Abstract
Objective
Identify the subcellular location and potential binding partners of two cerebellar degeneration‐related proteins, CDR2L and CDR2, associated with anti‐Yo‐mediated paraneoplastic cerebellar degeneration.
Methods
Cancer cells, rat Purkinje neuron cultures, and human cerebellar sections were exposed to cerebrospinal fluid and serum from patients with paraneoplastic cerebellar degeneration with Yo antibodies and with several antibodies against CDR2L and CDR2. We used mass spectrometry‐based proteomics, super‐resolution microscopy, proximity ligation assay, and co‐immunoprecipitation to verify the antibodies and to identify potential binding partners.
Results
We confirmed the CDR2L specificity of Yo antibodies by mass spectrometry‐based proteomics and found that CDR2L localized to the cytoplasm and CDR2 to the nucleus. CDR2L co‐localized with the 40S ribosomal protein S6, while CDR2 co‐localized with the nuclear speckle proteins SON, eukaryotic initiation factor 4A‐III, and serine/arginine‐rich splicing factor 2.
Interpretation
We showed that Yo antibodies specifically bind to CDR2L in Purkinje neurons of PCD patients where they potentially interfere with the function of the ribosomal machinery resulting in disrupted mRNA translation and/or protein synthesis. Our findings demonstrating that CDR2L interacts with ribosomal proteins and CDR2 with nuclear speckle proteins is an important step toward understanding PCD pathogenesis.
Introduction
Paraneoplastic neurological syndromes are rare autoimmune‐mediated diseases 1 , 2 characterized by the production of antibodies that target antigens expressed both by the tumor and endogenously in the central nervous system. 3 , 4 One of the most common forms of paraneoplastic neurological syndromes is paraneoplastic cerebellar degeneration (PCD). 5 In patients with PCD and breast or ovarian cancer, the dominant onconeural antibody, anti‐Yo, is detected in both serum and cerebrospinal fluid (CSF). 6 Anti‐Yo antibodies are directed against two proteins, cerebellar degeneration‐related protein 2 (CDR2) and CDR2‐like (CDR2L), which are endogenously expressed in Purkinje neurons of the cerebellum. 7 The interaction between anti‐Yo and CDR proteins is thought to mediate Purkinje neuron dysfunction and death. 5 A two‐step process has been proposed, with the internalization of Yo antibodies as the primary event, followed by the subsequent activation of cytotoxic T cells. 8 , 9 However, it has also been demonstrated that Yo antibodies can induce Purkinje neuron death in the absence of T lymphocytes. 8 , 10
Previously we showed that CDR2L is the major Yo antibody target in PCD. 7 However, we cannot exclude a functional role for CDR2 in anti‐Yo‐mediated PCD pathogenesis. These proteins display a high degree of homology with approximately 45% sequence identity, 11 , 12 and both are widely expressed in normal as well as malignant tissues. 3 , 13 Ovarian malignancy is the most frequent cancer type found in Yo‐mediated PCD, and both CDR2L and CDR2 are highly expressed in this type of cancer. 3 , 14
Earlier studies have suggested that CDR2L and CDR2 are cytoplasmic proteins. 3 , 13 However, detailed subcellular localization using antigen‐specific antibodies has not been performed. Current knowledge concerning the biologic function of CDR2L is limited. CDR2 has leucine zipper and zinc‐finger DNA binding domains, characteristic of transcriptional regulatory proteins 11 , 15 , 16 and occurrence of these domains in the predicted open reading frame suggests that CDR2 has a role in regulating gene expression. 11 , 17 CDR2 interacts with the serine/threonine protein kinase PKN and cell cycle‐related proteins MRG15 and MRGX; all involved in signal transduction or gene transcription. 15 , 18 , 19
In this study, we examined the subcellular locations of CDR2L and CDR2 and their protein‐protein interactions. Our findings suggest that CDR2L and CDR2 have different roles: CDR2L interacts with cytosolic ribosomes and appears to function in protein synthesis, while CDR2 associates with nuclear speckle proteins and appears to be involved in mRNA maturation.
Materials and Methods
Patient samples
Five sex‐ and age‐matched CSF samples from patients with Yo antibodies (PCD patients) and five without Yo antibodies and no neurological disease or underlying cancer (negative controls) were obtained from the Neurological Research Laboratory, Haukeland University Hospital (Regional Committees for Medical and Health Research Ethics, 2013/1480).
Cell culture
OvCar3 (American Type Culture Collection (ATCC), #HTB‐161) and HepG2 (ATCC, #HB‐8065) cancer cell lines were maintained and subcultivated on poly‐D‐lysine‐coated coverslips (Neuvitro, #GG‐18‐1.5‐pdl) according to the manufacturer’s protocol. Cells were washed twice with 0.1 M phosphate‐buffered saline (PBS), fixed (15 min, 4% paraformaldehyde in PBS, Thermo Fisher Scientific, #28908), and quenched (5 min, 50 mmol/L NH4Cl, Sigma‐Aldrich, #254134) prior to immunostaining.
Cerebellar tissue preparation
Cerebellar sections were cut from fresh frozen normal human tissue (REK, #2013/1503). Heat‐induced epitope retrieval was performed prior to immunostaining.
Rat Purkinje neuron cultures
All procedures were performed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals Norway (FOTS 20135149/20157494/20170001). Embryonic day 18 Wistar Hannover GLAST rat pups were used for neuronal culture preparation. The protocol has recently been described. 20
Immunochemistry
Fixed OvCar3 cells and cerebellar sections were permeabilized in 0.5% Triton X‐100‐PBS (Sigma‐Aldrich, #11332481001) for 5 min, washed in 0.5% gelatin‐PBS (Sigma‐Aldrich, #G7041) three times with 15 min each wash, blocked in 10% SEABLOCK (Thermo Fisher Scientific, #37527) in PBS for 30 min, and incubated with primary antibodies overnight at 4 °C. Following incubations, cells and sections were washed in gelatin‐PBS, incubated with secondary antibodies for 2 h at room temperature, and mounted using ProLong Diamond with DAPI (Thermo Fisher Scientific, #P36962). The following antibodies were used: rabbit anti‐CDR2 (Sigma‐Aldrich, #HPA018151), rabbit anti‐CDR2L (Protein Technology, #14563‐1‐AP), mouse anti‐rpS6 (Cell Signaling, #2317/Santa Cruz #sc‐74459), mouse anti‐SON (Santa Cruz, #sc398508), mouse anti‐eIF4A3 (Santa Cruz, #sc‐365549), mouse anti‐SRSF2 (Abcam, #ab11826), Alexa Fluor 488/594‐labeled goat anti‐human (Thermo Fisher Scientific, #A‐11013/#A11014), Alexa Fluor 488/594‐labeled goat anti‐rabbit (Thermo Fisher Scientific, #R37116/#R37117), rabbit anti‐STAR635P (Sigma‐Aldrich, #53399‐500UG), and Alexa Fluor 488/594‐labeled goat anti‐mouse (Thermo Fisher Scientific, #R37120/#R37121).
Super‐resolution microscopy
A Leica TCS SP8 Stimulated Emission Depletion (STED) 3X confocal microscope equipped with a 100x oil objective with a numerical aperture of 1.4 was used for imaging. The output of the excitation laser (up to 1.5 mW per line; pulsed) was kept between 1% and 20% and the STED laser (775 nm; up to 1.5 W) between 20% and 30%. Gating (between 1 and 6 ns) was applied for all channels as well as a minimum of three intensity averages. The lateral resolution was consistently measured to be between 40 and 50 nm.
Immunoprecipitation
OvCar3 and HepG2 cells were lysed in RIPA lysis buffer (Bioscience #786‐490) containing protease inhibitor cocktail (Sigma‐Aldrich #11873580001), 1 mmol/L phenylmethylsulfonyl fluoride (PMSF, Sigma‐Aldrich #P7626), 1 mmol/L sodium fluoride (NaF, Sigma‐Aldrich #S6776), and 1 mmol/L sodium orthovanadate (Na3VO4, Sigma‐Aldrich #450243). The lysate was centrifuged (22,000g, 4°C, 15 min) and the supernatant was collected.
Following the Bio‐Rad SureBeads immunoprecipitation protocol, the proteins were immunoprecipitated from OvCar3 and HepG2 cell lysates using Protein G Magnetic Beads (Dynabeads, Thermo Fischer Scientific, #1004D). Immunoprecipitated proteins were separated on a 10% TGX gel (Bio‐Rad, #456‐1035) and transferred to a polyvinylidene difluoride (PVDF) membrane using the Trans‐Blot Turbo Transfer kit (Bio‐Rad, #170‐4274). Western blot analysis was performed to detect proteins of interest using the following primary antibodies: rabbit anti‐CDR2L (Proteintech, #14563‐1‐AP), mouse anti‐rpS6 (Santa Cruz #sc‐74459), rabbit anti‐CDR2 (Sigma‐Aldrich, #018151), mouse anti‐CDR2 (Santa Cruz, #sc100320) mouse anti‐SON, mouse and rabbit anti‐eIF4A3 (Abcam, #ab32485). The secondary antibodies used were TidyBlot (Bio‐Rad, #STAR209PA) and horseradish peroxidase anti‐mouse IgG and anti‐rabbit IgG (Dako, #P0260 and #P0217). A negative control consisting of beads and cancer cell lysate was also included.
Proximity ligation assay
The proximity ligation assay was performed using the commercially available Duolink kit from Sigma‐Aldrich (#DUO92101). Fixed OvCar3 cells were permeabilized for 5 min using 0.5% Triton X‐100 diluted in PBS and blocked with 10% SEABLOCK in PBS. Primary antibodies against Hsp60 (EnCor Biotechnology, #CPCA‐HSP60), CDR2 (Sigma‐Aldrich, #018151), CDR2L (Proteintech, #14563‐1‐AP), SON, and SRSF2 were applied for 1 h (1:100 in blocking solution), followed by 3x 5‐minute washes with Wash Buffer A supplied with the kit. Probes (+ and −) were diluted in blocking solution (1:5) and added to the cells for 1 h (37 °C). The cells were washed 3x for 5 min each with Wash Buffer A and incubated with ligation buffer (1:5) and ligase enzyme (1:40) for 30 min (37°C). After 2x 5‐minute washes with Wash Buffer A amplification buffer (1:5) and the polymerase enzyme (1:80) were diluted in distilled water and applied to the cells for 100 min (37 °C, in the dark), followed by three 10‐minute washes with Wash Buffer B (supplied with the kit). Prolong Diamond with DAPI was used to mount the coverslips (overnight, 4 °C). Mounted cells were stored at −20 °C.
Mass spectrometry‐based proteomics analysis
Proteins of interest were immunoprecipitated from HepG2 or OvCar3 cell lysates using the antibodies listed in Table 1. A negative control consisting of beads and cancer cell lysate was also included. The samples were loaded on a 10% TGX gel and run approximately 1 cm into the resolving gel. Each lane was cut into cubes of approximately 1 mm2 and hydrated in Milli‐Q water (20 min, room temperature). Detergents (i.e. sodium dodecyl sulfate) and salts were removed by washing the gel in 25 mmol/L ammonium bicarbonate (Sigma‐Aldrich, #09830‐500G) and 50% acetonitrile (VWR, #34967‐2.5L). Cysteine reduction and alkylation were accomplished with a 45‐minute incubation in 10 mmol/L dithiothreitol (Amersham Biosciences, #171318‐02) at 56 °C followed by a 30‐minute incubation in 55 mmol/L iodoacetamide (VWR, #M216‐30G) at room temperature in the dark. After washing in 25 mmol/L ammonium bicarbonate and 50% acetonitrile, dried gel pieces were hydrated on ice for 20 min with a minimum volume of 6 ng/µL trypsin (sequencing‐grade modified, Promega, #V511A) in digestion buffer (20 mmol/L ammonium bicarbonate, 1 mmol/L calcium chloride (Sigma‐Aldrich,#C7902)), then covered with digestion buffer and incubated for 16 h at 37°C. Trypsin activity was quenched by acidification with trifluoracetic acid (VWR, #1.08218.0050), and samples were desalted using StageTip C18 columns (Empore disk‐C18, Agilent Life Sciences, #12145004) and the eluted peptides were dried and dissolved in 2% acetonitrile, 1% formic acid (VWR, #84865.260). 21
Table 1.
Antibody specificities determined by mass spectrometry analysis of CDR2L and CDR2 proteins immunoprecipitated from OvCar3 and HepG2 cell lysates.
| Target | Source/Supplier | Cat. no. | AA seq. | Cell line | #Peptides | Interaction |
|---|---|---|---|---|---|---|
|
Yo |
Yo positive CSF |
OvCar3 |
54 | CDR2L | ||
| Yo |
Yo positive CSF |
HepG2 |
‐ | ‐ | ||
| CDR2L | Sigma‐ Aldrich | HPA022015 | 395‐464 | OvCar3 | 56 | CDR2L |
|
CDR2L |
Proteintech |
14563‐1‐AP |
116‐465 |
OvCar3 |
68 |
CDR2L |
| CDR2L | Proteintech | 66791‐1‐Ig |
116‐465 |
OvCar3 | 69 |
CDR2L |
| CDR2 | Sigma‐Aldrich | HPA018151 | 270‐392 | HepG2 | 49 | CDR2 |
| CDR2 | Sigma‐Aldrich | HPA023870 | 112‐234 | HepG2 | 41 | CDR2 |
| CDR2 | Santa Cruz | Sc‐100320 | 296‐405 | HepG2 | 57 | CDR2 |
| CDR2 | LS Bio | C181958 | Full length | HepG2 | 51 | CDR2 |
AA seq., amino acid sequence.
About 0.5 µg tryptic peptides were loaded onto an Ultimate 3000 RSLC system (Thermo Fisher Scientific) connected online to a Q‐Exactive HF mass spectrometer (Thermo Fisher Scientific) equipped with EASY‐spray nano‐electrospray ion source (Thermo Fisher Scientific). All samples were loaded and desalted on a pre‐column (Acclaim PepMap 100, 2 cm x 75 µm ID nanoViper column, packed with 3 µm C18 beads) at a flow rate of 5 µL/min with 0.1% trifluoracetic acid. Peptides were separated during a biphasic acetonitrile gradient (flow rate of 200 nL/minute) on a 50‐cm analytical column (PepMap RSLC, 50 cm x 75 µm ID EASY‐spray column, packed with 2 µm C18 beads). Solvent A and B were 0.1% formic acid in water and 100% acetonitrile, respectively. The gradient composition was 5% B during trapping (5 min) followed by 5–7% B over 0.5 min, 7–22% B for the next 59.5 min, 22–35% B over 22 min, and 35–80% B over 5 min. Elution of very hydrophobic peptides and conditioning of the column was performed during a 10‐minute isocratic elution with 80% B and 15 min of isocratic conditioning with 5% B, respectively.
Charged peptides were analyzed by the Q‐Exactive HF, operating in the data‐dependent acquisition mode to automatically switch between full‐scan MS and MS/MS acquisition. Mass spectra were acquired in the scan range 375–1500 m/z with a resolution of 60,000 at m/z 200 after an accumulation of 3,000,000 charges (maximum trap time set at 50 ms in the C‐trap). The 12 peptides with the most intense signals above an intensity threshold of 50,000 counts and with charge states of 2 to 6 were sequentially isolated and accumulated to 100,000 charges (maximum trap time set at 110 ms) to a target value of 1 × 105 or a maximum trap time of 110 ms in the C‐trap with isolation width maintained at 1.6 m/z (offset of 0.3 m/z) before fragmentation in the higher energy collision dissociation cell. Fragmentation was performed with a normalized collision energy of 32%, and fragments were detected in the Q‐Exactive at a resolution of 60,000 at m/z 200 with first mass fixed at m/z 110. One MS/MS spectrum of a precursor mass was allowed before dynamic exclusion for 30 seconds with “exclude isotopes” on. Accurate mass measurements in MS mode were accomplished by enabling the lock‐mass internal calibration of the polydimethylcyclosiloxane ions generated in the electrospray process from ambient air (m/z 445.12003). 22
Database searching and criteria for protein identification
Tandem mass spectra data were extracted with Proteome Discoverer (version 2.3.0.523, Thermo Fisher Scientific) and were searched against human, reviewed protein sequences (SwissprotKB database, release 08‐2018) with Sequest HT and MS Amanda search engines. The following search criteria were used: carbamidomethylation of cysteine (fixed modification), oxidation of methionine and acetyl of the protein N‐terminus (variable modifications), a maximum of two missed trypsin cleavages, 0.02‐Da fragment ion mass tolerance, and 10‐ppm precursor ion tolerance. Search results from PD were loaded into Scaffold 4 (version 4.9.0, Proteome Software Inc.), and all spectra were searched with the X! Tandem search engine against identified proteins to identify nonspecific trypsin cleavages.
Peptide and protein identifications were filtered to achieve a false discovery rate < 1.0% (based on searching the reversed human database). Grouping of proteins sharing identical peptides was enabled. In order to evaluate the likelihood of the predicted interactions, the following criteria were established: (1) nonspecific bindings were removed based on the negative control (without primary antibodies); (2) the number of recognized peptides was set to at least two; (3) proteins that were identified by more than one of the antibodies to CDR2L or CDR2 were considered as more likely partners; (4) the likelihood of interaction was evaluated based on the predicted cellular location of each protein of interest. Protein‐protein interactions were analyzed using the STRING database. STRING implements all publicly available sources of known and predicted protein‐protein associations, together with computational analysis to evaluate potential connectivity networks. 23 , 24
Results
Antibody specificity
To evaluate antibody specificity, we immunoprecipitated CDR2L and CDR2 from cancer cell lysates and analyzed the precipitates using mass spectrometry‐based proteomics with the antibodies listed in Table 1. We found that the commercial antibodies raised against CDR2L and CDR2 were specific and recognized the expected antigens. Also, we confirmed our previous data showing that CDR2L is the major Yo antibody target. Analysis of lysates of OvCar3 cells, which expresses both CDR2L and CDR2, immunoprecipitated with Yo antibodies bound to magnetic beads showed that CDR2L, but not CDR2, was recognized by Yo antibodies. In similar experiments performed with a cell line that only expresses CDR2, HepG2 cells, Yo antibody did not precipitate CDR2.
CDR2L and CDR2 interaction partners identified by mass spectrometry analysis
Potential protein interaction partners were identified using mass spectrometry analysis of proteins immunoprecipitated with anti‐CDR2L and anti‐CDR2 antibodies from cancer cell lysates. Initially, several hundred hits were detected, and four criteria were established to determine the likelihood of the predicted interactions. Thereafter, we used the STRING database to evaluate the connectivity of the proteins that met our criteria. CDR2L was predicted to interact with 50 ribosomal proteins that were tightly connected (Fig. 1A). Of these 50 ribosomal proteins, 20 belong to the 40S subunit, and 30 belong to the 60S subunit. Proteins known to associate with nuclear speckles, eukaryotic initiation factor eIF4A3, SON, and the serine/arginine‐rich splicing factor SRSF2, were identified as potential interaction partners of CDR2. According to the STRING analysis eIF4A3 interacts with SON and SRSF2 (Fig. 1B), as well as with the 40S ribosomal subunit factor rpS6 (Fig. 1C).
Figure 1.
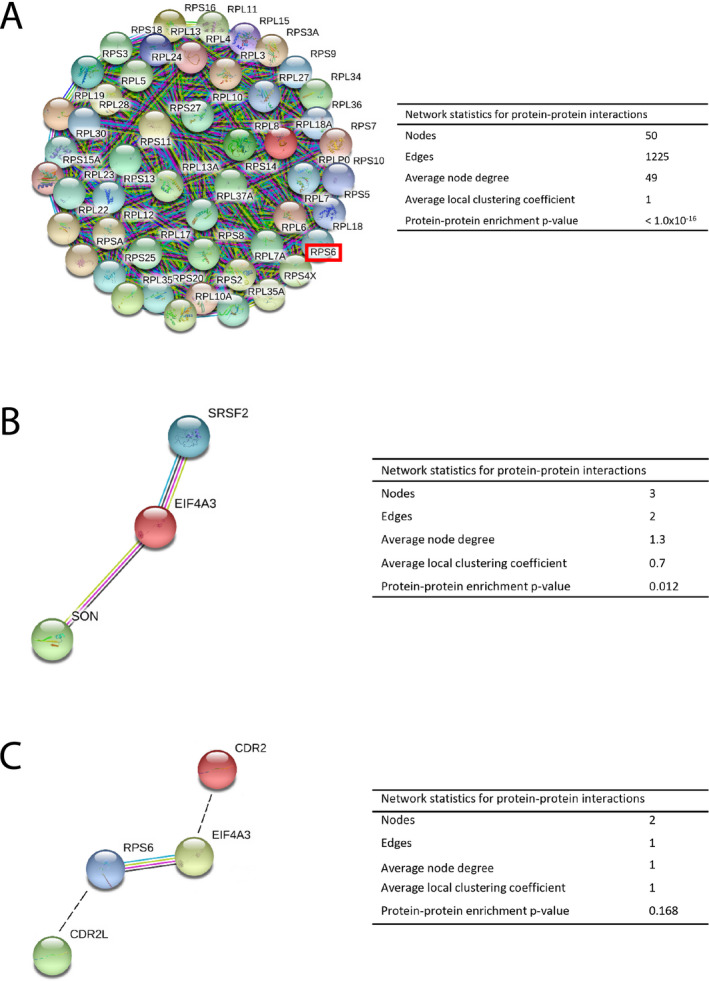
Protein‐protein interaction networks visualized by STRING. (A) CDR2L was predicted to interact with ribosomal proteins (rpS6, red box). The nodes indicate proteins, and the edges represent protein‐protein associations. (B) Protein‐protein interaction network of nuclear speckles proteins, SON, eIF4A3, and SRSF2, predicted to interact with CDR2. eIF4A3 (red) directly interacts with SON (light green) and SRSF2 (blue). (C) eIF4A3 (yellow) interacts with rpS6 (blue), indicated by colored edges. Predicted binding partners, CDR2L (green) and CDR2 (red), are manually gated (black, dotted lines). Color‐coded edges; light blue: curated databases, dark blue: gene co‐occurrence, pink: experimentally determined, green: text mining. Interactions with a medium score of 0.400 or more are shown.
CDR2L Co‐localizes with ribosomal proteins and CDR2 with nuclear speckle proteins in ovarian cancer cells
We used immunolabeling and proximity ligation assay to investigate the subcellular localization of CDR2L and CDR2. In OvCar3 cells, which express both CDR2L and CDR2, we found that CDR2L co‐localizes with rpS6, whereas CDR2 co‐localizes with nuclear speckle proteins SON, eIF4A3, and SRSF2 (Fig. 2A). These results were confirmed by proximity ligation assay in OvCar3 cells (Fig. 2B).
Figure 2.
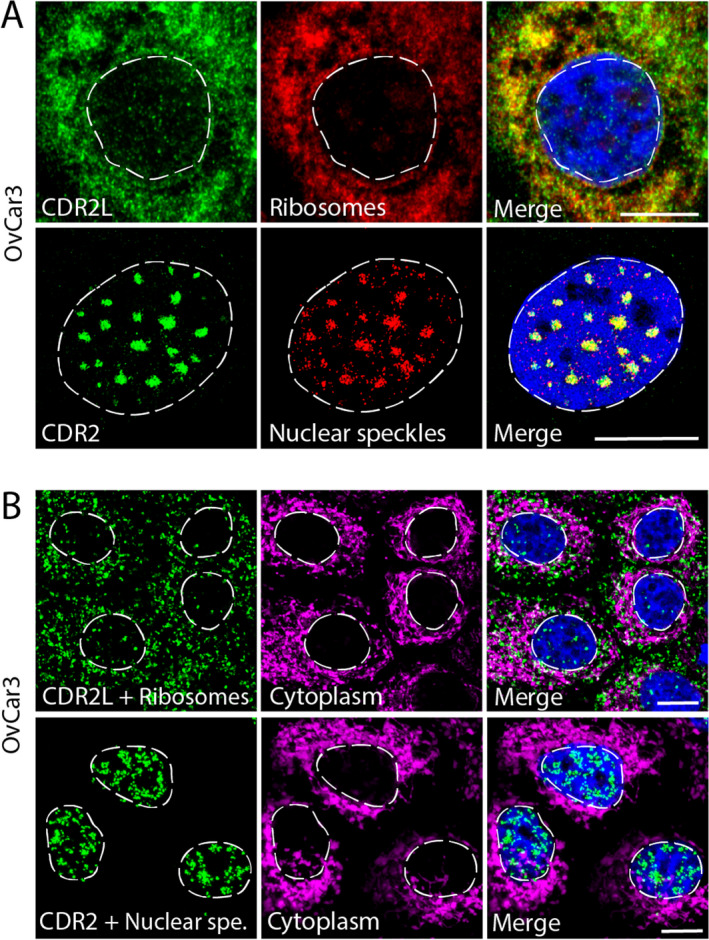
CDR2L co‐localizes with ribosomes and CDR2 with nuclear speckles in OvCar3 cells as shown using proximity ligation assay. (A) Upper row: Co‐localization of anti‐CDR2L (green) and ribosomes (rpS6; red) in the cytoplasm (yellow; merged image). Lower row: Co‐localization of anti‐CDR2 (green) and nuclear speckles (SRSF2; red) in the nucleus (yellow; merged image). (B) Upper row: Positive Duolink (green) between CDR2L and ribosomes (rpS6) in the cytoplasm (hsp60 in magenta was used to show the extent of the cell cytoplasm; merged image). Lower row: Positive Duolink (green) between CDR2 and nuclear speckle marker (SRSF2) in the nuclei; no co‐localization was observed with cytoplasmic marker hsp60 (magenta; merged image). DAPI was used as a marker for the nuclei (blue). Scare bars = 10 µm.
Co‐Immunoprecipitation of CDR2L and CDR2 from OvCar3 cells confirms protein‐protein interactions with ribosomal and nuclear speckle proteins
To analyze whether CDR2L directly interacts with rpS6, we performed co‐immunoprecipitation assays from OvCar3 cell lysates. CDR2L specifically co‐immunoprecipitated with rpS6, indicating that endogenous CDR2L forms a complex with rpS6 in cancer cells (Fig. 3A). Furthermore, we found that SON and eIF4A3 co‐immunoprecipitated with CDR2 from HepG2 cells, thus indicating a strong and stable interaction between these proteins and CDR2 (Fig. 3B).
Figure 3.
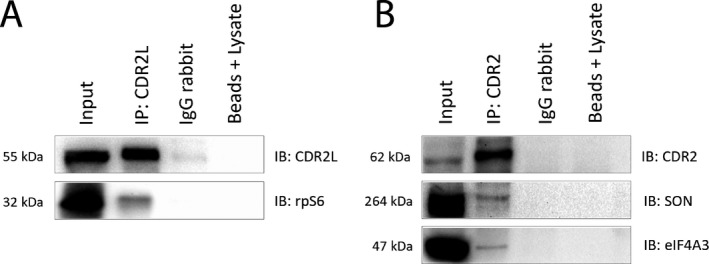
CDR2L co‐immunoprecipitates with ribosomal protein rpS6, whereas CDR2 co‐immunoprecipitates with nuclear speckle proteins SON and eIF4A3 in cancer cell lysates. (A) Immunoblot demonstrating the co‐immunoprecipitation of CDR2L and rpS6 from OvCar3 cell lysates. (B) Immunoblot demonstrating the co‐immunoprecipitation of CDR2, SON, and eIF4A3 from HepG2 cell lysates. Input = cancer cell lysates (OvCar3 or HepG2). Beads + lysate = samples that were not treated with primary antibody, and served as negative controls.
Co‐localizations of CDR2L with ribosomal proteins and of CDR2 with nuclear speckle proteins occurs in Purkinje neurons in Human cerebellum sections and in Purkinje neuron cultures
In human cerebellum sections, CDR2L and Yo antibodies stained the cytoplasm in regions that overlapped with regions stained for the ribosomal marker rpS6, whereas CDR2 showed nuclear staining that overlapped with nuclear speckle markers (eIF4A3, SON, and SRSF2; Fig. 4A). These results were replicated in cultured rat Purkinje neurons (Fig. 4B).
Figure 4.
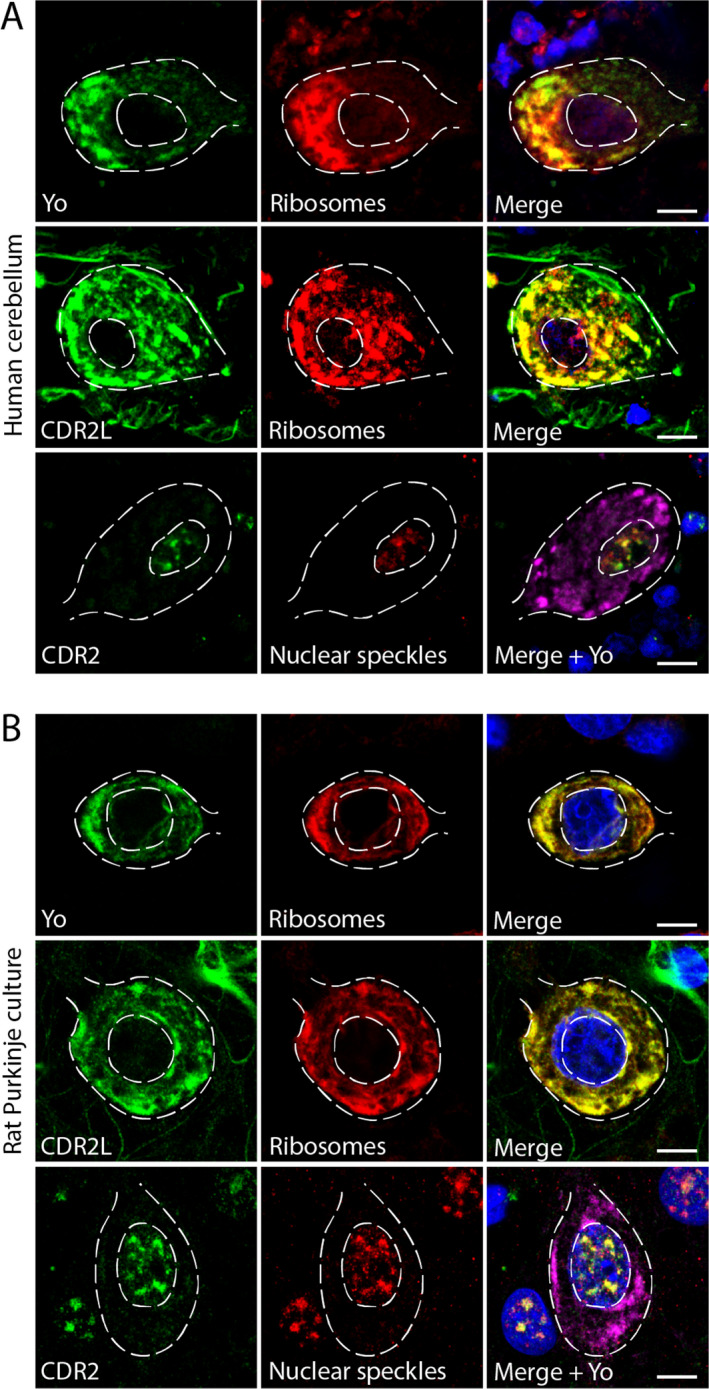
CDR2L and Yo co‐localize with ribosomal proteins and CDR2 co‐localizes with nuclear speckle proteins in cerebellar Purkinje neurons as shown by super‐resolution microscopy. (A) Upper row: Human cerebellar section stained with Yo‐CSF (green) and anti‐rpS6 (red); the proteins co‐localize in the cytoplasm (yellow; merged image). Middle row: Human cerebellar section stained with anti‐CDR2L (green) and ribosomal marker anti‐rpS6 (red); the proteins co‐localize in the cytoplasm (yellow; merged image). Lower row: Human cerebellar section stained with anti‐CDR2 (green) and nuclear speckle marker anti‐SRSF2 (red); the proteins co‐localize in the nucleus. No co‐localization was found with anti‐Yo (magenta; merged image). (B) Upper row: Rat Purkinje neuron cultures stained with anti‐Yo (CSF; green) and rpS6 (ribosomes; red); co‐localization was observed in the cytoplasm (yellow; merged image). Middle row: Rat Purkinje neuron cultures stained with anti‐CDR2L (green) and anti‐rpS6 (red); co‐localization was observed in the cytoplasm (yellow; merge image). Lower row: Rat Purkinje neurons stained with anti‐CDR2 (green), nuclear speckle protein (red), and anti‐Yo (magenta). CDR2 and the nuclear speckle protein co‐localize in the cell nucleus (yellow; merged image), whereas Yo does not. Scale bars = 10 µm.
Discussion
The pathogenesis of Yo‐mediated PCD remains incompletely understood, but it has been postulated that the Purkinje neuron loss is due to auto‐reactive T cells and a direct damaging effect of Yo antibodies. 3 , 4 , 6 , 25 We demonstrated previously that CDR2L, not CDR2, is the major target of the Yo antibody 7 : Yo antibodies bind both endogenous and recombinant CDR2L, but only recombinant CDR2, not the native form. In this study, we confirmed the CDR2L specificity of Yo antibodies by mass spectrometry‐based proteomics and showed that while CDR2L and CDR2 have differing localizations, it is possible to link their putative roles to ribosomal function.
The biological functions and precise subcellular localization of both CDR2L and CDR2 have been unresolved questions. Analysis of PCD patient sera has shown that Yo antibodies localize to the cytoplasm and associate with both membrane‐bound and free ribosomes. 26 , 27 In these studies, the Yo antigen is referred to as “CDR2.” However, based on our recent findings, we are confident that the main Yo antigen is indeed CDR2L. Here, we used available antibodies against CDR2L and CDR2, as well as anti‐Yo, to characterize the cellular localization of these proteins and their potential binding partners.
Immunolabeling cells with commercially available anti‐CDR2 antibodies result in various expression patterns, localizing CDR2 to both the cytoplasm and the nucleus. 7 , 13 Therefore, we first evaluated the specificity of the available CDR2L and CDR2 antibodies produced to recognize the full‐length protein or shorter sequences. Immunoprecipitation followed by mass spectrometry analysis confirmed antibody specificity. The previously reported inconsistent results for CDR2 may either stem from the antibody recognition of one of the four CDR2 isoforms (www.uniprot.org) or from the translocation of CDR2 between the cytoplasm and nucleus. Furthermore, previous studies also identified PKN, MRG15, and MRGX as CDR2 binding partners. Since these proteins function both in the cytoplasm and nucleus, this raises the possibility that CDR2 might facilitate the transport of these proteins or translocate itself. 15 , 18 , 19 , 28 In addition, no CDR2L‐CDR2 cross‐talk was observed, which supports our finding that there is no cross‐talk between CDR2L and CDR2 in their native forms. Furthermore, our immunoprecipitation‐mass spectrometry results showed that Yo antibodies only precipitated CDR2L and not CDR2 from cancer cells. This is in line with recent work, which shows that Yo antibodies bind to the CDR2L regions of least homology with CDR2. 29
In addition to confirming antibody specificity, the mass spectrometry analysis revealed potential interacting partners for CDR2L and CDR2. A number of ribosomal proteins, including rpS6, were identified as potential CDR2L binding partners. The most prominent CDR2 binding partners were three nuclear speckle proteins: SON, eIF4A3, and SRSF2. Next, we used super‐resolution microscopy and proximity ligation assay to evaluate co‐localization within a 40‐nm range in cancer cells and Purkinje neurons. CDR2L was found to co‐localize with rpS6, whereas CDR2 co‐localized with nuclear speckle proteins eIF4A3, SON, and SRSF2. Co‐immunoprecipitation analyses established that CDR2L directly interacts with rpS6 and that CDR2 directly interacts with eIF4A3 and SON.
Nuclear speckles are self‐assembled organelles consisting of around 200 proteins involved in pre‐mRNA processing including splicing, surveillance, and RNA export. 30 The speckles can vary in size and morphology within a single cell, but have been shown to be non‐random organizations of proteins and RNAs stabilized by favorable intermolecular interactions. 30 SRSF2 and SON localize to the core region of the speckle; both proteins have domains enriched with arginine and serine repeats that are crucial for speckle core formation. 30 , 31 Both proteins are also involved in mRNA splicing 32 , 33 and interact with the ATP‐dependent RNA helicase eIF4A3. 34 It has been suggested that eIF4A3 may provide a link between splicing and translation in the cytoplasm through its connection to rpS6 34 , 35 , which co‐localizes with CDR2L.
Translation in eukaryotes relies on the assembly of the small (40S) and the large (60S) ribosomal subunit into the 80S ribosomes. 36 Each subunit is composed of ribosomal proteins and RNAs that work together to catalyze protein synthesis using mRNA as a template. 37 , 38 Ribosomal proteins often undergo post‐translational modifications and rps6, the identified CDR2L binding partner, is regulated by phosphorylation. 39 , 40 Five phosphorylation sites have been identified and these phosphorylation events could participate in regulating the translation of specific subclasses of mRNA, synaptic plasticity and behavior. 41 Thus, rpS6 phosphorylation is often used to track neuronal activity. 40 , 41
Our findings linking CDR2 to nuclear speckles and CDR2L to ribosomes allow us to speculate that these two proteins may participate in a common pathway (Fig. 5). First, we show that CDR2 interacts with eIF4A3 in the nucleus. Second, eIF4A3, along with other initiation factors, facilitates mRNA binding to ribosomes. 42 Furthermore, eIF4A3 and rpS6 have been shown to interact based on affinity‐capture mass spectrometry analysis. 35 Third, we show that CDR2L interacts with the ribosomes through rpS6. These findings place CDR2 and CDR2L in the process of protein translation, one involved in mRNA maturation and the other directly with the synthesis of proteins.
Figure 5.
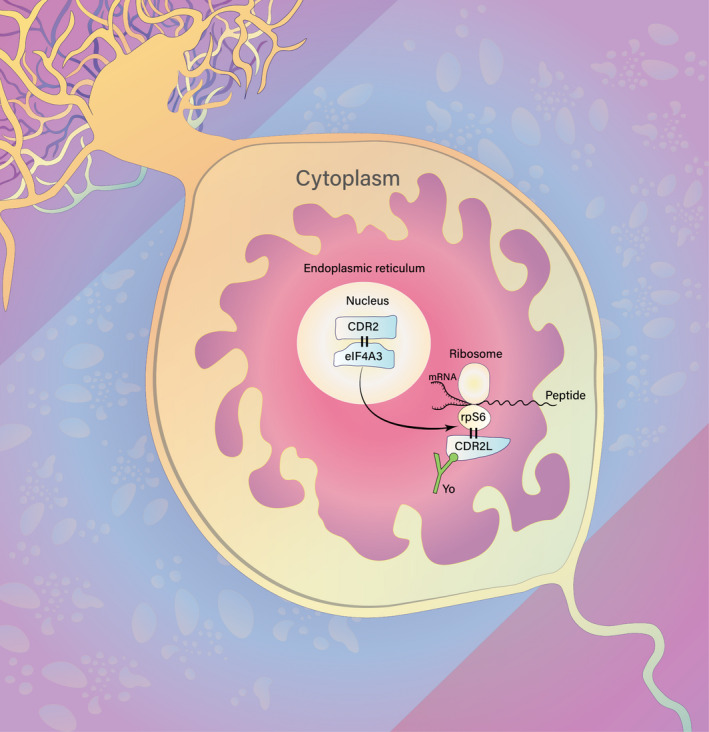
Hypothesis of CDR2L and CDR2 involvement in protein synthesis in Purkinje neurons. CDR2 localizes to the nucleus and directly interacts with nuclear speckle protein eIF4A3. eIF4A3, in conjugation with other cytoplasmic initiation factors, facilitates mRNA binding to the 40S ribosomal subunit. This event is important for mRNA maturation and translation, ultimately resulting in the synthesis of new proteins. CDR2L interacts with ribosomal subunit protein rpS6; therefore, we propose that CDR2L and CDR2 are both involved in the process of protein synthesis. Furthermore, Yo antibody (green) binding to CDR2L in Purkinje neurons of PCD patients may, therefore, interfere with the function of the ribosomal machinery, resulting in disrupted mRNA translation and/or protein synthesis.
Ensuring proper protein homeostasis is crucial to the cell. 36 , 38 We show that Yo antibodies specifically bind to CDR2L in Purkinje neurons of PCD patients where they potentially interfere with the function of the ribosomal machinery resulting in disrupted mRNA translation and/or protein synthesis. Taken together, our findings that CDR2L interacts with ribosomal proteins and CDR2 with nuclear speckle proteins is an important step toward understanding PCD pathogenesis. Future studies are needed to track the subcellular events in real‐time with the aim of addressing the dynamic interaction between the CDR2L and CDR2 molecules. This will be vital to understand whether there is a functional relationship between CDR2L and CDR2 in the Purkinje neuron deterioration that occurs in PCD.
Author Contributions
All authors contributed to the conception and design of the study; I.H. and T.K. performed the acquisition, data analysis, and prepared figures; I.H., T.K., M.S., and C.V. drafted the manuscript.
Conflicts of Interest
The authors have no conflicts of interest to report.
Acknowledgments
This work was supported by grants from Helse Vest, the University of Bergen, and Torbjørg Hauges Legacy. The confocal imaging was performed at the Molecular Imaging Center and was supported by the Department of Biomedicine and the Faculty of Medicine, University of Bergen, and its partners. The authors would like to thank Mette Haugen at the Department of Neurology, Haukeland University Hospital for valuable advice and discussion, and Anne Døskeland and Olav Mjaavatten at the Proteomics Unit (PROBE) at the University of Bergen for help and support related to the mass spectrometry analysis. Last but not least, we thank Dr. Laurence Bindoff for valuable discussion of our paper.
Funding Information
This work was supported by grants from Helse Vest, the University of Bergen, and Torbjørg Hauges Legacy. The confocal imaging was performed at the Molecular Imaging Center and was supported by the Department of Biomedicine and the Faculty of Medicine, University of Bergen, and its partners.
Funding Statement
This work was funded by Haukeland Universitetssjukehus grant F‐11533; Torbjørg Hauges Legacy grant ; Universitetet i Bergen grant #2016/7580‐RAKA.
Contributor Information
Ida Herdlevær, Email: idaherd@gmail.com.
Torbjørn Kråkenes, Email: torbjornkrakenes@gmail.com.
References
- 1. Peterson K, Rosenblum MK, Kotanides H, Posner JB. Paraneoplastic cerebellar degeneration. I. A clinical analysis of 55 anti‐Yo antibody‐positive patients. Neurol. 1992;42:1931–1937. [DOI] [PubMed] [Google Scholar]
- 2. Vedeler CA, Antoine JC, Giometto B, et al. Management of paraneoplastic neurological syndromes: report of an EFNS Task Force. European J Neurol. 2006;13:682–690. [DOI] [PubMed] [Google Scholar]
- 3. Raspotnig M, Haugen M, Thorsteinsdottir M, et al. Cerebellar degeneration‐related proteins 2 and 2‐like are present in ovarian cancer in patients with and without Yo antibodies. Cancer immunol immunother 2017;66:1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Darnell RB, Posner JB. Paraneoplastic Syndromes Involving the Nervous. System. 2003;349(16):1543–54. [DOI] [PubMed] [Google Scholar]
- 5. Rees JH. Paraneoplastic syndromes: when to suspect, how to confirm, and how to manage. J Neurol Neurosurg Psychiatry 2004;75 Suppl 2(Suppl 2):ii43‐ii50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eichler TW, Totland C, Haugen M, et al. CDR2L antibodies: a new player in paraneoplastic cerebellar degeneration. PloS one. 2013;8(6):e66002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krakenes T, Herdlevaer I, Raspotnig M, Haugen M, Schubert M, Vedeler CA. CDR2L is the major yo antibody target in paraneoplastic cerebellar degeneration. Annals of Neurol 2019;86:316–321. [DOI] [PubMed] [Google Scholar]
- 8. Schubert M, Panja D, Haugen M, Bramham CR, Vedeler CA. Paraneoplastic CDR2 and CDR2L antibodies affect Purkinje cell calcium homeostasis. Acta Neuropathol 2014;128(6):835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albert ML, Darnell JC, Bender A, Francisco LM, Bhardwaj N, Darnell RB. Tumor‐specific killer cells in paraneoplastic cerebellar degeneration. Nature Med 1998;4:1321–1324. [DOI] [PubMed] [Google Scholar]
- 10. Greenlee JE, Clawson SA, Hill KE, Wood BL, Tsunoda I, Carlson NG. Purkinje cell death after uptake of anti‐Yo antibodies in cerebellar slice cultures. J Neuropathol Exp Neurol 2010;69:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sato S, Inuzuka T, Nakano R, et al. Antibody to a zinc finger protein in a patient with paraneoplastic cerebellar degeneration. Biochem Biophys Res Commun 1991;178:198–206. [DOI] [PubMed] [Google Scholar]
- 12. Corradi JP, Yang C, Darnell JC, Dalmau J, Darnell RB. A Post‐transcriptional regulatory mechanism restricts expression of the paraneoplastic cerebellar degeneration antigen cdr2 to Immune Privileged Tissues. J Neurosci. 1997;17:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Totland C, Aarskog NK, Eichler TW, et al. CDR2 antigen and Yo antibodies. Cancer Immunology Immunother 2011;60:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Small M, Treilleux I, Couillault C, et al. Genetic alterations and tumor immune attack in Yo paraneoplastic cerebellar degeneration. Acta Neuropathol 2018;135:569–579. [DOI] [PubMed] [Google Scholar]
- 15. Sakai K, Shirakawa T, Li Y, Kitagawa Y, Hirose G. Interaction of a paraneoplastic cerebellar degeneration‐associated neuronal protein with the nuclear helix‐loop‐helix leucine zipper protein MRG X. Mol Cell Neurosci 2002;19:477–484. [DOI] [PubMed] [Google Scholar]
- 16. Okano HJ, Park WY, Corradi JP, Darnell RB. The cytoplasmic Purkinje onconeural antigen cdr2 down‐regulates c‐Myc function: implications for neuronal and tumor cell survival. Genes Dev 1999;13:2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fathallah‐Shaykh H, Wolf S, Wong E, Posner JB, Furneaux HM. Cloning of a leucine‐zipper protein recognized by the sera of patients with antibody‐associated paraneoplastic cerebellar degeneration. Proc Natl Acad Sci USA 1991;88:3451–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takanaga H, Mukai H, Shibata H, Toshimori M, Ono Y. PKN interacts with a paraneoplastic cerebellar degeneration‐associated antigen, which is a potential transcription factor. Exp Cell Res 1998;241:363–372. [DOI] [PubMed] [Google Scholar]
- 19. Sakai K, Kitagawa Y, Saiki S, Saiki M, Hirose G. Effect of a paraneoplastic cerebellar degeneration‐associated neural protein on B‐myb promoter activity. Neurobiol Dis 2004;15:529–533. [DOI] [PubMed] [Google Scholar]
- 20. Uggerud IM, Krakenes T, Hirai H, Vedeler CA, Schubert M. Development and optimization of a high‐throughput 3D rat Purkinje neuron culture. bioRxiv. 2020;105858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Analytical Chem 2003;75:663–670. [DOI] [PubMed] [Google Scholar]
- 22. Olsen JV, de Godoy LM, Li G, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C‐trap. Mol Cell Proteomics 2005;4:2010–2021. [DOI] [PubMed] [Google Scholar]
- 23. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res 2017;45(D1):D362–d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–d13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Storstein A, Krossnes BK, Vedeler CA. Morphological and immunohistochemical characterization of paraneoplastic cerebellar degeneration associated with Yo antibodies. Acta Neurol Scand 2009;120:64–7. [DOI] [PubMed] [Google Scholar]
- 26. Rodriguez M, Truh LI, O'Neill BP, Lennon VA. Autoimmune‐paraneoplastic‐cerebellar‐degeneration‐Ultrastructural‐localization‐of‐antibody‐binding‐sites‐in‐purkinje‐cells. Neurology 1988;38:1380. [DOI] [PubMed] [Google Scholar]
- 27. Hida C, Tsukamoto T, Awano H, Yamamoto T. Ultrastructural localization of anti‐Purkinje cell antibody‐binding sites in paraneoplastic cerebellar degeneration. Arch Neurol 1994;51:555–558. [DOI] [PubMed] [Google Scholar]
- 28. Mukai H, Miyahara M, Sunakawa H, et al. Translocation of PKN from the cytosol to the nucleus induced by stresses. Proc Natl Acad Sci USA. 1996;93(19):10195–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O’Donovan B, Mandel‐Brehm C, Vazquez SE, et al. High resolution epitope mapping of anti‐Hu and anti‐Yo autoimmunity by programmable phage display. Brain Communications. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fei J, Jadaliha M, Harmon TS, et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 2017;130:4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin S, Coutinho‐Mansfield G, Wang D, Pandit S, Fu X‐D. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 2008;15:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahn E‐Y, DeKelver Russell C, Lo M‐C, et al. son controls cell‐cycle progression by coordinated regulation of RNA Splicing. Molecular Cell 2011;42:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan CC, Dostie J, Diem MD, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh G, Kucukural A, Cenik C, et al. The cellular EJC interactome reveals higher‐order mRNP structure and an EJC‐SR protein nexus. Cell. 2012;151:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J Cell Biol 2018;217:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012;19:568–576. [DOI] [PubMed] [Google Scholar]
- 38. Lehmkuhl EM, Zarnescu DC. Lost in Translation: Evidence for Protein Synthesis Deficits in ALS/FTD and Related Neurodegenerative Diseases. Adv Neurobiol 2018;20:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 2006;31:342–348. [DOI] [PubMed] [Google Scholar]
- 40. Biever A, Valjent E, Puighermanal E. Ribosomal protein S6 phosphorylation in the nervous system. From Regulation to Function. Front Mol Neurosci 2015;8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Puighermanal E, Biever A, Pascoli V, et al. Ribosomal protein S6 phosphorylation is involved in novelty‐induced locomotion, synaptic plasticity and mRNA translation. Front Mol Neurosci 2017;10:419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rogers GW Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 2001;276:30914–30922. [DOI] [PubMed] [Google Scholar]


