Abstract
Recent declines in the health of the honey bee have startled researchers and lay people alike as honey bees are agriculture’s most important pollinator. Honey bees are important pollinators of many major crops and add billions of dollars annually to the US economy through their services. One factor that may influence colony health is the microbial community. Indeed, the honey bee worker digestive tract harbors a characteristic community of bee-specific microbes, and the composition of this community is known to impact honey bee health. However, the honey bee is a superorganism, a colony of eusocial insects with overlapping generations where nestmates cooperate, building a hive, gathering and storing food, and raising brood. In contrast to what is known regarding the honey bee worker gut microbiome, less is known of the microbes associated with developing brood, with food stores, and with the rest of the built hive environment. More recently, the microbe Bombella apis was identified as associated with nectar, with developing larvae, and with honey bee queens. This bacterium is related to flower-associated microbes such as Saccharibacter floricola and other species in the genus Saccharibacter, and initial phylogenetic analyses placed it as sister to these environmental bacteria. Here, we used comparative genomics of multiple honey bee-associated strains and the nectar-associated Saccharibacter to identify genomic changes that may be associated with the ecological transition to honey bee association. We identified several genomic differences in the honey bee-associated strains, including a complete CRISPR/Cas system. Many of the changes we note here are predicted to confer upon Bombella the ability to survive in royal jelly and defend themselves against mobile elements, including phages. Our results are a first step toward identifying potential function of this microbe in the honey bee superorganism.
Keywords: microbiome, evolution, horizontal gene transfer, Bombella apis
Significance
The honey bee is the world’s most important pollinator and its microbial symbionts impact its health and vitality. One important symbiont of the bee is Bombella apis, recently discovered to protect developing bees from fungal pathogens. Here, we use comparative genomics to identify differences in genetic content between B. apis and the closely related flower-associated Saccharibacter clade. We find that in comparison to the outgroup, Bombella harbors many loci potentially involved in carbohydrate use, host interaction, microbe–microbe interaction, and in virulence and defense. These results are the foundation to understanding the function of B. apis in a honey bee colony.
Introduction
The honey bee (Apis mellifera) is extremely important economically because of the pollination services it provides to numerous agricultural crops. As a result, there is increasing interest in determining how the microbiome supports and influences bee function. Although a honey bee colony is made up of bees with diverse roles, or castes, the majority of studies on bee microbiomes have focused on workers specifically. The microbial community of worker bees consists of eight to ten core bacterial species (Martinson et al. 2011, 2012; Moran et al. 2012; Sabree et al. 2012; Moran 2015). The characterization of these groups led to research into their role in honey bee health and we now know these microbes provision nutrients (Moran 2015) and assist in the breakdown of plant-derived carbohydrates (Lee et al. 2018), as is the case in other insect–microbe interactions (McCutcheon and Moran 2007; Douglas 2013). There has also been speculation as to the role of the microbiome in resistance to pathogens, as microbial communities have been shown to protect the bumble bee (Bombus terristris) from the parasite Crithidia bombi (Koch and Schmid-Hempel 2011). Honey bee-associated microbes interact with each other in diverse ways both in vitro and in vivo, suggesting that they may interact syntrophically within workers (Martinson et al. 2012; Rokop et al. 2015). Although these studies focused on honey bee workers are intriguing, less is known about the microbes found associated with the honey bee queen, the hive environment, with developing brood, or with food stores. Amplicon studies suggest that this community is depauperate and more transient (Anderson et al. 2013; Rokop et al. 2015; Tarpy et al. 2015). However, one microbe is found across all of these environments: Parasaccharibacter apium (proposed to be reclassified as B. apis; Corby-Harris et al. 2016; Yun et al. 2017; Bonilla-Rosso et al. 2019; Smith, Anderson et al. 2020). Bombella apis is in the family Acetobacteraceae and occupies defined niches within the hive, including: queen guts, nurse hypopharyngeal glands, nurse crops, and royal jelly, and is only rarely found outside of these areas (Anderson et al. 2013; Vojvodic et al. 2013; Corby-Harris et al. 2014). Evidence suggests that it might play a role in protecting developing larvae and worker bees from fungal pathogens (Corby-Harris et al. 2016; Miller et al. 2020), although direct antagonism against Nosema has yet to be rigorously tested. Given that B. apis makes up a large proportion of the queen gut microbiome, it is possible that it plays important roles in queen health and physiology, as well (Tarpy et al. 2015; Anderson et al. 2018), although this has yet to be determined and it is equally likely that B. apis transits through queen gut via her diet of royal jelly.
Bombella apis is part of a clade of acetic acid bacteria (a group within the family Acetobacteraceae) that contains both free-living and bee-associated members. Comparative genomics, then, can give us insights into genomic changes associated with the transition to honey bee association in this clade. This comparison can also help elucidate what sets B. apis apart from closely related species and the role(s) it might be playing in the hive environment. To that end, we used the genomes of eight B. apis strains (Chouaia et al. 2014; Veress et al. 2017; Corby-Harris and Anderson 2018; Smith et al. 2019), a Bombella sp. genome assembly, as well as five genomes of the closely related genus Saccharibacter (Jojima et al. 2004; Smith, Vuong et al. 2020) and a genome of the bumblebee symbiont, Bombella intestini (Li et al. 2015, 2016), to begin to tease apart the unique capabilities of B. apis (table 1). Insights gained here could prove critical in determining the factors responsible for maintaining queen health in colonies and could ultimately lead to the development of interventions to improve queen health and mitigate the detrimental impacts of queen failure on this economically critical species.
Table 1.
Genome Names, Accession Number, and Isolation Sources for Genomes Used in These Analyses
| Genome | GenBank Accession | Isolation Source | Genome Size (Mb) | Contigs | %GC | Number of Genes | % N sequence | Ref | BUSCO String |
|---|---|---|---|---|---|---|---|---|---|
| Bombella apis G7_7_3c | GCA_002079945.1 | Apis mellifera hindgut | 2.01 | 1 | 59.42 | 1,873 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis SME1 | GCA_009362775.1 | Apis mellifera honey | 2.09 | 11 | 59.51 | 1,984 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis A29 | GCA_002917995.1 | Apis mellifera larva | 2.01 | 27 | 59.39 | 1,846 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis B8 | GCA_002917945.1 | Apis mellifera larva | 2.01 | 29 | 59.38 | 1,854 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis C6 | GCA_002917985.1 | Apis mellifera larva | 2.01 | 34 | 59.38 | 1,852 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis AM169 | GCA_000723565.1 | Apis mellifera stomach | 1.98 | 9 | 59.32 | 1,816 | 0 | C: 95.9%[S: 95.9%, D: 0.0%], F: 2.7%, M: 1.4%, n: 221 | |
| Bombella apis 3.A.1 | GCA_002150125.1 | Honey | 2.01 | 24 | 59.41 | 1,864 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Bombella apis M18 | GCA_002150105.1 | Apis mellifera stomach | 2.01 | 11 | 59.35 | 1,972 | 0 | C: 97.7%[S: 97.7%, D: 0.0%], F: 0.9%, M: 1.4%, n: 221 | |
| Bombella intestini R-52487 | GCA_002003665.1 | Bombus lapidarius crop | 2.02 | 12 | 54.94 | 1,910 | 0 | C: 98.6%[S: 98.6%, D: 0.0%], F: 0.9%, M: 0.5%, n: 221 | |
| Bombella sp. AS1 | GCA_002592045.1 | Apis mellifera larva | 1.85 | 13 | 52.64 | 1,738 | 0 | C: 96.4%[S: 96.4%, D: 0.0%], F: 0.9%, M: 2.7%, n: 221 | |
| Saccharibacter sp. EH611 | GCA_009834765.1 | Anthophora sp. Crop | 2.28 | 25 | 51.47 | 2,253 | 0 | C: 99.1%[S: 99.1%, D: 0.0%], F: 0.0%, M: 0.9%, n: 221 | |
| Saccharibacter sp. EH60 | GCA_009834775.1 | Anthophora sp. Crop | 2.32 | 30 | 51.47 | 2,304 | 0 | C: 99.1%[S: 99.1%, D: 0.0%], F: 0.0%, M: 0.9%, n: 221 | |
| Saccharibacter sp. EH70 | GCA_009834795.1 | Anthophora sp. Crop | 2.29 | 26 | 50.47 | 2,215 | 0 | C: 99.1%[S: 99.1%, D: 0.0%], F: 0.0%, M: 0.9%, n: 221 | |
| Saccharibacter floricola DSM15669 | GCA_000378165.1 | Flower | 2.38 | 43 | 51.22 | 2,352 | 0.0018 | C: 99.1%[S: 99.1%, D: 0.0%], F: 0.0%, M: 0.9%, n: 221 | |
| Saccharibacter sp. 17.LH.SD | GCA_009834805.1 | Melissodes sp. Crop | 2.10 | 13 | 48.61 | 1,996 | 0 | C: 98.2%[S: 98.2%, D: 0.0%], F: 0.9%, M: 0.9%, n: 221 | |
| Gluconobacter oxydans H24 | GCA_000311765.1 | Industrial sample | 3.82 | 2 | 56.24 | 3,742 | 0 | C: 98.6%[S: 97.7%, D: 0.9%], F: 0.5%, M: 0.9%, n: 221 |
Note.—Bombella apis G7_7_3c is the B. apis reference genome. BUSCO string abbreviations are as follows: Complete: X%[Singleton: X%, Duplicated: X%]; Fragmented: 0.0%; Missing: X%; number of genes in the reference set: 221. C, complete; S, singleton (of the complete reference genes, what % are singleton in the genome being tested); D, duplicated (of the complete reference genes, what % are duplicated in the genome being tested); F, fragmented; M, missing; n, number of genes in reference set.
Materials and Methods
Phylogenetic Relationship of Bombella and Saccharibacter
To determine the phylogenetic placement of Bombella and Saccharibacter strains used here, we initially used the 16S rRNA gene sequences from the Silva database (Quast et al. 2012; Yilmaz et al. 2014; Glockner et al. 2017) that met the following criteria: 1) from a species belonging to either Bombella or Saccharibacter, 2) of length at least 1,200 bases, and 3) sequence quality >90. Additionally, the 16S rRNA gene sequence for Gluconobacter oxydans was included as an outgroup. The 16S rRNA sequences for B. intestini (Li et al. 2015) and B. apis (Yun et al. 2017) were included. We used BLAST to find the 16S rRNA gene sequences in the Bombella and Saccharibacter genomes (table 1) to pull out their respective 16S rRNA sequences for use in this phylogeny. All sequences were aligned using the SINA aligner (Pruesse et al. 2012); parameters used were set using the –auto option. A maximum likelihood phylogeny was constructed using RAxML with the GTRGAMMA substitution model and 1,000 bootstrap replicates (v8.2.11; Stamatakis 2006). The final tree was visualized using FigTree (v1.4.2, http://tree.bio.ed.ac.uk/software/figtree/).
Orthology Analysis
To facilitate downstream analyses, we clustered genes from all genomes in table 1—plus G. oxydans H24 as an outgroup—into groups of orthologous genes (GOGs) using OrthoMCL (v.2.0.9; Li et al. 2003) (supplementary table S1, Supplementary Material online). Amino acid sequences were downloaded from NCBI and clustering was performed using default OrthoMCL parameters, namely percentMatchCutoff = 50 and evalueExponentCutoff=−5. These clusters were then classified as single-copy orthologs (defined as containing exactly one representative from each genome), variable (defined as missing a representative from at least one genome and having varying numbers of representatives from each of the other genomes), multicopy ortholog (containing at least one representative from each genome, but multiple copies from at least one genome), or genome-specific (containing at least two genes that all came from the same genome) using an in-house Perl script.
Bombella and Saccharibacter Core Ortholog Phylogeny
We constructed a phylogeny using concatenated amino acid alignments of all single-copy GOGs. The amino acid sequences were aligned using the MAFFT L-INS-I algorithm (v7.310; Katoh et al. 2002), and alignments were then concatenated, and used to construct a maximum likelihood phylogeny using RAxML with substitution model PROTGAMMALGF and 1,000 bootstrap replicates (v8.2.11; Stamatakis 2006). The final tree was visualized using FigTree (v1.4.2, http://tree.bio.ed.ac.uk/software/figtree/).
Calculation of Genomic Similarity
To determine relatedness and species assignment, we calculated genome-wide Average Nucleotide Identity (gANI) and aligned fraction (AF) for each pairwise comparison using ANIcalculator (Varghese et al. 2015). Predicted transcript sequences for each pairwise comparison were passed to the software, which output gANI and AF in each direction for the pairwise comparison. As gANI and AF can vary depending on the direction of comparison due to differences in genome length, we report the average of the pairwise calculations in each direction. These values were used with genus and species cutoff values to determine taxonomic identify of our sequenced genomes.
Synteny Analysis
Before our analysis of synteny, we subjected all of our assemblies to Quast v. 5.0.2 run with default parameters on contigs from our genomes compared with the circularized genome of B. apis strain G7_7_3 using the –pe1 and –pe2 flags to include raw reads and calculate genome completeness as well as determine potential misassemblies. Few potential within-contig misassemblies were identified in each genome, with most having no flagged misassemblies. We identified some within-contig misassemblies for the following strains: nine for B. apis A29, and seven for B. apis C6 and B. apis SME1. Importantly, we cannot rule out misassembly of the G7_7_3 genome or natural genomic inversions and rearrangements between these strains over time. In addition, no misassemblies were identified in regions hosting horizontal gene transfers (HGTs) mentioned below. We then used Mauve (Darling et al. 2004, 2010) to determine the syntenic regions between the Bombella spp genomes. The B. apis G7_7_3c reference genome is resolved to a single chromosome, so it was used as the reference sequence in Mauve’s “move contigs” tool, and the likely order and orientation of contigs in the other genomes was determined. To facilitate downstream analyses, the output of Mauve’s “move contigs” tool was used to order, orient, and concatenate contigs into single pseudochromosomes for each genome. Structural rearrangements were then visualized using Mauve’s built-in graphical interface.
Annotation of CRISPR Arrays and Phage Sequences
Pseudochromosomes for each genome were uploaded to CRISPRFinder to determine location and sequence of CRISPR arrays (Grissa et al. 2007). To assess whether CRISPR arrays differ genome-to-genome (an indication that the arrays were incorporated after the strains diverged), we used an in-house Perl script to determine the maximum intergenomic percent identity of spacer sequences by aligning each spacer from a given genome to every spacer in every other genome and calculating percent identity (available via github here: https://github.com/esmith1032/Bombella_apis_evol/blob/master/README.md). For example, when we aligned two spacers, we took the shorter of the two and made linear, local alignments to every possible position in the longer of the two spacers, such that the entirety of the smaller spacer could still be aligned. For each of these alignments, we calculated the percent identity for the alignment and the maximum percent identity was retained as the percent identity for the two spacers. We also used the SEA-PHAGES data based to identify phages from which these spacers might be derived (supplementary table S6, Supplementary Material online). We used PHAge Search Tool Enhanced Release (PHASTER) (Zhou et al. 2011; Arndt et al. 2016) to define phage-like regions. Any region determined to be “questionable” or “intact” by PHASTER was labeled as likely to be of phage origin.
Determination of Bee-Associated Bacteria-Specific Orthologs
We identified all GOGs that contained at least one gene from each genome of Bombella spp. and no genes from Saccharibacter spp. We then took the B. apis G7_7_3c genome representative for each of these GOGs and got KEGG annotations for as many as possible using BlastKOALA (Kanehisa et al. 2016). Any hit that was given a definitive KO number by BlastKOALA was considered valid. For those genes that we were not able to get KEGG annotations, we used NCBI’s BLAST to aid in determining potential function of these bee-associated bacteria-specific genes (supplementary table S2, Supplementary Material online). This list of genes and their potential functions was then manually inspected to hypothesize genes that may have allowed for the transition to bee association.
Analysis of HGTs
To determine whether or not genes in any of the Bombella spp. genomes were horizontally transferred, we employed a combination of sequence-composition, phylogenetic, and synteny approaches. We mapped genes of particular interest (e.g., genes unique to certain clades, species, or strains) to their locations on the linear pseudochromosomes constructed during synteny analysis. Additionally, we calculated the %GC for each gene. We then determined how many SDs each gene was from its genome-wide mean %GC. The third prong of this analysis involved identifying genes that were phylogenetically aberrant. To do this, we used Darkhorse (Podell and Gaasterland 2007) to calculate the lineage probability index (LPI) for each gene. LPI measures the likelihood that a particular gene was inherited vertically, from the ancestor of the species of interest. Higher LPIs indicate a higher likelihood that the gene is ancestral and has not been horizontally transferred into the resident genome, whereas lower LPIs indicate that HGT may have occurred. Darkhorse calculates LPI using the taxonomy string for the best BLAST hits for each gene. We ran LPI twice, once including BLAST hits to Bombella and Saccharibacter subject sequences and once excluding such hits from the analysis. In doing so, genes with orthologs in close relatives of the Bombella/Saccharibacter clade (likely vertically inherited) will have high LPIs in both analyses, whereas genes without orthologs in close relatives of the clade (potentially horizontally transferred) will have a high LPI in the analysis that included Bombella and Saccharibacter BLAST hits, but a low LPI in the analysis that excluded those BLAST hits. For each gene, then, we can calculate the difference in LPI between the two analyses to determine how far from the genome-wide mean LPI difference (in SDs) each gene is. In doing so, genes that are likely to be horizontally transferred will have a larger discrepancy between LPI values than genes that were vertically inherited. We then classified regions as likely to be HGTs if they met the following criteria: 1) a block of at least three syntenic genes that show interesting phylogenetic distributions (e.g., unique to clade, species, or strain) where 2) a majority of genes in the region are at least 1 SD from the mean %GC or LPI difference (or both).
Domain Annotation of Genes of Interest
We used HHpred (https://toolkit.tuebingen.mpg.de/#/tools/hhpred; Soding et al. 2005), accessed on 8/2018, to determine domain architecture and gain an understanding of potential function of the genes in each HGT. For genes of interest that were part of a GOG, all members of the GOG were first aligned using the MAFFT L-INS-I algorithm (v7.310; Katoh et al. 2002). These multiple sequence alignments (or single amino acid sequences in the case of strain-unique genes) were then uploaded to HHpred’s online tool and homology was determined using HMMs in the COG_KOG_v1.0, Pfam-A_v31.0, and SMART_v6.0 databases; only domains scoring >60% probability are discussed here. Gene models for each region of interest were then constructed and visualized using the HHpred results and in-house R scripts to “draw” the gene models. A BLAST search using the nucleotide sequence of this gene against the NCBI nr database was used to determine a putative function.
Results
Saccharibacter and Bombella Are Sister Clades
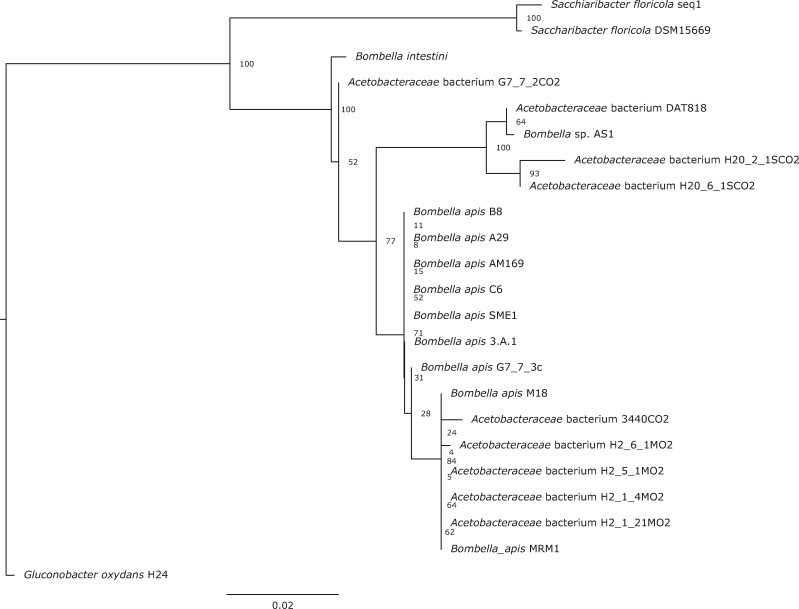
To robustly determine the relationship between Bombella and Saccharibacter spp., we constructed a maximum likelihood phylogeny using 16S rRNA sequences. Our final tree largely agrees with previously published phylogenies for this group (fig. 1) (Corby-Harris et al. 2014; Miller et al. 2020). Sequences were largely grouped into two monophyletic clades by genus. Bombella plus Saccharibacter comes out as sister to the Gluconobacter outgroup. The taxonomic nomenclature of this entire group has recently been revised but clearly Bombella and Saccharibacter are separate clades (Li et al. 2015; Yun et al. 2017). This conclusion is also supported by a recent publication by Bonilla-Rosso et al. (2019).
Fig. 1.
Maximum likelihood phylogenetic tree of Bombella and Saccharibacter species constructed from full-length 16S rRNA sequences, Gluconobacter oxydans as an outgroup. Bootstrap scores are indicated at each node.
Core Ortholog Phylogeny of Bombella and Saccharibacter Strains
We used OrthoMCL (v2.0.9; Li et al. 2003) to define GOGs using the Bombella and Saccharibacter genomes listed in table 1; G. oxydans H24 was used as an outgroup. In total, 3,017 GOGs were defined, with an average of 10.3 genes per GOG. Of these, 1,209 GOGs were present as single copies in every genome in the analysis, whereas an additional 34 GOGs were present in every genome, but in varying numbers in each genome. There were 1,526 GOGs that were variable (i.e., missing a representative from at least one genome, and present in varying numbers in the other genomes); and 248 GOGs that consisted of at least two genes that all came from the same genome (supplementary table S1, Supplementary Material online).
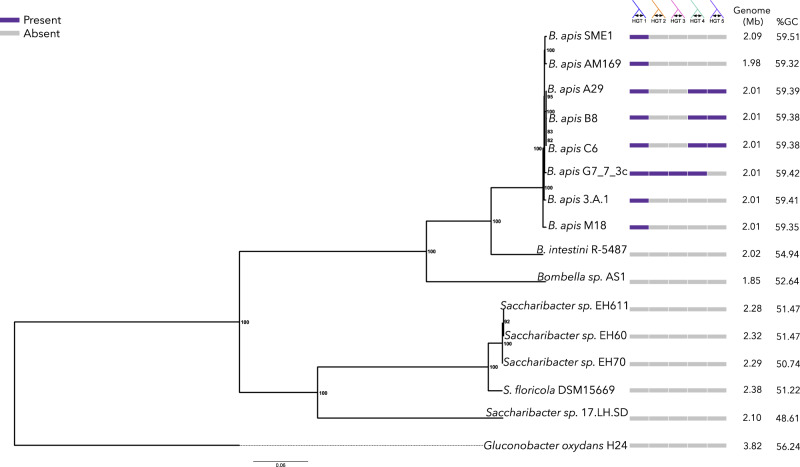
To better resolve the phylogenetic relationships between Bombella spp. and Saccharibacter spp., we constructed a second maximum likelihood phylogeny using aligned and concatenated amino acid sequences of the 1,209 single-copy GOGs (fig. 2). This robustly supported amino acid phylogeny broadly agrees with our previously constructed 16S phylogeny. In the core ortholog tree, B. intestini interrupts the monophyly of honey bee-associated Bombella genomes. Notably, this tree groups B. intestini more closely to the majority of the B. apis strains, whereas Bombella sp. AS1 is more distantly related, and possibly a different species, as suggested by Bonilla-Rosso et al. (2019). Similar to the 16S tree, we again see quite short branch lengths within the honey bee-associated acetic acid bacteria, particularly among those in the clade including all B. apis genomes (fig. 2).
Fig. 2.
Maximum likelihood phylogenetic tree of the Bombella/Saccharibacter clades constructed from concatenated amino acid alignment of 1,259 single-copy orthologous genes. Bootstrap scores are indicated at each node. Colored boxes represent the presence (purple) or absence (gray) of each of seven genomic regions of interest. Genome size and %GC are also displayed.
Identification of Sequences of Phage Origin
Movement and insertion of bacteriophage sequences in a genome can have profound effects on the evolution of that genome (Casas and Maloy 2011; Koskella and Brockhurst 2014; Harrison et al. 2017). Mobile genetic elements can also provide insight into the lifestyle of a bacterium, as the fraction of mobile DNA varies significantly with host ecology (Newton and Bordenstein 2011). We identified two regions of phage origin among the 15 genomes analyzed. One region in S. floricola was identified as “intact,” stretching ∼60 kb, and containing 89 proteins, all of which are identified by BLAST hits as being of phage origin or hypothetical. Synteny alignments indicate that this region is unique to S. floricola and not contained in any other genome. Likewise, OrthoMCL did not cluster any of the genes within this region with any other genes in our analysis, further supporting the idea that these genes are unique to S. floricola. The second region is in the B. apis G7_7_3c genome and was identified as “questionable” (one step below “intact”). This phage region is ∼31-kb long and contains 40 proteins, all of which were identified as either being of phage origin or hypothetical. Like Phage 1, synteny mapping and OrthoMCL clustering showed that this region is unique to B. apis G7_7_3c and shows no homology to any of the other genomes (fig. 2).
Signatures of Honey Bee Association in the B. apis Genomes
To identify genes associated with the transition to honey bee association, we identified GOGs that contained at least one gene from each Bombella spp. and were also missing in Saccharibacter spp. There were a total of 1,542 GOGs containing at least one gene from each of the aforementioned genomes, but only 74 were also missing in all Saccharibacter spp. We determined the putative functions of these genes using the B. apis G7_7_3c genome representative for each GOG (supplementary table S2, Supplementary Material online). It should be noted that all annotations discussed from here forward are putative and require further functional characterization.
Several bee-associated unique genes stood out as particularly interesting, the first being gluconolactonase. Lactonases, such as gluconolactonase, reversibly catalyze the hydrolysis of lactones (such as gluconolactone) to the hydroxyl acid form (such as gluconic acid). Gluconolactone is found in both honey and royal jelly and is thought to be partially responsible for the antibacterial properties of both compounds (Sagona et al. 2015). In water, this compound can be hydrolyzed into gluconic acid, acidifying the environment and preventing bacterial growth (Li et al. 2007; Schonleben et al. 2007; Furusawa et al. 2008). The presence of this gene, encoding an enzyme capable of reversing this acidification—at least locally—may explain how B. apis is able to thrive in the presence of royal jelly (Ramachandran et al. 2006; Corby-Harris et al. 2014). Alternatively, it is possible that B. apis is contributing to the production of gluconic acid in this environment. BLAST searches of the metatranscriptomes and metagenomes of bacteria in the “core” honey bee microbiome (Moran 2015; Lee et al. 2018) resulted in zero hits, indicating that none of the “core” microbiome members possesses a homolog of this gene. The presence of gluconolactonase may help explain the unique distribution of B. apis within the hive. Another bee-associated unique gene is an HdeD family acid-resistance protein, which in Escherichia coli participates in resistance to acids at high cell densities (Mates et al. 2007). The presence of this gene in B. apis may indicate an adaptation to living in low pH environments—such as the queen bee digestive tract or royal jelly (Anderson et al. 2011).
An AI-2 E family transporter was identified as unique to Bombella spp. AI-2 is an auto-inducer responsible for activating cascades associated with quorum sensing. Although B. apis does not contain any AI-2 synthesis genes, the presence of an AI-2 E family transporter indicates that it may be responding to exogenous AI-2 produced by other bacteria, possibly in a competitive interaction. Indeed, honey bee microbes are known to produce AI-2 (Miller et al. 2018). It is possible, also, that B. apis simply consumes this metabolite, using it as a source of carbon and energy. Bolstering the competition hypothesis is the presence of fusaric acid resistance (FUSC) genes in B. apis. Fusaric acid and its analogs can be quorum sensing inhibitors (Tung et al. 2017), so the presence of FUSC genes might be an adaptation that allows B. apis to evade quorum sensing inhibition attempts by other microbes. Alternatively, these FUSC genes may play a role in competition with fungal species. Fusaric acid is produced by several species of fungus and is antibacterial (Crutcher et al. 2017). Therefore, the FUSC genes may play a role in B. apis’s protection of honey bee larvae from fungal infection by allowing it to tolerate antibacterial capabilities of fungi and exert its antifungal properties (Corby-Harris et al. 2016; Miller et al. 2020). Interestingly, none of the canonical honey bee gut symbionts encodes FUSC genes, further suggesting a unique role for this gene in B. apis among honey bee symbionts.
The final set of genes of particular interest in this analysis is a complete Type I-E CRISPR/Cas cassette, found only in Bombella strains and not Saccharibacter. To determine if this CRISPR/Cas cassette was active, we annotated the genomes for the presence of CRISPR arrays, and found that all of the genomes that have this CRISPR/Cas cassette contain multiple CRISPR arrays. It is possible that these CRISPR arrays were present in the most recent common ancestor of the Bombella clade and have simply remained in these current genomes; if that were the case, we would expect the spacers in these CRISPR arrays to be highly similar between all strains. However, if these arrays are part of an active CRISPR/Cas system, we would expect the spacers to differ from strain to strain, reflecting unique challenges encountered by each strain. To rule out the possibility that these arrays are ancestral, we aligned each spacer sequence from a given genome to all other spacer sequences from the other genomes and calculated the percent identity. The minimum best intergenomic match for any spacer was 40%, whereas the maximum was just 65% identical over the length of the spacer, indicating that the spacer sequences are unique from genome to genome and the CRISPR/Cas systems identified here are likely active and/or were incorporated after the B. apis strains diverged from one another. Importantly, the spacers also do not match the existing prophage genomes in the Saccharibacter or Bombella genomes and have homology to known, sequenced phages (supplementary table S6, Supplementary Material online). Top SEA-PHAGES hits for each spacer in each genome are quite distinct and reflect the fact that each of these sequenced B. apis strains has likely interacted with a different pool of phages.
Bombella intestini was isolated from a bumble bee gut, so we also looked at genes that were unique to this bacterium. There were a total of 65 genes that were unique to B. intestini, including a complete type IV secretion system (T4SS) and several genes involved in antibiotic production or resistance. Putative annotations of these 65 genes are in supplementary table S3, Supplementary Material online.
Identification of Horizontally Transferred Gene Regions
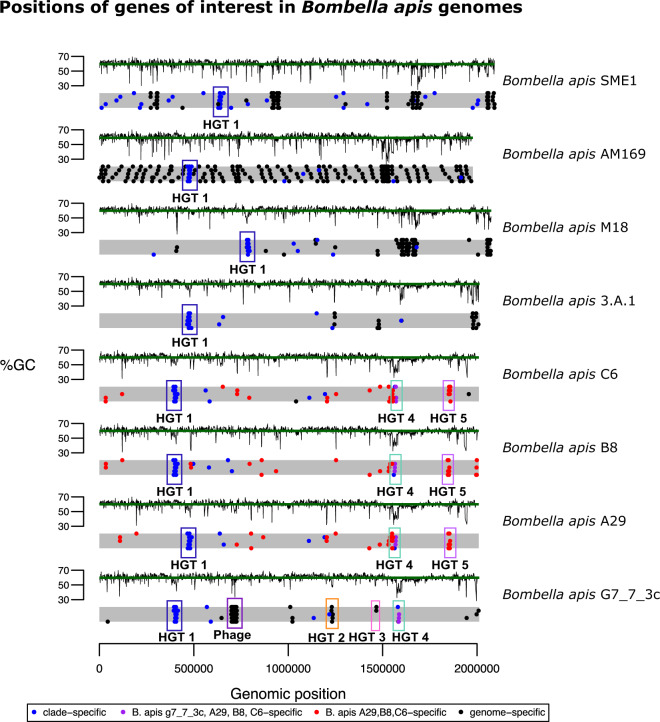
Horizontal transfer of DNA between unrelated bacteria is a commonly known mechanism by which bacteria can acquire new traits and adapt to novel environments (Gogarten et al. 2002; Nakamura et al. 2004; Wiedenbeck and Cohan 2011; Roberts and Kreth 2014). We identified two regions of phage origin, one in S. floricola and one in B. apis G7_7_3c (discussed above, fig. 2). To determine whether the bacteria in the Bombella clade have undergone other potential HGT events, we determined the spatial distribution of genes of particular interest (e.g., clade-specific, species-specific, or strain-specific genes) across the bacterial genomes (fig. 3). Some of the genes specific to different clades occur in clusters, an indication that they may have originated elsewhere and been horizontally inherited as a chunk of contiguous DNA. We then looked for anomalies in sequence composition (%GC) and phylogeny to determine whether they were putatively horizontally transferred. Using this combination of methods, we identified a total of five HGT regions in the Bombella clade, which we have numbered 1–5 (see supplementary table S4, Supplementary Material online, for %GC and LPI-difference deviations for each gene in each HGT).
Fig. 3.
Genomic locations of genes of interest in Bombella and Saccharibacter genomes. Each gray bar is a representation of the genome, with each dot representing the location of a gene in each of four categories (see legend). Regions of interest mentioned in the text are highlighted and labeled. %GC for every gene is plotted above each genome representation, with the green line indicating the genome-wide average %GC.
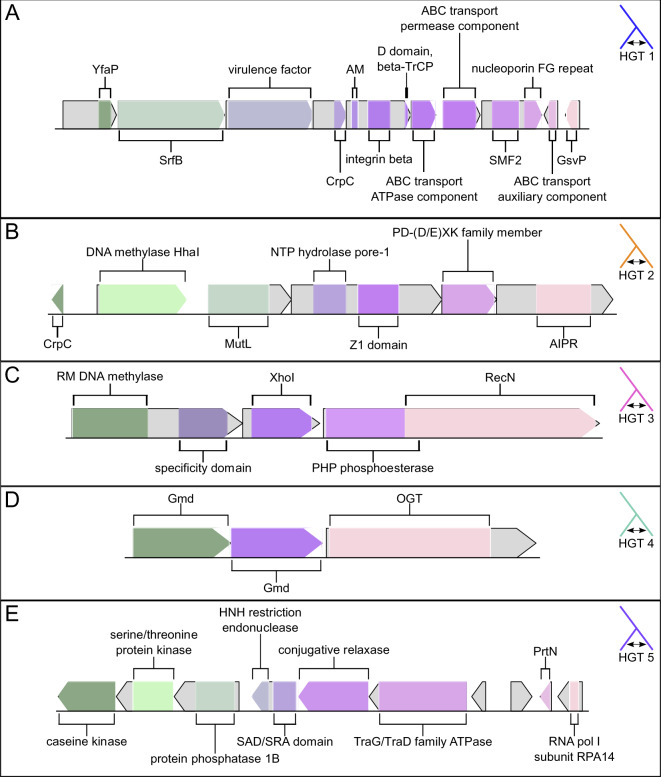
HGT1 (fig. 4A) is present in all genomes in the Bombella clade, and contains ten genes, although B. apis C6 is missing one of the genes (the second-to-last gene at the 3′ end of the HGT, annotated as an ABC transport auxiliary component). The three most 5′ genes show homology to YfaP (an uncharacterized conserved protein), SrfB (part of the surfactin antibiotic synthesis machinery), and an uncharacterized bacterial virulence factor. The genes in the 3′ half of this HGT contain a number of domains involved in membrane transport. We hypothesize that the two halves of this HGT work together to synthesize and export antibiotics as a form of defense or regulation of competing bacteria. Lending support to the hypothesis that this HGT is involved in defense or immunity is the fact that a CRISPR array lies immediately 5′ of this HGT in each genome (supplementary table S5, Supplementary Material online). Bacterial defense mechanisms tend to occur in clusters of “defense islands” (Koonin et al. 2017; Doron et al. 2018), where CRISPR/Cas systems are found in proximity to restriction–modification (R–M) systems so the presence of this CRISPR array is perhaps a further indication of this HGTs role in bacterial immunity.
Fig. 4.
Gene models for each of five genomic regions of interest. Gene models are drawn to scale within each panel, but not across panels. (A) HGT1. Abbreviations are: CrpC, cysteine-rich protein C; AM, automated matches; SMF2, sulfatase modifying factor 2; GsvP, gas vesicle protein C. (B) HGT2. Abbreviations are: CrpC, cysteine-rich protein C; AIPR, abortive infection phage resistance protein. (C) HGT3. Abbreviations are: RM, restriction–modification. (D) HGT 4. Abbreviations are: Gmd, GDP-d-mannose dehydratase; OGT, O-linked N-acetylglucosamine transferase OGT. (E) HGT5. Abbreviations are: SAD/SRA, SET and Ring finger Associated; PrtN, pyocin-activator protein.
HGTs 2 and 3 (fig. 4B and C) are restricted solely to B. apis G7_7_3c and are both bacterial R–M systems. Bacterial R–M systems are a defense against invading DNA (i.e., bacteriophage). They act by methylating host DNA at specific sites; invading DNA with the same recognition site will be unmethylated, recognized as foreign, and targeted for degradation (Rodic et al. 2017). HGT2 contains six genes, which make up the core components of a bacterial (R–M) system. Interestingly, the domain architecture in this R–M system has been recognized as an evolutionary precursor to eukaryotic defenses against transposable elements (Iyer et al. 2011). HGT3 (fig. 4C) consists of three genes comprising five domains; the 5′-most gene consists of a predicted R–M DNA methylase coupled to a specificity domain, the middle gene is predicted to be an XhoI restriction enzyme, and the 3′-most gene is a PHP phosphoesterase coupled to a RecN DNA repair ATPase. Taken together, it appears that HGTs 2 and 3 are responsible for recognition of and defense against foreign DNA.
HGT4 (fig. 4D) is present in B. apis strains G7_7_3c, A29, B8, and C6 and contains three genes: two GDP-d-mannose dehydratases (GMD) and an O-linked N-acetylglucosamine transferase (OGT). GMD plays a role in the metabolism of mannose and fructose, sugars commonly found in nectar and pollen (Freeman and Madrono 1985). The presence of GMD in B. apis genomes might allow for the consumption of nectar or nectar components by these bacteria. OGT, on the other hand, plays a role in posttranslational modification of thousands of identified proteins (Love and Hanover 2005). However, although OGT-mediated posttranslational modification is common in eukaryotes, it is far more rare in bacteria (Ostrowski et al. 2015). To date, only a handful of prokaryotic OGTs have been identified, and the targets of these OGTs remain unclear (Shen et al. 2006; Sokol and Olszewski 2015). Given the role OGTs play in eukaryotic posttranslational modification and the fact that many bacterial effector proteins show homology to eukaryotic proteins (Galan 2009), it is possible that the presence of OGT in B. apis represents a pathway for host–microbe interaction and symbiont-mediated protein modification.
HGT5 (fig. 4E) is unique to B. apis strains A29, B8, and C6, all strains that had been isolated from honey bee larvae. The fifth HGT identified (HGT5) occurs at the junction of two contigs in the linear pseudochromosomes we constructed. The abutting ends of each contig have annotations for partial pseudogenes, such that when they are joined a complete gene is created. Like HGTs 1–3, HGT5 contains genes that may play a role in protection against foreign DNA. There are four genes in the 5′ section of HGT5, three of which are kinases, and the fourth contains a SAD/SRA domain in its 5′ end, and an HNH endonuclease domain in its 3′ end. In bacteria, the SAD/SRA domain is often found associated with an HNH domain (Makarova et al. 2011) and it is thought that the two domains act together to recognize and cleave foreign DNA (Iyer et al. 2011). The 3′ section of HGT5 consists of a conjugative relaxase, a TraG/TraD family ATPase (a coupling protein involved in bacterial conjugation and/or T4SS), a homolog of the pyocin-activator protein PrtN, a homolog of a yeast RNA polymerase I subunit, and two additional genes with no annotations. The presence of a PrtN homolog is particularly interesting, as in Pseudomonas aeruginosa pyocins are antibacterial agents, often acting to depolarize the membrane of target cells (Jacob 1954; Michel-Briand and Baysse 2002). Interestingly, one of the two unannotated genes in the 3′ region of HGT5 shows weak homology to a phage shock protein, which are proteins involved in the response to stress that may weaken the energy status of the cell (Flores-Kim and Darwin 2016). This protein, then, may play a part in immunity to membrane depolarization. Given the presence in HGT5 of: an HNH endonuclease coupled to a SAD/SRA domain, a conjugative relaxase, a TraG/TraD family ATPase, a pyocin-activator protein, and a protein with at least some homology to a phage shock protein, we hypothesize that it may play a role in pathogenesis or defense.
Discussion
Here, we used the genomes of ten Bombella spp. and five Saccharibacter spp. to gain insight into the genomic changes associated with the transition to honey bee symbiosis in this group. We note several genomic differences—some of which were horizontally acquired—between bee-associated bacteria and the flower-associated Saccharibacter that may have allowed for the expansion of B. apis into previously unoccupied niches within the honey bee colony. These differences can be classified as changes that introduce: 1) novel metabolic capabilities, 2) defense and/or virulence mechanisms, and 3) mechanisms for interaction with other microbes and/or the host.
Metabolic genes identified here include gluconolactonase, which may allow for the deacidification of royal jelly (Li et al. 2007; Schonleben et al. 2007; Furusawa et al. 2008; Sagona et al. 2015), and two copies of GMD, a gene that plays a role in the metabolism of mannose and fructose, components of nectar and honey (Freeman and Madrono 1985). Distinct defense and/or virulence mechanisms were identified, including: a CRISPR/Cas system, two R–M systems, and an HGT with some homology to known virulence mechanisms. Interestingly, the R–M systems were identified in the only strain in the clade that also contains a phage sequence (B. apis G7_7_3c). R–M systems, like phages, can act as selfish genetic elements (Kobayashi 2001), so their presence in this genome may indicate that it was historically more permissive to invading DNA. These R–M systems may also have been coopted by the prophage to prevent super-infection with additional phages (Van Etten et al. 1988).
Genes involved in the interaction with other microbes and/or the host that we identified include: an AI-2 family transporter, fusaric acid resistance genes, and ogt. Given that B. apis does not encode any of the canonical genes for the production of quorum-sensing molecules, it seems likely that B. apis is responding to exogenous AI-2 (and/or fusaric acid and its analogs) produced by other members of the bee microbiome (Miller et al. 2018). The ogt genes provide routes for interaction with the host, as ogt is known to modify thousands of eukaryotic proteins (Love and Hanover 2005). Taken together, we hypothesize that the novel combination of metabolic, quorum-sensing, defense/virulence, and eukaryotic interaction genes in the Bombella clade genomes allowed for the utilization of a unique food source and protection from an onslaught of previously unencountered challenges and facilitated the transition to honey bee association in this clade.
Bombella apis has been shown to benefit honey bee larval development and provide protection against fungal pathogens (Corby-Harris et al. 2016; Miller et al. 2020). Some of the genes identified here, while allowing B. apis to transition to honey bee symbiosis, may also be related to its ability to protect the bee host from infection with fungi or other pathogens. If indeed these genes are responsible for the transition to, and maintenance of, honey bee symbiosis, we would expect to see a modified evolutionary trajectory relative to those genes not involved in the symbiosis. We currently lack sufficient sampling of nonbee-associated bacteria in this clade to do such analyses; however, future studies addressing this question should allow for further elucidation of the genes involved in the transition to honey bee association. Those analyses, coupled with functional characterization of the genes of interest identified here, should lay the foundation for the development of beneficial intervention strategies in this economically critical insect.
Supplementary Material
Acknowledgments
We thank Amelia R.I. Lindsey for feedback on initial versions of this article. E.A.S. was supported by a Faculty Research Support grant from Indiana University and by a USDA NIFA Postdoctoral Fellowship. This work was supported by NSF IOS 2005306 to I.L.G.N.
Literature Cited
- Anderson KE, et al. 2013. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8(12):e83125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, et al. 2018. The queen’s gut refines with age: longevity phenotypes in a social insect model. Microbiome 6(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffman G. 2011. An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis meliffera). Insect Soc. 58(4):431–444. [Google Scholar]
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Rosso G, et al. 2019. Acetobacteraceae in the honey bee gut comprise two distant clades with diverging metabolism and ecological niches. 861260: bioRxiv.
- Casas V, Maloy S. 2011. Role of bacteriophage-encoded exotoxins in the evolution of bacterial pathogens. Future Microbiol. 6(12):1461–1473. [DOI] [PubMed] [Google Scholar]
- Chouaia B, et al. 2014. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol Evol. 6(4):912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, Anderson KE. 2018. Draft genome sequences of four Parasaccharibacter apium strains isolated from honey bees. Genome Announc. 6(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, et al. 2014. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl Environ Microbiol. 80(24):7460–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, et al. 2016. Parasaccharibacter apium, gen. nov., sp. nov., improves honey bee (Hymenoptera: Apidae) resistance to Nosema. J Econ Entomol. 109(2):537–543. [DOI] [PubMed] [Google Scholar]
- Crutcher FK, et al. 2017. Microbial resistance mechanisms to the antibiotic and phytotoxin fusaric acid. J Chem Ecol. 43(10):996–1006. [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, et al. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359(6379):eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 2013. Microbial brokers of insect-plant interactions revisited. J Chem Ecol. 39(7):952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Kim J, Darwin AJ. 2016. The phage shock protein response. Annu Rev Microbiol. 70(1):83–101. [DOI] [PubMed] [Google Scholar]
- Freeman CE, Madrono RW. 1985. Some floral nectar-sugar compositions of species from southeastern Arizona and southwestern New Mexico. Madrono 32:78–86. [Google Scholar]
- Furusawa T, et al. 2008. Comprehensive royal jelly (RJ) proteomics using one- and two-dimensional proteomics platforms reveals novel RJ proteins and potential phospho/glycoproteins. J Proteome Res. 7(8):3194–3229. [DOI] [PubMed] [Google Scholar]
- Galan JE. 2009. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 5:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner FO, et al. 2017. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 261:169–176. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 19(12):2226–2238. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35(Web Server):W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E, Hall JPJ, Paterson S, Spiers AJ, Brockhurst MA. 2017. Conflicting selection alters the trajectory of molecular evolution in a tripartite bacteria-plasmid-phage interaction. Mol Ecol. 26(10):2757–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L. 2011. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 101:25–104. [DOI] [PubMed] [Google Scholar]
- Jacob F. 1954. Induced biosynthesis and mode of action of a pyocine, antibiotic produced by Pseudomonas aeruginosa. Ann Inst Pasteur (Paris). 86(2):149–160. [PubMed] [Google Scholar]
- Jojima Y, et al. 2004. Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int J Syst Evol Microbiol. 54(Pt 6):2263–2267. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428(4):726–731. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29(18):3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A. 108(48):19288–19292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Wolf YI. 2017. Evolutionary genomics of defense systems in archaea and bacteria. Annu Rev Microbiol. 71(1):233–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38(5):916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Miller KI, McKinlay JB, Newton ILG. 2018. Differential carbohydrate utilization and organic acid production by honey bee symbionts. FEMS Microbiol Ecol. 94(8). doi: 10.1093/femsec/fiy113. [DOI] [PubMed] [Google Scholar]
- Li J, Wang T, Zhang Z, Pan Y. 2007. Proteomic analysis of royal jelly from three strains of western honeybees (Apis mellifera). J Agric Food Chem. 55(21):8411–8422. [DOI] [PubMed] [Google Scholar]
- Li L, et al. 2015. Bombella intestini gen. nov., sp. nov., an acetic acid bacterium isolated from bumble bee crop. Int J Syst Evol Microbiol. 65(Pt 1):267–273. [DOI] [PubMed] [Google Scholar]
- Li L, et al. 2016. Whole-genome sequence analysis of Bombella intestini LMG 28161T, a novel acetic acid bacterium isolated from the crop of a red-tailed bumble bee, Bombus lapidarius. PLoS One 11(11):e0165611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA. 2005. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005(312):re13. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Snir S, Koonin EV. 2011. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 193(21):6039–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, et al. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 20(3):619–628. [DOI] [PubMed] [Google Scholar]
- Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 78(8):2830–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol. 189(7):2759–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 104(49):19392–19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84(5–6):499–510. [DOI] [PubMed] [Google Scholar]
- Miller DL, Smith EA, Newton ILG. 2020. A bacterial symbiont protects honey bees from fungal disease. bioRxiv. [DOI] [PMC free article] [PubMed]
- Miller KI, Franklin CD, Mattila HR, Newton IL. 2018. Social communication between microbes colonizing the social honey bee Apis mellifera. bioRxiv. doi: 10.1101/287995. [Google Scholar]
- Moran NA. 2015. Genomics of the honey bee microbiome. Curr Opin Insect Sci. 10:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7(4):e36393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 36(7):760–766. [DOI] [PubMed] [Google Scholar]
- Newton IL, Bordenstein SR. 2011. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 62(1):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA, van Aalten DM. 2015. Evidence for a functional O-linked N-acetylglucosamine (O-GlcNAc) system in the thermophilic bacterium Thermobaculum terrenum. J Biol Chem. 290(51):30291–30305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T. 2007. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 8(2):R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, et al. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(D1):D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Fontanille P, Pandey A, Larroche C. 2006. Gluconic acid: properties, applications, and microbial production. Food Technol Biotechnol. 44:185–195. [Google Scholar]
- Roberts AP, Kreth J. 2014. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front Cell Infect Microbiol. 4:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic A, Blagojevic B, Zdobnov E, Djordjevic M, Djordjevic M. 2017. Understanding key features of bacterial restriction-modification systems through quantitative modeling. BMC Syst Biol. 11(S1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokop ZP, Horton MA, Newton IL. 2015. Interactions between cooccurring lactic acid bacteria in honey bee hives. Appl Environ Microbiol. 81(20):7261–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree ZL, Hansen AK, Moran NA. 2012. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One 7(7):e41250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona S, Turchi B, Fratini F, Guiusti M, Bull BT. 2015. Preliminary evaluation of glucose oxidase and its products in vitro antimicrobial activities on Paenibacillus larvae ATCC9545 vegetative form. Bullet Insectol. 68:233–237. [Google Scholar]
- Schonleben S, Sickmann A, Mueller MJ, Reinders J. 2007. Proteome analysis of Apis mellifera royal jelly. Anal Bioanal Chem. 389(4):1087–1093. [DOI] [PubMed] [Google Scholar]
- Shen A, Kamp HD, Grundling A, Higgins DE. 2006. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20(23):3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Anderson KE, Corby-Harris V, McFrederick QS, Newton IL. 2020. Reclassification of seven honey bee symbiont strains as Bombella apis. bioRxiv. doi: 10.1101/2020.05.06.081802. [DOI] [PubMed]
- Smith EA, Martin-Eberhardt SA, Miller DL, Parish AJ, Newton ILG. 2019. Draft genome sequence of a Bombella apis strain isolated from honey bees. Microbiol Resour Announc. 8:e01329-19. doi: 10.1128/MRA.01329-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Vuong HQ, et al. 2020. Draft genome sequences of four Saccharibacter sp. strains isolated from native bees. Microbiol Resour Announc.9:e00022-20. doi: 10.1128/MRA.00022-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server):W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol KA, Olszewski NE. 2015. The putative eukaryote-like O-GlcNAc transferase of the cyanobacterium Synechococcus elongatus PCC 7942 hydrolyzes UDP-GlcNAc and is involved in multiple cellular processes. J Bacteriol. 197(2):354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Tarpy DR, Mattila HR, Newton IL. 2015. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl Environ Microbiol. 81(9):3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung TT, et al. 2017. Fusaric acid and analogues as Gram-negative bacterial quorum sensing inhibitors. Eur J Med Chem. 126:1011–1020. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Xia YN, Burbank DE, Narva KE. 1988. Chlorella viruses code for restriction and modification enzymes. Gene 74(1):113–115. [DOI] [PubMed] [Google Scholar]
- Varghese NJ, et al. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43(14):6761–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veress A, Wilk T, Kiss J, Olasz F, Papp PP. 2017. Draft genome sequences of Saccharibacter sp. strains 3.A.1 and M18 isolated from honey and a honey bee (Apis mellifera) stomach. Genome Announc. 5(30). doi:10.1128/genomeA.00744-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojvodic S, Rehan SM, Anderson KE. 2013. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One 8(8):e72106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 35(5):957–976. [DOI] [PubMed] [Google Scholar]
- Yilmaz P, et al. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42(D1):D643–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JH, Lee JY, Hyun DW, Jung MJ, Bae JW. 2017. Bombella apis sp. nov., an acetic acid bacterium isolated from the midgut of a honey bee. Int J Syst Evol Microbiol. 67(7):2184–2188. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39(Suppl):W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.