results show that TL can significantly improve the performance of decoding models across subjects/sessions and can reduce the calibration time of brain–computer interface (BCI) systems. This review summarizes the current practical suggestions and performance outcomes in the hope that it will provide guidance and help for EEG research in the future.
Keywords: EEG, transfer learning, review, decoding, classification
1. Introduction
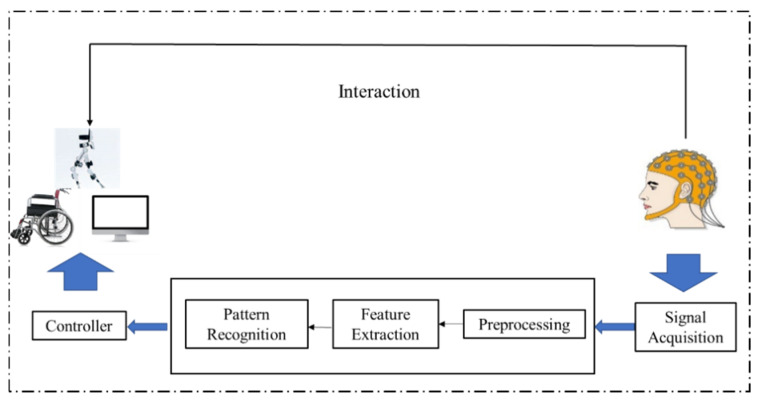
A brain–computer interface (BCI) is a communication method between a user and a computer that does not rely on the normal neural pathways of the brain and muscles [1]. According to the methods of electroencephalography (EEG) signal collection, BCIs can be divided into three types, namely, non-invasive, invasive, and partially-invasive BCIs. Among them, non-invasive BCIs realize the control of external equipment via EEG and by transforming EEG recordings into a command, which have been widely used due to their convenient operation. Figure 1 shows a typical non-invasive BCI system framework based on EEG, which usually consists of three parts: EEG signal acquisition, signal decoding, and external device control. During this process, signal decoding is the key step to ensure the operation of the whole system.
Figure 1.
Framework of an electroencephalography (EEG)-based brain–computer interface (BCI) system.
The representation of EEG typically takes the form of a high-dimensional matrix, which includes the information of sampling points, channels, trials, and subjects [2]. Meanwhile, the most common features of EEG-based BCIs include spatial filtering, band power, time points, and so on. Recently, machine learning (ML) has shown its powerful ability for feature extraction in EEG-based BCI tasks [3,4].
BCI technology based on EEG has made great progress, but the challenges of weak robustness and low accuracy greatly hinder the application of BCIs in practice [5]. From the perspective of signal decoding, the reasons are as follows: First, one of the main assumptions of ML is that training and test data belong to the same feature space and are subject to the same probability distribution. However, this assumption is often violated in the field of bioelectric signal processing, because differences in physiological structure and psychological states may cause obvious variation in EEG. Therefore, signals from different sessions/subjects on the same task show different features and distribution.
Second, EEG signals are extremely weak and are always accompanied by unrelated artifacts from other areas of the brain, which potentially mislead discriminant results and decrease the classification accuracy. Third, the strict requirements for the experimental conditions of BCI systems make it difficult to obtain large and high-quality datasets in practice. It is difficult for a classification model based on small-scale samples to obtain strong robustness and high classification accuracy. However, large-scale and high-quality datasets are the basis for guaranteeing the decoding accuracy of models.
One promising approach to solve these problems is transfer learning (TL). The principle of TL is realizing the knowledge transfer from different but related tasks, i.e., using existing knowledge learned from accomplished tasks to help with new tasks. The definition of TL is as follows: A given domain D consists of a feature space X and a marginal probability distribution P(X). A task T consists of a label space y and a prediction function . A source domain and a target domain may have different feature spaces or different marginal probability distributions, i.e., or . Meanwhile, tasks and are subject to different label spaces. The aim of TL is to help improve the learning ability of the target predictive function (·) in using the knowledge in and [6].
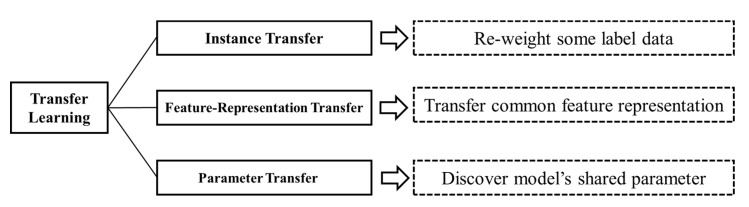
There are two main scenarios in EEG-based BCIs, namely, cross-subject transfer and cross-session transfer. The goal of TL is to find the similarity between new and original tasks and then to realize the discriminative and stationary information transfer across domains [7]. In this study, we attempted to summarize the transferred knowledge for EEG based on following three types: Knowledge of instance, knowledge of feature representation, and knowledge of model parameters.
This review of TL applications for EEG classification attempted to address the following critical questions: What problems does TL solve for EEG decoding? (Section 3.1); which paradigms of EEG are used for TL analysis? (Section 3.2); what kind of datasets can we refer to in order to verify the performance of these methods? (Section 3.3); what types of TL frameworks are available? (Section 3.4).
First, the search methods for the identification of studies are introduced in Section 2. Then, the principle and classification criteria of TL are analyzed in Section 3. Next, the TL algorithms for EEG from 2010 to 2020 are described in Section 4. Finally, the current challenges of TL in EEG decoding are discussed in Section 5.
2. Methodology
A wide literature search from 2010 to 2020 was conducted, resorting to the main databases, such as Web of Science, PubMed, and IEEE Xplore. The keywords used for the electronic search were TL, electroencephalogram, brain–computer interface, inter-subject, and covariate shift. Table 1 lists the collection criteria for inclusion or exclusion.
Table 1.
Inclusion and exclusion criteria.
| Inclusion Criteria | Exclusion Criteria |
|---|---|
|
|
|
|
|
\ |
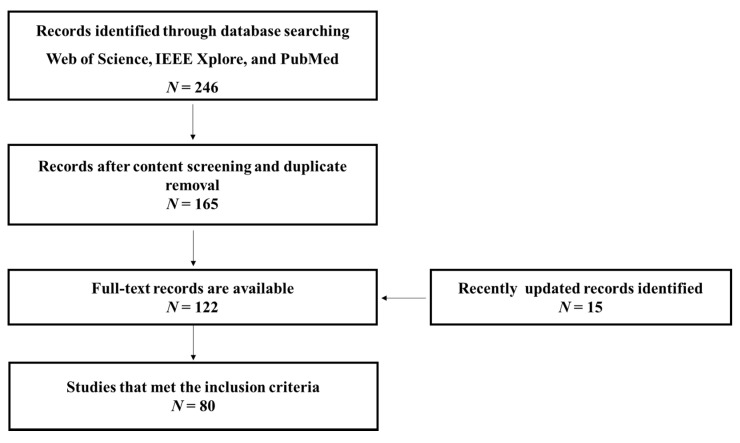
The search method of this review is shown in Figure 2, which was used to identify and to narrow down the collection of TL-based studies, resulting in a total of 246 papers. Duplicates between all datasets and studies without full-text links were excluded. Finally, 80 papers that meet the inclusion criteria were included.
Figure 2.
The search method for identifying relevant studies.
3. Results
3.1. What Problems Does Transfer Learning Solve?
This review of the literature on TL applications for EEG attempted to address the following critical questions:
3.1.1. The Problem of Differences across Subjects/Sessions
Although advanced methods such as machine learning have been proven to be a critical tool in EEG processing or analysis, they still suffer from some limitations that hinder their wide application in practice. Consistency of the feature space and probability distribution of training and test data is an important prior condition of machine learning. However, in the field of biomedical engineering, such as EEG based on BCIs, this hypothesis is often violated. Obvious variation in feature distribution typically occurs in representations of EEG across sessions/subjects. This phenomenon results in a scattered distribution of EEG signal features, an increase in the difficulty of feature extraction, and a reduction in the performance of the classifier.
3.1.2. The Problem of Small Sample Size
In recent years, machine learning and deep neural networks have provided good results for the classification of linguistic features, images, sounds, and natural texts. A main reason for its success is that their massive amount of data guarantees the performance of the classifier. However, in practical applications of BCI, it is difficult to collect high-quality and large EEG datasets due to the limitations of strict requirements for the experimental environment and available subjects. The performance of these methods is highly sensitive to the number of samples; a small sample size tends to lead to overfitting during model training, which adversely affects the classification accuracy [8].
3.1.3. The Problem of Time-Consuming Calibration
A large amount of data are required to calibrate a BCI system when a subject performs a specific EEG task. This requirement commonly takes a long calibration session, which is inevitable for a new user. For example, when a subject performs a steady-state visually evoked potential (SSVEP) speller task, the various commands cause a long calibration time. However, collecting calibration data is time-consuming and laborious, which reduces the efficiency of the BCI system.
3.2. EEG Paradigms for Transfer Learning
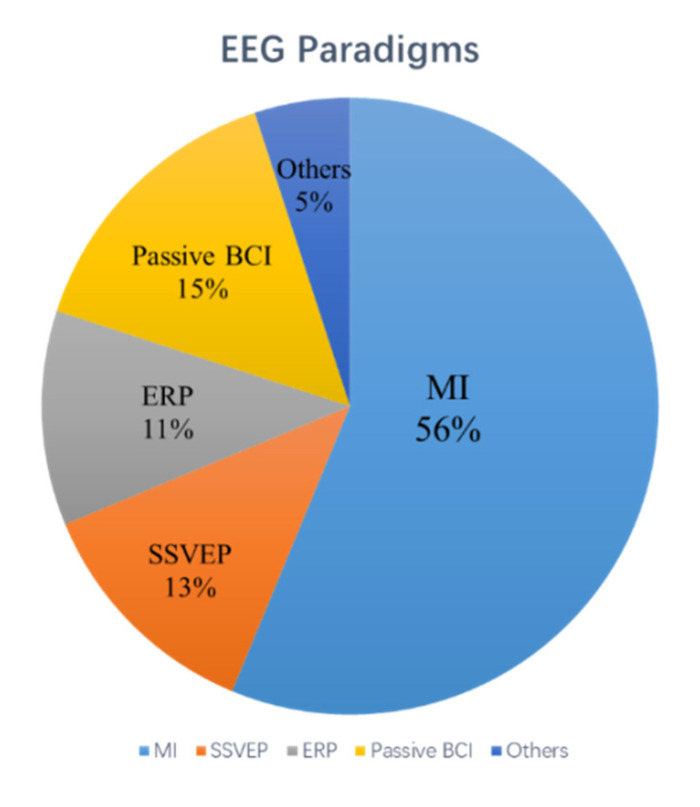
There are four paradigms of EEG-BCIs discussed in this paper and the percentage of these paradigms across collected studies are shown in Figure 3.
Figure 3.
The percentage of different EEG pattern strategies across collected studies.
3.2.1. Motor Imagery
Motor imagery (MI) is a mental process that imitates motor intention without real motion output [9], which activates the neural potential in primary sensorimotor areas. Different imagery tasks will induce potential activity in different regions of the brain. Thus, this response can be converted into a classification task. The feature of MI signals is often expressed in the form of frequency or band energy [10]. Due to task objectives and various feature representations, a variety of machine learning algorithms (e.g., deep learning and Riemannian geometry) can be applied to the decoding of MI [11,12].
3.2.2. Steady-State Visually Evoked Potentials
When a human receives a fixed frequency of flashing visual stimuli, the potential activity of the cerebral cortex is modulated to produce a continuous response related to the frequency (same or multiples) of these stimuli. This physiological phenomenon is referred to a SSVEP [13]. Due to their stable and obvious representation of signals, BCI systems based on SSVEP are widely used to control equipment such as mobile devices, wheelchairs, and spellers.
3.2.3. Event-Related Potentials
Event-related potentials are responses for multiple or diverse stimuli corresponding to specific meanings [14]. P300 is the most representative type of ERP, which occurs about 300 ms after a visual or auditory stimulus. A feature classification model can be used for decoding P300.
3.2.4. Passive BCIs
A passive BCI is a form of interaction that does not rely on external stimuli. It achieves a brain control task by encoding the mental activity from different states of the brain [15]. Common types of passive BCI tasks include driver drowsiness, emotion recognition, mental workload assessment, and epileptic detection [16], which can be decoded by regression and classification models [17,18].
3.3. Case Studies on a Shared Dataset
Analysis between different datasets is not valid because they use different equipment or communication protocols. In addition, different mental tasks and collecting procedures also bring great differences to EEG. Therefore, the reviewed studies mainly concentrate on the TL across subjects/sessions in the same dataset. In Table 2, we briefly summarize the publicly available EEG dataset in this review.
Table 2.
Dataset.
| Datasets | Task | Subject | Channel | Amount of Data (Per Subject) | Sampling Rate | Reference |
|---|---|---|---|---|---|---|
| BCIC-II-IV | 2 MI classes | 1 | 28 | 3 sessions/416 trials | 1000 Hz | [19] |
| BCIC-III-II | P300 | 2 | 64 | 5 sessions | 240 Hz | [20] |
| BCIC-III-IVa | 2 MI classes | 5 | 118 | 4 sessions/280 trials | 1000 Hz | [21] |
| BCIC-IV-2a | MI | 9 | 25 | 2 sessions/288 trials | 250 Hz | [22] |
| BCIC-IV-2b | 2 MI classes | 9 | 6 | 720 trials | 250 Hz | [23] |
| P300 speller | P300 | 8 | 8 | 5 sessions/20 trials | 256 Hz | [24] |
| DEAP | ER | 32 | 40 | 125 trials | 128 Hz | [25] |
| BCIC -III- IVC | MI | 1 | 118 | 630 trials | 200 Hz | [26] |
| SEED | ER | 15 | 64 | 3 sessions/15 trials | 200 Hz | [27] |
| OpenMIIR | Music Imagery | 10 | 64 | 5 sessions/12 trials in four tasks | 512 Hz | [28] |
| CHB-MIT | ED | 22 | 23 | 844 hours’ collection | 256 Hz | [29] |
3.4. Transfer Learning Architecture
In this review, we summarize previous studies according to “what knowledge should be transferred in EEG processing.” Multi-step processing for EEG across subjects/sessions results in discriminative information in different steps. Therefore, determining what should be transferred is the key problem according to different EEG tasks. Pan et al. [6] proposed authoritative classification approaches based on “what to transfer.” All papers collected in this review were classified according to this method (Figure 4). In the following sections, we have selected several representative methods for analysis.
Figure 4.
Different approaches to transfer learning.
3.4.1. Transfer Learning Based on Instance Knowledge
It is often assumed that we can easily obtain large amounts of markup data from a source domain, but this data cannot be directly reused. Instance transfer approaches re-weight some source domain data as a supplement for the target domain. Based on instance transfer, the majority of the literature utilized the measurement method to evaluate the similarity between data from the source and target domains. The similarity metric was then converted into the transfer weight coefficient, which was directly used to instance transfer by re-weighting the source domain data [30,31,32]. Herein, we have listed a few typical methods based on instance transfer.
Reference [33] proposed an instance TL method based on K–L divergence measurements. They measured the similarity of the normal distribution between two domains and transformed this similarity into a transfer weight coefficient for the target subject.
Suppose that the normal distribution from the two datasets and can be expressed as:
| (1) |
where and are the mean value and variance (i = 1/0), respectively. The K–L divergence of the two distributions can be expressed as:
| (2) |
where K denotes the dimension of the data, represents the mean value, and is the variance, det represents calculation of the determinant.
The similarity weight can be calculated by:
| (3) |
where is the balancing coefficient and is the summed divergence of the distribution characteristics of the target subjects. The results show that instance transfer can effectively reduce the calibration time and can significantly improve the average classification accuracy of MI tasks.
Li et al. [34] proposed importance-weighted linear discriminant analysis (IWLDA) with bootstrap aggregation. They defined the ratio of test and training input densities as transfer weight:
| (4) |
where and represent the marginal probability distribution of the training set and the test set, respectively.
Then, they optimized the parameters of the LDA model by adding a regularization coefficient and transfer weights:
| (5) |
where refers to the target labels corresponding to the feature vectors for i-th trials. Parameter is learned by least-squares.
| (6) |
where
| (7) |
The least-squares solution can be obtained by:
| (8) |
where λ (≥0) is the regularization parameter, D is the diagonal matrix with the i-th diagonal element, I is the identity matrix and is the least-squares solution. They also combined the bagging method that independently constructs accurate and diverse base learners to improve the classification accuracy and to reduce the variance. The weighted parameters of the LDA model in the target domain can thus be optimized.
Covariate shift [35] is a common phenomenon in EEG processing across subjects/sessions. It is defined as follows: Given an input space X and an output space Y, the marginal distribution of is inconsistent with , i.e., . However, the conditional distribution of the two domains is the same, . Covariate shift obviously affects the unbiasedness of a model in standard model selection, which reduces the generalization ability of the machine model during EEG decoding [30].
To address this issue, research has proposed covariate shift adaptation. For example, Raza et al. [36] proposed a transductive learning model based on the k-nearest neighbor principle. They initialized the classifier using data from the calibration stage and trained the optimal classification boundary. Then, adaptation was executed to update the classifier. The updated rules are as follows:
First, the Euclidean distance is used to measure unlabeled and labeled data:
| (9) |
where p and q refer to the unlabeled and labeled data points, respectively, and dist is the Euclidean distance. Then, the k-nearest neighbors are selected based on the Euclidean distance. Next, this distance is converted to inverse form , which represents the corresponding pattern in the training database that is closer to the current unlabeled feature set.
| (10) |
where i is the label and is the bias. To decide if the current trial’s features and estimated label should be added to the existing knowledge base, a confidence ratio CR is calculated:
| (11) |
The CR index is calculated to predict the label for the unlabeled test data. The predicted test data are then added into the knowledge database, following which the decision boundary is recalculated to realize the update.
3.4.2. Transfer Learning Based on Feature Representation
TL based on feature representation can be achieved by reducing the difference between two domains by feature transformation or projecting the feature from two domains into the uniform feature space [37,38,39]. Unlike instance transfer, feature representation TL aims to encode the shared information across subjects/sessions into a feature representation. For example, spatial filtering and time–frequency transformation are used to transform the raw data into feature representations.
Nakanish et al. proposed a spatial filtering approach called the task-related component analysis (TRCA) method to enhance the reproducibility during SSVEP tasks and to improve the performance of an SSVEP-based BCI [40].
Suppose that two domain signals consist of two parts: A task-related signal and a task-unrelated signal . A multichannel signal from can be calculated as:
| (12) |
where i represents the number of channels and a refers to the project coefficients; 1 and 2 represent labels.
| (13) |
where refers to the target data, and the optimization goal is to solve and . The covariance between the and the trials is described as:
| (14) |
All combinations of the trials are summed as:
| (15) |
where j represents the number of trials and refers to the spatial filters. Matrix s is defined as:
| (16) |
The variance of is constrained to obtain a finite solution:
| (17) |
The optimization is calculated as:
| (18) |
where is the optimal spatial filter. Finally, the correlation coefficient is calculated by Pearson’s correlation analysis between the data from the two domains. In their study, spatial filters as a feature representation were transferred to the target domain. The results showed that this method significantly improves the information transfer rates and classification accuracy. Based on this research, Tanaka [41] improved the TRCA method by maximizing the similarity across group of subjects, and they named this novel method group TRCA. The results showed that the group representation calculated by the group TRCA method achieve high consistency between two domains and offer effective data supplementation during brain–computer interaction.
CSP is a popular method for feature extraction, which is often used for MI classification. During calculation, a spatial filter is adopted to maximize the separation between the class variances of EEG. However, heterogeneous data across subjects/sessions causes poor classification performance of the model in the training stage. One feasible approach to solve the limitation is regularization. Lotte [42] presented regularized CSP to improve the classification accuracy across subjects. In their study, they discussed two strategies. One of them was regularizing the covariance matrix estimated. They can be, respectively, expressed as:
| (19) |
| (20) |
where represents the initial spatial covariance matrix for class i, is the regularized estimate, I is the identity matrix, is a constant scaling parameter, and represents the generic covariance matrix. The regularization parameters can be defined as and . This strategy aims to optimize the covariance matrix by transforming other subjects’ data into covariance combined with the regularization parameters and by transferring this feature to the target subject.
Another approach is regularizing the CSP objective function. CSP uses spatial filters to extremize the function:
| (21) |
where is spatial covariance matrix from class i. This approach optimizes CSP algorithms by regularizing the CSP objective function itself:
| (22) |
where represents a penalty function for the measurement distance between the spatial filter and the prior information. The goal of the objective function is to maximize and to minimize . is a user-defined regularization parameter. The prior information from the source domain provides a good solution to guide the optimization direction of the estimation of spatial filters.
In addition, adaptation regularization is a typical feature TL method based on the structural risk minimization principle and the regularization theory. Cross-domain feature transfer is mainly operated by three methods: (1) Utilize the structural risk minimization principle and minimize the structural risk functional; (2) minimize the distribution difference between the joint probability distributions; (3) maximize the manifold consistency underlying the marginal distributions [43]. In recent research, Chen et al. [44] developed an efficient cross-subject TL framework for driving status detection. They used adaptation regularization to measure and reduce the difference of the features from the two domains and to extract the features by filtering algorithms. The results showed that this framework can achieve high recognition accuracy and good transfer ability.
3.4.3. Transfer Learning Based on Model Parameters
The assumption of model parameter TL is that individual models across subjects/sessions should share some parameters. The key step of this approach is to find shared parameter information and to realize knowledge transfer. The domain adaption (DA) of a classifier is the common method of model parameter transfer. The knowledge of the parameter information from is reused and adjusted according to the prior distribution of [45]. A DA method, named adaptive extreme learning machine (ELM), was proposed by Bamdadian et al. [46]. ELM is a single-hidden layer feedforward neural network, which determines the output weights by operating the inverse operation of the hidden layer weight matrices [47]. This method has two steps: First, the classifier is initialized by data from the calibration session. Then, the update rule for the output weight based on least-square minimization is calculated. The update rule is calculated as follows:
The initial output weight can be defined as:
| (23) |
where H is the output matrix of hidden layer, and refer to the Moore–Penrose pseudo-inverse of H, and T represents the label category. The updated weight is calculated as:
| (24) |
| (25) |
| (26) |
where k is k-th hidden node, is orthogonal matrix calculated by . The experiential results showed that adaptive ELM can significantly improve the classification accuracy in MI classification across subjects.
Another strategy is ensemble learning, which combines multiple weak classifiers from the source domain into a strong classifier. Dalhoumi et al. [48] proposed a novel ensemble strategy based on Bayesian model averaging. They calculated the probability of having a class label given a feature vector :
| (27) |
where is the logarithmic variance feature vector, is a set of hypotheses from the source domain, and T is the test set. The hypothesis prior is estimated in the following method:
| (28) |
| (29) |
where is the projection of the feature vector x on the spatial filters of subject k. The learned ensemble classifier can be used to predict labels for the target user:
| (30) |
The results showed that this ensemble strategy can improve the classification performance in small-scale EEG data by evaluation on a real dataset.
In recent years, deep neural networks have provided good results for the processing of EEG signals [49,50]. Due to their end-to-end model structure and automatic feature extraction ability, deep neural networks minimize the interference of redundant information and improve the classification performance. Inspired by computer vision, a deep neural network learns generic feature representations by lower layers of the model. Specific feature representations with the relevant specific subjects or sessions are learned by the high layer [51]. Therefore, freezing lower layers and fine-tuning higher layers is a good way to realize model parameter transfer based on deep learning.
Zhao et al. [52] proposed an end-to-end deep convolution network for MI classification. To avoid the limitation of a small sample and overfitting, they utilized the data from to pre-train the source network and to transfer the parameters of several layers to initialize the target network. First, the network was pre-trained using data from the source domain. Then, they used the M source subjects to initialize the nth layer’s target network by a weight average:
| (31) |
where represents the strength of the source network and refers to the connecting weights of the nth layer to the next layer. The next stage is to fine-tune the target initialized network by data from . The results showed that the parameter transfer strategy can reduce the calibration time for new subjects and can help the deep convolution network to obtain better classification performance.
Raghu et al. used CNN combined with TL to recognize epileptic seizures [53]. They proposed two different transfer methods: To finetune a pre-trained network and then extract image features by said pre-trained network, and to classify the status of brain using an SVM. Popular networks such as Alexnet, VGG16net, VGG19net, and Squeezenet, were used to verify the performance of the proposed framework.
The summary of collected studies is shown in Table 3.
Table 3.
Summary of transfer learning for EEG decoding.
| Pattern | Reference | Type | Transfer Method | Feature Extraction | Datasets | Results |
|---|---|---|---|---|---|---|
| MI | [33] | ITL | Similarity measurement with KL divergence | LR+CSP | 19 subjects’ data, BCIC-IV-2a, and BCIC-III-4a | 70.3% and 75% and 75% |
| MI | [54] | ITL | Informative subspace transferring and selective ITL with active learning | LDA | BCIC-IV-2b | \ |
| MI | [55] | FTL | Ensemble learning and adaptive learning | LDA | NIPS | 68.1% |
| MI | [56] | ITL | Similarity measurement with Jensen Shannon ratio and rule adaptation TL | CSP+LDA | BCIC-IV-2a | 77% |
| MRP | [57] | ITL | MMD and regularized discriminative spatial pattern | Linear RR | BCIC-I-1 and BCIC-II-4 | \ |
| P300 | [58] | ITL | Ensemble learning generic information | Bayesian LDA | 8 participants’ data | 62.5% |
| SSVEP | [59] | ITL | Variability assessment Fisher’s discriminant ratios | Cluster | 8 subjects’ data | \ |
| P300 | [60] | ITL | Dynamically adjusts weights of instances | Liner SVM | BCIC-III-2 and dataset of P300 speller | 74.9% |
| MI | [61] | ITL | Selective informative with normalized entropy | LDA | BCIC-IV-2b | 75.8% |
| MI | [62] | ITL | Selective informative expected decision boundary | LDA | BCIC-IV-2a | 75.6% |
| MI | [63] | MTL | Domain adaptation parallel transport on the cone manifold of SPD | Linear SVM | BCIC-IV-2a | \ |
| DD | [64] | ITL | Selective transfer learning | Linear regression | 15 subjects’ data | 66% |
| MI | [65] | ITL | Composite local temporal correlation CSP Frobenius distance | Liner quadratic Mahalanobis | BCIC-III-IVa | 89.21% |
| SSVEP | [66] | ITL | Ensemble learning and similarity measurement with mutual information | LDA | 10 healthy subjects’ data | \ |
| Cognitive detection | [67] | ITL | Similarity measurement by Pearson’s correlation coefficient | SVM | 28 subjects’ data | 87.6% |
| ERP | [68] | FTL | Probabilistic zero framework | Unsupervised adaptation | Akimpech dataset | \ |
| MI | [69] | FTL | DA with power spectral density | CNN | BCIC-IV-2a | \ |
| MI | [70] | FTL | Many-objective optimization | Linear SVM | BCIC-III-IVa | 75.8% |
| VEP | [71] | FTL | Active semi-supervised TL | SVM | 14 subjects’ experiments | \ |
| MI | [72] | FTL | Adaptive Selective CSP | Discriminant analysis | 6 participant experiments | 61% |
| MI | [34] | FTL | CSA | Importance-weighted LDA | BCIC-III dataset | 79.1% |
| MI | [73] | ITL | Instance TL based p-hash | CNN | BCIC-IV-2b | |
| MI | [74] | FTL | Informative TL with AL | LDA | BCIC-IV-2a | 67.7% |
| MI | [75] | MTL | Modifications of CSP | SVM | BCIC-III-IVa | \ |
| MI | [76] | MTL | k-nearest neighbors principle | SVM+LDA | BCIC-IV-b | \ |
| ER | [77] | FTL | Transfer recursive feature elimination | Least-squares SVM | DEAP dataset | 78% |
| SSVEP | [78] | MTL | Least-squares transformation | \ | 8 participant experiments | 82.1% |
| MI | [79] | FTL | Domain transfer multiple kernel boosting | SVM |
BCIC-III-Iva 5 subjects’ data |
81.6% 76% |
| SSVEP | [80] | FTL | Spatial filtering transfer | TRCA | 10 subjects’ data | \ |
| SSVEP | [81] | FTL | Reference template transfer | MestCCA | 10 subjects’ data | \ |
| SSVEP | [82] | FTL | Reference template transfer | Transfer template-CCA | 12 subjects’ data | 85% |
| SSVEP | [83] | FTL | Reference template transfer | Adaptive combined-CCA | 10 subjects’ data | 83% |
| MI | [84] | FTL | Fuzzy TL based on generalized hidden-mapping RR | SVM | BCIC-IV-2a | 89.3% |
| MI | [46] | MTL | Adaptive extreme learning machine | SVM+ELM | 12 subjects’ data | 71.8% |
| MI | [48] | MTL | Classifier ensemble | LDA | BCIC-IV-2a | \ |
| MI | [13] | FTL | Regularized CSP with TL | LDA | BCIC-III-IVa | 78.9% |
| Multitask | [85] | FTL | Geometrical transformations on Riemannian Procrustes analysis | / | 8 publicly available BCI datasets | \ |
| ERP | [86] | FTL | Spectral transfer using information geometry | MDRM | 15 subjects’ data | 62% |
| MI | [87] | FTL | Space adaptation | LDA | BCIC-IV-2a | 77.5% |
| MI | [88] | FTL | Feature space transformation | LDA | PhysioNet datasets | 72% |
| MI | [89] | FTL | Tangent space-based TL | LDA | BCIC-IV-2a | \ |
| ER | [90] | FTL | Transfer component analysis and kernel principle component analysis | SVM | SEED | 77.96% |
| MI | [91] | FTL | Transfer kernel CSP | SVM | BCIC-III-IVa | 81.14% |
|
MI/
ERP |
[92] | FTL | Affine transform | Minimum distance mean and Bayesian classifier | BCIC-IV-2a and Brain Invaders experiment | \ |
| SSVEP | [93] | ITL | Riemannian similarities | Bootstrapping | 12 subjects’ data | 80.9% & 81.3% |
| MI | [94] | FTL | Multitask learning | RR+SVM | 10 healthy subjects’ data and an ALS subject’s data | 85% |
| Imagined speech | [95] | ITL | Inductive transfer learning | Naïve Bayesian classifier | 27 subjects’ data | 68.9% |
| MI | [96] | FTL |
Transferable discriminative dimensionality reduction |
KNN+SVM | 5 subjects’ data | 74.4% |
| MI | [7] | FTL | Nonstationary information transfers | LDA | 5 subjects’ data and BCIC-III-IVa | 80.4% |
| MI | [97] | MTL | Fine-tuned based on VGG16 | CNN | BCIC-IV-2b | 74.2% |
| MI | [98] | MTL | Fine-tuned based on pre-trained network | CNN | BCIC-IV-2a | 69.71% |
| ErrPs | [99] | MTL | Fine-tuned based on pre-trained network | CNN | 15 epilepsy patients’ data | 81.50% |
| Music Imagination | [100] | MTL | Fine-tuned based on AlexNet | LSTM | OpenMIIR dataset | 30.83% |
| ErrPs | [101] | MTL | Fine-tuned based on pre-trained network | CNN | 31 subjects’ data | 84.1% |
| DD | [102] | ITL | Source domain selection | Weighted adaptation regularization | 16 subjects ‘data | \ |
|
Attention detection |
[103] | ITL | Subject adaptation | CNN | 8 subjects ‘data | 84.17% |
| MI | [52] | ITL | Subject transfer | CNN |
BCIC-IV2a and BCIC-IV-2b |
0.56 and 0.65 (MK) |
| MI | [104] | MTL | Fine-tuned based on pre-trained network | RBM |
BCIC-IV2a and 12 subjects’ data |
88.9% |
| P300 | [105] | MTL | Fine-tuned based on pre-trained network | CNN | BCIC-III-2 | 90.5% |
| MI | [106] | MTL | Fine-tuned based on pre-trained network | CNN | BCIC-IV-2b | 0.57 (MK) |
| MI | [107] | MTL | Fine-tuned based on pre-trained network | Conditional variational autoencoder | PhysioNet datasets | 73% |
| Music Imagination | [108] | FTL | Cross-domain encoder | Attention decoder-RNN | OpenMIIR datasets | 37.9% |
| MI | [36] | ITL | Covariate shift detection and adaptation | Linear SVM |
BCIC-IV-2a BCIC-IV-2b |
73.8%
and
69.7% |
| MWA | [109] | FTL | Cross-subject statistical shift | Random forest | 9 subjects‘ data | \ |
| MI | [110] | MTL | Fine-tuned based on multiple network | CNN |
BCIC-IV-2a and BCIC-IV-2b |
\ |
| MI | [111] | MTL | Four-strategy model transfer learning | Deep neural network | BCIC-IV-2a | \ |
| MI | [112] | ITL | Subject–subject transfer | CNN | 3 subjects’ data | \ |
| P300 | [113] | ITL | Subject–subject transfer | Linear SVM | 22 subjects’ data | 68.7% |
| ER | [114] | ITL | Measurement on Riemannian geometry instance transfer | SVM | MDME and SDMN datasets | \ |
| DD | [40] | FTL | Adaptation regularization based TL | Multiple classifier |
23 subjects’ data and NIH physio bank |
89.59% |
| ED | [53] | MTL | Fine-tuned based on pre-trained network | GoogLeNet and Inception v3 | TUH open-source database |
82.85%
and
88.30% |
| ED | [115] | MTL | Fine-tuned based on pre-trained network | CNN and bidirectional LSTM | CHB-MIT EEG dataset | 99.6% |
| ED | [116] | MTL | Fine-tuned based on pre-trained network | CNN | CHB-MIT EEG dataset | 92.7% |
| MI | [117] | FTL | Spatial filtering transfer and matrix decomposition | ELM |
BCIC-III-IVa and BCIC-IV-1 |
89% 62% |
| SSVEP | [41] | FTL | Spatial filtering transfer | Group TRCA | Benchmark dataset | \ |
| MI | [118] | MTL | Adversarial inference | CNN | 52 subjects’ data | \ |
| ER | [119] | FTL | Power spectral density feature | Polynomial/Gaussian kernels/ naïve Bayesian SVM | DEAP, MAHNOB-HCI, and DREAMER | \ |
| MWA | [120] | FTL | Ensemble learning | Stacked denoising autoencoder | 8 subjects’ data | 92% |
| MI | [121] | FTL | Data mapping and ensemble learning | LDA | BCIC-IV-2a | 0.58 (MK) |
| MI | [122] | FTL | Center-based discriminative feature learning | CNN | BCIC-III-IVa | 76% |
“\” represents that there is no specific description or else multiple descriptions for the results.
4. Discussion
Based on the numerous papers surveyed herein, we briefly summarized the development of the application of TL to EEG decoding. This will help researchers scan the status of this field and receive useful guidance in future work.
According to the various studies surveyed in this paper, it is not hard to determine the points of interest that researchers focus on. As shown in Figure 2, more studies have focused on active BCI (i.e., MI, SSVEP, and ERP) among these different EEG paradigms. One possible explanation is that the goal of these mental activity decoding studies is to categorize EEG from different classes. This would allow many machine learning methods to be applied to this paradigm. From Table 2, it can be seen that the application scenarios of TL in the existing literature have focused almost only on classification and regression tasks.
The method of model parameter transfer is not applicable to only a few subjects with initially low BCI performance. The feature of EEG from these subjects exhibits inseparability in feature space. Therefore, the parameter optimization of the classifier does not significantly improve the classification results. It is worth noting that the adaptive strategy of the classifier should be considered a supplement to achieve the goal of a calibration-free mode of operation [123]. The combination of TL and the adaptive strategy may receive increasing attention in future studies.
It is also worth noting that TL showed good results across subjects/experiments, but the detail of variability across sessions/subjects was unclear. Some studies proposed that the Bayesian model is a promising approach to capture variability. This model is built based on multitask learning, and variation in some features is often extracted, such as spectral and spatial [124,125].
Due to its end-to-end structure and competitive performance, deep learning has been successful in processing EEG data [126]. However, the computational power and small-scale data are a limitation during practical operation. A hybrid structure based on TL and deep learning is a promising way to address this issue. For example, one of the methods is fine-tuning the pre-trained network, which has proven to be effective. With the development of deep learning technology, the research for such a hybrid structure is still a hot topic for future research.
As reported in the above-cited studies, TL is instrumental in EEG decoding across subjects/sessions. However, knowledge transfer across tasks/device is still a blank field. This issue is worth exploring and will make EEG-based BCI systems much more practical.
5. Conclusions
In this paper, we reviewed the research on TL for EEG decoding that was published between 2010 and 2020. We discussed numerous approaches that can be divided into three categories: Instance transfer, feature representation transfer, and parameter of classifier transfer. Based on the summary of their results, we can conclude that TL can effectively improve the decoding performance in classification and regression tasks. In addition, TL provides adequate performance in initializing BCI systems for a new subject, which reduces the length of time of the calibration process. Although there are some limitations for using TL for EEG decoding, such as the scope of application of TL and suboptimal performance on some occasions, TL shows strong robustness. Overall, TL is instrumental in EEG decoding across subjects/sessions. In addition, achieving a calibration-free model of operation and higher accuracy of decoding are worthy of further research.
Acknowledgments
The authors would like to thank the support by Guangdong Institute of Medical Instruments & National Engineering Research Center for Healthcare Devices.
Abbreviations: List of Acronyms
| AL | Active learning |
| ALS | Amyotrophic Lateral Sclerosis |
| ATL | Active Transfer Learning |
| BCIC | Brain Computer Interface Competition |
| CCA | Canonical Correlation Analysis |
| CNN | Convolution Neural Network |
| CSA | Covariate Shift Adaptation |
| CSP | Common Spatial Pattern |
| DA | Domain Adaptation |
| DD | Drowsiness Detection |
| ELM | Extreme Learning Machine |
| ED | Epileptic Detection |
| ER | Emotion Recognition |
| ERP | Event-Related Potential |
| ErrPs | Electroencephalography -measured error-related potentials |
| FTL | Feature Transfer Learning |
| ITL | Instance Transfer Learning |
| JSR | Jensen Shannon Ratio |
| KL | Kullback–Leibler |
| KNN | K-Nearest Neighbor |
| LDA | Linear Discriminate Analysis |
| LR | Logistic Regression |
| LSTM | Long Short-Term Memory |
| MDRM | Minimum Distance to Riemannian Mean classifiers |
| MI | Motor Imagery |
| MMD | Maximum mean discrepancy |
| MK | Means Kappa value |
| MTL | Model Transfer Learning |
| MWA | Mental-Workload Assessment |
| MRP | Movement Related Potentials |
| RNN | Recurrent Neural Network |
| RR | Ridge Regression |
| RBM | Restricted Boltzmann Machine |
| SMR | Sensory Motor Rhythm |
| SVM | Support Vector Machine |
| SSVEP | Steady State Visual Evoked Potential |
| SELM | Sigmoid Extreme Learning Machine |
| SPD | Symmetric Positive Definite |
| TL | Transfer Learning |
| TRCA | Task-Related Component Analysis |
| VEP | Visual Evoked Potential |
| VGG | Visual Geometry Group |
Author Contributions
K.Z. and G.X. designed the study. K.Z. wrote the manuscript, X.Z., H.L. collected the relevant papers. S.Z., Y.Y., R.L. prepared the figures. All authors have read and agreed to the published version of the manuscript.
Funding
Research supported by National Key Research & Development Plan of China (Grant No.2017YFC1308500), GDAS’ Project of Science and Technology Development (Grant No. 2019GDASYL-0502002) and Key Research & Development Plan of Shaanxi Province (Grant No. 2018ZDCXL-GY-06-01).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yuan H., He B. Brain–Computer Interfaces Using Sensorimotor Rhythms: Current State and Future Perspectives. IEEE Trans. Biomed. Eng. 2014;61:1425–1435. doi: 10.1109/TBME.2014.2312397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subasi A., Gursoy M.I. EEG signal classification using PCA, ICA, LDA and support vector machines. Expert Syst. Appl. 2010;37:8659–8666. doi: 10.1016/j.eswa.2010.06.065. [DOI] [Google Scholar]
- 3.Lotte F., Bougrain L., Cichocki A., Clerc M., Congedo M., Rakotomamonjy A., Yger F. A review of classification algorithms for EEG-based brain-computer interfaces: A 10 year update. J. Neural Eng. 2018;15:031005. doi: 10.1088/1741-2552/aab2f2. [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y., Zhang Y., Chen X., Yin E., Jin J., Wang X., Cichocki A. Sparse Group Representation Model for Motor Imagery EEG Classification. IEEE J. Biomed. Health Inf. 2018;23:631–641. doi: 10.1109/JBHI.2018.2832538. [DOI] [PubMed] [Google Scholar]
- 5.Krepki R., Blankertz B., Curio G., Müller K.-R. The Berlin Brain Computer Interface (BBCI)—Towards a new communication channel for online control in gaming applications. Multimed. Tools Appl. 2017;33:73–90. doi: 10.1007/s11042-006-0094-3. [DOI] [Google Scholar]
- 6.Pan S.J., Yang Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2009;22:1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 7.Samek W., Meinecke F., Muller K.-R. Transferring Subspaces between Subjects in Brain-Computer Interfacing. IEEE Trans. Biomed. Eng. 2013;60:2289–2298. doi: 10.1109/TBME.2013.2253608. [DOI] [PubMed] [Google Scholar]
- 8.Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 9.Bonassi G., Biggio M., Bisio A., Ruggeri P., Bove M., Avanzino L. Provision of somatosensory inputs during motor imagery enhances learning-induced plasticity in human motor cortex. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-09597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfurtscheller G., Neuper C. Motor imagery and direct brain-computer communication. Proc. IEEE. 2001;89:1123–1134. doi: 10.1109/5.939829. [DOI] [Google Scholar]
- 11.Ahn M., Jun S.C. Performance variation in motor imagery brain–computer interface: A brief review. J. Neurosci. Methods. 2015;243:103–110. doi: 10.1016/j.jneumeth.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K., Xu G., Han Z., Ma K., Zheng X., Chen L., Duan N., Zhang S. Data Augmentation for Motor Imagery Signal Classification Based on a Hybrid Neural Network. Sensors. 2020;20:4485. doi: 10.3390/s20164485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng M., Gao X., Gao S., Xu D. Design and Implementation of a Brain-Computer Interface With High Transfer Rates. IEEE Trans. Biomed. Eng. 2002;49:1181–1186. doi: 10.1109/TBME.2002.803536. [DOI] [PubMed] [Google Scholar]
- 14.Handy T.C., editor. Event-Related Potentials: A Methods Handbook. The MIT Press; Boston, MA, USA: 2005. [Google Scholar]
- 15.Zander T.O., Kothe C. Towards passive brain–computer interfaces: Applying brain–computer interface technology to human–machine systems in general. J. Neural Eng. 2011;8:025005. doi: 10.1088/1741-2560/8/2/025005. [DOI] [PubMed] [Google Scholar]
- 16.Arico P., Borghini G., Di Flumeri G., Sciaraffa N., Colosimo A., Babiloni F. Passive BCI in Operational Environments: Insights, Recent Advances, and Future Trends. IEEE Trans. Biomed. Eng. 2017;64:1431–1436. doi: 10.1109/TBME.2017.2694856. [DOI] [PubMed] [Google Scholar]
- 17.Cui Y., Xu Y., Wu D. EEG-Based Driver Drowsiness Estimation Using Feature Weighted Episodic Training. IEEE Trans. Neural Syst. Rehabi. Eng. 2019;27:2263–2273. doi: 10.1109/TNSRE.2019.2945794. [DOI] [PubMed] [Google Scholar]
- 18.Muehl C., Allison B., Nijholt A., Chanel G. A survey of affective brain computer interfaces: Principles, state-of-the-art, and challenges. Brain-Comput. Interfaces. 2014;1:66–84. doi: 10.1080/2326263X.2014.912881. [DOI] [Google Scholar]
- 19.Blankertz B., Curio G., Müller K. Classifying Single Trial EEG: Towards Brain Computer Interfacing. In: Diettrich T.G., Becker S., Ghahramani Z., editors. Advances in Neural Information Processing Systems 14 (NIPS 01) The MIT Press; Cambridge, MA, USA: 2002. [Google Scholar]
- 20.Wolpaw J.R., Birbaumer N., McFarland D.J., Pfurtscheller G., Vaughan T.M. Brain–computer interfaces for communication and control. Clin. Neurophysiol. 2002;113:767–791. doi: 10.1016/S1388-2457(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 21.Dornhege G., Blankertz B., Curio G., Müller K. Boosting bit rates in non-invasive EEG single-trial classifications by feature combination and multi-class paradigms. IEEE Trans. Biomed. Eng. 2004;51:993–1002. doi: 10.1109/TBME.2004.827088. [DOI] [PubMed] [Google Scholar]
- 22.Naeem M., Brunner C., Leeb R., Graimann B., Pfurtscheller G. Seperability of four-class motor imagery data using independent components analysis. J. Neural Eng. 2006;3:208–216. doi: 10.1088/1741-2560/3/3/003. [DOI] [PubMed] [Google Scholar]
- 23.Leeb R., Lee F., Keinrath C., Scherer R., Bischof H., Pfurtscheller G. Brain–Computer Communication: Motivation, Aim, and Impact of Exploring a Virtual Apartment. IEEE Trans. Neural Syst. Rehabil. Eng. 2007;15:473–482. doi: 10.1109/TNSRE.2007.906956. [DOI] [PubMed] [Google Scholar]
- 24.Riccio A., Esimione L., Eschettini F., Epizzimenti A., Inghilleri M., Belardinelli M.E., Mattia D., Cincotti F. Attention and P300-based BCI performance in people with amyotrophic lateral sclerosis. Front. Hum. Neurosci. 2013;7:732. doi: 10.3389/fnhum.2013.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koelstra S., Muhl C., Soleymani M., Lee J.-S., Yazdani A., Ebrahimi T., Pun T., Nijholt A., Patras I. DEAP: A Database for Emotion Analysis; Using Physiological Signals. IEEE Trans. Affect. Comput. 2011;3:18–31. doi: 10.1109/T-AFFC.2011.15. [DOI] [Google Scholar]
- 26.Li Y., Koike Y., Sugiyama M. A Framework of Adaptive Brain Computer Interfaces; Proceedings of the International Conference on Biomedical Engineering & Informatics; Tianjin, China. 17–19 October 2009. [Google Scholar]
- 27.Zheng W.-L., Lu B.-L. Investigating Critical Frequency Bands and Channels for EEG-Based Emotion Recognition with Deep Neural Networks. IEEE Trans. Auton. Ment. Dev. 2015;7:162–175. doi: 10.1109/TAMD.2015.2431497. [DOI] [Google Scholar]
- 28.Stober S., Sternin A., Owen A.M., Grahn J.A. Towards music imagery information retrieval: Introducing the openmiir dataset of eeg recordings from music perception and imagination; Proceedings of the 16th ISMIR Conference; Malaga, Spain. 26–30 October 2015; pp. 763–769. [Google Scholar]
- 29.Shoeb A. Ph.D. Thesis. Massachusetts Institute of Technology; Cambridge, MA, USA: 2009. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. [Google Scholar]
- 30.Masashi S., Krauledat M., MÞller K. Covariate shift adaptation by importance weighted cross validation. J. Mach. Learn. Res. 2007;8:985–1005. [Google Scholar]
- 31.Khan M., Heisterkamp D.R. Adapting instance weights for unsupervised domain adaptation using quadratic mutual information and subspace learning; Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR); Cancun, Mexico. 4–8 December 2016; pp. 1560–1565. [Google Scholar]
- 32.Zadrozny B. Proceedings of the Twenty-First International Conference on Machine Learning—ICML ’04. Association for Computing Machinery (ACM); New York, NY, USA: 2004. Learning and evaluating classifiers under sample selection bias; p. 114. [Google Scholar]
- 33.Azab A.M., Mihaylova L., Ang K.K., Arvaneh M. Weighted Transfer Learning for Improving Motor Imagery-Based Brain–Computer Interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2019;27:1352–1359. doi: 10.1109/TNSRE.2019.2923315. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Kambara H., Koike Y., Sugiyama M. Application of covariate shift adaptation techniques in brain–computer interfaces. IEEE Trans. Biomed. Eng. 2010;57:1318–1324. doi: 10.1109/TBME.2009.2039997. [DOI] [PubMed] [Google Scholar]
- 35.Shimodaira H. Improving predictive inference under covariate shift by weighting the log-likelihood function. J. Stat. Plan. Inference. 2000;90:227–244. doi: 10.1016/S0378-3758(00)00115-4. [DOI] [Google Scholar]
- 36.Raza H., Cecotti H., Li Y., Prasad G. Adaptive learning with covariate shift-detection for motor imagery-based brain–computer interface. Soft Comput. 2016;20:3085–3096. doi: 10.1007/s00500-015-1937-5. [DOI] [Google Scholar]
- 37.Liu J., Shah M., Kuipers B., Savarese S. Cross-view action recognition via view knowledge transfer; Proceedings of the CVPR 2011; Providence, RI, USA. 20–25 June 2011; pp. 3209–3216. [Google Scholar]
- 38.Zheng V.W., Pan S.J., Yang Q., Pan J.J. Transferring ulti-device localization models using latent multi-task learning; Proceedings of the Twenty-Third AAAI Conference on Artificial Intelligence; Chicago, IL, USA. 13–17 July 2008; pp. 1427–1432. [Google Scholar]
- 39.Pan S.J., Tsang I.W., Kwok J.T., Yang Q. Domain Adaptation via Transfer Component Analysis. IEEE Trans. Neural Netw. 2010;22:199–210. doi: 10.1109/TNN.2010.2091281. [DOI] [PubMed] [Google Scholar]
- 40.Nakanishi M., Wang Y., Chen X., Wang Y.T., Gao X., Jung T.P. Enhancing Detection of SSVEPs for a High-Speed Brain Speller Using Task-Related Component Analysis. IEEE Trans. Biomed. Eng. 2017;65:104–112. doi: 10.1109/TBME.2017.2694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka H. Group task-related component analysis (gTRCA): A multivariate method for inter-trial reproducibility and inter-subject similarity maximization for EEG data analysis. Sci. Rep. 2020;10:1–17. doi: 10.1038/s41598-019-56962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotte F., Guan C. Regularizing Common Spatial Patterns to Improve BCI Designs: Unified Theory and New Algorithms. IEEE Trans. Biomed. Eng. 2011;58:355–362. doi: 10.1109/TBME.2010.2082539. [DOI] [PubMed] [Google Scholar]
- 43.Long M., Wang J., Ding G., Pan S.J., Yu P.S. Adaptation Regularization: A General Framework for Transfer Learning. IEEE Trans. Knowl. Data Eng. 2013;26:1076–1089. doi: 10.1109/TKDE.2013.111. [DOI] [Google Scholar]
- 44.Chen L., Zhang A., Lou X. Cross-subject driver status detection from physiological signals based on hybrid feature selection and transfer learning. Expert Syst. Appl. 2019;137:266–280. doi: 10.1016/j.eswa.2019.02.005. [DOI] [Google Scholar]
- 45.Zhang B., Lu J., Peitao W., Tang Z. A review on transfer learning for brain-computer interface classification; Proceedings of the 5th International Conference on Information Science and Technology (ICIST); Changsha, China. 24–26 April 2015. [Google Scholar]
- 46.Bamdadian A., Guan C., Ang K.K., Xu J. Improving session-to-session transfer performance of motor imagery-based BCI using adaptive extreme learning machine; Proceedings of the 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Osaka, Japan. 3–7 July 2013; pp. 2188–2191. [DOI] [PubMed] [Google Scholar]
- 47.Huang G.-B., Zhu Q.-Y., Siew C.-K. Extreme learning machine: Theory and applications. Neurocomputing. 2006;70:489–501. doi: 10.1016/j.neucom.2005.12.126. [DOI] [Google Scholar]
- 48.Dalhoumi S., Dray G., Montmain J. Knowledge Transfer for Reducing Calibration Time in Brain-Computer Interfacing; Proceedings of the IEEE International Conference on Tools with Artificial Intelligence—ICTAI; Limassol, Cyprus. 10–12 November 2014. [Google Scholar]
- 49.Dai M., Zheng D., Na R., Wang S., Zhang S. EEG Classification of Motor Imagery Using a Novel Deep Learning Framework. Sensors. 2019;19:551. doi: 10.3390/s19030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu N., Li T., Ren X. A Deep Learning Scheme for Motor Imagery Classification based on Restricted Boltzmann Machines. IEEE Trans. Neural Syst. Rehabil. Eng. 2016;25:566–576. doi: 10.1109/TNSRE.2016.2601240. [DOI] [PubMed] [Google Scholar]
- 51.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM. :2012. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 52.Zhao D., Tang F., Si B., Feng X. Learning joint space–time–frequency features for EEG decoding on small labeled data. Neural Netw. 2019;114:67–77. doi: 10.1016/j.neunet.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Raghu S., Sriraam N., Temel Y., Rao S.V., Kubben P.L. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw. 2020;124:202–212. doi: 10.1016/j.neunet.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Hossain I., Khosravi A., Nahavandhi S. Active transfer learning and selective instance transfer with active learning for motor imagery based BCI; Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN); Vancouver, BC, Canada. 24–29 July 2016. [Google Scholar]
- 55.Tu W., Sun S. A subject transfer framework for EEG classification. Neurocomputing. 2012;82:109–116. doi: 10.1016/j.neucom.2011.10.024. [DOI] [Google Scholar]
- 56.Giles J., Ang K.K., Mihaylova L.S., Arvaneh M. A Subject-to-subject Transfer Learning Framework Based on Jensen-shannon Divergence for Improving Brain-computer Interface; Proceedings of the ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); Brighton, UK. 12–17 May 2019; pp. 3087–3091. [Google Scholar]
- 57.Wang P., Lu J., Lu C., Tang Z. An algorithm for movement related potentials feature extraction based on transfer learning; Proceedings of the 2015 5th International Conference on Information Science and Technology (ICIST); Changsha, China. 24–26 April 2015; pp. 309–314. [Google Scholar]
- 58.Adair J., Brownlee A., Daolio F., Ochoa G. International Workshop on Machine Learning, Optimization, and Big Data. Springer; Cham, Switzerland: 2017. Evolving training sets for improved transfer learning in brain computer interfaces; pp. 186–197. [Google Scholar]
- 59.Wei C.-S., Nakanishi M., Chiang K.-J., Jung T.-P. Exploring Human Variability in Steady-State Visual Evoked Potentials; Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Miyazaki, Japan. 7–10 October 2018; pp. 474–479. [Google Scholar]
- 60.Hou J., Li Y., Liu H., Wang S. Improving the P300-based brain-computer interface with transfer learning; Proceedings of the2017 8th International IEEE/EMBS Conference on Neural Engineering (NER); Shanghai, China. 25–28 May 2017; pp. 485–488. [Google Scholar]
- 61.Hossain I., Khosravi A., Hettiarachchi I.T., Nahavandhi S. Informative instance transfer learning with subject specific frequency responses for motor imagery brain computer interface; Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Banff, AB, Canada. 5–8 October 2017; pp. 252–257. [Google Scholar]
- 62.Hossain I., Khosravi A., Hettiarachchi I., Nahavandi S. Multiclass informative instance transfer learning framework for motor imagery-based brain-computer interface. Comput. Intell. Neurosci. 2018;2018:6323414. doi: 10.1155/2018/6323414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yair O., Ben-Chen M., Talmon R. Parallel Transport on the Cone Manifold of SPD Matrices for Domain Adaptation. IEEE Trans. Signal Process. 2019;67:1797–1811. doi: 10.1109/TSP.2019.2894801. [DOI] [Google Scholar]
- 64.Wei C.-S., Lin Y.-P., Wang Y.-T., Jung T.-P., Bigdely-Shamlo N., Lin C.-T. Selective Transfer Learning for EEG-Based Drowsiness Detection; Proceedings of the 2015 IEEE International Conference on Systems, Man, and Cybernetics; Kowloon, China. 9–12 October 2015; pp. 3229–3232. [Google Scholar]
- 65.Hatamikia S., Nasrabadi A.M. Subject transfer BCI based on composite local temporal correlation common spatial pattern. Comput. Boil. Med. 2015;64:1–11. doi: 10.1016/j.compbiomed.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Sybeldon M., Schmit L., Akcakaya M. Transfer Learning for SSVEP Electroencephalography Based Brain–Computer Interfaces Using Learn++. NSE and Mutual Information. Entropy. 2017;19:41. doi: 10.3390/e19010041. [DOI] [Google Scholar]
- 67.Wei C.S., Lin Y.P., Wang Y.T., Lin C.T., Jung T.P. Transfer learning with large-scale data in brain-computer interfaces; Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Orlando, FL, USA. 16–20 August 2016; pp. 4666–4669. [DOI] [PubMed] [Google Scholar]
- 68.Kindermans P.J., Tangermann M., Müller K.R., Schrauwen B. Integrating dynamic stopping, transfer learning and language models in an adaptive zero-training ERP speller. J. Neural Eng. 2014;11:035005. doi: 10.1088/1741-2560/11/3/035005. [DOI] [PubMed] [Google Scholar]
- 69.Jeon E., Ko W., Suk H.I. Domain Adaptation with Source Selection for Motor-Imagery based BCI; Proceedings of the 2019 7th International Winter Conference on Brain-Computer Interface (BCI); Gangwon, Korea. 18–20 February 2019; pp. 1–4. [Google Scholar]
- 70.Pal M., Bandyopadhyay S., Bhattacharyya S. A Many Objective Optimization Approach for Transfer Learning in EEG Classification. arXiv. 20191904.04156 [Google Scholar]
- 71.Wu D., Lance B., Lawhern V. Transfer learning and active transfer learning for reducing calibration data in single-trial classification of visually-evoked potentials; Proceedings of the 2014 IEEE International Conference on Systems, Man, and Cybernetics (SMC); San Diego, CA, USA. 5–8 October 2014; pp. 2801–2807. [Google Scholar]
- 72.Jin Y., Mousavi M., de Sa V.R. Adaptive CSP with subspace alignment for subject-to-subject transfer in motor imagery brain-computer interfaces; Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI); GangWon, Korea. 15–17 January 2018; pp. 1–4. [Google Scholar]
- 73.Zhang K., Xu G., Chen L., Tian P., Han C., Zhang S., Duan N. Instance Transfer Subject-Dependent Strategy for Motor Imagery Signal Classification Using Deep Convolutional Neural Networks. Comput. Math. Meth. Med. 2020;2020:1683013. doi: 10.1155/2020/1683013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hossain I., Khosravi A., Hettiarachchi I., Nahavandi S. Calibration Time Reduction Using Subjective Features Selection Based Transfer Learning For Multiclass BCI; Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Miyazaki, Japan. 7–10 October 2018; pp. 491–498. [Google Scholar]
- 75.Kang H., Nam Y., Choi S. Composite common spatial pattern for subject-to-subject transfer. IEEE Signal Process. Lett. 2009;16:683–686. doi: 10.1109/LSP.2009.2022557. [DOI] [Google Scholar]
- 76.Raza H., Prasad G., Li Y., Cecotti H. Covariate shift-adaptation using a transductive learning model for handling non-stationarity in EEG based brain-computer interfaces; Proceedings of the 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Belfast, UK. 2–5 November 2014; pp. 230–236. [Google Scholar]
- 77.Yin Z., Wang Y., Liu L., Zhang W., Zhang J. Cross-subject EEG feature selection for emotion recognition using transfer recursive feature elimination. Front. Neurorobot. 2017;11:19. doi: 10.3389/fnbot.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang K.J., Wei C.S., Nakanishi M., Jung T.P. Cross-Subject Transfer Learning Improves the Practicality of Real-World Applications of Brain-Computer Interfaces; Proceedings of the 2019 9th International IEEE/EMBS Conference on Neural Engineering (NER); San Francisco, CA, USA. 20–23 March 2019; pp. 424–427. [Google Scholar]
- 79.Dai M., Wang S., Zheng D., Na R., Zhang S. Domain transfer multiple kernel boosting for classification of EEG motor imagery signals. IEEE Access. 2019;7:49951–49960. doi: 10.1109/ACCESS.2019.2908851. [DOI] [Google Scholar]
- 80.Nakanishi M., Wang Y.T., Wei C.S., Chiang K.J., Jung T.P. Facilitating Calibration in High-Speed BCI Spellers via Leveraging Cross-Device Shared Latent Responses. IEEE Trans. Biomed. Eng. 2019;67:1105–1113. doi: 10.1109/TBME.2019.2929745. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y.U., Zhou G., Jin J., Wang X., Cichocki A. Frequency recognition in SSVEP-based BCI using multiset canonical correlation analysis. Int. J. Neural Syst. 2014;24:1450013. doi: 10.1142/S0129065714500130. [DOI] [PubMed] [Google Scholar]
- 82.Yuan P., Chen X., Wang Y., Gao X., Gao S. Enhancing performances of SSVEP-based brain–computer interfaces via exploiting inter-subject information. J. Neural Eng. 2015;12:046006. doi: 10.1088/1741-2560/12/4/046006. [DOI] [PubMed] [Google Scholar]
- 83.Waytowich N.R., Faller J., Garcia J.O., Vettel J.M., Sajda P. Unsupervised adaptive transfer learning for Steady-State Visual Evoked Potential brain-computer interfaces; Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Budapest, Hungary. 9–12 October 2016; pp. 004135–004140. [Google Scholar]
- 84.Salami A., Khodabakhshi M.B., Moradi M.H. Fuzzy transfer learning approach for analysing imagery BCI tasks; Proceedings of the 2017 Artificial Intelligence and Signal Processing Conference (AISP); Shiraz, Iran. 20–27 October 2017; pp. 300–305. [Google Scholar]
- 85.Rodrigues P.L.C., Jutten C., Congedo M. Riemannian Procrustes Analysis: Transfer Learning for Brain–Computer Interfaces. IEEE Trans. Biomed. Eng. 2018;66:2390–2401. doi: 10.1109/TBME.2018.2889705. [DOI] [PubMed] [Google Scholar]
- 86.Waytowich N.R., Lawhern V.J., Bohannon A.W., Ball K.R., Lance B.J. Spectral Transfer Learning Using Information Geometry for a User-Independent Brain-Computer Interface. Front. Neurosci. 2016;10:430. doi: 10.3389/fnins.2016.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arvaneh M., Robertson I., Ward T.E. Subject-to-subject adaptation to reduce calibration time in motor imagery-based brain-computer interface; Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; Chicago, IL, USA. 26–30 August 2014; pp. 6501–6504. [DOI] [PubMed] [Google Scholar]
- 88.Heger D., Putze F., Herff C., Schultz T. Subject-to-subject transfer for CSP based BCIs: Feature space transformation and decision-level fusion; Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Osaka, Japan. 3–7 July 2013; pp. 5614–5617. [DOI] [PubMed] [Google Scholar]
- 89.Gaur P., McCreadie K., Pachori R.B., Wang H., Prasad G. Tangent Space Features-Based Transfer Learning Classification Model for Two-Class Motor Imagery Brain–Computer Interface. Int. J. Neural Syst. 2019;29:1950025. doi: 10.1142/S0129065719500254. [DOI] [PubMed] [Google Scholar]
- 90.Zheng W.L., Zhang Y.Q., Zhu J.Y., Lu B.L. Transfer components between subjects for EEG-based emotion recognition; Proceedings of the 2015 International Conference on Affective Computing and Intelligent Interaction (ACII); Xi’an, China. 21–24 September 2015; pp. 917–922. [Google Scholar]
- 91.Dai M., Zheng D., Liu S., Zhang P. Transfer kernel common spatial patterns for motor imagery brain-computer interface classification. Comput. Math. Meth. Med. 2018;2018:9871603. doi: 10.1155/2018/9871603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zanini P., Congedo M., Jutten C., Said S., Berthoumieu Y. Transfer learning: A Riemannian geometry framework with applications to brain–computer interfaces. IEEE Trans. Biomed. Eng. 2017;65:1107–1116. doi: 10.1109/TBME.2017.2742541. [DOI] [PubMed] [Google Scholar]
- 93.Kalunga E.K., Chevallier S., Barthélemy Q. Transfer learning for SSVEP-based BCI using Riemannian similarities between users; Proceedings of the 2018 26th European Signal Processing Conference (EUSIPCO); Rome, Italy. 3–7 September 2018; pp. 1685–1689. [Google Scholar]
- 94.Jayaram V., Alamgir M., Altun Y., Scholkopf B., Grosse-Wentrup M. Transfer learning in brain-computer interfaces. IEEE Comput. Intell. Mag. 2016;11:20–31. doi: 10.1109/MCI.2015.2501545. [DOI] [Google Scholar]
- 95.García-Salinas J.S., Villaseñor-Pineda L., Reyes-García C.A., Torres-García A.A. Transfer learning in imagined speech EEG-based BCIs. Biomed. Signal Process. Control. 2019;50:151–157. doi: 10.1016/j.bspc.2019.01.006. [DOI] [Google Scholar]
- 96.Tu W., Sun S. Transferable discriminative dimensionality reduction; Proceedings of the 2011 IEEE 23rd International Conference on Tools with Artificial Intelligence; Boca Raton, FL, USA. 7–9 November 2011; pp. 865–868. [Google Scholar]
- 97.Xu G., Shen X., Chen S., Zong Y., Zhang C., Yue H., Liu M., Chen F., Che W. A Deep Transfer Convolutional Neural Network Framework for EEG Signal Classification. IEEE Access. 2019;7:112767–112776. doi: 10.1109/ACCESS.2019.2930958. [DOI] [Google Scholar]
- 98.Sakhavi S., Guan C. Convolutional neural network-based transfer learning and knowledge distillation using multi-subject data in motor imagery BCI; Proceedings of the 2017 8th International IEEE/EMBS Conference on Neural Engineering (NER); Shanghai, China. 25–28 May 2017; pp. 588–591. [Google Scholar]
- 99.Behncke J., Schirrmeister R.T., Volker M., Hammer J., Marusic P., Schulze-Bonhage A., Burgard W., Ball T. Cross-Paradigm Pretraining of Convolutional Networks Improves Intracranial EEG Decoding; Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Miyazaki, Japan. 7–10 October 2018; pp. 1046–1053. [Google Scholar]
- 100.Tan C., Sun F., Zhang W. Deep Transfer Learning for EEG-Based Brain Computer Interface; Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); Calgary, AB, Canada. 15–20 April 2018; pp. 916–920. [Google Scholar]
- 101.Völker M., Schirrmeister R.T., Fiederer L.D.J., Burgard W., Ball T. Deep transfer learning for error decoding from non-invasive EEG; Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI); GangWon, Korea. 15–17 January 2018; pp. 1–6. [Google Scholar]
- 102.Wu D., Lawhern V.J., Gordon S., Lance B.J., Lin C.-T. Driver Drowsiness Estimation From EEG Signals Using Online Weighted Adaptation Regularization for Regression (OwARR) IEEE Trans. Fuzzy Syst. 2016;25:1522–1535. doi: 10.1109/TFUZZ.2016.2633379. [DOI] [Google Scholar]
- 103.Fahimi F., Zhang Z., Goh W.B., Lee T.S., Ang K.K., Guan C. Inter-subject transfer learning with an end-to-end deep convolutional neural network for EEG-based BCI. J. Neural Eng. 2019;16:026007. doi: 10.1088/1741-2552/aaf3f6. [DOI] [PubMed] [Google Scholar]
- 104.Kobler R.J., Scherer R. Restricted Boltzmann Machines in Sensory Motor Rhythm Brain-Computer Interfacing: A study on inter-subject transfer and co-adaptation; Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC); Budapest, Hungary. 9–12 October 2016; pp. 000469–000474. [Google Scholar]
- 105.Liu Y., Yang C., Li Z. The Application of Transfer Learning in P300 Detection; Proceedings of the 2018 IEEE International Conference on Cyborg and Bionic Systems (CBS); Shenzhen, China. 25–27 October 2018; pp. 412–417. [Google Scholar]
- 106.Parvan M., Ghiasi A.R., Rezaii T.Y., Farzamnia A. Transfer Learning based Motor Imagery Classification using Convolutional Neural Networks; Proceedings of the 2019 27th Iranian Conference on Electrical Engineering (ICEE); Yazd, Iran. 30 April–2 May 2019; pp. 1825–1828. [Google Scholar]
- 107.Özdenizci O., Wang Y., Koike-Akino T., Erdoğmuş D. Learning invariant representations from EEG via adversarial inference. IEEE Access. 2020;8:27074–27085. doi: 10.1109/ACCESS.2020.2971600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tan C., Sun F., Kong T., Fang B., Zhang W. Attention-based Transfer Learning for Brain-computer Interface; Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); Brighton, UK. 12–17 May 2019; pp. 1154–1158. [Google Scholar]
- 109.Albuquerque I., Monteiro J., Rosanne O., Tiwari A., Gagnon J.F., Falk T.H. Cross-Subject Statistical Shift Estimation for Generalized Electroencephalography-based Mental Workload Assessment; Proceedings of the 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC); Bari, Italy. 6–9 October 2019; pp. 3647–3653. [Google Scholar]
- 110.Wu H., Li F., Li Y., Fu B., Shi G., Dong M., Niu Y. A Parallel Multiscale Filter Bank Convolutional Neural Networks for Motor Imagery EEG Classification. Front. Neurosci. 2019;13:1275. doi: 10.3389/fnins.2019.01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uran A., van Gemeren C., van Diepen R., Chavarriaga R., Millán J.D. Applying Transfer Learning To Deep Learned Models for EEG Analysis. arXiv. 20191907.01332 [Google Scholar]
- 112.Craik A., Kilicarslan A., Contreras-Vidal J.L. Classification and Transfer Learning of EEG during a Kinesthetic Motor Imagery Task using Deep Convolutional Neural Networks; Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Berlin, Germany. 23–27 July 2019; pp. 3046–3049. [DOI] [PubMed] [Google Scholar]
- 113.Onishi A., Nakagawa S. Comparison of Classifiers for the Transfer Learning of Affective Auditory P300-Based BCIs; Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Berlin, Germany. 23–27 July 2019; pp. 6766–6769. [DOI] [PubMed] [Google Scholar]
- 114.Lin Y.P. Constructing a Personalized Cross-day EEG-based Emotion-Classification Model Using Transfer Learning. IEEE J. Biomed. Health Inf. 2019;24:1255–1264. doi: 10.1109/JBHI.2019.2934172. [DOI] [PubMed] [Google Scholar]
- 115.Daoud H., Bayoumi M.A. Efficient epileptic seizure prediction based on deep learning. IEEE Trans. Biomed. Circuits Syst. 2019;13:804–813. doi: 10.1109/TBCAS.2019.2929053. [DOI] [PubMed] [Google Scholar]
- 116.Wang Y., Cao J., Wang J., Hu D., Deng M. Epileptic Signal Classification with Deep Transfer Learning Feature on Mean Amplitude Spectrum; Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Berlin, Germany. 23–27 July 2019; pp. 2392–2395. [DOI] [PubMed] [Google Scholar]
- 117.Fauzi H., Shapiai M.I., Khairuddin U. Transfer Learning of BCI Using CUR Algorithm. J. Signal Process. Syst. 2020;92:109–121. doi: 10.1007/s11265-019-1440-9. [DOI] [Google Scholar]
- 118.Özdenizci O., Wang Y., Koike-Akino T., Erdoğmuş D. Transfer learning in brain-computer interfaces with adversarial variational autoencoders; Proceedings of the 2019 9th International IEEE/EMBS Conference on Neural Engineering (NER); San Francisco, CA, USA. 20–23 March 2019; pp. 207–210. [Google Scholar]
- 119.Arevalillo-Herráez M., Cobos M., Roger S., García-Pineda M. Combining Inter-Subject Modeling with a Subject-Based Data Transformation to Improve Affect Recognition from EEG Signals. Sensors. 2019;19:2999. doi: 10.3390/s19132999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang S., Yin Z., Wang Y., Zhang W., Wang Y., Zhang J. Assessing cognitive mental workload via EEG signals and an ensemble deep learning classifier based on denoising autoencoders. Comput. Boil. Med. 2019;109:159–170. doi: 10.1016/j.compbiomed.2019.04.034. [DOI] [PubMed] [Google Scholar]
- 121.Zou Y., Zhao X., Chu Y., Zhao Y., Xu W., Han J. An inter-subject model to reduce the calibration time for motion imagination-based brain-computer interface. Med. Boil. Eng. Comput. 2019;57:939–952. doi: 10.1007/s11517-018-1917-x. [DOI] [PubMed] [Google Scholar]
- 122.Hang W., Feng W., Du R., Liang S., Chen Y., Wang Q., Liu X. Cross-Subject EEG Signal Recognition Using Deep Domain Adaptation Network. IEEE Access. 2019;7:128273–128282. doi: 10.1109/ACCESS.2019.2939288. [DOI] [Google Scholar]
- 123.Congedo M., Sherlin L. EEG Source Analysis. Elsevier BV; Amsterdam, The Netherlands: 2011. pp. 25–433. [Google Scholar]
- 124.Alamgir M., Grosse-Wentrup M., Altun Y. Multitask learning for brain–computer interfaces; Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics; Sardinia, Italy. 13–15 May 2010; pp. 17–24. [Google Scholar]
- 125.Kang H., Choi S. Bayesian common spatial patterns for multi-subject EEG classification. Neural Netw. 2014;57:39–50. doi: 10.1016/j.neunet.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 126.Craik A., He Y., Contreras-Vidal J.L. Deep Learning for Electroencephalogram (EEG) Classification Tasks: A Review. J. Neural Eng. 2019;16:031001. doi: 10.1088/1741-2552/ab0ab5. [DOI] [PubMed] [Google Scholar]