Abstract
It has been observed that some colorectal cancer patients due to germline or double somatic pathogenic variants in the mismatch repair (MMR) genes may have intact protein expression observed in their tumors by immunohistochemistry (IHC). This has been speculated to occur more frequently in Lynch Syndrome (LS) cases due to pathogenic missense mutations leading to expression of a full-length but nonfunctional protein with retained antigenicity. Our goals were to study the frequency of unexpected MMR expression in colorectal cancers among LS patients with missense mutations, LS patients with truncating mutations, as well as cases with double somatic MMR mutations, and evaluate if the unexpected MMR expression is more common in certain categories. IHC slides were available on 82 patients with MMR deficiency without methylation, including 56 LS cases and 26 double somatic MMR mutation cases. 16 of 82 MMR-defective cases showed unexpected MMR expression, with 10 cases showing tumor staining weaker than control and 6 cases (7%) with intact staining. Unexpected MMR expression was most commonly seen with LS cases with missense mutations (4 of 9, 44%), followed by MMR double somatic mutation cases (7 of 26, 27%), and lastly by LS cases with truncating mutations (5 of 47, 11%). Cautious interpretation of MMR IHC is advised when dealing with tumor staining that is weaker than control regardless of percentage of tumor staining, as these cases may harbor pathogenic MMR gene mutations. Missense mutations may account for some LS cases that may be missed by IHC alone. Strict adherence to proper interpretation of IHC with attention to staining intensity and the status of heterodimer partner protein will prevent many potential misses.
Keywords: Colorectal cancer, mismatch repair protein, immunohistochemistry, missense mutation, double somatic mutation, unexpected staining, MMR IHC
Introduction
Assessment of mismatch repair (MMR) expression by immunohistochemistry (IHC) is the cornerstone of universal screening for Lynch syndrome (LS) at most institutions. We expect expression of MMR protein will be lost for any MMR gene that has a pathogenic germline mutation with a second hit, double somatic mutations, or, in the case of MLH1, methylation of the promoter. While most cases encountered during daily practice are straight-forward with the expected staining pattern and intensity of MMR proteins, occasional cases exist that may show weak tumor staining compared to internal control or show discordant IHC findings compared to microsatellite instability (MSI) testing. Some cases even show retained intact IHC expression in the presence of known pathogenic mutations of the corresponding MMR gene. This phenomenon has been speculated to be more frequent in LS cases due to pathogenic or hypofunctional missense mutations, which lead to expression of a nonfunctional protein with retained antigenicity [1–4]. In addition, we hypothesize that double somatic mutation cases could also have unexpected MMR expression, because these mutations are often at low variant allele fractions indicating that a subpopulation of cells without both somatic mutations may retain expression. However, little is known regarding the frequency of retained staining due to missense mutation and double somatic MMR mutations as compared to tumors due to other pathogenic MMR mutations in LS. In this study, our goals were to 1) study the frequency of unexpected MMR expression in colorectal cancers (CRC) among LS patients with missense mutations, LS patients with truncating mutations, as well as cases with double somatic MMR mutations, and 2) evaluate if the unexpected MMR expression is more common in any of these scenarios.
Materials and Methods
3312 adults with colorectal cancer diagnosed from 1/1/2013 to 12/31/2016 were evaluated in our statewide initiative (Ohio Colorectal Cancer Prevention Initiative, ClinicalTrials.gov identifier: NCT01850654). Institutional Review Board approval and written informed consent were obtained. Mismatch repair-deficient tumors were identified by MMR IHC and/or MSI PCR (Promega MSI Analysis System (Version 1.2) with five repeat markers (BAT-25, BAT-26, NR-21, NR-24, MONO-27). Tumors with ≥ 2 of 5 markers showing instability were classified as MSI-high (MSI-H). If MSI-H or absent MLH1 staining, MLH1 methylation was analyzed by pyrosequencing (with ≥15% methylation classified as positive).
MMR immunostaining performed at The Ohio State University Wexner Medical Center used the following commercial antibodies: MLH1 (Leica/Novacastra, Clone ES05, Buffalo Grove, IL), PMS2 (BD Pharmingen, Clone A16–4, San Jose, CA), MSH2 (Calbiochem, Clone FE11, Basel-Land, Switzerland), MSH6 (Epitomics, Clone EP49, Burlingame, CA). MMR IHC slides were available for review of 82 patients with MMR deficiency (without methylation) and germline Next Generation Sequencing (NGS) results. A subset (26 cases) also have tumor sequencing results indicating the presence of double somatic MMR gene mutations. MMR IHC slides on these 82 cases were reviewed without the knowledge of mutation status. The MMR protein expression level (strength and percentage of tumor cells stained) was quantified manually. Background lymphocytes and basal crypt epithelial cells served as internal control for assessing staining intensity in the tumor nuclei. For this study, intact staining was defined as tumor staining equal to or greater than internal positive control, in ≥ 5% of tumor nuclei. Staining that was present in < 5% of tumor nuclei was considered absent/loss of staining. If tumor cells stained weaker than the control and the staining was present in ≥ 5% of tumor nuclei, the case was flagged as abnormal staining pattern.
Patients with MMR deficiency without MLH1 methylation underwent germline NGS panel testing (25 to 66 cancer genes, ColoSeq or BROCA, University of Washington). Genomic regions were captured using biotinylated RNA oliognucleotides (SureSelect) and sequenced on an Illumina HiSeq2000 instrument [5]. Large rearrangements were detected [6]. If no germline MMR mutation was found in these cases, tumor sequencing (ColoSeqTumor or Oncoplex) of the MMR genes (MLH1, PMS2, MSH2, MSH6, EPCAM) was performed. Data was created by the UW NGS Laboratory and Analytics group.
Statistical analysis was performed using Fisher’s Exact test and unpaired t test.
Results
Patient Characteristics
Of the 3312 CRC cases, we included a total of 82 cases that had MMR IHC slides available for review, including 56 cases with pathogenic germline MMR mutations and 26 cases with double somatic MMR mutations identified via tumor sequencing. Average age for the 82 patients was 52 years (20 to 84 years). There is no gender predilection, with a male to female ratio of 1.1 to 1. Forty-five of eighty-two (55%) of the CRCs were located in the right colon versus thirty-seven (45%) in the left colon.
MMR Immunohistochemistry and Molecular Characteristics
Among the 82 patients evaluated for this study, 56 had pathogenic germline MMR gene mutations and 26 had double somatic MMR gene mutations. 16 of 82 (20%) MMR-defective cases showed unexpected MMR expression. Detailed case data are summarized in Tables 1 and 2. Specifically, unexpected MMR expression with defective MMR was more common in LS cases with missense mutations (4 of 9, 44%) than LS cases with truncating mutations (5 of 47, 11%, p=0.0287). Of note, double somatic MMR mutation group (7 of 26, 27%) also showed a trend of more unexpected MMR expression than LS cases with truncating mutations although not statistically significant (p=0.1006). There was no statistically significant difference when comparing the rate of unexpected MMR expression in LS with missense mutation group versus double somatic MMR mutation group (p=0.4159).
Table 1.
Expression of Mismatch Repair (MMR) Proteins by Immunohistochemistry in 82 MMR-Deficient Colorectal Cancers without MLH1 Methylation.
| Cases (n) | MLH1 | PMS2 | MSH2 | MSH6 | |
|---|---|---|---|---|---|
| Lynch syndrome, Missense Mutations (9) | Unexpected Staining (4) | 0 | 2 | 0 | 2 |
| Expected Staining (5) | 0 | 0 | 4 | 1 | |
| Lynch syndrome, Truncating Mutations (47) | Unexpected Staining (5) | 1 | 1 | 0 | 3 |
| Expected Staining(42) | 6 | 8 | 21 | 7 | |
| Double Somatic MMR Mutations (26) | Unexpected Staining (7) | 6 | 0 | 1 | 0 |
| Expected Staining(19) | 14 | 0 | 3 | 2 | |
Table 2.
Unexpected MMR Expression in MMR-Deficient Colorectal Cancers without MLH1 Methylation.
| Cases (n) | Cases with Retained MMR Expression n (%) | # | Age (yr) | Sex | Site | MSI | MMR Gene Mutation | Unexpected Expression of Afflicted MMR Gene by IHC* (% staining) | Expression of Corresponding Partner Gene by IHC (% staining) |
|---|---|---|---|---|---|---|---|---|---|
| Lynch Syndrome with Missense Mutations (9) | 4 (44%) | 1 | 48 | F | Rectum | MSS* | PMS2 c.137G>T, p.S46I; VUS (c.-7T>C) | PMS2 Intact* | MLH1 Intact |
| 2 | 44 | M | Descending | MSI-H* | PMS2 c.2113G>A, E705K | PMS2 WTC* (75%) | MLH1 Intact | ||
| 3 | 49 | M | Sigmoid | MSI-H | MSH6 c.1109T>C, p.L370S | MSH6 Intact | MSH2 Intact | ||
| 4 | 47 | M | Sigmoid | MSI-H | MSH6 c.1109T>C, p.L370S | MSH6 WTC (30%) | MSH2 Intact | ||
| Lynch Syndrome with Truncating Mutations (47) | 5 (11%) | 5 | 46 | F | Rectum | MSI-H | MLH1 c.2252_2253delAA, p.K751Sfs*3; MSH2 VUS (c.80C>T, p.P27L) | MLH1 Intact | PMS2 WTC (5%) |
| 6 | 29 | M | Rectum | MSS | PMS2 c.1281del, p.H428Tfs*20 | PMS2 Intact | MLH1 Intact | ||
| 7 | 51 | M | Rectum | MSI-H | MSH6 del promoter-exon 1 | MSH6 Intact | MSH2 Intact | ||
| 8 | 51 | M | Cecum | MSI-H | MSH6 c.3840_3846del, p.E1281Lfs*44 | MSH6 WTC (20%) | MSH2 Intact | ||
| 9 | 34 | M | Rectum | MSI-H | MSH6 c.3840_3846del, p.E1281Lfs*44 | MSH6 WTC (50%) | MSH2 WTC (80%) | ||
| Double Somatic MMR Mutations (26) | 7 (27%) | 10 | 59 | F | Ascending | MSI-H | MLH1 Hit 1: c.95T>A, p.I32N; Hit 2: LOH | MLH1 WTC (10%) | PMS2 Loss |
| 11 | 61 | M | Cecum | MSI-H | MLH1 Hit 1: c.2054C>A, p.S685Y; Hit 2: LOH | MLH1 WTC (10%) | PMS2 Loss | ||
| 12 | 61 | M | Ascending | MSI-H | MLH1 Hit 1: c.199G>A, p.G67R; Hit 2: LOH | MLH1 WTC (10%) | PMS2 WTC (30%) | ||
| 13 | 39 | M | Ascending | MSI-H | MLH1 Hit 1: c.2135G>A, p.W712X; Hit 2: c.2041G>A, p.A681T | MLH1 WTC (20%) | PMS2 Loss | ||
| 14 | 62 | M | Sigmoid | MSS | MLH1 Hit 1: c.298C>T, p.R100X; Hit 2: LOH | MLH1 WTC (75%) | PMS2 Loss | ||
| 15 | 60 | F | Transverse | MSI-H | MLH1 Hit 1: c.100G>A, p.E34K; Hit 2: LOH | MLH1 Intact | PMS2 Intact | ||
| 16 | 54 | M | Descending | MSI-H | MSH2 Hit 1: c.2038C>T, p.R680X; Hit 2: LOH | MSH2 WTC (>95%) | MSH6 WTC (80%) | ||
IHC = Immunohistochemistry; Intact = Diffuse Strong Staining Equal to or Greater than Control; LOH=loss of heterozygosity; MMR=Mismatch Repair Protein; MSI-H= Microsatellite Instable-High; MSS=Microsatellite Stable; WTC=Weaker Than Control.
Interestingly, in the MMR double somatic mutation group that had unexpected staining, the majority cases (86%, 6 of 7) had loss of heterozygosity (LOH) as the second hit, whereas only one case had two sequencing changes. However, this was not statistically different when compared to the double somatic mutation cases with expected staining (p=1.000). Among the 19 double somatic mutation cases with expected staining, 74% (14 of 19) had LOH as the second hit.
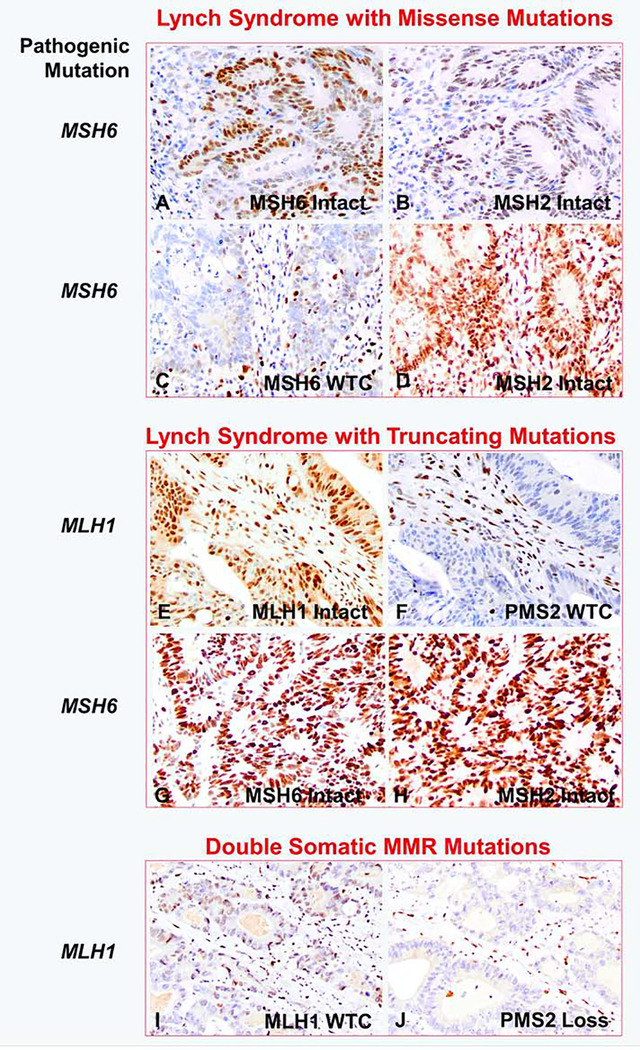
Among the 16 MMR-defective cases with unexpected MMR expression, the staining intensity and percentage of tumor cells stained were variable (Figure 1). 63% (10 of 16) cases demonstrated tumor staining weaker than control and 37% (6 of 16) cases showed intact (diffuse strong) staining. The ten cases with tumor staining weaker than control had a range of percent tumor cells stained (10% to >95%). In the clinical setting, such cases with tumor staining weaker than control may have been interpreted as abnormal or indeterminate by most pathologists and further workup may have been suggested, for example Figure 1C and 1F (weaker than control staining in 30% and 5% tumor nuclei respectively).
Figure 1.
Examples of unexpected protein expression in colorectal cancers with pathogenic mismatch repair gene mutation. (A-B) Lynch syndrome with germline MSH6 missense mutation (case #3), showing intact MSH6 and MSH2 staining; (C-D) Lynch syndrome with germline MSH6 missense mutation (case #4), showing retained but weaker than control MSH6 staining in 30% tumor, MSH2 staining is intact; (E-F) Lynch syndrome with germline MLH1 truncating mutation (case #5), showing intact MLH1 staining, but weaker than control PMS2 staining in 5% tumor; (G-H) Lynch syndrome with germline MSH6 truncating mutation (case #7), showing intact MSH6 and MSH2 staining; (I-J) Sporadic colorectal cancer with double somatic MLH1 mutations (case #10), showing weaker than control MLH1 staining in 10% tumor and loss of PMS2 expression. WTC: weaker than internal control. MMR, mismatch repair Original magnification: 400x.
The unexpected MMR protein expression was found in cases with pathogenic mutations in all four MMR genes. Of MLH1, PMS2, MSH2, MSH6 pathogenic mutations (including all germline and double somatic mutation cases), 26%, 27%, 3%, and 33% had unexpected immunoreactivity respectively. Of note, among the four LS cases with missense mutations and unexpected staining, two cases were associated with a recurrent mutation in MSH6 c.1109T>C, p.L370S.
Discussion
The phenomenon of retained expression of protein with certain molecular alterations of the corresponding gene is uncommon but well documented. Retained MLH1 staining has been identified in some MLH1 mutation or hypermethylation cases [2,4,7–9]. It has been suggested that this is due to missense mutation that lead to a still expressed, albeit non-functional, protein. Missense mutation has been speculated to be the most common genetic alteration associated with this phenomenon [1,3,4]. In addition, we hypothesize that double somatic mutation cases could also have unexpected MMR expression, since such mutations often occur at low variant allele fractions indicating that a subpopulation of cells without both somatic mutations may retain expression. Rarely, apparent ‘normal’ expression of protein can also be seen with truncating mutations. It has been reported that TP53 truncating mutations with c-terminal stopgain can result in detectable but non-functional p53 protein yielding a normal wild-type staining pattern [10]. We hypothesize that similar mechanism could explain the retained expression in defective MMR gene with truncating mutation, i.e. despite the truncating mutation a small fragment of protein is still made, which contains an intact antibody-binding site for the immunohistochemistry to work. Lastly, it is also possible that tumor with unexpectedly retained expression from a LS patient is an incidental cancer that is MMR proficient and unrelated to the germline mutation.
Here we demonstrated that among 82 CRC with LS or double somatic MMR mutations, 16 cases had unexpectedly retained MMR protein expression. This finding of some degree of nuclear staining despite pathogenic mutations was statistically more common in LS cases with missense mutations (44%, 4 of 9) than LS cases with truncating mutations (11%, 5 of 47, p=0.0287). There is a trend of more unexpected MMR expression in the double somatic MMR mutation group (27%, 7 of 26) than the truncating germline LS group (p=0.1006), and there is no statistically significant difference between the LS with missense mutation group and double somatic MMR mutation group (p=0.4159), suggesting that double somatic MMR mutation cases may also be more likely to have unexpected IHC staining patterns. In a recent study by Hechtman et al. [11], the authors also found that retained MMR expression was more commonly seen in tumors from patients with missense mutations of the MMR genes in a cohort composed of CRC, endometrial cancer, and various other cancer types. They found that 69% (25) of MSI-high/MMR-IHC discordant cases harbored either a pathogenic germline or somatic MMR missense mutation, while only 16% (6) of the control group (MSI-high/MMR IHC-deficient cases) harbored a pathogenic germline or somatic MMR missense mutation (p=0.0001).
Unexpected staining with pathogenic mutations were found with all four MMR genes in the 16 cases in this study, and all these IHC stains were performed on resections, except one biopsy of liver metastasis (case#7), in house at the Ohio State University. Of MLH1, PMS2, MSH2, MSH6 pathogenic mutations (including all germline mutation and double somatic mutation cases), 26%, 27%, 3%, and 33% had unexpected staining respectively. In an interobserver interpretation variability study of MMR IHC, Klarskov et al [12] observed that CRC cases with tumor staining weaker than control was the main source of reduced consensus. Such weak staining was present in all MMR proteins, from the most to the least frequently seen - MLH1, MSH6, PMS2, and MSH2. Mangold et al. [8] reported that 34% tumors from MLH1 germline mutation carriers exhibit weak or partial MLH1 staining. One plausible reason is that a high proportion (more than one third) of all MLH1 and MSH6 pathogenic genetic alterations are missense mutations, despite the fact that the majority of LS-associated MMR gene alterations are frameshift or nonsense mutations with resultant truncated proteins [13], but this cannot explain the high rate of unexpected staining we found among PMS2 cases in our cohort.
The levels of MMR protein expression in the presence of mutations, as indicated by staining intensity on IHC, can be highly variable from weak in 10% tumor nuclei to strong diffuse staining in almost all tumor nuclei. This variation of staining intensity could be related to how much the antigenic epitopes are altered and the residual binding ability to antibodies. Variable degrees of immunostaining have also been described in other genes with missense mutation, for example, Fumarate hydratase in uterine leiomyomata [14]. However, we should point out that rare aberrant staining patterns such as granular/speckled nuclear staining and nuclear membrane staining have been reported with MMR deficient cases, and these aberrant staining patterns are thought to be related to technical issues, and should not be interpreted as evidence of preserved staining [15]. None of our cases with unexpectedly retained staining showed the above aberrant nuclear staining pattern. The effects of chemoradiation have been previously shown to impact MMR IHC by decreased or loss of staining [15–18]. While IHC was performed in the post-therapy setting for all 5 rectal cancers with unexpected staining, retained MMR staining in the presence of molecular abnormality is not expected; therefore the retained staining in our rectal cases are unlikely to be therapy-related artifact.
This variable retained staining seen with defective MMR cases begs the questions: ‘What is considered intact MMR staining?’ and ‘How can we better identify defective MMR cases that masquerade with retained staining?’ The cutoff value for what is considered intact MMR staining ranges in the literature from any convincing staining [19], 1%, 5% [20], up to 10% [15, 21]. Regardless of what the cutoff value is, one key factor is the evaluation of the staining intensity – MMR staining in the tumor nuclei must be equal to or stronger than internal control to be considered ‘intact’. In this study, we found that 63% (10 of 16) MMR deficient cases with unexpected staining demonstrated tumor staining weaker than control. If staining weaker than control is confirmed on a repeat staining, the result should be interpreted as abnormal and additional studies are warranted [22]. However, not all defective MMR tumors with retained protein expression show weaker than control staining; strong diffuse staining can be seen. Clinical suspicion (family or personal history of cancer and patient age) as well as the staining of the heterodimer partner, may help suggest that additional tests are needed to identify a mutation. For instance, MLH1 mutation cases (especially with missense mutations) may show retained and strong MLH1 staining; but examination of its heterodimer partner PMS2 often reveals isolated loss of PMS2 staining, which should prompt additional studies thus avoiding missing such cases (case #10, Figure 1 I&J) [23,24].
As we and others have previously demonstrated, both MMR IHC and MSI are reliable screening tests for Lynch syndrome, although they are not perfect and occasional cases will be missed by both methods [7, 25–28]. Unexpectedly retained MMR staining could contribute to missed Lynch syndrome cases when using MMR IHC screening alone. Fortunately, understanding the nuances of MMR IHC interpretation as discussed above may help avoid pitfalls in most cases. As shown in this study, 9 of 56 (16%) LS cases had unexpected staining. Among these 9 cases, 4 exhibited weaker staining than control and one case showed abnormal staining in the partner gene (case #5), so these 5 cases should have been flagged as abnormal and should not have been missed. For the remaining 4 of 56 (7%) LS cases with retained diffuse strong intact staining (cases #1, 3, 6, 7), 3 patients were younger than 50 years old and additional workup would be suggested according to current guidelines for early onset CRC [29,30].
These observations highlight the importance of tumor staining intensity in comparison to control during MMR IHC interpretation, and the evaluation of which should be a prerequisite even before determining if there is certain percentage staining. At our institution, if tumor nuclei stain weaker than control, the case is interpreted as abnormal and further workup is needed. Tumor sequencing by multigene panel with genetic counseling is suggested, particularly for patients with a strong family history or if the age of diagnosis is less than 50 years [29–31]. A limitation of this study is the small number of CRC cases that had unexpectedly retained staining, future studies with more cases may be helpful to avoid sampling bias.
Conclusions
Unexpected expression of MMR proteins may occur in both LS cases and cases with double somatic MMR mutation, a potential pitfall in the screening process when using IHC only. Such MMR protein expression is most commonly seen with pathogenic germline missense mutations, but it is not limited to these mutations. In addition, it may be more common in double somatic MMR mutation cases where one of the mutations is loss of heterozygosity. Cautious interpretation of MMR IHC is advised when dealing with tumor staining weaker than control regardless of percentage of tumor staining, as these cases may harbor pathogenic MMR gene mutations. Missense mutations appear to account for some, but not all CRC cases that may be missed in LS screening by IHC alone. Strict adherence to proper interpretation of IHC with attention to staining intensity and the status of heterodimer partner protein will prevent many potential misses.
Acknowledgements
The research was approved by Ohio State University Institutional Review Board (IRB). The data reported here were derived from the Ohio Colorectal Cancer Prevention Initiative, which is supported by a grant from Pelotonia, an annual cycling event in Columbus, Ohio that supports cancer research at The Ohio State University Comprehensive Cancer Center – James Cancer Hospital and Solove Research Institute. This study was supported in part by grant P30 CA016058, National Cancer Institute, Bethesda, MD.
The authors would like to thank Shawn Scully for his assistance with image production. Part of this work was presented as a poster at the United States & Canadian Academy of Pathology (USCAP) 2019 Annual Meeting in National Harbor, Maryland
Footnotes
Conflict of Interests:
Ms. Hampel is on the scientific advisory board for InVitae Genetics, Genome Medical and Promega, has conducted collaborative research with Myriad Genetics Laboratories, Inc, Ambry Genetics, and InVitae Genetics, and has stock in Genome Medical. Ms. Pearlman has done collaborative research with Ambry Genetics, Myriad Genetics Laboratories, Inc, and InVitae Genetics. Drs. Chen, Jones, Zhao, Alsomali, Frankel, and Ms. Knight have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dieumegard B, Grandjouan S, Sabourin JC, et al. Extensive molecular screening for hereditary non-polyposis colorectal cancer, Br J Cancer 2000;82:871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shia J, Holck S, Depetris G, Greenson JK, Klimstra DS. Lynch syndrome-associated neoplasms: a discussion on histopathology and immunohistochemistry. Fam Cancer 2013;12:241–60. [DOI] [PubMed] [Google Scholar]
- [3].Thibodeau SN, French AJ, Roche PC, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 1996;56:4836–40. [PubMed] [Google Scholar]
- [4].Wahlberg SS, Schmeits J, Thomas G, et al. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res 2002;62:3485–92. [PubMed] [Google Scholar]
- [5].Pritchard CC, Smith C, Salipante SJ et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing, J Mol Diagn 2012;14:357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data, BMC genomics 2011;12:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].de JPong AE, van Puijenbroek M, Hendriks Y, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer, Clin Cancer Res 2004:10:972–80. [DOI] [PubMed] [Google Scholar]
- [8].Mangold E, Pagenstecher C, Friedl W, et al. Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining, J Pathol 2005:207(4):385–95. [DOI] [PubMed] [Google Scholar]
- [9].Salahshor S, Koelble K, Rubio C, Lindblom A. Microsatellite Instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer, Lab Invest 2001;81:535–41. [DOI] [PubMed] [Google Scholar]
- [10].Köbel M, Ronnett BM, Singh N, et al. Interpretation of P53 Immunohistochemistry in Endometrial Carcinomas: Toward Increased Reproducibility. Int J Gynecol Pathol 2019:38 Suppl 1(Iss 1 Suppl 1): S123–s131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hechtman JF, Rana S, Middha S, et al. Retained mismatch repair protein expression occurs in approximately 6% of microsatellite instability-high cancers and is associated with missense mutations in mismatch repair genes, Mod Pathol 2020;33:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Klarskov L, Ladelund S, Holck S, et al. Interobserver variability in the evaluation of mismatch repair protein immunostaining.” Hum Pathol 2010;41(10): 1387–1396. [DOI] [PubMed] [Google Scholar]
- [13].Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database, Disease markers 2004;20: 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Joseph NM, Solomon DA, Frizzell N, Rabban JT, Zaloudek C, Garg K. Morphology and Immunohistochemistry for 2SC and FH Aid in Detection of Fumarate Hydratase Gene Aberrations in Uterine Leiomyomas From Young Patients, Am J Surg Pathol 2015;39:1529–39. [DOI] [PubMed] [Google Scholar]
- [15].Pai RK, Pai RK. A Practical Approach to the Evaluation of Gastrointestinal Tract Carcinomas for Lynch Syndrome, Am J Surg Pathol 2016;40:e17–34. [DOI] [PubMed] [Google Scholar]
- [16].Bao F, Panarelli NC, Rennert H, Sherr DL, Yantiss RK. Neoadjuvant therapy induces loss of MSH6 expression in colorectal carcinoma. Am J Surg Pathol. 2010;34(12):1798–1804. [DOI] [PubMed] [Google Scholar]
- [17].Radu OM, Nikiforova MN, Farkas LM, Krasinskas AM. Challenging cases encountered in colorectal cancer screening for Lynch syndrome reveal novel findings: nucleolar MSH6 staining and impact of prior chemoradiation therapy. Hum Pathol. 2011;42(9):1247–1258. [DOI] [PubMed] [Google Scholar]
- [18].Vilkin A, Halpern M, Morgenstern S, et al. How reliable is immunohistochemical staining for DNA mismatch repair proteins performed after neoadjuvant chemoradiation? Hum Pathol. 2014;45(10):2029–2036. [DOI] [PubMed] [Google Scholar]
- [19].Bartley AN, Hamilton SR, Alsabeh R, et al. Template for reporting results of biomarker testing of specimens from patients with carcinoma of the colon and rectum, Arch Pathol Lab Med 2014;138:166–70. [DOI] [PubMed] [Google Scholar]
- [20].Markow M, Chen W, Frankel WL. Immunohistochemical Pitfalls: Common Mistakes in the Evaluation of Lynch Syndrome, Surg Pathol Clin 2017;10:977–1007. [DOI] [PubMed] [Google Scholar]
- [21].Sarode VR, Robinson L. Screening for Lynch Syndrome by Immunohistochemistry of Mismatch Repair Proteins: Significance of Indeterminate Result and Correlation With Mutational Studies, Arch Pathol Lab Med 2019;143:1225–1233. [DOI] [PubMed] [Google Scholar]
- [22].Chen W, Frankel WL. A practical guide to biomarkers for the evaluation of colorectal cancer, Mod Pathol 2019;32(Suppl 1):1–15. [DOI] [PubMed] [Google Scholar]
- [23].Dudley B, Brand RE, Thull D, Bahary N, Nikiforova MN, Pai RK. Germline MLH1 Mutations Are Frequently Identified in Lynch Syndrome Patients With Colorectal and Endometrial Carcinoma Demonstrating Isolated Loss of PMS2 Immunohistochemical Expression, Am J Surg Pathol 2015;39:1114–20. [DOI] [PubMed] [Google Scholar]
- [24].Rosty C, Clendenning M, Walsh MD, et al. Germline mutations in PMS2 and MLH1 in individuals with solitary loss of PMS2 expression in colorectal carcinomas from the Colon Cancer Family Registry Cohort, BMJ Open 2016;6:e010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352(18): 1851–1860. [DOI] [PubMed] [Google Scholar]
- [26].Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 2008; 26(35): 5783–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sachak T, Pearlman R, Hampel H, et al. Next generation sequencing sheds light on Lynch syndrome screening using immunohistochemistry vs. microsatellite instability by polymerase chain reaction. USCAP 2018 Abstracts: Gastrointestinal Pathology. Lab Invest 2018;98:300. [Google Scholar]
- [28].Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20(4):1043–1048. [DOI] [PubMed] [Google Scholar]
- [29].National Comprehensive Cancer Network clinical practice guidelines in oncology (NCCN guidelines): Genetic/Familial high-risk assessment: Colorectal. Version 3.2019 – December 13, 2019 Accessed 6/26/2020 https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf
- [30].Pearlman R, Frankel WL, Swanson B et al. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol 2017; 3: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hampel H, Pearlman R, Beightol M, et al. Assessment of tumor sequencing as a replacement for lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 2018; 4(6): 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]