Abstract
Mechanisms for somatic chromosomal mosaicism (SCM) and chromosomal instability (CIN) are not completely understood. During molecular karyotyping and bioinformatic analyses of children with neurodevelopmental disorders and congenital malformations (n = 612), we observed colocalization of regular chromosomal imbalances or copy number variations (CNV) with mosaic ones (n = 47 or 7.7%). Analyzing molecular karyotyping data and pathways affected by CNV burdens, we proposed a mechanism for SCM/CIN, which had been designated as “chromohelkosis” (from the Greek words chromosome ulceration/open wound). Briefly, structural chromosomal imbalances are likely to cause local instability (“wreckage”) at the breakpoints, which results either in partial/whole chromosome loss (e.g., aneuploidy) or elongation of duplicated regions. Accordingly, a function for classical/alpha satellite DNA (protection from the wreckage towards the centromere) has been hypothesized. Since SCM and CIN are ubiquitously involved in development, homeostasis and disease (e.g., prenatal development, cancer, brain diseases, aging), we have metaphorically (ironically) designate the system explaining chromohelkosis contribution to SCM/CIN as the cytogenomic “theory of everything”, similar to the homonymous theory in physics inasmuch as it might explain numerous phenomena in chromosome biology. Recognizing possible empirical and theoretical weaknesses of this “theory”, we nevertheless believe that studies of chromohelkosis-like processes are required to understand structural variability and flexibility of the genome.
Keywords: chromosome, copy number variations, chromosome instability, chromosomal mosaicism, chromosomal imbalances, aneuploidy, disease, aging, pathways, cytogenomics
1. Introduction
Despite the fact that mechanisms of chromosomal instability (CIN) and somatic chromosomal mosaicism (SCM) remain to be further explored [1,2,3], CIN-associated genome behavior (chromothripsis, chromoanasynthesis, chromoanagenesis) has already been described [4,5,6]. Uncovering underlying processes of the commonest types of SCM/CIN (e.g., aneuploidy) is generally less successful [7,8,9]. Still, somatic aneuploidy (CIN manifested as aneuploidy) has been found to result from alterations to a number of molecular/cellular pathways, gene mutations [10,11], and/or genetic–environmental interactions [12]. In the postgenomic context, it appears important to evaluate the contribution of heritable/sporadic cytogenomic variations (i.e., genomic variations at chromosomal and subchromosomal levels) to the formation of CIN and SCM using whole-genome analyses and systems biology approaches. Currently, studies of pathways from regular chromosomal imbalances and/or copy number variations (CNV) to CIN/SCM are rare.
Genome architecture at the DNA sequence level has long been recognized as playing a key role in formation of structural genome variations [13,14]. A large series of studies using a panel of molecular, cytogenomic, and bioinformatic techniques have shown that structural genomic variants (chromosomal rearrangements and CNV) frequently occur through mechanisms involving repeat sequences at the breakpoints as well as DNA recombination-based and replication-based processes [15,16,17,18,19]. At the chromosomal/subchromosomal levels, related phenomena have not been systematically addressed. However, chromosome segregation errors have been indicated to form a wide spectrum of somatic genome rearrangements [20]. These results promise the success of forthcoming studies of interplays between genome behavior at chromosomal (subchromosomal) level, CIN and SCM. Therefore, whole-genome and bioinformatic analyses of co-occurring non-mosaic and mosaic chromosomal (subchromosomal) variations or CIN in an individual may help to uncover previously unrecognized mechanisms for somatic genome variations at the chromosomal level.
In the present contribution, we describe an SNP (single nucleotide polymorphism)-array study of colocalized regular/mosaic chromosome imbalances in a cohort of children with neurodevelopmental disorders and congenital malformations. Analysis of the structural genomic variations allowed us to propose a previously unknown pathway from regular chromosomal imbalances or CNV to SCM (CIN). Furthermore, we have introduced a kind of theory, which might be relevant to numerous areas of chromosome research, cytogenomics (molecular cytogenetics), and medical cytogenetics.
2. Results and Discussion
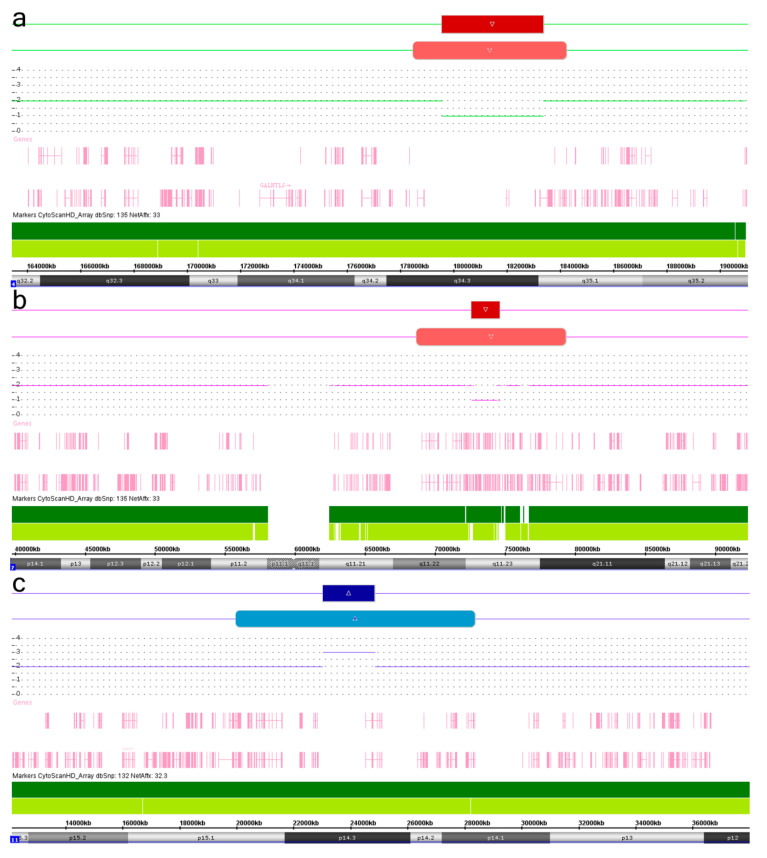
Molecular karyotyping of the Russian cohort of children with neurodevelopmental disorders and congenital malformations [21,22], which currently includes 612 individuals, has been performed by high resolution SNP-array using Affymetrix CytoScan platform (HD). During the analysis, we observed concomitant regular and mosaic chromosome imbalances located at the same chromosomal locus or encompassed similar genomic regions in 47 individuals (7.7%). These colocalized structural chromosome abnormalities manifested as deletions or duplications with mosaic imbalances presenting mainly larger than regular ones. Accordingly, we have suggested that the colocalizations are not coincidental, representing, therefore, a snapshot of a possible dynamic process, which starts as a regular chromosomal imbalance (CNV) and proceeds with the formation of mosaic imbalances by a kind of “wreckage” at both breakpoints of the regular one. Hence, regular structural genomic variants are likely to be the first step initiating genomic instability (GIN) at adjacent chromosomal loci (genomic regions). The instability is likely to produce a larger rearrangement through defective DNA damage response and/or reparation (duplication—elongation by erroneous reparation; deletion—loss of chromosome parts). To define these colocalized chromosome abnormalities in an individual and to name the process producing SMC and CIN/GIN from regular genomic/chromosomal changes (CNV) by a single term, we have introduced the neologism “chromohelkosis”, which literally means “chromosome ulceration or ulcer” (from the Greek words “chromo” designating “chromosome” and “helkosis” derived from helkos (ἕλκος), which means “ulceration”, “ulcer”, or “open wound”). Consequently, the process is designated as chromohelkosis, whereas colocalized regular and mosaic chromosomal changes are designated as chromohelkosis imbalances. We intentionally composed the word “chromohelkosis” to mimic chromothripsis (chromosome shattering characterized by extensive rearrangements and an oscillating pattern of DNA copy numbers), chromoanasynthesis (local chromosome shattering associated with a random restitching of chromosomal fragments), and chromoanagenesis (complex rearrangements at one/several chromosomal loci produced by a catastrophic event) [4,5,6], inasmuch as it also seems to be a common mechanism for somatic chromosome rearrangements and CIN. Figure 1 shows three examples of chromosomal loci affected by chromohelkosis.
Figure 1.
SNP-array analysis of colocalized regular/mosaic chromosome imbalances in an individual (chromosomal loci suggested to be affected by chromohelkosis or chromohelkosis imbalances): (A) regular/mosaic deletions at 4q34.3q35.1; (B) regular/mosaic deletions at 7q11.23/7q11.22q21.11; (C) regular/mosaic duplications at 11p14.3. Dark red—regular deletions; light red—mosaic deletions; dark blue—regular duplication; light blue—mosaic duplication.
Chromohelkosis imbalances have been found to affect loci of almost all chromosomes apart from chromosomes 6, 8, 18–21, and Y. There have been detected 26 duplications and 21 deletions (55/45). Sizes of regular chromosome imbalances have been found to be highly variable from small CNV (tens to hundreds of kbp) to partial trisomies and monosomies (large chromosome deletions/duplications, e.g., >20 Mb). Mosaic chromosome imbalances have been generally more than 2 Mb, except one case (0.87 Mb). Details on the chromohelkosis imbalances are given in Table 1.
Table 1.
Colocalized regular/mosaic chromosome imbalances and copy number variations (CNV) marking chromosomal loci affected by chromohelkosis (chromohelkosis imbalances).
| Chromosomal Localization | Deletion (del)/Duplication (dup) | Genomic Localization 1 | Size (Mb), Regular | Size (Mb), Mosaic | Mosaicism Rate (%) 2 |
||
|---|---|---|---|---|---|---|---|
| Regular | Mosaic | Regular | Mosaic | ||||
| 1p36.32 | 1p36.33p36.32 | Dup | 2,793,846–3,123,524 | 849,466–3,586,707 | 0.329 | 2.737 | 20 |
| 1p36.33 | 1p36.33p36.32 | Dup | 1,134,091–1,207,463 | 849,466–5,278,786 | 0.073 | 4.429 | 25 |
| 1q21.1q21.2 | 1p12q21.2 | Dup | 146,003,044–147,398,560 | 118,506,747–149,965,913 | 1.395 | 31.459 | 20 |
| 2p25.3 | 2p25.3 | Dup | 1,611,691–1,861,548 | 12,770–3,007,511 | 0.249 | 2.994 | 25 |
| 2q22.2 q22.3 | 2q22.1q23.3 | Del | 143,410,303–145,299,945 | 140,410,739–150,635,360 | 1.889 | 10.224 | 50 |
| 2q23.1q23.3 | 2q22.2q24.1 | Del | 148,851,963–151,316,465 | 143,753,727–155,408,790 | 2.464 | 11.655 | 40 |
| 2q23.1q23.3 | 2q22.2q24.1 | Del | 149,073,384–151,886,100 | 144,007,224–156,393,001 | 2.812 | 12.385 | 45 |
| 2q24.1 | 2q23.3q24.2 | Del | 155,684,576–157,919,431 | 151,497,654–162,200,234 | 2.234 | 10.702 | 35 |
| 3p26.1 | 3p26.3p26.1 | Dup | 4,311,166–7,256,278 | 2,788,170–8,587,443 | 2.945 | 5.799 | 40 |
| 3p26.3p26.2 | 3p26.3p26.1 | Dup | 1,839,722–3,372,758 | 61,891–4,693,249 | 1.533 | 4.631 | 60 |
| 3q26.2q26.31 | 3q26.1q26.31 | Del | 170,316,791–171,650,195 | 165,957,466–175,300,706 | 1.333 | 9.343 | 30 |
| 4q34.3q35.1 | 4q34.3q35.1 | Del | 179,568,373–183,377,810 | 178,503,425–184,251,370 | 3.809 | 5.747 | 55 |
| 5q35.2q35.3 | 5q35.1q35.3 | Del | 175,029,372–177,324,736 | 171,538,904–180,719,789 | 3.395 | 9.180 | 30 |
| 7p22.1p15.2 | 7p22.1p15.2 | Dup | 4,783,314–26,275,210 | 4,790,968–26,522,153 | 21.491 | 21.731 | 80 |
| 7p22.2p21.3 | 7p22.3p21.3 | Del | 3,235,409–7,970,015 | 43,360–8,320,635 | 4.734 | 8.277 | 20 |
| 7q11.23 | 7q11.22q21.11 | Del | 72,612,042–74,610,673 | 68,665,592–79,305,748 | 1.998 | 10.640 | 40 |
| 9p24.3 | 9p24.3p24.2 | Dup | 203,861–823,845 | 203,861–2,593,900 | 0.619 | 2.390 | 30 |
| 9p24.3 | 9p24.3 | Del | 203,861–410,357 | 203,861–1,074,830 | 0.206 | 0.870 | 25 |
| 9q22.31q22.33 | 9q22.31q22.33 | Del | 96,109,697–99,973,789 | 95,891,880–100,145,863 | 3.864 | 4.253 | 70 |
| 9q34.3 | 9q34.13q34.3 | Dup | 139,053,501–139,435,356 | 134,317,328–141,020,389 | 0.381 | 6.703 | 30 |
| 9q34.3 | 9q34.13q34.3 | Del | 139,784,913–141,020,389 | 135,282,452–141,020,389 | 1.235 | 5.737 | 40 |
| 10q21.1 | 10q11.23q21.1 | Dup | 53,156,807–57,931,080 | 52,693,425–58,936,553 | 4.774 | 6.243 | 75 |
| 10q23.1q23.2 | 10q23.1q23.2 | Del | 86,412,180–88,502,670 | 85,638,142–89,465,109 | 2.090 | 3.826 | 50 |
| 11p14.3 | 11p14.3 | Dup | 23,032,300–24,850,872 | 19,983,179–28,380,051 | 1.819 | 8.396 | 25 |
| 12q24.33 | 12q24.33 | Dup | 129,804,153–130,492,863 | 129,577,575–133,777,902 | 0.688 | 4.200 | 20 |
| 12q24.33 | 12q24.33 | Dup | 129,803,493–130,485,474 | 130,035,491–133,777,902 | 0.681 | 3.742 | 20 |
| 13q12.11 | 13q11q12.11 | Dup | 21,683,950–22,155,929 | 19,436,287–22,422,460 | 0.471 | 2.986 | 20 |
| 13q34 | 13q34 | Del | 114,085,478–115,107,733 | 110,963,086–115,107,733 | 1.022 | 4.144 | 40 |
| 14q32.2 | 14q32.13q32.2 | Dup | 99,153,952–101,024,454 | 95,563,168–100,095,249 | 1.870 | 4.532 | 20 |
| 15q11.2 | 15q11.2 | Dup | 22,770,421–23,082,328 | 22,770,421–25,083,880 | 0.311 | 2.313 | 20 |
| 15q11.2 | 15q11.2 | Dup | 22,770,421–23,288,350 | 22,770,421–25,318,376 | 0.517 | 2.547 | 25 |
| 15q11.2q13.1 | 15q11.2q13.1 | Del | 22,770,421–29,021,034 | 22,770,421–28,373,187 | 5.732 | 5.602 | 85 |
| 15q13.2q13.3 | 15q13.1q14 | Del | 30,913,573–32,914,239 | 28,394,840–36,544,674 | 2.518 | 8.149 | 25 |
| 16p11.2q11.2 | 16p11.2q12.1 | Dup | 32,038,693–46,463,769 | 34,448,198–51,124,520 | 14.425 | 16.676 | 20 |
| 16q23.1 | 16q22.3q23.3 | Dup | 77,496,014–78,916,839 | 73,357,720–82,335,001 | 1.420 | 8.977 | 30 |
| 16q24.3 | 16q24.2q24.3 | Del | 89,683,742–90,155,062 | 87,157,300–90,155,062 | 0.471 | 2.997 | 25 |
| 17p12 | 17p13.1p11.2 | Dup | 14,082,944–15,479,940 | 10,219,298–17,108,606 | 1.396 | 6.889 | 20 |
| 17p13.3 | 17p13.3p13.2 | Del | 525–1,323,904 | 525–4,375,742 | 1.323 | 4.375 | 40 |
| 17q25.3 | 17q25.3 | Del | 80,396,463–81,041,938 | 77,947,778–81,041,938 | 0.645 | 3.094 | 30 |
| 22q11.21 | 22q11.1q11.22 | Dup | 18,974,541–21,800,797 | 17,398,811–23,374,206 | 2.826 | 5.975 | 45 |
| 22q11.21 | 22q11.1q11.21 | Dup | 18,649,189–20,311,810 | 16,888,899–22,034,665 | 1.662 | 5.145 | 40 |
| 22q11.21 | 22q11.1q11.22 | Dup | 18,979,345–21,465,659 | 16,888,899–23,410,418 | 2.486 | 6.521 | 50 |
| 22q11.21 | 22q11.1q11.23 | Dup | 18,916,842–21,465,659 | 16,888,899–23,535,339 | 2.548 | 6.646 | 50 |
| 22q13.2q13.31 | 22q13.2q13.31 | Dup | 43,337,317–46,575,998 | 42,018,242–44,860,024 | 3.238 | 2.841 | 50 |
| Xp22.31 | Xp22.32p22.2 | Del | 6,784,550–7,495,395 | 4,931,788–9,634,138 | 0.710 | 4.702 | 25 |
| Xp21.1 | Xp21.1p11.4 | Del | 32,881,263–35,187,430 | 31,875,672–38,716,579 | 2.306 | 6.840 | 50 |
| Xq28 | Xq28 | Dup | 153,747,685–153,761,134 | 147,843,549–152,036,631 | 0.013 | 4.193 | 20 |
1 GRCh37/hg19. 2 mosaicism rates according to Chromosome Analysis Suite (ChAS) software (Affymetrix).
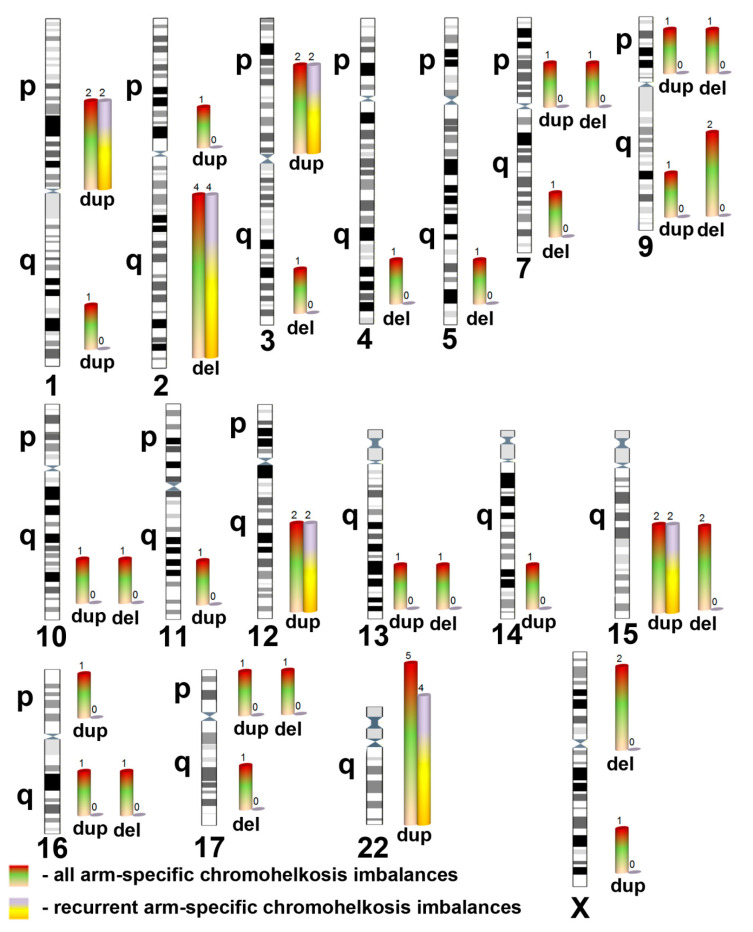
Five chromohelkosis imbalances were recurrent, i.e., non-random variations at same genomic regions (Figure 2). This observation allowed us to conclude that specific genome architecture is likely to be involved in chromohelkosis as in germline and somatic variations mediating genomic disorders [8,13,14,15,16]. Additionally, reciprocal chromohelkosis imbalances (i.e., chromohelkosis deletions and duplications at the same chromosomal locus/genomic region) at 9p24.3, 9q34.13q34.3, and 15q11.2 have been revealed (Table 1). This observation additionally suggests that specificity of local genomic architecture makes the loci susceptible to genomic/chromosomal rearrangements) [13,14]. However, to provide further evidence and a molecular/cellular basis for chromohelkosis, additional analyses of molecular karyotyping data appeared to be required.
Figure 2.
Spectrum of chromohelkosis imbalances classified according to chromosome arms and recurrence at the same chromosomal locus, encompassing the same genomic region (del—deletion; dup—duplication). Two differently colored bars show number of chromohelkosis imbalances per chromosome. The scale of bar length is the same for all chromosomes, whereas the scale of chromosomes is conserved.
To characterize the spectrum of chromohelkosis imbalances, we used the ratio between sizes of regular and mosaic chromosome abnormalities. According to this parameter, cases were divided in quartiles (Q1—the smallest ratio/the longest distance between breakpoints of regular and mosaic imbalances; Q4—the largest ratio/the shortest distance between breakpoints of regular and mosaic imbalances). The first quartile (Q1) comprised 27 (57%) chromohelkosis imbalances (1p (n = 2), 1q, 2p, 2q (n = 4), 3q(del), 7q, reciprocal 9p(del), and 9p(dup), reciprocal 9q(del) and 9q(dup), 11p, 12q (n = 2), 13q, 15q(dup) (n = 2), 16q(del), 16q(del), 17p(dup), 17q, Xp, Xq), whereas the remaining quartiles (Q2–Q4) comprised 20 chromohelkosis imbalances (3p (n = 2), 4q, 5q, reciprocal 7p(del), and 7p(dup), 9q(del), 10q (n = 2), 14q, 15q(del) (n = 2), 16p, 22q (n = 5), Xp). The distribution of chromohelkosis imbalances demonstrates a large segregation towards abnormalities with dramatically increased sizes of mosaic ones. Q1-imbalances are, therefore, more likely to be associated with a higher rate of local GIN both at the breakpoints and at the affected chromosomal loci. Correlations between the ratio and patient age, chromosomal localization, or imbalance specificity (deletion/duplication) have not been found. Thus, individual susceptibility to chromohelkosis has been proposed as the mechanism of the occurrence and distribution in favor of Q1. As described previously, such a kind of susceptibility may be a result of alterations to molecular and cellular pathways safeguarding genome stability [12,23,24]. Recently, a model for pathway-based classification of genomic burden (CNV burden) resulting in CIN/GIN and SCM was proposed [25]. Using this model and a novel bioinformatic method for pathway-based CNV/gene prioritization [26], we evaluated CNV burden effect on pathways required for genome stability maintenance (e.g., DNA reparation and damage response, programmed cell death, cell cycle regulation, cancer-related pathways, etc.). Accordingly, we retrieved ontologies of genes affected by regular CNV and chromosome imbalances in the genome stability context and identified Ipp (index of pathway prioritization): Ipp(Q1) = 2.4 and 46 pathways; Ipp(Q2–Q4) = 1.7 and 27 pathways (for more details, see [26]). We observed a statistically significant difference between Q1 and Q2–Q4 in enrichment of genome stability maintenance pathways (Z-test, p < 0.001). As a result, we concluded that non-random distribution of chromohelkosis imbalances is likely to be caused by individual susceptibility to GIN/CIN produced by alterations to pathways safeguarding genome stability. In other words, a kind of saturation in CNV encompassing genes involved in these pathways may result in chromohelkosis. Moreover, a correlation between a higher CNV burden and a measure of chromohelkosis progression appears to exist. It is highly likely that the measure of chromohelkosis progression may be connected to repeat DNA, which is involved in genome organization and stability [27] (discussed hereafter).
The appreciable difference between the regular and mosaic (colocalized) rearrangements is reflected in gene content of a chromohelkosis imbalance (Supplementary Table S1). For instance, three individuals exhibited chromohelkosis imbalances characterized by the lack of functionally annotated genes in regions affected by regular rearrangements (chromohelkosis imbalance at 11p and twice at 2q). In addition, numerous cases demonstrated phenotypic outcomes associated with mosaic rearrangements but not with regular ones (e.g., rearrangements at 1p, 9q, 15q, 16q, 17p, and 22q). However, it is noteworthy that these phenotypic manifestations are milder than those observed in recognizable chromosomal (microdeletion or microduplication) syndromes. These observations might be used to explain extensive variability of SCM phenotypes, as suggested previously [8,28,29,30]. Here, it is recognized that chromohelkosis may have a diagnostic relevance comparable to chromothripsis, chromoanasynthesis, and chromoanagenesis.
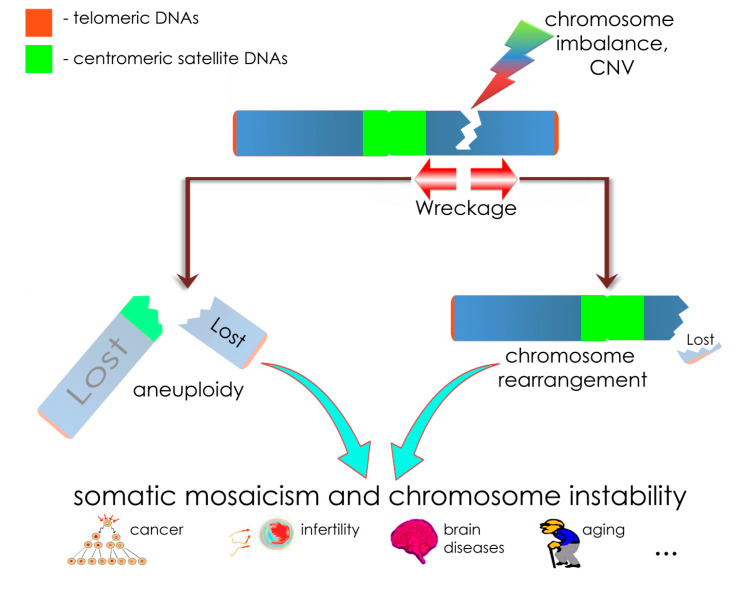
Among others, chromohelkosis may represent a model of somatic genome variability (i.e., SMC, GIN, and CIN). We have suggested that chromohelkosis deletions produce local instability at the breakpoints leading to progressive loss of adjacent chromosomal regions. As a result, mosaic deletions are larger than regular ones. This mechanism is similar to some extent to those proposed for explaining pathways from chromosome fragile sites to somatic chromosomal aberrations and CIN to some extent [31,32]. Moreover, fragile sites affect replication timing producing DNA flexibility peaks and stress inducible asynchrony at the breakage to produce GIN or CIN [31]. This process mimics “wreckages” at the breakpoints of chromohelkosis imbalances. However, chromohelkosis is able to lead to the loss of a larger chromosomal region due to the dynamic nature. Chromohelkosis affecting the centromere would result in a loss of the whole chromosome (mosaic aneuploidy/monosomy). Alterations to centromere stability (scission) have been recently shown to be a mechanism for GIN/CIN [33]. It is also known that CIN and GIN may be mediated by pathways involving centromeric DNA (e.g., classical/alpha satellite DNA) [3,20,33,34]. Although functional significance of centromeric satellite DNA (constitutive heterochromatin) remains a matter of further investigation, numerous studies indicate the involvement in granting genome/chromosome stability [35,36,37]. Satellite DNA is suggested either to mediate GIN or to protect from alterations to chromosome structure [27]. Here, we propose a role of centromeric satellite DNA similar to telomeric DNA [38], which seems to play a role in CIN caused by chromohelkosis, as well. Thus, centromeric satellite DNA protects centromere from chromohelkosis “wreckage”. Similarly, telomeric DNA is proposed to play the canonical role of protecting chromosomal ends from chromohelkosis “wreckage”. The failure of centromeric satellite DNA to protect centromere from the “wreckage” would result in chromosomal loss (aneuploidy). The failure of telomeric DNA to protect chromosomal ends from “wreckage” would result in chromosomal rearrangements. Figure 3 schematically shows suggested mechanisms of chromohelkosis mediated by deletions.
Figure 3.
Schematic depiction of chromohelkosis due to a deletion and suggested outcomes relevant to the cytogenomic “theory of everything” (discussed hereafter). Chromosomal imbalances or CNV manifesting as deletions are able to cause instability at the sequence level (GIN) at the breakpoints. Consequently, wreckage may occur through altered DNA damage response and reparation, which cause progressive loss of chromosomal DNA localized at the breakpoints. According to the theory, (centromeric) satellite DNA protects chromosomes from propagation of chromohelkosis in the same way as telomeres do. However, the protection may fail to initiate two scenarios: (i) the distal chromosome part is wrecked and lost, when telomeres are affected; as a result, somatic chromosome rearrangements are formed (on the right-hand side); (ii) the proximal chromosome part is wrecked and, if centromeric satellite DNA fails to protect the centromere, the whole chromosome is lost (i.e., aneuploidy/monosomy) (on the left-hand side). This cascade of CIN/GIN processes results in somatic mosaicism and CIN, which are mechanisms for cancer, infertility, brain diseases, aging, and, probably, other morbid conditions. To depict a biological basis of chromohelkosis, we have used parts of figures from our previous articles distributed under the terms of the Creative Commons Attribution License [28,39].
The elongation of duplicated regions might be mediated by fails of chromosomal DNA reparation at the breakpoints progressively occurring during each fail. Related mechanisms for CIN have been already described in cancers [20,23]. These duplications may also involve repeat (satellite) DNA as a driving force for chromohelkosis progression [27]. Probably, the consequences of chromohelkosis mediated by deletions may be used to explain a slight preponderance of duplications over deletions. More precisely, mosaic deletion is more likely to disappear, becoming aneuploidy (chromosome loss), whereas mosaic duplication is likely to become a larger one.
SMC and CIN are important contributors to development, homeostasis, interindividual diversity, and disease. To be more exact, SMC and CIN are integrated parts of human prenatal development [40,41,42], aging [43,44], cancer [24,45,46], interindividual/intercellular genome diversification, and disease [40,47,48,49,50,51,52]. SMC and CIN are considered a major focus of basic and diagnostic research for providing therapeutic opportunities in disease and aging [29,52,53,54]. Therefore, it is hard to overestimate the role of SMC and CIN. Our current observation hallmarks a common mechanism of somatic genome variability produced by chromosomal imbalances and CNV. Taking into account a kind of omnipresence of chromosomal variations and CNV [7,28,47,53], the chromohelkosis-based pathway to SMC/CIN might underlie a cytogenomic theory, which would be relevant to numerous areas of genetics, genomics, and chromosome research. In our opinion, such a theory resembles the elusive theory of everything described by Stephen Hawking and Leonard Mlodinow [55], because both create a temptation to explain almost everything in a given research area. To decrease the temptation, we used metaphoric (ironic) designation “the cytogenomic theory of everything” for the system explaining chromohelkosis contribution to SCM/CIN. Finally, recognizing empirical and theoretical weaknesses of our contribution, we nevertheless insist that chromohelkosis is a process to explore for understanding structural variability and flexibility of the genome.
3. Materials and Methods
3.1. Patients and Samples
Molecular karyotyping was performed to identify chromosomal abnormalities and CNV in the Russian cohort of children with neurodevelopmental disorders (intellectual disability, autism, epilepsy) and congenital anomalies (612 individuals), which has been clinically described in previous studies [21,22]. This study was approved by the Ethics Committee of the Veltischev Research and Clinical Institute for Pediatrics of the Pirogov Russian National Research Medical University, Ministry of Health of Russian Federation, Moscow, Russia (AAAA—A18—118051590122—7 # 6, 19 June 2019). Written informed consent was obtained from the patients’ parents.
3.2. SNP-Array
Molecular karyotyping was performed by CytoScan HD Arrays (Affymetrix, Santa Clara, CA, USA) consisting of about 2.7 million markers. All the procedures have been repeatedly described in detail previously [21,22,56,57]. Cytogenomic variations were visualized using the Affymetrix ChAS (Chromosome Analysis Suite) software (CytoScan® HD Array Version 4.1.0.90/r29400). The reference sequence was GRCh37/hg19.
3.3. Bioinformatic Analysis
An original approach to prioritization of CNV, candidate genes, and processes using molecular karyotyping data was carried out as described earlier [58]. The procedure is an ontology-based gene filtering/ranking with fusion of data acquired from databases dedicated to clinical genetics, genomics, epigenetics (gene expression), proteomics (interactome), and metabolome. Additionally, genomic data were analyzed by obtaining the ratio between the size of regular and mosaic imbalances. Using this ratio, imbalances were divided in quartiles. The data have been also used for pathway analysis.
Pathway Analysis
Pathway analysis was performed using CNVariome concept and data laundering protocol, which were recently described in detail [25,26]. The enrichment by genome stability maintenance pathways was additionally analyzed using statistical Z-test.
4. Conclusions
In summary, we observed co-occurrence of regular and mosaic imbalances (deletions/duplications) at same chromosomal loci during molecular karyotyping. Data analysis allowed us to propose a mechanism for SMC and CIN — chromohelkosis (chromosome ulceration/open wound). Further exploration of this mechanism has led to suggesting a function for classical/alpha satellite DNA (protection from the wreckage towards the centromere) and cytogenomic “theory of everything”, a metaphorically termed system explaining chromohelkosis contribution to SCM and CIN. Chromohelkosis is proposed to be the result of a cumulative effect of chromosomal imbalance/CNV (or, probably, other alterations to chromosomal structure) and mutational/CNV burden, which alters pathways of genome stability maintenance. These combinations of genomic variations would be critical for chromohelkosis dynamics and the levels of SCM and CIN. Thus, forthcoming studies are to uncover the spectrum of genomic variations leading to chromohelkosis to show whether this process is restricted to specific chromosomal imbalances or CNV.
Acknowledgments
The authors thank Irina A Demidova, Alexei D Kolotii and Victor S Kravets.
Abbreviations
| ChAS | Chromosome Analysis Suite |
| CIN | Chromosomal instability |
| CNV | Copy number variations |
| Del | deletion (tables & figures) |
| dup | duplication (tables & figures) |
| GIN | Genome instability |
| IPP | Index of pathway prioritization |
| SCM | Somatic chromosomal mosaicism |
| SNP | Single nucleotide polymorphism |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/21/8328/s1.
Author Contributions
Conceptualization, I.Y.I., S.G.V. and Y.B.Y.; methodology, I.Y.I., S.G.V., Y.B.Y. and M.A.Z.; formal analysis, I.Y.I., S.G.V. and Y.B.Y.; investigation, I.Y.I., S.G.V., Y.B.Y., M.A.Z., O.S.K. and K.S.V.; data curation, I.Y.I.; writing—original draft preparation, I.Y.I.; writing—review and editing, I.Y.I., S.G.V. and M.A.Z.; visualization, I.Y.I., S.G.V., Y.B.Y., M.A.Z., O.S.K. and K.S.V.; project administration, I.Y.I., S.G.V. and S.I.K.; funding acquisition, I.Y.I., S.G.V. and S.I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially supported by RFBR and CITMA according to the research project №18-515-34005; prof. IY Iourov’s lab is supported by the Government Assignment of the Russian Ministry of Science and Higher Education, Assignment no. AAAA-A19–119040490101-6; prof. SG Vorsanova’s lab is supported by the Government Assignment of the Russian Ministry of Health, Assignment no. AAAA-A18–118051590122-7.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thompson S.L., Bakhoum S.F., Compton D.A. Mechanisms of Chromosomal Instability. Curr. Biol. 2010;20:285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heng H.H.Q., Bremer S.W., Stevens J.B., Horne S.D., Liu G., Abdallah B.Y., Ye K.J., Ye C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013;32:325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 3.Iourov I.Y., Vorsanova S.G., Yurov Y.B., Kutsev S.I. Ontogenetic and Pathogenetic Views on Somatic Chromosomal Mosaicism. Genes. 2019;10:379. doi: 10.3390/genes10050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leibowitz M.L., Zhang C.-Z., Pellman D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu. Rev. Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 5.Ly P., Cleveland D.W. Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends Cell Biol. 2017;27:917–930. doi: 10.1016/j.tcb.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellestor F. Chromoanagenesis: Cataclysms behind complex chromosomal rearrangements. Mol. Cytogenet. 2019;12:1–12. doi: 10.1186/s13039-019-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iourov I., Vorsanova S.G., Yurov Y.B. Somatic Genome Variations in Health and Disease. Curr. Genom. 2010;11:387–396. doi: 10.2174/138920210793176065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell I.M., Shaw C.A., Stankiewicz P., Lupski J.R. Somatic mosaicism: Implications for disease and transmission genetics. Trends Genet. 2015;31:382–392. doi: 10.1016/j.tig.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye C.J., Sharpe Z., Heng H.H. Origins and Consequences of Chromosomal Instability: From Cellular Adaptation to Genome Chaos-Mediated System Survival. Genes. 2020;11:1162. doi: 10.3390/genes11101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 11.Taylor A.M., Shih J., Ha G., Gao G.F., Zhang X., Berger A.C., Schumacher S.E., Wang C., Hu H., Liu J., et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33:676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iourov I.Y., Vorsanova S.G., Yurov Y.B. Somatic Cell Genomics of Brain Disorders: A New Opportunity to Clarify Genetic-Environmental Interactions. Cytogenet. Genome Res. 2013;139:181–188. doi: 10.1159/000347053. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho C.M.B., Lupski J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: The long and the short of it. Genome Biol. 2019;20:1–14. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukami M., Shima H., Suzuki E., Ogata T., Matsubara K., Kamimaki T. Catastrophic cellular events leading to complex chromosomal rearrangements in the germline. Clin. Genet. 2017;91:653–660. doi: 10.1111/cge.12928. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Wang J., Zhang C., Li D., Carvalho C.M., Ji H., Xiao J., Wu Y., Zhou W., Wang H., et al. Efficient CNV breakpoint analysis reveals unexpected structural complexity and correlation of dosage-sensitive genes with clinical severity in genomic disorders. Hum. Mol. Genet. 2017;26:1927–1941. doi: 10.1093/hmg/ddx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balachandran P., Beck C.R. Structural variant identification and characterization. Chromosom. Res. 2020;28:31–47. doi: 10.1007/s10577-019-09623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Q., Maurais E.G., Ly P. Cellular and genomic approaches for exploring structural chromosomal rearrangements. Chromosom. Res. 2020;28:19–30. doi: 10.1007/s10577-020-09626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umbreit N.T., Zhang C.-Z., Lynch L.D., Blaine L.J., Cheng A.M., Tourdot R., Sun L., Almubarak H.F., Judge K., Mitchell T.J., et al. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368:0712. doi: 10.1126/science.aba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ly P., Brunner S.F., Shoshani O., Kim D.H., Lan W., Pyntikova T., Flanagan A.M., Behjati S., Cleveland D.W., Campbell P.J., et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019;51:705–715. doi: 10.1038/s41588-019-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iourov I.Y., Vorsanova S.G., Korostelev S.A., Zelenova M.A., Yurov Y.B. Long contiguous stretches of homozygosity spanning shortly the imprinted loci are associated with intellectual disability, autism and/or epilepsy. Mol. Cytogenet. 2015;8:77. doi: 10.1186/s13039-015-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iourov I.Y., Vorsanova S.G., Zelenova M.A., Vasin K.S., Kurinnaia O.S., Korostelev S.A., Yurov Y.B. Epigenomic variations manifesting as a loss of heterozygosity affecting imprinted genes represent a molecular mechanism of autism spectrum disorders and intellectual disability in children. Zh. Nevrol. Psikhiatr. Im. S. S. Korsakova. 2019;119:91–97. doi: 10.17116/jnevro201911905191. [DOI] [PubMed] [Google Scholar]
- 23.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry H.H. New data collection priority: Focusing on genome-based bioinformation. Res. Result. Biomed. 2020;6:5–8. [Google Scholar]
- 25.Iourov I.Y., Vorsanova S.G., Yurov Y.B. The variome concept: Focus on CNVariome. Mol. Cytogenet. 2019;12:52–56. doi: 10.1186/s13039-019-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelenova M.A., Yurov Y.B., Vorsanova S.G., Iourov I.Y. Laundering CNV data for candidate process prioritization in brain disorders. Mol. Cytogenet. 2019;12:54–56. doi: 10.1186/s13039-019-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padeken J., Zeller P., Gasser S.M. Repeat DNA in genome organization and stability. Curr. Opin. Genet. Dev. 2015;31:12–19. doi: 10.1016/j.gde.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Iourov I.Y., Vorsanova S.G., Yurov Y.B. Chromosomal mosaicism goes global. Mol. Cytogenet. 2008;1:26. doi: 10.1186/1755-8166-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorsanova S.G., Yurov Y.B., Soloviev I., Iourov I. Molecular Cytogenetic Diagnosis and Somatic Genome Variations. Curr. Genom. 2010;11:440–446. doi: 10.2174/138920210793176010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potter H.P. Beyond Trisomy 21: Phenotypic Variability in People with Down Syndrome Explained by Further Chromosome Mis-segregation and Mosaic Aneuploidy. J. Down Syndr. Chromosom. Abnorm. 2016;2:1–4. doi: 10.4172/2472-1115.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma K., Qiu L., Mrasek K., Zhang J., Liehr T., Quintana L.G., Li Z. Common Fragile Sites: Genomic Hotspots of DNA Damage and Carcinogenesis. Int. J. Mol. Sci. 2012;13:11974–11999. doi: 10.3390/ijms130911974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puliti A., Rizzato C., Conti V., Bedini A., Gimelli G., Barale R., Sbrana I. Low-copy repeats on chromosome 22q11.2 show replication timing switches, DNA flexibility peaks and stress inducible asynchrony, sharing instability features with fragile sites. Mutat. Res. Mol. Mech. Mutagen. 2010;686:74–83. doi: 10.1016/j.mrfmmm.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Yadav V., Sun S., Coelho M.A., Heitman J. Centromere scission drives chromosome shuffling and reproductive isolation. Proc. Natl. Acad. Sci. USA. 2020;117:7917–7928. doi: 10.1073/pnas.1918659117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iourov I.Y., Vorsanova S.G., Zelenova M.A., Korostelev S.A., Yurov Y.B. Genomic Copy Number Variation Affecting Genes Involved in the Cell Cycle Pathway: Implications for Somatic Mosaicism. Int. J. Genom. 2015;2015:1–7. doi: 10.1155/2015/757680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 36.McNulty S.M., Sullivan B.A. Alpha satellite DNA biology: Finding function in the recesses of the genome. Chromosom. Res. 2018;26:115–138. doi: 10.1007/s10577-018-9582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miga K.H. Centromeric Satellite DNAs: Hidden Sequence Variation in the Human Population. Genes. 2019;10:352. doi: 10.3390/genes10050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolzán A.D. Using telomeric chromosomal aberrations to evaluate clastogen-induced genomic instability in mammalian cells. Chromosom. Res. 2020:1–18. doi: 10.1007/s10577-020-09641-2. [DOI] [PubMed] [Google Scholar]
- 39.Yurov Y.B., Vorsanova S.G., Iourov I.Y. Chromosome Instability in the Neurodegenerating Brain. Front. Genet. 2019;10:892. doi: 10.3389/fgene.2019.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurov Y.B., Iourov I.Y., Vorsanova S.G., Liehr T., Kolotii A.D., Kutsev S.I., Pellestor F., Beresheva A.K., Demidova I.A., Kravets V.S., et al. Aneuploidy and Confined Chromosomal Mosaicism in the Developing Human Brain. PLoS ONE. 2007;2:558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor T.H., Gitlin S.A., Patrick J.L., Crain J.L., Wilson J.M., Griffin D.K. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum. Reprod. Update. 2014;20:571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 42.Rohrback S., Siddoway B., Liu C.S., Chun J. Genomic mosaicism in the developing and adult brain. Dev. Neurobiol. 2018;78:1026–1048. doi: 10.1002/dneu.22626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurov Y.B., Vorsanova S.G., Iourov I. Ontogenetic Variation of the Human Genome. Curr. Genom. 2010;11:420–425. doi: 10.2174/138920210793175958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijg J. Somatic mutations, genome mosaicism, cancer and aging. Curr. Opin. Genet. Dev. 2014;26:141–149. doi: 10.1016/j.gde.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machiela M.J. Mosaicism, aging and cancer. Curr. Opin. Oncol. 2019;31:108–113. doi: 10.1097/CCO.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonetti G., Bruno S., Padella A., Tenti E., Martinelli G. Aneuploidy: Cancer strength or vulnerability? Int. J. Cancer. 2018;144:8–25. doi: 10.1002/ijc.31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iourov I.Y., Vorsanova S.G., Yurov Y.B. Chromosomal Variation in Mammalian Neuronal Cells: Known Facts and Attractive Hypotheses. Int. Rev. Cytol. 2006;249:143–191. doi: 10.1016/s0074-7696(06)49003-3. [DOI] [PubMed] [Google Scholar]
- 48.Iourov I.Y., Vorsanova S.G., Yurov Y.B. Single Cell Genomics of the Brain: Focus on Neuronal Diversity and Neuropsychiatric Diseases. Curr. Genom. 2012;13:477–488. doi: 10.2174/138920212802510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liehr T., Al-Rikabi A. Mosaicism: Reason for Normal Phenotypes in Carriers of Small Supernumerary Marker Chromosomes with Known Adverse Outcome. A Systematic Review. Front. Genet. 2019;10:10. doi: 10.3389/fgene.2019.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potter H., Chial H.J., Caneus J., Elos M., Elder N., Borysov S., Granic A. Chromosome Instability and Mosaic Aneuploidy in Neurodegenerative and Neurodevelopmental Disorders. Front. Genet. 2019;10:1092. doi: 10.3389/fgene.2019.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorpe J., Osei-Owusu I.A., Avigdor B.E., Tupler R., Pevsner J. Mosaicism in Human Health and Disease. Annu. Rev. Genet. 2020;54:1–24. doi: 10.1146/annurev-genet-041720-093403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vorsanova S.G., Yurov Y.B., Iourov I.Y. Dynamic nature of somatic chromosomal mosaicism, genetic-environmental interactions and therapeutic opportunities in disease and aging. Mol. Cytogenet. 2020;13:1–6. doi: 10.1186/s13039-020-00488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hochstenbach R., Buizer-Voskamp J., Vorstman J., Ophoff R. Genome Arrays for the Detection of Copy Number Variations in Idiopathic Mental Retardation, Idiopathic Generalized Epilepsy and Neuropsychiatric Disorders: Lessons for Diagnostic Workflow and Research. Cytogenet. Genome Res. 2011;135:174–202. doi: 10.1159/000332928. [DOI] [PubMed] [Google Scholar]
- 54.Schang A.-L., Saberan-Djoneidi D., Mezger V. The impact of epigenomic next-generation sequencing approaches on our understanding of neuropsychiatric disorders. Clin. Genet. 2017;93:467–480. doi: 10.1111/cge.13097. [DOI] [PubMed] [Google Scholar]
- 55.Hawking S., Mlodinow L. The (elusive) theory of everything. Sci. Am. 2010;303:68–71. doi: 10.1038/scientificamerican1010-68. [DOI] [PubMed] [Google Scholar]
- 56.Mao J., Wang H., Li H., Song X., Wang T., Xiang J., Li H. Genetic analysis of products of conception using a HLPA/SNP-array strategy. Mol. Cytogenet. 2019;12:1–7. doi: 10.1186/s13039-019-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue H., Huang H.L., Wang Y., An G., Zhang M., Xu L., Lin Y. Molecular cytogenetic identification of small supernumerary marker chromosomes using chromosome microarray analysis. Mol. Cytogenet. 2019;12:13. doi: 10.1186/s13039-019-0425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iourov I.Y., Vorsanova S.G., Yurov Y.B. In silico molecular cytogenetics: A bioinformatic approach to prioritization of candidate genes and copy number variations for basic and clinical genome research. Mol. Cytogenet. 2014;7:98. doi: 10.1186/s13039-014-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.