Abstract
The three-dimensional organization of the genome within the cell nucleus has come into sharp focus over the last decade. This has largely arisen because of the application of genomic approaches that have revealed numerous levels of genomic and chromatin interactions, including topologically associated domains (TADs). The current review examines how these domains were identified, are organized, how their boundaries arise and are regulated, and how genes within TADs are coordinately regulated. There are many examples of the disruption to TAD structure in cancer and the altered regulation, structure and function of TADs are discussed in the context of hormone responsive cancers, including breast, prostate and ovarian cancer. Finally, some aspects of the statistical insight and computational skills required to interrogate TAD organization are considered and future directions discussed.
Keywords: topological associated domain, prostate cancer, breast cancer, ovarian cancer, CTCF, cohesin, enhancer
1. Topologically Associated Domains in gene regulation.
1.1. The three-dimensional organization of the nucleus
The seventeenth century development of microscopes enabled observation of biological structures, such as the eye of a fly, at a previously unprecedented level of detail. These observations led to Matthias Schleiden and Theodor Schwann in 1838 to spearhead the development of cell theory (reviewed in(Franke, 1988)), which proposed that cells were the basic unit of structure of all organisms, and that they were produced form preexisting cells. With ever-increasing microscope power, cells were revealed to contain numerous sub-structures, both in the cytoplasm and the nucleus, and that indeed the nucleus was a functionally-vital structure within the cell. For example, although there is considerable variability in the size, shape and longevity of cells in the human body, the nucleus they contain is remarkably consistent, being approximately 6 μM in diameter, representing approximately 10% of the cell volume. Of course, of key importance, the nucleus contains the same genetic material.
Given its constant shape and ubiquitous nature, the nucleus has been the subject of intense study for the last several centuries, and led to the identification of sub-structures and organization. For example, analyses of the cells of sea urchin and other readily accessible model organisms revealed that the nucleus which had previously been perceived as a dark sea of unknown function, was in fact richly detailed with sub-features. These newly identified structures included the so-called “A/B regions” which contained the so-called “Active” central part of the nucleus and the B inactive region at the periphery; the nucleolus; various “speckled features” including the olfactory region; and chromosomal territories. Thus, the nucleus actually contained structural components that were not static, but instead the chromosomes and other features moved on currents through the cell cycle and during differentiation. Determining how these nuclear structures and movements relate to gene regulatory processes has been a central research focus for much of the last 100 years.
Charting the topography of the nucleus was profoundly catalyzed by the publication in 2001 of the draft human genome (Wright et al., 2001, Wolfsberg et al., 2001, Olivier et al., 2001), which ushered in the genomics era of research. Genomic-centered research necessitated and was further catalyzed by the development of bioinformatics and statistical approaches, combined with sequencing technologies of ever-increasing precision and throughput. These statistical approaches, computational developments and technology advancements all combined to develop an unprecedented understanding of the content and structure of the nucleus, and the diversity and regulation of nucleic acids. Arising from these interdisciplinary efforts has been the concept the three-dimensional (3D) genome (reviewed in (Rowley et al., 2018, Spielmann et al., 2018, Gibcus et al., 2013)).
1.2. Identification of Topologically Associated Domains
Following on from the discovery of these physical structures within the nucleus and their movements, many workers focused on understanding gene regulation mechanisms with the goal to understand how genic features, such as gene enhancer regions bring about coordinated gene expression, and of which gene(s).
A well-studied and revealing gene regulatory scenario has been the analyses of the immunoglobulin (IgG) region that includes the beta and gamma-globin genes; β-globulin is initially transcribed in the fetal liver but becomes fully active during erythropoiesis in the adult bone marrow, whereas γ-globulin is strongly expressed in fetal liver and very lowly expressed in the adult bone marrow. The IgG region has become a well-examined model to investigate enhancer-gene regulation relationships, due to their essential functions and disruptions in various leukemias (reviewed in(Chenoweth et al., 2015, Heijnen et al., 1997, Unkeless, 1989)). Initial studies reasoned that open chromatin sites within the locus, identified by DNAase hypersensitivity, would be accessible to transcription factors and were used to identify the key features required for these dynamic gene expression patterns(Hanscombe et al., 1991, Blom van Assendelft et al., 1989). Subsequently, studies by Fraser and colleagues extended this concept to reveal that enhancer-gene communication existed across large genomic distances(Carter et al., 2002, Dillon et al., 1997).
The concept of inter and intra-chromosomal functional interactions was supported by earlier studies in sea urchin that had observed chromosomal co-segregation and physical contact in an ordered manner (Arceci et al., 1980, Wang et al., 1979, Kojima, 1960). Subsequently, non-homologous chromosomal contacts were identified in the ribosome, which includes ~ 300 genes in the nucleolus from five different chromosomes. Similarly, hundreds of olfactory genes from multiple chromosomes converge often in heterochromatic loci and are coordinately regulated(Olender et al., 2016, Gilad et al., 2003).
However, to catalog the potential for large scale and dynamic interactions within and between chromosomes required new technologies and development of statistical approaches. Specifically, chromosome cross-talk was comprehensively addressed by the development of chromatin capture techniques (reviewed in(Fu et al., 2018, Chang et al., 2018, Denker et al., 2016)). These approaches leverage the possibly of capturing the physical proximity of widely spaced regions and applying next generation sequencing technologies to define the location and extent of these associations.
In this manner, and with improvements in computational power and refinements to statistical modelling, ever more complex maps of chromatin loop formation were generated. In turn this gave rise to the concept that these loop structures were contained within topologically associated domains (TADs)(Nora et al., 2012, de Laat et al., 2013, Dixon et al., 2012). In parallel, other approaches revealed that lamin-associated domains (LADs) were also organized and were associated with movement of chromosomes to the outer lamina membrane of the nucleus. Remarkably, TAD structure appears to be essentially invariant across cells within a given organism, and in humans they range from ~ 100 kb to ~ 1 mb, and across cell types there are somewhere between 500 – 1500 TADs that can cover much of the genome. However, between cell types there is significant variation in the activity of the contents of each TAD (reviewed in (Wang et al., 2018a, Krumm et al., 2018, Roy et al., 2018)).
There is evidence for TAD structure to be conserved between Drosophila, mice and humans. Indeed, TAD structure is highly conserved over syntenic chromosomal regions and it is tempting to speculate that over evolution this has influenced how evolutionary pressures have selected genomic changes within and outside TADs as alterations are more common outside of TADs than inside, suggesting they are selected against when they occur(Krefting et al., 2018, Gong et al., 2018, Harmston et al., 2017).
The precise function and significance of TAD structure is still a topic of significant investigation but a consensus is that this is the unit of organization for the genome, and has arisen as it gives a number of biological advantages. Namely, it allows for coordination of gene regulation programs, and the integration of transcriptional signals (Dixon et al., 2016) (Figure 1A). That is there are many more enhancers across the genome than there are genes to be regulated; it is estimated that there are potentially as many as 10 enhancers for every gene, and that these enhancers can be often exceedingly distal from target genes (Furlong et al., 2018, Ron et al., 2017, Murakawa et al., 2016, Schmidl et al., 2014). By enclosing multiple genes within a regulatory TAD region, promiscuous regulation is limited, and also for those genes within the TAD coordinated regulation is more readily established. It has also emerged that TADs are hierarchical and contain smaller regions of looping within them(Racko et al., 2019, Hansen et al., 2018).
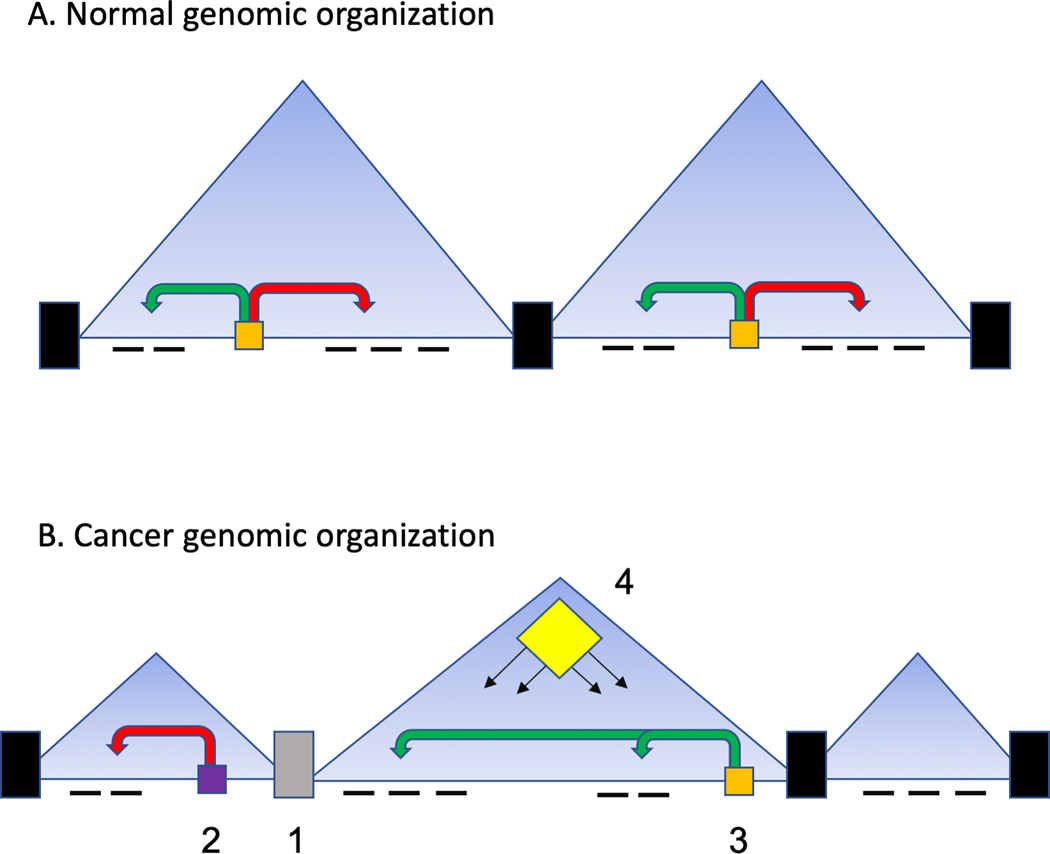
Figure 1: TAD organization in health and disease.
A. In normal cells TADs are organized such that boundaries arise by the action of multi-protein complexes centered on CTCF and cohesin complexes. These act to insulate the contents of the TAD which range in size from 100 kb to 1–2 mb. Within a TAD, enhancers (in orange) regulate target genes through further looping events and exert positive and negative impacts on gene expression. Often these gene expression patterns are highly coordinated and governed by multiple enhancer interactions.
B. In cancer cells there many examples of these process being corrupted. There is good evidence that boundary function is altered (1) with loss and gain of boundaries and changing the TAD structure. Generally, it appears the cancer genome gains more TADs of shorter length. This change of TAD structure can change enhancer function (2, 3) such that previously insulated genes are subsequently regulated by new enhancers. This is further distorted by structural variations impacting boundary function (1) and enhancer responses (2,3). The coordinated gene regulation within a TAD also allows for emergent changes in epigenetic regulation (4). That is, mutations in epigenetic regulators such as EZH2 can modestly change the regulation of individual genes in a TAD, but the collective effect on all genes is pronounced if all the genes are on a pathway for example.
Thus, in general terms the TAD structure allows for the coordinated regulation of genes, although as with all complex biological phenomena, there is diversity in the level of organization and response. That is, the TAD structure is not the mammalian equivalent of a bacterial operon. There are strong examples of co-regulated genes within a TAD, and specific TADs are beginning to be characterized by investigators. For example, the regulation of ~1 Mb TAD on chr 5 containing the proto-cadherins has emerged as a well-investigated example (Jiang et al., 2017, Symmons et al., 2014, Dixon et al., 2012, Nora et al., 2012). Whilst these individual loci are informative, it is clear there remain areas of ambiguity in terms of how TADs are formed and regulated that suggest other mechanisms participate in gene regulation(Luo et al., 2018, Barutcu et al., 2018, Rodriguez-Carballo et al., 2017).
Therefore, the view has emerged that the TAD can be considered as the functional unit of the genome being strongly conserved between cell types. A significant focus of TAD research has been to establish what mechanisms drive TAD boundary formation, and how these processes are disrupted in diseases, including cancer.
1.3. CCCTC-Binding Factor roles in TAD structure and genomic functions.
Principal amongst the proteins that organize TAD boundaries and therefore define TAD structure is the DNA binding factor CCCTC-Binding Factor (CCTF), which contains 11 zinc finger motifs and binds across the genome. Human CTCF was cloned by Filippova and colleagues in 1996 by examining regulatory sequences proximal to the MYC gene, and used these regions as bait to identify binding proteins(Filippova et al., 1996). This revealed the significant relationship between CTCF and MYC, in which CTCF is able to repress MYC expression. Analyses of the multiple CTCF zinc finger motifs that revealed differential binding abilities, and suggested significant dexterity in binding and function(Filippova et al., 2002). Of course, this intimate relationship with MYC directly implicated CTCF in cancer development, and catalyzed an intense arena of subsequent investigation.
Again, the β-globin gene locus offered further clues to the function of CTCF binding identified sites and supported the concept that CTCF could regulate gene expression, but also act to insulate and therefore control regulation (Bell et al., 1999). These discoveries suggested that these CTCF functions maybe common and conserved through vertebrates and necessary for precise gene regulation. Subsequently, Chromatin Conformation Capture (3C) technologies applied to the histocompatibility complex genes also revealed the coordination and insulation roles that were dependent on CTCF(Majumder et al., 2010, Majumder et al., 2008). Chromatin immunoprecipitation and microarray approaches (ChIP-chip) initially demonstrated that at promoters chromatin states and CTCF binding sites were highly similar across cell types, whereas binding at enhancer regions was highly variable and correlated with cell type specific gene expression(Heintzman et al., 2009, Smith et al., 2009).
These findings supported the concept that CTCF functions were in part dependent upon the genomic context of binding. Indeed, CTCF was shown to interact with numerous other proteins, and principal amongst these are members of the Cohesin complex(Katainen et al., 2015, Guo et al., 2012, Wendt et al., 2008). Cohesin-dependent TAD folding can bring distant enhancers into play with genes, and many known genomic binding factors including the Polycomb complex and YY1 all play a role in these functions(Donohoe et al., 2007). One process where CTCF has been extensively examined is during X inactivation(Donohoe et al., 2009), which has revealed that CTCF also interacts with noncoding lncRNA including TSIX and XIST to bring about silencing in part mediated by the repressor YY1. Indeed, YY1 heterozygote mice have altered TSIX and XIST expression which was pheno-copied by knockdown of CTCF(Yang et al., 2015, Spencer et al., 2011, Pugacheva et al., 2005). Similarly, FIRR lncRNA plays a role in X-chromosome inactivation and shown to be dependent on NCOR2 and SHARP binding(McHugh et al., 2015). CTCF also demonstrates very significant flexibility in terms of the interactions it undergoes with a wide array of other proteins(Shin, 2018, Ghirlando et al., 2016). Furthermore, there are multiple CTCF isoforms ranging from 55 to 130 kD and can modify each other’s behavior(Li et al., 2019b), and furthermore they are the target of a variety of post-translational modifications(de Wit et al., 2015, Holwerda et al., 2013, Del Rosario et al., 2019, Wang et al., 2012b). Analyses of CTCF genomic binding patters and chromatin organization has identified several patterns of looping(Handoko et al., 2011) including active and repressive loops that are functionally dependent upon size.
Together these findings support models of diverse interactions between CTCF and its isoforms, other repressors and lncRNA in a manner that is heavily influenced by genomic context. The complexity of these interactions, and their potential genomic impact, is heightened by the discoveries that the human genome contains approximately 30000 to 40000 CTCF binding sites(Narendra et al., 2015, Sanyal et al., 2012). Therefore, CTCF is biologically impactful as a result of the frequency and diversity of its genomic interactions.
In parallel to studies in cell models, transgenic murine studies have been undertaken and have revealed that homozygous CTCF deletion is embryonically lethal, whereas heterozygotes are viable but poor breeders. In CTCF heterozygote animals there is clear evidence for changes in TAD structure that impact enhancer promoter interactions(Kemp et al., 2014, Splinter et al., 2006). For example, one of phenotypes in these mice is disruption of glucose homeostasis due to loss of insulation around the Pax6 locus (Tsui et al., 2012).
The boundary function of CTCF was conclusively demonstrated in 2015 by Narendra and colleagues by examining regulation on the HOX locus, and established that CTCF insulates between regions of opposite HOX gene activity during development leading to activation of normally repressed members(Narendra et al., 2015, Ing-Simmons et al., 2015). In the context of this locus, CTCF insulates heterochromatin from euchromatin. More generally, it appears that TAD boundaries limit spread of activity across the genome, and in this way limit transcriptional noise from bidirectional enhancers. More recently, genome-editing approaches have allowed incremental removal of CTCF binding sites associated with the well-established Sox9–Kcnj2 TAD on chr 11 of the murine genome. These approaches demonstrated consistent gene regulation, but subsequent gene inversions and repositioning of boundaries led to disease phenotypes suggesting that boundary erosion is required but not always sufficient for aberrant gene expression and phenotypic consequences(Despang et al., 2019). Therefore, the early proposition for TAD structure to limit the spread of repressive heterochromatin is probably not a universal function. Rather it now appears that the TAD structure constrains whatever is contained, be it active or repressive chromatin, and for aberrant transcriptional function other genomic distortion mechanisms may need to occur.
1.4. Mechanisms of TAD boundary formation
Although ChIP-Seq reveals that ~90% of TAD boundaries contain CTCF binding, there are many more CTCF binding sites that are not contained within boundaries(Zhang et al., 2018, Lobanenkov et al., 2018), and TAD boundary seems to be initiated and maintained by multiple proteins(Huang et al., 2018, Marques et al., 2015, Wang et al., 2012a). A caveat to this is that all these structures are identified in large bulk cell populations, and single cell studies will need be applied (and technologies refined and developed) to address this issue accurately.
Although genetic and biochemical approaches have implicated different proteins in the formation of TAD boundaries, questions remain over how the protein complexes form, how are they guided, and how do they sustain boundary structure. Various models have been proposed to address this challenge including so-called “handcuff” and “extrusion” models(Dixon et al., 2016, Racko et al., 2018, Hansen et al., 2018). In the handcuff model CTCF and Cohesin have a common association, as is established with ChIA-Pet, which shows CTCF spanning TAD boundaries. That is, the Cohesin ring encircles chromatin fibers in a 30–40 nm ring and in this way, holds sister chromatids together.
Alternatively, a loop extrusion model has been proposed whereby DNA binding complexes scan along DNA whilst also being tethered to an anchor CTCF site and do not close until it finds another CTCF motif in a convergent orientation. This process would explain why CTCF motif orientation at boundaries are convergent(de Wit et al., 2015) and that loss of one CTFC motif interacting and TAD bounds shift to next one and in this manner gives a degree of inherent stability. However, this model is also incomplete. For example, fundamentally, it remains unknown what protein complex is able to scan large genomic distances of 100s of kb in opposite directions and what happens to the supercoils that would be arise in the wake.
Most likely, there is contribution from all these forces with looping from enhancer to promoter that is stabilized by protein complexes and topological changes trigger activation. Loops can be further coordinated in transcription factor hubs, which has been appreciated for several years. Pre-looped topologies give hubs that provide liquid phase transitions(Strom et al., 2017) to promote stabilize the hub. In this manner, the enhancer-promoter interactions have a higher diffusion rate and faster transcriptional responses.
Finally, it is interesting to note that TAD boundaries are often enriched for highly active housekeeping genes, and the nucleosome spacing is shorter resulting in greater stiffness(Hug et al., 2017). It is possible that changing the flexibility can impact boundary and the affinity of CTCF, and that these physical properties help to illuminate CTCF-independent boundaries.
2. Disruption to TAD structure and CTCF in hormone responsive cancers
2.1. Altered TAD structure and enhancer usage in hormone responsive cancers.
Given these important relationships between genomic organization and coordinate gene regulation, it is not surprising that disruption to TAD structure is strongly implicated in a wide variety of diseases. One of the clearest and earliest identified examples was of the WNT6/IHH/EPHA4/PAX3 TAD on chr 2. Disruption and variation in boundary regions can result in the ectopic expression of these powerful developmental transcription factors in developing limb buds and leading to profound phenotypes in mice and humans (Fabre et al., 2017, Koch, 2016, Lupianez et al., 2015, Cutrupi et al., 2018, Wu et al., 2017).
Similarly, epigenetic mechanisms can impact boundary formation. One of the earlier clues that de-regulated CTCF function was profoundly impactful was revealed through the altered DNA methylation at CTCF sites. Specifically, CpG methylation can impede CTCF binding and therefore remove insulator function as a driver of myotonic dystrophy (Lopez Castel et al., 2011, Filippova et al., 2001).
Altered CTCF status and function is strongly associated with cancer(Kemp et al., 2014, Splinter et al., 2006, Filippova et al., 2002). Murine modelling of its actions revealed that hemizygous deletion predisposes to cancer(Kemp et al., 2014). Indeed, CTCF deletion is associated with deregulated and increased levels of CpG methylation, and therefore directly links changes in CTCF function to both genomic and epigenomic disruption. Given the wide diversity of genomic and epigenomic alterations that occur in cancer it is not surprising that they can combine with mechanisms that control TAD boundary formation and can act as cancer-drivers (Figure 1B). Loss of the cohesin complex member, RAD21, leads to reduced intra-TAD chromatin interactions, and as a result increases TAD volume (Poterlowicz et al., 2017, Rodriguez-Carballo et al., 2017, Fudenberg et al., 2016). Targeted disruption of CTCF leads changes TAD sizes and is antagonized by cohesin changes(Sima et al., 2019, Luo et al., 2018, Barutcu et al., 2018).
There are strong examples of so-called “enhancer hijacking” in which genomic disruptions to TAD boundaries allows previously insulated enhancers access to neighboring genes and changes in expression(Haller et al., 2019, Martin-Garcia et al., 2019, Cuartero et al., 2018, Zimmerman et al., 2018, Ryan et al., 2015, Northcott et al., 2014). This has been observed on chromosome 3 in AML where changes in CTCF expression weakens boundary and allow oncogenes to be regulated by nearby enhancers(Luo et al., 2018). Deletion of the CTCF motif at a boundary can shift the TAD structure and change gene-regulation for example as has been shown for the HOX locus(Luo et al., 2018). Similar examples of such oncogenic hijacking of transcription factors is seen with NOTCH signaling in breast cancer(Petrovic et al., 2019), the nuclear receptor NR4A3 in acinic cell carcinomas (Haller et al., 2019) and in a similar manner Cyclin D highjacks enhancers from IgG locus in mantle cell lymphoma(Martin-Garcia et al., 2019). In other hijacking events a ~ 1.8 Mb TAD of chr 8 containing the MYC gene (Hyle et al., 2019) is extended to recruit enhancer regions that are themselves amplified in pediatric neuroblastoma (Zimmerman et al., 2018) by mechanisms that include insulator erosion. The net result is that enhancers associated with the lncRNA PVT1, downstream of MYC, are activated (Cho et al., 2018, Ren et al., 2019, Parolia et al., 2018). Pan-cancer approaches have identified altered regions containing enhancers that changed genes expression and provided integrative framework to begin to classify these events (Weischenfeldt et al., 2017).
TADs are enriched for specific histone modifications such as H3K37me3, and mutations to EZH2 change its ability to govern this histone modification within a TAD. As a result, they occur more frequently than expected by chance, and therefore a single mutation in an epigenetic regulatory protein can impact the regulation of multiple genes within a TAD. This has been demonstrated in lymphoma cells with wild-type or mutant EZH2, in EZH2 should normally target inactive TADs, and limit multiple genes to control differentiation but mutant EZH2 selectively target these genes and therefore disrupts this capacity(Donaldson-Collier et al., 2019).
Amongst hormone responsive cancers there are examples of altered TAD regulation and structure. One of the most extensively studied is in the case of prostate cancer (PCa). In normal murine prostate development there is clear evidence that the developmental transcription factor, TBx18, is important for prostate development (Negi et al., 2019) and the use Circularized Chromosome Conformation Capture (4C) was used to find interacting regions that were stage- and tissue-specific in normal prostate development. In this manner the authors were able to show enhancer interactions within a 1.5 Mb TAD on chr 9 that controlled TBx18, and reveal how dynamically these interactions change during differentiation.
To identify aggressive PCa phenotypes investigators in the Cancer Genome Atlas (TCGA)(Cancer Genome Atlas Research et al., 2013, Cancer Genome Atlas Research Network. Electronic address et al., 2015) consortium and other groups(Fraser et al., 2017) have applied integrative genomic approaches. These and earlier studies(Tomlins et al., 2005) identified common translocations between TMPRRS2 and ETS genes in PCa (Sanda et al., 2017, Tomlins et al., 2005). The TMPRSS2 gene is androgen responsive, and its translocation leads to androgen-driven overexpression of the ETS transcription factor ERG, or other ETS family members, and acts as a cancer-driver(Ulz et al., 2016, Wang et al., 2014, Wang et al., 2011). TMPRSS2:ERG fusions are being assessed as biomarkers(Tomlins et al., 2015) for precision medicine efforts that combine genomic information in algorithms to define disease state and predict treatment responses. This frequent translocation of ERG in prostate leads to significant changes in chromatin organization(Rickman et al., 2012, Rickman et al., 2010).
Susan Clark and coworkers have focused on dissecting how this translocation and other events drive PCa. In the first instance they demonstrated changes in enhancer interactions as a result of the TMPRSS2 translocation that may re-wire cancer signaling and lead to dysregulation of gene expression programs(Taberlay et al., 2014). Subsequently they used the chromatin conformation capture technique Hi-C to capture all genomic-genomic interactions in prostate models (PrEC, LNCaP, PC-3) coupled with analyses of CTCF boundaries, and other genomic histone modification data. In this manner they were able to develop a highly-textured annotation of epigenomic context and TAD associations in non-malignant and malignant cell models(Taberlay et al., 2016). This approach was able to demonstrate in PCa cell models that TAD boundaries were shifted, and new ones created, which overlapped with H3K4me3. That is, large TADs were divided into smaller TADs. As a result, 317 TADs were identified in PrEC whereas in the cancer cell models this increased to 622 (PC-3) and 1111 (LNCaP). Thus, it appears that larger TADs are normal, and divided in cancer systems that most likely impact enhancer usage (Figure 1B). There was also evidence that copy number variation impacts these structures. Finally, the anchor point associations of TADs correlated with changes in expression of adjacent genes, and remarkably these patterns of differential expression were identified both in cell lines and primary tumor data.
Other workers have begun to reveal the coordinated regulation of lncRNA and androgen receptor regulated target genes were more likely within the same TAD, than different TADs (daSilva et al., 2018), again supporting the idea that TAD are organizational units in the cell.
In breast cancer, TAD mapping has been undertaken as part of analyses of RUNX1, and the impact of RUNX1 on TAD structure measured using Hi-C. Whilst RUNX1 binding sites were enriched at TAD boundaries, there was no significant change in the boundary structure from knockdown of RUNCX1, which probably reflects the complex and stable nature of TAD boundaries. That is, they arise from many proteins interacting, but few of the protein alone can significantly change how they form(Barutcu et al., 2016).
2.2. Changes to CTCF function in hormone-responsive cancers.
Across cancers, altered CTCF and its downstream consequences are much studied. Many workers in breast cancer have focused on mapping disease-associated enhancer access. For example, Luca Magnani and co-workers(Patten et al., 2018) have mapped enhancers from primary patient material and overlapped with GWAS identified SNPs enriched in enhancers. They also demonstrated relationships between CTCF-dependent boundaries, enhancer strength and correlation with changes in mRNA suggesting that the interplay between boundaries, enhancer access and gene expression are all measurable in primary tumors. Another approach has been to delete CTCF binding sites within proximity to estrogen receptor alpha (ERα) regulated target genes using genome-editing and identified that some, but not all, ERα enhancer-gene relationships are altered(Korkmaz et al., 2019). These studies reflects those of others also in breast cancer where CTCF was shown to insulate the ERα pioneer factor, FOXA1 (Zhang et al., 2010), and to repress oncogenic signals downstream of ERB2 (Brix et al., 2019).
Indeed many ERα regulated genes associate with CTCF and can be captured in association with enhancer regions that loop to the lamina associated with gene suppression (Fiorito et al., 2016), and associate with periodic expression through the cell cycle, and disruptions to these relationships associate with worse patient outcome (Dominguez et al., 2016, Yamamoto et al., 2014). Together these findings suggest that CTCF actions are a significant regulator of ERα responses (Chan et al., 2008) related to specific histology (Razavi et al., 2018) and overall outcomes.
In prostate cancer, CTCF sites associated with outcome identified by GWAS and CRISPR deletion of sites leads to increased gene expression of genes within loops and that FOXA1, AR and in same CTCF block (Taslim et al., 2012).
Workers have also examined how CTCF mutations and expression changes impacts the epigenome in ovarian cancer, and established relationships to disease progression risks(Woloszynska-Read et al., 2011). Furthermore, the CTCF paralog, BORIS (Brother of the Regulator of Imprinted Sites) is altered in ovarian cancer and changes enhancer access and promotes invasive phenotypes(Hillman et al., 2019).
In pancreatic cancer a focus has been on the regulation of the PAX6 gene which is required for islet development and it has been revealed that CTCF regulates the locus(Buckle et al., 2018), reflecting the phenotype of the CTCF heterozygote deletion in mice(Tsui et al., 2012). Pancreatic cancer has also been a cancer in which the interplay between CTCF and repression of MYC has been seen to associated with progression and outcomes(Peng et al., 2019). Bioinformatic approaches have also been applied to integrate GWAS data with CTCF binding sites and therefore identify a critical impact of a risk variant with altered boundary function and reduced expression of the putative anti-oncogene MFSD13A(Mei et al., 2019).
In testes the CTCF/BORIS regulation of gene programs is expressed in primary spermatocytes but then silenced by DNA methylation. Reactivation in cancer cells is apparent making it a cancer testes- antigen gene expression and a potential target for immunotherapy(Martin-Kleiner, 2012). Reactivation of BORIS can bind hTERT and increase expression in ovarian and testicular cancers(Renaud et al., 2011). Finally, in the pituitary the human growth hormone cluster has been investigated and shown long range genomic interactions(Tsai et al., 2016, Tsai et al., 2014), but it remains to be established how important this is in pituitary cancer.
2.3. A survey of CTCF and interacting components in hormone-responsive cancers.
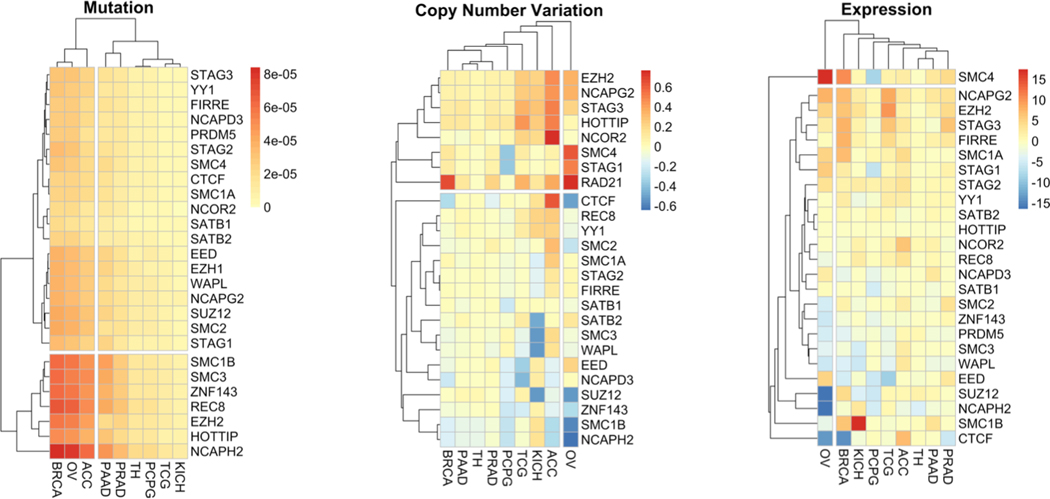
To complement this review TCGA data from the adrenal, breast, kidney, ovarian, pancreatic, parathyroid, prostate, testis and thyroid cancer cohorts has been examined to reveal the mutation, copy number variation and expression of CTCF and genes for the various interacting proteins (Figure 2). By mutational status, breast, ovarian and adrenocortical tumors cluster together driven by common mutations to cohesin complex (e.g. SMC3) and other chromosomal structural proteins (e.g. NCAPH2 and REC8). The lncRNA HOTTIP is also frequently mutated in these tumors. By contrast, kidney, parathyroid and testicular cancers are essentially free from mutation in any of the genes examined. CTCF is not commonly mutated.
Figure 2: Mutation, copy number variation and expression of genes involved in boundary formation in hormone responsive cancers.
Gene mutation, copy number variation and expression z-score pan-cancer data for breast (BRCA), ovarian (OV), pancreatic (PAAD), prostate (PRAD), thyroid (TH), pheochromocytoma (PCPG), Testicular Germ Cell (TCG), Adrenocortical (ACC) were downloaded using the R platform for statistical computing using the cgdsr package. The genes selected were chosen by the studies highlighted in the text, as well as choosing paralogs identified by Human Genome Nomenclature. Gene mutation and copy number variation data was normalized to the number of tumors within the cohort, and also normalized to gene length for the mutation data. The normalized mutation, copy number variation and expression was then visualized as a heatmap (pheatmap) using average Manhattan clustering.
This is somewhat echoed by changes in copy number variation with NCAPH2 and SMC1B showing copy number loss in ovarian and adrenal cancer. CTCF has common loss in ovarian and breast cancer, but gain in adrenal cancer. However, there are other clear differences, with kidney cancer for example showing clear copy number loss for SMC3 but not mutation. Notable gains in copy number included RAD21 and STAG1 in ovarian, and RAD21 also in adrenal, testicular and breast cancer. STAG3 is also amplifies in adrenal, kidney and testicular cancer. Similarly, STAG1 has loss of copy number variation and reduced expression uniquely in Pheochromocytoma. These findings reflect an emerging appreciation of the role of STAG family members across cancers(Romero-Perez et al., 2019, Mondal et al., 2019, Cheng et al., 2015). Whilst not mutated in any of the cancer NCOR2 is highly amplified in adrenal cancer, as are HOTTIP, NCAPG2 and EZH2, which are all mutated in the same cancer. Expression changes also distinguish ovarian cancer with reduced expression of CTCF, NCAPH2 and upregulation of SMC4. These patterns are somewhat echoed by breast and kidney cancer. The amplification of NCOR2 in adrenal cancer is reflected by modest upregulation of the mRNA. Similarly, SMC1B has a modest gain of copy number in kidney cancer but strong mRNA upregulation.
3. Statistical and computational challenges in the analyses of TADs
Statistical and computational frameworks were essential for the capture, analyses and interpretation of chromatin conformation data, and the development of these frameworks finally enabled the genome wide analyses in Hi-C data 2012(Nora et al., 2012, Dixon et al., 2012). These approaches apply complex genomic analyses and there are several areas where variance can impact the data interpretation, from wet-lab experimental parameters to dry-lab analyses. An ongoing challenge is to harmonize these techniques and analytical pipelines. These include cross-linking parameters, choice of restriction enzymes and library preparation. Similarly, the statistical approaches differ in their assumptions, for example how to model the distribution of the data, how to consider the impact of distance between events, and how best to determine differential TAD events(Stansfield et al., 2019, Mora et al., 2016).
There are multiple algorithms to identify TADs. For example, Dali and Blanchette(Dali et al., 2017) recently compared seven algorithms. Interestingly, the requirements of RAM to run algorithms whilst significant (~ 20 Gb), depending on the resolution e.g. 100 kb compared to 25 kb, is now such that once the raw read data are aligned, the analyses can be undertaken on most desktop computers. Perhaps, the optimal resolution to consider TAD analyses is around 50 kb and requires 500 million reads. Naturally, with higher resolution, more TADs are predicted and the mean size decreases. Amongst most of the commonly used algorithms there ~ 75% sites convergence predicted in at least two tools e.g. with HiCSeg(Levy-Leduc et al., 2014), TopDom (Shin et al., 2016). Tools probably are very conservative, and under-estimate by missing 25% of manually annotated TADs(Stansfield et al., 2019) (Table 1).
Table 1.
Representative computational packages for analyses of chromatin capture experiments. Each package is implemented in the R language for statistical computing and the focus of the data type and function of the analytical approache alongside the relevant publications are indicated.
| Package | Focus | Function | Publication |
|---|---|---|---|
| sevenC | CTCF ChIP-Seq | Predicts chromatin loops from CTCF ChIP-Seq data | |
| diffloop | ChIA-PET and RNA-Seq | Identify differential chromatin topology between cell conditions and annotate with gene expression | (165) |
| R3Cseq | 3C | Identify genomic loops between two fixed points | (166) |
| CHiCAGO | Capture-Hi-C | Analyses of HiC data that are enriched for genomic features of interest | (167) |
| HiTC | Hi-C | Normalization and visualization of Hi-C data, TAD detection | (168) |
| multiHiCcompare | Hi-C | Normalization and visualization of Hi-C data, TAD detection | (144) |
| HiCRep HiCseg | Hi-C | Assess reproducibility in Hi-C data, and ormalization and visualization of Hi-C data, TAD detection | (169) |
| TopDom | Hi-C | Normalization and visualization of Hi-C data, TAD detection | (148) |
Of course, a challenge for the field in both wet-lab design and dry-lab analyses is that the work to-date is from bulk culture and there is a need to undertake single cell or purified cell populations to ensure that TAD structure is not an emergent property from cell population averaging. Recently, this issue has been addressed by complementary approaches in single cells using partition ~30 kb across a region of chromosome 21. Interestingly, this revealed TAD structure and associations with CTCF and cohesin preference; cohesin depletion did not impact structure at single cell suggesting its CTCF-cohesion that is important(Bintu et al., 2018). In parallel, others are optimizing single cell approaches that combine Hi-C and imaging and suggest that there areas of significant heterogeneity between cells in terms of genomic and allele-specific organization (Finn et al., 2019).
However, perhaps, the greatest challenge will be the data integration approaches, which will be designed, implemented and interpreted by bioinformatically trained researchers. We now stand approximately 50 years after the conception of bioinformatics (reviewed in (Hogeweg, 2011)) and, in comparison to the 50-year widespread application of molecular biology (Weaver, 1970, Danna et al., 1971, Saiki et al., 1988, Hunkapiller et al., 1991), it is far from clear that bioinformatics is on a trajectory to also become democratized. Currently, it is far from clear how the research community stands in terms of the widespread application of bioinformatics. The unprecedented insight generated by the large-scale biological meta-projects such as The Human Genome Project(Roberts et al., 2001), ENCODE(Birney, 2012, Consortium et al., 2007), RoadMap Epigenome(Roadmap Epigenomics et al., 2015), FANTOM(Sanli et al., 2013), IHEC (Bujold et al., 2016, Chen et al., 2016) and TCGA (Cancer Genome Atlas Research et al., 2013) was achieved in large part through the success of the intensive data analytic initiatives within these projects. Unfortunately, at present, there is a worrying possibility that, given the ever-increasing computational and statistical requirements to map genomic topology, these remarkable achievements will themselves become islands of bioinformatically-driven genomic insight in oceans of reductionist biology. This will profoundly impede dissection and exploitation of how TAD structure governs hormonal signaling and is corrupted in cancer.
4. Summary and Future Challenges.
The visual density of the nucleus impeded early workers from appreciating the diversity of the structure-function relationships that were dynamically regulated across the life-cycle of the cell. Advancements in microscopy and the development of molecular biology techniques enabled significant developments in understanding, but the major catalyst for deciphering genomic organization and function was sequencing the human genome, and the development of statistical approaches and computational resources to undertake complex genomic analyses. This led to the first attempts to capture the 3D genome in 2012 and the identification of TADs(Dixon et al., 2012, Nora et al., 2012). Subsequently, it has become clear that the TAD structure is both biologically important and provides a major conceptual level for understanding genomic organization and gene regulation.
In terms of understanding how TAD structure can impact hormone-responsive cancers, there are both opportunities and challenges. The opportunity exists to integrate the understanding of TAD structure and organization into a comprehensive understanding of cancer and progression risks. For example, the choices of genomic bindings sites (cistrome) for either the ERα in breast cancer or the AR in prostate cancer are highly diverse. Understanding is emerging of the forces that shape these cistromes and how they determine the magnitude and amplitude of transcriptional signals (transcriptome), but many questions remain. It is unclear how TAD structure truly determines these cistrome-transcriptome relationships, and which mechanisms distort these actions in cancer progression and therapy resistance. There are clear examples of TAD boundary erosion and enhancer hijacking, but it is largely unknown at the genome-wide level how germline variation, somatic structural variation and epigenomic mechanisms may each contribute to these processes of boundary erosion and determining enhancer promiscuity.
In many ways, this is a readily achievable research goal at least within cell models; 3D genomic approaches are well-developed as are cistromic and transcriptomic methodologies. However, this now raises the prospect that hormone responsive cancer research will require 3D-cistrome-transcriptome data capture and analyses. Collectively, this is neither a trivial experimental challenge nor a financially modest endeavor. However, significant efforts have been made by leaders in the field to generate 3D genome browsers that will potentially enable more rapid assimilation of this information to the wider research community(Li et al., 2019a, Morita et al., 2019, Wang et al., 2018b, Yauy et al., 2018, Robinson et al., 2018).
Transition into patient samples is always more challenging, but there is a realistic possibility that greater understanding in the basic biology of TAD structure and function and how germline variation, somatic structural variation and epigenomic mechanisms converge selectively on specific TAD will lead to a personalized prediction of individual risk of disease and progression. For example, analyses of peripheral blood cells will reveal germline interplay with TAD boundary structure, and analyses of circulating tumor cells will similarly reveal how these boundaries are distorted. Computational approaches can impute or predict TAD structure, and intra-TAD configuration(Matthews et al., 2018, Sauerwald et al., 2017).
In turn, mathematical modelling of these approaches within a systems biology framework will allow predictive modelling of hormonal responsiveness, perhaps in the first instance for key cancer pre-disposing TADs such as the one on chr 8 containing MYC. It is highly likely that analyses of how germline variation, somatic structural variation and epigenomic mechanisms impacts TAD structure and in turn governs hormone signaling and will be essential to develop a more complete understanding of how hormone responsive cancers arise in humans, propagate and respond to therapies.
Acknowledgments
Funding:
This work was funded by the Prostate program of the Department of Defense Congressionally Directed Medical Research Programs [W81XWH-14–1-0608] and from the National Cancer Institute (NCI) grant awarded to the OSUCCC The James, CCSG P30CA016058
Footnotes
Declaration of Interest:
I declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- ARCECI RJ & GROSS PR 1980. Histone gene expression: progeny of isolated early blastomeres in culture make the same change as in the embryo. Science, 209, 607–9. [DOI] [PubMed] [Google Scholar]
- BARUTCU AR, et al. 2016. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta, 1859, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARUTCU AR, MAASS PG, LEWANDOWSKI JP, WEINER CL & RINN JL 2018. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat Commun, 9, 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL AC, WEST AG & FELSENFELD G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–96. [DOI] [PubMed] [Google Scholar]
- BINTU B, MATEO LJ, SU JH, SINNOTT-ARMSTRONG NA, PARKER M, KINROT S, YAMAYA K, BOETTIGER AN & ZHUANG X 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIRNEY E 2012. The making of ENCODE: Lessons for big-data projects. Nature, 489, 49–51. [DOI] [PubMed] [Google Scholar]
- BLOM VAN ASSENDELFT G, HANSCOMBE O, GROSVELD F & GREAVES DR 1989. The beta-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell, 56, 969–77. [DOI] [PubMed] [Google Scholar]
- BRIX DM, et al. 2019. Release of transcriptional repression via ErbB2-induced, SUMO-directed phosphorylation of myeloid zinc finger-1 serine 27 activates lysosome redistribution and invasion. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCKLE A, NOZAWA RS, KLEINJAN DA & GILBERT N 2018. Functional characteristics of novel pancreatic Pax6 regulatory elements. Hum Mol Genet, 27, 3434–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUJOLD D, et al. 2016. The International Human Epigenome Consortium Data Portal. Cell Syst, 3, 496–499 e2. [DOI] [PubMed] [Google Scholar]
- CANCER GENOME ATLAS RESEARCH, N., et al. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet, 45, 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANCER GENOME ATLAS RESEARCH NETWORK. ELECTRONIC ADDRESS, S. C. M. O. & CANCER GENOME ATLAS RESEARCH, N. 2015. The Molecular Taxonomy of Primary Prostate Cancer. Cell, 163, 1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARTER D, CHAKALOVA L, OSBORNE CS, DAI YF & FRASER P 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet, 32, 623–6. [DOI] [PubMed] [Google Scholar]
- CHAN CS & SONG JS 2008. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res, 68, 9041–9. [DOI] [PubMed] [Google Scholar]
- CHANG P, GOHAIN M, YEN MR & CHEN PY 2018. Computational Methods for Assessing Chromatin Hierarchy. Comput Struct Biotechnol J, 16, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN L, et al. 2016. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell, 167, 1398–1414 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENG F, LIU C, LIN CC, ZHAO J, JIA P, LI WH & ZHAO Z 2015. A Gene Gravity Model for the Evolution of Cancer Genomes: A Study of 3,000 Cancer Genomes across 9 Cancer Types. PLoS Comput Biol, 11, e1004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENOWETH AM, TRIST HM, TAN PS, WINES BD & HOGARTH PM 2015. The high-affinity receptor for IgG, FcgammaRI, of humans and non-human primates. Immunol Rev, 268, 175–91. [DOI] [PubMed] [Google Scholar]
- CHO SW, et al. 2018. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell, 173, 1398–1412 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONSORTIUM, E. P., et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUARTERO S & MERKENSCHLAGER M 2018. Three-dimensional genome organization in normal and malignant haematopoiesis. Curr Opin Hematol, 25, 323–328. [DOI] [PubMed] [Google Scholar]
- CUTRUPI AN, BREWER MH, NICHOLSON GA & KENNERSON ML 2018. Structural variations causing inherited peripheral neuropathies: A paradigm for understanding genomic organization, chromatin interactions, and gene dysregulation. Mol Genet Genomic Med, 6, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALI R & BLANCHETTE M 2017. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res, 45, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANNA K & NATHANS D 1971. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A, 68, 2913–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DASILVA LF, BECKEDORFF FC, AYUPE AC, AMARAL MS, MESEL V, VIDEIRA A, REIS EM, SETUBAL JC & VERJOVSKI-ALMEIDA S 2018. Chromatin Landscape Distinguishes the Genomic Loci of Hundreds of Androgen-Receptor-Associated LincRNAs From the Loci of Non-associated LincRNAs. Front Genet, 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE LAAT W & DUBOULE D 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 502, 499–506. [DOI] [PubMed] [Google Scholar]
- DE WIT E, et al. 2015. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell, 60, 676–84. [DOI] [PubMed] [Google Scholar]
- DEL ROSARIO BC, KRIZ AJ, DEL ROSARIO AM, ANSELMO A, FRY CJ, WHITE FM, SADREYEV RI & LEE JT 2019. Exploration of CTCF post-translation modifications uncovers Serine-224 phosphorylation by PLK1 at pericentric regions during the G2/M transition. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENKER A & DE LAAT W 2016. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev, 30, 1357–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DESPANG A, et al. 2019. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet, 51, 1263–1271. [DOI] [PubMed] [Google Scholar]
- DILLON N, TRIMBORN T, STROUBOULIS J, FRASER P & GROSVELD F 1997. The effect of distance on long-range chromatin interactions. Mol Cell, 1, 131–9. [DOI] [PubMed] [Google Scholar]
- DIXON JR, GORKIN DU & REN B 2016. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell, 62, 668–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIXON JR, SELVARAJ S, YUE F, KIM A, LI Y, SHEN Y, HU M, LIU JS & REN B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMINGUEZ D, TSAI YH, GOMEZ N, JHA DK, DAVIS I & WANG Z 2016. A high-resolution transcriptome map of cell cycle reveals novel connections between periodic genes and cancer. Cell Res, 26, 946–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONALDSON-COLLIER MC, et al. 2019. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet, 51, 517–528. [DOI] [PubMed] [Google Scholar]
- DONOHOE ME, SILVA SS, PINTER SF, XU N & LEE JT 2009. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature, 460, 128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONOHOE ME, ZHANG LF, XU N, SHI Y & LEE JT 2007. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell, 25, 43–56. [DOI] [PubMed] [Google Scholar]
- FABRE PJ, LELEU M, MORMANN BH, LOPEZ-DELISLE L, NOORDERMEER D, BECCARI L & DUBOULE D 2017. Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol, 18, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FILIPPOVA GN, FAGERLIE S, KLENOVA EM, MYERS C, DEHNER Y, GOODWIN G, NEIMAN PE, COLLINS SJ & LOBANENKOV VV 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol, 16, 2802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FILIPPOVA GN, et al. 2002. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res, 62, 48–52. [PubMed] [Google Scholar]
- FILIPPOVA GN, THIENES CP, PENN BH, CHO DH, HU YJ, MOORE JM, KLESERT TR, LOBANENKOV VV & TAPSCOTT SJ 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet, 28, 335–43. [DOI] [PubMed] [Google Scholar]
- FINN EH, PEGORARO G, BRANDAO HB, VALTON AL, OOMEN ME, DEKKER J, MIRNY L & MISTELI T 2019. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell, 176, 1502–1515 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIORITO E, et al. 2016. CTCF modulates Estrogen Receptor function through specific chromatin and nuclear matrix interactions. Nucleic Acids Res, 44, 10588–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKE WW 1988. Matthias Jacob Schleiden and the definition of the cell nucleus. Eur J Cell Biol, 47, 145–56. [PubMed] [Google Scholar]
- FRASER M, et al. 2017. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. [DOI] [PubMed] [Google Scholar]
- FU S, ZHANG L, LV J, ZHU B, WANG W & WANG X 2018. Two main stream methods analysis and visual 3D genome architecture. Semin Cell Dev Biol. [DOI] [PubMed] [Google Scholar]
- FUDENBERG G, IMAKAEV M, LU C, GOLOBORODKO A, ABDENNUR N & MIRNY LA 2016. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep, 15, 2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FURLONG EEM & LEVINE M 2018. Developmental enhancers and chromosome topology. Science, 361, 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHIRLANDO R & FELSENFELD G 2016. CTCF: making the right connections. Genes Dev, 30, 881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIBCUS JH & DEKKER J 2013. The hierarchy of the 3D genome. Mol Cell, 49, 773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILAD Y & LANCET D 2003. Population differences in the human functional olfactory repertoire. Mol Biol Evol, 20, 307–14. [DOI] [PubMed] [Google Scholar]
- GONG Y, LAZARIS C, SAKELLAROPOULOS T, LOZANO A, KAMBADUR P, NTZIACHRISTOS P, AIFANTIS I & TSIRIGOS A 2018. Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat Commun, 9, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO Y, MONAHAN K, WU H, GERTZ J, VARLEY KE, LI W, MYERS RM, MANIATIS T & WU Q 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A, 109, 21081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALLER F, et al. 2019. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun, 10, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANDOKO L, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet, 43, 630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANSCOMBE O, WHYATT D, FRASER P, YANNOUTSOS N, GREAVES D, DILLON N & GROSVELD F 1991. Importance of globin gene order for correct developmental expression. Genes Dev, 5, 1387–94. [DOI] [PubMed] [Google Scholar]
- HANSEN AS, CATTOGLIO C, DARZACQ X & TJIAN R 2018. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus, 9, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARMSTON N, ING-SIMMONS E, TAN G, PERRY M, MERKENSCHLAGER M & LENHARD B 2017. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat Commun, 8, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEIJNEN IA & VAN DE WINKEL JG 1997. Human IgG Fc receptors. Int Rev Immunol, 16, 29–55. [DOI] [PubMed] [Google Scholar]
- HEINTZMAN ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature, 459, 108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILLMAN JC, et al. 2019. BORIS expression in ovarian cancer precursor cells alters the CTCF cistrome and enhances invasiveness through GALNT14. Mol Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOGEWEG P 2011. The roots of bioinformatics in theoretical biology. PLoS Comput Biol, 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLWERDA SJ & DE LAAT W 2013. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci, 368, 20120369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG J, LI K, CAI W, LIU X, ZHANG Y, ORKIN SH, XU J & YUAN GC 2018. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun, 9, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUG CB, GRIMALDI AG, KRUSE K & VAQUERIZAS JM 2017. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell, 169, 216–228 e19. [DOI] [PubMed] [Google Scholar]
- HUNKAPILLER T, KAISER RJ, KOOP BF & HOOD L 1991. Large-scale and automated DNA sequence determination. Science, 254, 59–67. [DOI] [PubMed] [Google Scholar]
- HYLE J, et al. 2019. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer-promoter looping. Nucleic Acids Res, 47, 6699–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ING-SIMMONS E, SEITAN VC, FAURE AJ, FLICEK P, CARROLL T, DEKKER J, FISHER AG, LENHARD B & MERKENSCHLAGER M 2015. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res, 25, 504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG Y, et al. 2017. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet, 49, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATAINEN R, et al. 2015. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet, 47, 818–21. [DOI] [PubMed] [Google Scholar]
- KEMP CJ, et al. 2014. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep, 7, 1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOCH L 2016. Chromatin: Going a TAD out on a limb. Nat Rev Genet, 17, 717. [DOI] [PubMed] [Google Scholar]
- KOJIMA MK 1960. Acceleration of the cleavage of sea urchin eggs by vital staining with neutral red. Exp Cell Res, 20, 565–73. [DOI] [PubMed] [Google Scholar]
- KORKMAZ G, et al. 2019. A CRISPR-Cas9 screen identifies essential CTCF anchor sites for estrogen receptor-driven breast cancer cell proliferation. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KREFTING J, ANDRADE-NAVARRO MA & IBN-SALEM J 2018. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol, 16, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRUMM A & DUAN Z 2018. Understanding the 3D genome: Emerging impacts on human disease. Semin Cell Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVY-LEDUC C, DELATTRE M, MARY-HUARD T & ROBIN S 2014. Two-dimensional segmentation for analyzing Hi-C data. Bioinformatics, 30, i386–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI D, HSU S, PURUSHOTHAM D, SEARS RL & WANG T 2019a. WashU Epigenome Browser update 2019. Nucleic Acids Res, 47, W158–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, et al. 2019b. An alternative CTCF isoform antagonizes canonical CTCF occupancy and changes chromatin architecture to promote apoptosis. Nat Commun, 10, 1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOBANENKOV VV & ZENTNER GE 2018. Discovering a binary CTCF code with a little help from BORIS. Nucleus, 9, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOPEZ CASTEL A, NAKAMORI M, TOME S, CHITAYAT D, GOURDON G, THORNTON CA & PEARSON CE 2011. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet, 20, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO H, et al. 2018. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood, 132, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUPIANEZ DG, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAJUMDER P & BOSS JM 2010. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol, 30, 4211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAJUMDER P, GOMEZ JA, CHADWICK BP & BOSS JM 2008. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med, 205, 785–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARQUES M, HERNANDEZ RP & WITCHER M 2015. Analysis of changes to mRNA levels and CTCF occupancy upon TFII-I knockdown. Genom Data, 4, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN-GARCIA D, et al. 2019. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1(−) mantle cell lymphoma. Blood, 133, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN-KLEINER I 2012. BORIS in human cancers -- a review. Eur J Cancer, 48, 929–35. [DOI] [PubMed] [Google Scholar]
- MATTHEWS BJ & WAXMAN DJ 2018. Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. Elife, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCHUGH CA, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature, 521, 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEI S, et al. 2019. A functional variant in the boundary of a topological association domain is associated with pancreatic cancer risk. Mol Carcinog. [DOI] [PubMed] [Google Scholar]
- MONDAL G, STEVERS M, GOODE B, ASHWORTH A & SOLOMON DA 2019. A requirement for STAG2 in replication fork progression creates a targetable synthetic lethality in cohesin-mutant cancers. Nat Commun, 10, 1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORA A, SANDVE GK, GABRIELSEN OS & ESKELAND R 2016. In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform, 17, 980–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORITA M, et al. 2019. ViBrism DB: an interactive search and viewer platform for 2D/3D anatomical images of gene expression and co-expression networks. Nucleic Acids Res, 47, D859–D866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURAKAWA Y, YOSHIHARA M, KAWAJI H, NISHIKAWA M, ZAYED H, SUZUKI H, FANTOM C & HAYASHIZAKI Y 2016. Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights into Diseases. Trends Genet, 32, 76–88. [DOI] [PubMed] [Google Scholar]
- NARENDRA V, ROCHA PP, AN D, RAVIRAM R, SKOK JA, MAZZONI EO & REINBERG D 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science, 347, 1017–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEGI S, BOLT CC, ZHANG H & STUBBS L 2019. An extended regulatory landscape drives Tbx18 activity in a variety of prostate-associated cell lineages. Dev Biol, 446, 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORA EP, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature, 485, 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORTHCOTT PA, et al. 2014. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature, 511, 428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLENDER T, et al. 2016. The human olfactory transcriptome. BMC Genomics, 17, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLIVIER M, et al. 2001. A high-resolution radiation hybrid map of the human genome draft sequence. Science, 291, 1298–302. [DOI] [PubMed] [Google Scholar]
- PAROLIA A, CIESLIK M & CHINNAIYAN AM 2018. Competing for enhancers: PVT1 fine-tunes MYC expression. Cell Res, 28, 785–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATTEN DK, et al. 2018. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat Med, 24, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENG WX, HE RZ, ZHANG Z, YANG L & MO YY 2019. LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene. [DOI] [PubMed] [Google Scholar]
- PETROVIC J, et al. 2019. Oncogenic Notch Promotes Long-Range Regulatory Interactions within Hyperconnected 3D Cliques. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POTERLOWICZ K, et al. 2017. 5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells. PLoS Genet, 13, e1006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUGACHEVA EM, TIWARI VK, ABDULLAEV Z, VOSTROV AA, FLANAGAN PT, QUITSCHKE WW, LOUKINOV DI, OHLSSON R & LOBANENKOV VV 2005. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum Mol Genet, 14, 953–65. [DOI] [PubMed] [Google Scholar]
- RACKO D, BENEDETTI F, DORIER J & STASIAK A 2018. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res, 46, 1648–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RACKO D, BENEDETTI F, DORIER J & STASIAK A 2019. Are TADs supercoiled? Nucleic Acids Res, 47, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAZAVI P, et al. 2018. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell, 34, 427–438 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REN X, et al. 2019. High Expression of long non-coding RNA PVT1 predicts metastasis in Han and Uygur Patients with Gastric Cancer in Xinjiang, China. Sci Rep, 9, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RENAUD S, LOUKINOV D, ALBERTI L, VOSTROV A, KWON YW, BOSMAN FT, LOBANENKOV V & BENHATTAR J 2011. BORIS/CTCFL-mediated transcriptional regulation of the hTERT telomerase gene in testicular and ovarian tumor cells. Nucleic Acids Res, 39, 862–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICKMAN DS, et al. 2010. ERG cooperates with androgen receptor in regulating trefoil factor 3 in prostate cancer disease progression. Neoplasia, 12, 1031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICKMAN DS, et al. 2012. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci U S A, 109, 9083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROADMAP EPIGENOMICS, C., et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTS L, DAVENPORT RJ, PENNISI E & MARSHALL E 2001. A history of the Human Genome Project. Science, 291, 1195. [DOI] [PubMed] [Google Scholar]
- ROBINSON JT, TURNER D, DURAND NC, THORVALDSDOTTIR H, MESIROV JP & AIDEN EL 2018. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst, 6, 256–258 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUEZ-CARBALLO E, et al. 2017. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev, 31, 2264–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMERO-PEREZ L, SURDEZ D, BRUNET E, DELATTRE O & GRUNEWALD TGP 2019. STAG Mutations in Cancer. Trends Cancer, 5, 506–520. [DOI] [PubMed] [Google Scholar]
- RON G, GLOBERSON Y, MORAN D & KAPLAN T 2017. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun, 8, 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROWLEY MJ & CORCES VG 2018. Organizational principles of 3D genome architecture. Nat Rev Genet, 19, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROY SS, MUKHERJEE AK & CHOWDHURY S 2018. Insights about genome function from spatial organization of the genome. Hum Genomics, 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYAN RJ, et al. 2015. Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov, 5, 1058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAIKI RK, GELFAND DH, STOFFEL S, SCHARF SJ, HIGUCHI R, HORN GT, MULLIS KB & ERLICH HA 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–91. [DOI] [PubMed] [Google Scholar]
- SANDA MG, et al. 2017. Association Between Combined TMPRSS2:ERG and PCA3 RNA Urinary Testing and Detection of Aggressive Prostate Cancer. JAMA Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANLI K, KARLSSON FH, NOOKAEW I & NIELSEN J 2013. FANTOM: Functional and taxonomic analysis of metagenomes. BMC Bioinformatics, 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANYAL A, LAJOIE BR, JAIN G & DEKKER J 2012. The long-range interaction landscape of gene promoters. Nature, 489, 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAUERWALD N, ZHANG S, KINGSFORD C & BAHAR I 2017. Chromosomal dynamics predicted by an elastic network model explains genome-wide accessibility and long-range couplings. Nucleic Acids Res, 45, 3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMIDL C, et al. 2014. Transcription and enhancer profiling in human monocyte subsets. Blood, 123, e90–9. [DOI] [PubMed] [Google Scholar]
- SHIN H, SHI Y, DAI C, TJONG H, GONG K, ALBER F & ZHOU XJ 2016. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res, 44, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN HY 2018. The structural and functional roles of CTCF in the regulation of cell type-specific and human disease-associated super-enhancers. Genes Genomics. [DOI] [PubMed] [Google Scholar]
- SIMA J, et al. 2019. Identifying cis Elements for Spatiotemporal Control of Mammalian DNA Replication. Cell, 176, 816–830 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH ST, et al. 2009. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol, 328, 518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCER RJ, DEL ROSARIO BC, PINTER SF, LESSING D, SADREYEV RI & LEE JT 2011. A boundary element between Tsix and Xist binds the chromatin insulator Ctcf and contributes to initiation of X-chromosome inactivation. Genetics, 189, 441–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPIELMANN M, LUPIANEZ DG & MUNDLOS S 2018. Structural variation in the 3D genome. Nat Rev Genet, 19, 453–467. [DOI] [PubMed] [Google Scholar]
- SPLINTER E, HEATH H, KOOREN J, PALSTRA RJ, KLOUS P, GROSVELD F, GALJART N & DE LAAT W 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev, 20, 2349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANSFIELD JC, CRESSWELL KG & DOZMOROV MG 2019. multiHiCcompare: joint normalization and comparative analysis of complex Hi-C experiments. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STROM AR, EMELYANOV AV, MIR M, FYODOROV DV, DARZACQ X & KARPEN GH 2017. Phase separation drives heterochromatin domain formation. Nature, 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SYMMONS O, USLU VV, TSUJIMURA T, RUF S, NASSARI S, SCHWARZER W, ETTWILLER L & SPITZ F 2014. Functional and topological characteristics of mammalian regulatory domains. Genome Res, 24, 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TABERLAY PC, et al. 2016. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res, 26, 719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TABERLAY PC, STATHAM AL, KELLY TK, CLARK SJ & JONES PA 2014. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res, 24, 1421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TASLIM C, CHEN Z, HUANG K, HUANG TH, WANG Q & LIN S 2012. Integrated analysis identifies a class of androgen-responsive genes regulated by short combinatorial long-range mechanism facilitated by CTCF. Nucleic Acids Res, 40, 4754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMLINS SA, et al. 2015. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMLINS SA, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science, 310, 644–8. [DOI] [PubMed] [Google Scholar]
- TSAI YC, COOKE NE & LIEBHABER SA 2014. Tissue specific CTCF occupancy and boundary function at the human growth hormone locus. Nucleic Acids Res, 42, 4906–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI YC, COOKE NE & LIEBHABER SA 2016. Long-range looping of a locus control region drives tissue-specific chromatin packing within a multigene cluster. Nucleic Acids Res, 44, 4651–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUI S, GAO J, WANG C & LU L 2012. CTCF mediates effect of insulin on glucagon expression. Exp Cell Res, 318, 887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ULZ P, et al. 2016. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun, 7, 12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNKELESS JC 1989. Human Fc receptors for IgG. Int Rev Immunol, 5, 165–71. [DOI] [PubMed] [Google Scholar]
- WANG DC, WANG W, ZHANG L & WANG X 2018a. A tour of 3D genome with a focus on CTCF. Semin Cell Dev Biol. [DOI] [PubMed] [Google Scholar]
- WANG H, et al. 2012a. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res, 22, 1680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, CAI Y, SHAO LJ, SIDDIQUI J, PALANISAMY N, LI R, REN C, AYALA G & ITTMANN M 2011. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res, 71, 1325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, WANG Y & LU L 2012b. De-SUMOylation of CCCTC binding factor (CTCF) in hypoxic stress-induced human corneal epithelial cells. J Biol Chem, 287, 12469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG S, et al. 2014. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci U S A, 111, 4251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Y, et al. 2018b. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol, 19, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG YL & TAYLOR DL 1979. Distribution of fluorescently labeled actin in living sea urchin eggs during early development. J Cell Biol, 81, 672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEAVER W 1970. Molecular biology: origin of the term. Science, 170, 581–2. [DOI] [PubMed] [Google Scholar]
- WEISCHENFELDT J, et al. 2017. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet, 49, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WENDT KS, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature, 451, 796–801. [DOI] [PubMed] [Google Scholar]
- WOLFSBERG TG, MCENTYRE J & SCHULER GD 2001. Guide to the draft human genome. Nature, 409, 824–6. [DOI] [PubMed] [Google Scholar]
- WOLOSZYNSKA-READ A, et al. 2011. Coordinated cancer germline antigen promoter and global DNA hypomethylation in ovarian cancer: association with the BORIS/CTCF expression ratio and advanced stage. Clin Cancer Res, 17, 2170–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WRIGHT FA, et al. 2001. A draft annotation and overview of the human genome. Genome Biol, 2, RESEARCH0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU P, et al. 2017. 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat Commun, 8, 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO S, et al. 2014. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell, 25, 762–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG F, et al. 2015. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol, 16, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAUY K, GATINOIS V, GUIGNARD T, SATI S, PUECHBERTY J, GAILLARD JB, SCHNEIDER A & PELLESTOR F 2018. Looking for Broken TAD Boundaries and Changes on DNA Interactions: Clinical Guide to 3D Chromatin Change Analysis in Complex Chromosomal Rearrangements and Chromothripsis. Methods Mol Biol, 1769, 353–361. [DOI] [PubMed] [Google Scholar]
- ZHANG R, WANG Y, YANG Y, ZHANG Y & MA J 2018. Predicting CTCF-mediated chromatin loops using CTCF-MP. Bioinformatics, 34, i133–i141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, LIANG J, LI Y, XUAN C, WANG F, WANG D, SHI L, ZHANG D & SHANG Y 2010. CCCTC-binding factor acts upstream of FOXA1 and demarcates the genomic response to estrogen. J Biol Chem, 285, 28604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIMMERMAN MW, et al. 2018. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov, 8, 320–335. [DOI] [PMC free article] [PubMed] [Google Scholar]