Abstract
Isolated reports of new-onset diabetes in individuals with COVID-19 have led to the hypothesis that SARS-CoV-2 is directly cytotoxic to pancreatic islet β cells. This would require binding and entry of SARS-CoV-2 into β cells via co-expression of its canonical cell entry factors, angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2); however, their expression in human pancreas has not been clearly defined. We analyzed six transcriptional datasets of primary human islet cells and found that ACE2 and TMPRSS2 were not co-expressed in single β cells. In pancreatic sections, ACE2 and TMPRSS2 protein was not detected in β cells from donors with and without diabetes. Instead, ACE2 protein was expressed in islet and exocrine tissue microvasculature and in a subset of pancreatic ducts, whereas TMPRSS2 protein was restricted to ductal cells. These findings reduce the likelihood that SARS-CoV-2 directly infects β cells in vivo through ACE2 and TMPRSS2.
Keywords: SARS-CoV-2, COVID-19, ACE2, TMPRSS2, islet, beta cell, pericyte, microvasculature, duct, pancreas
Graphical Abstract
Highlights
-
•
ACE2 and TMPRSS2 mRNA are expressed in the human pancreas
-
•
ACE2 protein is expressed in some islet and exocrine capillaries and pericytes
-
•
ACE2 and TMPRSS2 proteins are co-expressed in some pancreatic ducts
-
•
ACE2 and TMPRSS2 protein is not detected in β cells of the human pancreas
Coate et al. examined expression of canonical SARS-CoV-2 entry proteins ACE2 and TMPRSS2 in the human pancreas and report ACE2 expression in the microvasculature, including islet pericytes, whereas both ACE2 and TMPRSS2 are expressed in some ducts. Conversely, neither protein is detected in β cells, arguing against direct β cell viral infection in vivo.
Introduction
In coronavirus disease of 2019 (COVID-19), elevated body mass index (BMI) and plasma glucose levels, with or without pre-existing diabetes, have been identified as independent risk factors of morbidity and mortality with severe acute respiratory syndrome-associated coronavirus-2 (SARS-CoV-2) infection (Barron et al., 2020; Cariou et al., 2020; Holman et al., 2020; Riddle et al., 2020; Wang et al., 2020b; Zhou et al., 2020). Isolated cases of new-onset diabetes and diabetic emergencies such as ketoacidosis and hyperosmolar hyperglycemia have been reported with COVID-19 (Chee et al., 2020; Goldman et al., 2020; Hollstein et al., 2020; Kim et al., 2020; Li et al., 2020a; Rafique and Ahmed, 2020; Unsworth et al., 2020), leading to the hypothesis that SARS-CoV-2 has a diabetogenic effect mediated by direct cytotoxicity to pancreatic islet β cells (Koch et al., 2020).
In vitro studies have shown that SARS-CoV-2 entry into human host cells requires binding to the cell surface receptor angiotensin-converting enzyme 2 (ACE2), as well as proteolytic cleavage of the viral spike (S) protein by transmembrane serine protease 2 (TMPRSS2) (Hoffmann et al., 2020; Lan et al., 2020; Shang et al., 2020; Wiersinga et al., 2020). In 2010, Yang et al. (2010) examined autopsy samples from a single deceased patient infected by SARS-CoV-1, which uses similar machinery for binding and cellular entry, and reported expression of ACE2 in pancreatic islet cells. Though the identity of these islet cells was not assessed, the authors suggested that binding of ACE2 by SARS-CoV-1 damages islets and causes acute diabetes, which could be reversed after viral recovery (Yang et al., 2010). Further, there have been occasional reports of other viral infections eliciting a diabetogenic effect (reviewed in Filippi and von Herrath, 2008). More recently, Yang and colleagues reported that β-like cells derived from human pluripotent stem cells (hPSCs) as well as β cells of primary human islets express ACE2, raising the possibility of direct infection and cytotoxicity of β cells by SARS-CoV-2 (Yang et al., 2020). Importantly, neither of these prior studies (Yang et al., 2010, 2020) characterized the expression and localization of TMPRSS2, an obligate co-factor for SARS-CoV-2 cellular entry. Thus, a more detailed analysis of both ACE2 and TMPRSS2 expression and localization in human pancreatic tissue from normal donors and those with diabetes is urgently needed. The purpose of this study was to test the hypothesis that native pancreatic islet β cells possess the cellular machinery that could render them direct targets of SARS-CoV-2. Importantly, we found that ACE2 and TMPRSS2 proteins are not detectable in human islet endocrine cells from normal donors or those with diabetes, making a direct diabetogenic effect of SARS-CoV-2 via ACE2 and TMPRSS2 unlikely.
Results and Discussion
ACE2 and TMPRSS2 mRNA Expression Is Minimal in Human α or β Cells
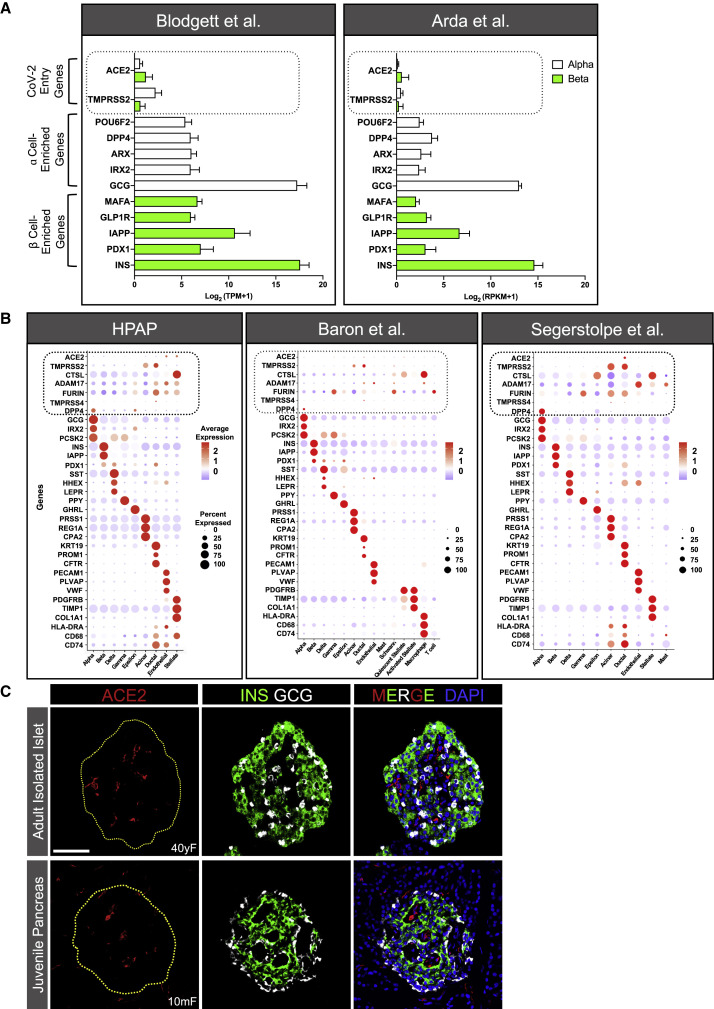
We first evaluated mRNA expression of ACE2 and TMPRSS2 from two existing bulk RNA sequencing (RNA-seq) datasets (Arda et al., 2016; Blodgett et al., 2015), where human islet α and β cells were enriched by fluorescence-activated cell sorting, and compared their expression to that of key islet-enriched genes, some of which are normally expressed at relatively low levels in islet cells (e.g., transcription factors). Median expression level of ACE2 and TMPRSS2 mRNA was much less than transcripts of such key islet-enriched genes in human α and β cells (∼84% and 92% lower than α and β cell-enriched transcripts, respectively) (Figure 1 A; n = 7–8 adult donors per study). In addition, analysis of four single-cell (sc) RNA-seq datasets of human pancreatic cells (Baron et al., 2016; Camunas-Soler et al., 2020; Kaestner et al., 2019; Segerstolpe et al., 2016) revealed that, in aggregate, less than 1.5% of β cells expressed ACE2 or TMPRSS2, and each transcript was minimally expressed or undetectable in all other endocrine cell subsets (Figures 1B and S1A; Table S1). We note that this included analysis of the robust Human Pancreas Analysis Program (HPAP) dataset that contained more than 25,000 cells from 11 normal donors (Kaestner et al., 2019), the findings of which were confirmed in the analyses of three previously reported, but smaller, datasets (Figures 1B and S1A). Furthermore, given that ACE2 and TMPRSS2 co-expression is required for canonical SARS-CoV-2 host cell entry (Hoffmann et al., 2020), we also evaluated this occurrence but found that no β cells co-expressed ACE2 and TMPRSS2 in any of these four datasets (Table S1).
Figure 1.
ACE2 and TMPRSS2 Expression in Isolated Human Islet Cells and Juvenile Pancreas
(A) Relative expression of ACE2 and TMPRSS2 compared with select α (white bars) and β (green bars) cell-type-enriched genes in sorted human islet α and β cells from previously published bulk RNA-seq datasets, reported as transcript per million mapped reads (TPM; n = 7; Blodgett et al., 2015) or reads per kilobase of transcript per million mapped reads (RPKM; n = 8; Arda et al., 2016). Mean expression values are presented as log2 (TPM+1) or log2 (RPKM+1) to account for negative values. Dotted line highlights ACE2 and TMPRSS2 expression.
(B) Dot plots of ACE2, TMPRSS2, CTSL, ADAM17, FURIN, TMPRSS4, and DPP4 expression compared with cell-type-enriched genes from three single-cell (sc) RNA-seq datasets (Baron et al., 2016; Kaestner et al., 2019; Segerstolpe et al., 2016). Dot size indicates percentage of cells in a given population expressing the gene; dot color represents scaled average expression. Dotted lines highlight expression of putative SARS-CoV-2 entry machinery. Percentages of β cells expressing and co-expressing these genes are available in Table S1.
(C) Representative images of an isolated islet from an adult human donor (top panels; n = 6 total images analyzed) and a pancreatic section from a juvenile human donor (bottom panels; n = 10 total images analyzed) stained for ACE2 (red; antibody ab15348), insulin (INS; green), glucagon (GCG; white), and DAPI (blue). Dotted yellow line denotes islet area. Human pancreatic donor information is available in Table S2 (donors I1–I3 and J1–J5). See also Figures S1, S2, and S4. Scale bar, 50 μm.
As for non-endocrine cells, the HPAP dataset (Kaestner et al., 2019) revealed that a small subset (<5%) of endothelial and stellate cells (which include pericytes) expressed moderate to high levels of ACE2, whereas only ∼1%–3% of either population expressed ACE2 in the datasets by Baron et al. (2016) and Segerstolpe et al. (2016) (Figure 1B). This difference most likely stemmed from the number of cells analyzed; the HPAP dataset contains ∼2.5- and 20-fold more total cells than that of Baron et al. (2016) or Segerstolpe et al. (2016), respectively. In addition, pooled analysis of these datasets (Baron et al., 2016; Camunas-Soler et al., 2020; Kaestner et al., 2019) showed that less than 1% of acinar or ductal cells expressed ACE2, whereas ∼35% of both cell types expressed TMPRSS2. In the dataset by Segerstolpe et al. (2016), ACE2 was expressed in ∼20% of acinar and ductal cells, whereas TMPRSS2 was expressed in greater than 75% of both populations (Figure 1B). Evaluation of ACE2 and TMPRSS2 co-expression in non-endocrine cells revealed that on average, less than 1% of acinar, ductal, endothelial, and stellate cells co-expressed both transcripts in these datasets (Baron et al., 2016; Camunas-Soler et al., 2020; Kaestner et al., 2019). In the Segerstolpe et al. (2016) dataset, ∼5% of acinar cells and 15% of ductal cells co-expressed ACE2 and TMPRSS2, but this difference could represent unintended cell selection bias as this study analyzed the fewest number of cells.
Altogether, these gene expression findings reduce the likelihood that SARS-CoV-2 can bind and enter human β cells via the canonical pathway involving ACE2 and TMPRSS2. However, they do not exclude the possibility of viral entry via non-canonical pathways involving other suggested effector proteases of SARS-CoV-2, such as Cathepsin L (CTSL), ADAM metallopeptidase domain 17 (ADAM17), FURIN, and TMPRSS4 (Breidenbach et al., 2020; Schreiber et al., 2020; Seyedpour et al., 2020; Zang et al., 2020). We evaluated the expression of these transcripts across all four scRNA-seq datasets and found that with the exception of TMPRSS4, each is ubiquitously expressed to varying degrees in all cell types (Figures 1B and S1A; Table S1). In aggregate, however, less than 1.3% of β cells co-expressed ACE2 with CTSL, ADAM17, FURIN, or TMPRSS4 (Table S1). An outstanding question is whether expression of these transcripts in even 1% of β cells is sufficient to confer permissiveness to SARS-CoV-2 infection in vivo. Furthermore, heparan sulfate in the extracellular glycocalyx (Clausen et al., 2020) and motile cilia (present on islet endocrine cells) (Lee et al., 2020) are mechanisms proposed to assist ACE2-mediated viral entry. Future studies addressing these possibilities in β cells are urgently needed. Notwithstanding, CTSL, ADAM17, FURIN, and TMPRSS4 were more highly expressed in exocrine, endothelial, and stellate cells, as was ACE2 (Figure 1B). These gene expression patterns raise the possibility that infection of certain pancreatic cell types by SARS-CoV-2 indirectly impacts β cell function.
Obesity is a key metabolic risk factor associated with mortality with COVID-19. A recent report found a positive correlation between BMI and TMPRSS2, but not ACE2, expression (Taneera et al., 2020). We evaluated the expression of ACE2, TMPRSS2, and ADAM17 in β cells from 11 donors according to their BMI in our largest scRNA-seq dataset (Kaestner et al., 2019). We did not observe changes in the expression of ACE2 or TMPRSS2 expression with higher BMI but observed a trend toward higher ADAM17 expression (Figure S1B). However, this dataset included only one patient with a BMI in the obese range. Studies with a larger number of donor islets are needed to address the role of obesity and other pancreas-resident SARS-CoV-2 effector molecules on β cell function in the context of COVID-19.
ACE2 and TMPRSS2 Proteins Are Not Detected in Adult or Juvenile Human Islet α or β Cells
Recently, Yang et al. (2020) reported that ACE2 protein was present in α and β cells from primary human islets and in β-like cells derived from hPSCs. To investigate ACE2 expression, we immunostained cryosections of intact isolated human islets using the same ACE2 antibody as used by Yang et al. (2020) (antibody validation studies in Figure S2; mouse [Figure S2A] and human [Figures S2J, S2O, and S2T] kidney tissues were utilized as positive controls for ACE2 immunostaining). In accordance with the transcriptomic data reported above, ACE2 was not detected in insulin-positive β cells or glucagon-positive α cells in our samples; instead, ACE2 signal was prominent in surrounding cells such as those of the microvasculature (Figure 1C, top panels).
Since hPSC-derived β-like cells differentiated in vitro are considered “juvenile-like” and not functionally mature β cells (Nair et al., 2019), one possibility is that ACE2 is expressed in less differentiated cells, or in the cultured, immortalized β cells (EndoCβH1) studied by Fignani et al. (2020). We investigated such a possibility by staining for ACE2 in pancreatic sections from juvenile human donors (age range 5 days to 5 years) using the same ACE2 antibody as used by Yang et al. (2020). However, ACE2 was not detected in these juvenile β or α cells (Figure 1C, bottom panels), making this explanation less likely.
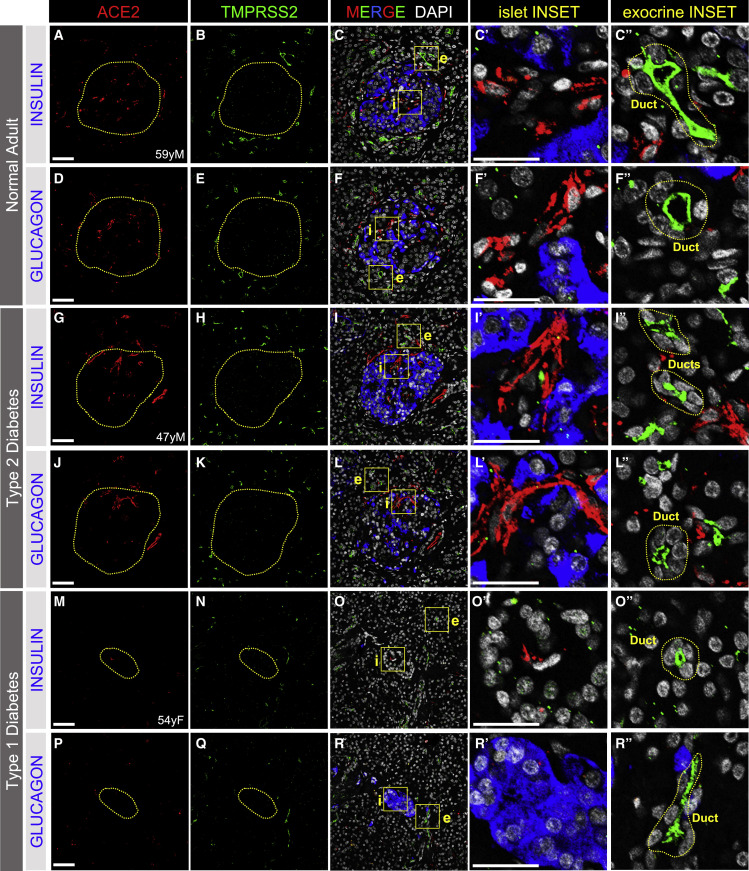
To further characterize ACE2 and TMPRSS2 protein expression in the native pancreas, we next analyzed human pancreatic tissue sections from normal donors (ND, n = 14; age range 18–59 years) and those with type 2 diabetes (T2D, n = 12; age range 42–66 years) or type 1 diabetes (T1D, n = 11; age range 13–63 years) for these proteins on the same sections. ACE2 protein did not co-localize with markers of α or β cells but was detected within the distinct islet areas indicative of microvascular structures in all ND individuals (Figures 2A–2F″ and S3A–S3D) as well as those with T2D (Figures 2G–2L″ and S3I–S3L) or T1D (Figures 2M–2R″ and S3O–S3R). We noted similar findings with three additional ACE2-directed antibodies, including the same antibodies and dilutions reported by Yang et al. (2020) and Fignani et al., (2020), and one antibody validated by the Human Protein Atlas (HPA) (Hikmet et al., 2020) (Figure S2). Three of these antibodies detect epitopes corresponding to the extracellular domain of ACE2 (AF933, MAB933, and HPA000288), while one detects an epitope in the C-terminal domain (ab15348). Staining patterns were similar across all antibodies (Figure S2). Furthermore, ACE2 antibody specificity (ab15348) was confirmed via peptide competition with the commercially available immunizing peptide (Figures S2F–S2I).
Figure 2.
ACE2 and TMPRSS2 Protein in Human Islets and Exocrine Tissue of Adult Pancreas
(A–F″) Immunostaining of SARS-CoV-2 cell entry markers ACE2 (red; antibody AF933; A, C, C,′ D, F, and F′) or TMPRSS2 (green; B, C, C″, E, F, and F″) in islet β cells (insulin, blue; C and C′) and islet α cells (glucagon, blue; F and F′) in native pancreatic sections from adult normal donors without diabetes (n = 14 total images analyzed).
(G–L″) Immunostaining of SARS-CoV-2 cell entry markers ACE2 (red; antibody AF933; G, I, I′, J, L, and L′) or TMPRSS2 (green; H, I, I″, K, L, and L″) in islet β cells (insulin, blue; I and I′) and islet α cells (glucagon, blue; L and L′) in native pancreatic sections from adult donors with type 2 diabetes (n = 8 total images analyzed).
(M–R″) Immunostaining of SARS-CoV-2 cell entry markers ACE2 (red; antibody AF933; M, O, O′, P, R, and R′) or TMPRSS2 (green; N, O, O″, Q, R, and R″) in islet β cells (insulin, blue; O and O′) and islet α cells (glucagon, blue; R and R′) in native pancreatic sections from adult donors with type 1 diabetes (n = 4 total images analyzed).
Dotted yellow lines denote islets. Islet (i) and exocrine (e) inset areas are marked by yellow boxes in MERGE column with DAPI counterstain. Pancreatic ducts, identified structurally by rosette pattern of DAPI-labeled nuclei (white), are shown within dotted yellow lines in exocrine INSET column. Human pancreatic donor information is available in Table S2 (A–F″, donor N9; G–L″, donor 2H; M–R″, donor 1C). See also Figures S2–S4. Scale bars, 50 μm (A–R) and 25 μm (insets, C′–R″).
TMPRSS2 protein was not detected within islets in ND (Figures 2A–2F″ and S3E–S3H), T2D (Figures 2G–2L″, S3M, and S3N), or T1D donors (Figures 2M–2R″ and S3S–S3V). The TMPRSS2-positive structures in exocrine tissue resembled intercalated and larger ducts as indicated by the typical organization of their nuclei (Figures 2C″, 2F″, 2I″, 2L″, 2O″, and 2R″). We did not detect differences in the signal intensity or spatial distribution of ACE2 or TMPRSS2 between ND and T2D tissue (Figures 2 and S3), but labeling for both proteins appeared to be reduced in T1D pancreatic sections (Figures 2M–2R″ and S3O–S3V). Altogether, these observations indicate that the cell surface proteins required for canonical SARS-CoV-2 host cell entry were not detected on islet endocrine cells from normal or diabetic donors.
Recent in silico analyses by Vankadari and Wilce (2020) and Li et al. (2020b) suggested that human dipeptidyl peptidase 4 (DPP4) may interact with SARS-CoV-2 and facilitate its entry into host cells. Therefore, we examined the distribution of DPP4 in human pancreas and found that it localized to α cells, but not β cells, of ND, T2D, and T1D islets (Figures S4A–S4I). This finding is consistent with our analysis of four scRNA-seq datasets showing DPP4-enriched α cells (Figures 1B and S1A). Given that we did not detect TMPRSS2 within islets, these data suggest that DPP4 is an unlikely mediator of β cell entry by SARS-CoV-2 (Drucker, 2020).
The discrepancy between our findings and those of recent studies identifying ACE2 and TMPRSS2 in human β cells (Fignani et al., 2020; Yang et al., 2010, 2020) is likely explained by important differences in experimental approaches and contexts. Yang et al. (2010) showed ACE2 staining in a single donor islet but did not identify its cellular identity with endocrine markers. In addition, the ACE2 antibody used in their study was not reported, which precluded replication of their finding. Furthermore, Yang et al. (2020) examined ACE2 expression in isolated dispersed human islet cells, whereas our study examined ACE2 in isolated intact islets and in human islets in native pancreas in situ. It is possible that dispersion of pancreatic cells alters the expression of ACE2 in this context (van den Brink et al., 2017). Fignani et al., (2020) reported ACE2 protein in some islet endocrine cells from seven non-diabetic donors; however, ACE2 was primarily seen in subcellular compartments rather than the expected cell surface location. Intriguingly, Fignani et al., (2020) observed differential immunostaining patterns in human pancreas and islets upon testing three ACE2 antibodies that recognize different epitopes of the full-length ACE2 protein, leading them to conclude that a recently discovered short isoform of ACE2 (Blume et al., 2020; Ng et al., 2020; Onabajo et al., 2020) is expressed in human β cells. However, at least two of the four ACE2 antibodies (ab15348 and AF933) we used in our study are predicted to recognize both the short and long isoforms of ACE2, yet we did not detect either form of the ACE2 protein in human islet β cells with either of these reagents. One of these antibodies (ab15348) was also used by Fignani et al., (2020). Furthermore, the short isoform of ACE2 appears to be unstable (Ng et al., 2020) and, importantly, lacks the residues required for SARS-CoV-2 spike glycoprotein binding (Blume et al., 2020; Ng et al., 2020; Onabajo et al., 2020), making it an unlikely receptor for SARS-CoV-2. Sources of variability in immunostaining techniques can stem from differences in tissue fixation procedures, antigen retrieval methods, antibodies used, and their dilutions. Notwithstanding, the remarkable concordance of our findings with those from a recent independent effort in native human pancreas (Kusmartseva et al., 2020) suggests that differences in experimental conditions may explain the discordance with other studies.
ACE2 Is Localized to Islet and Exocrine Tissue Capillaries
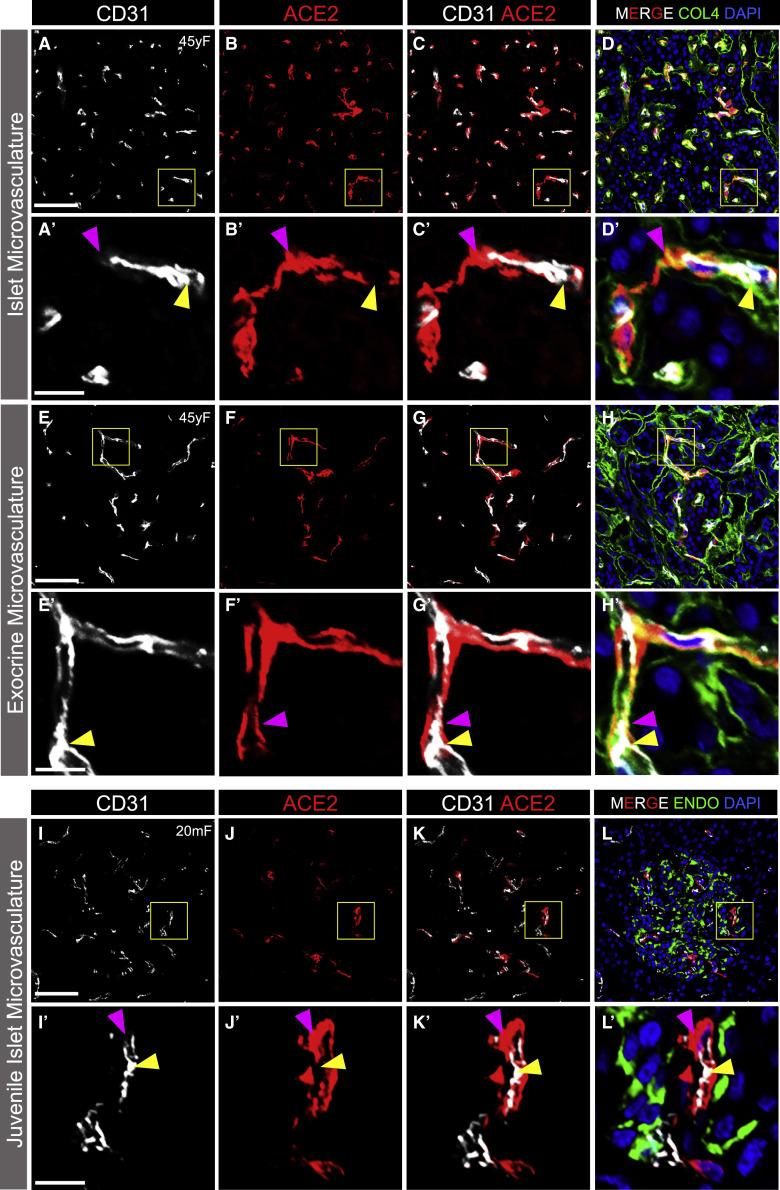
The non-endocrine cell staining pattern of ACE2 in islets prompted us to examine whether ACE2 was expressed in the microvasculature, as described in other organs (Hamming et al., 2004). Indeed, staining of adult and juvenile human pancreatic tissue sections with CD31, an endothelial cell marker, revealed that ACE2 labeling was localized to the perivascular compartment of islet capillaries (Figures 3A–3D′, 3I–3L′, and S5A–S5H′). In addition, exocrine tissue capillaries, similar to those in islets, showed perivascular ACE2 labeling (Figures 3E–3H′ and S5I–S5P′). As ACE2-positive cells were scarce in T1D pancreatic tissues (Figures 2M, 2O′, 2O″, 2P, 2R′, 2R″, and S3O–S3R), the presence of ACE2 labeling in the perivascular compartment was relatively rare in both islet (Figures S5E–S5H′) and exocrine (Figures S5M–S5P′) T1D tissues. The structure and close relationship between ACE2-positive perivascular cells and CD31-positive endothelial cells in islet and exocrine tissue capillaries raised the possibility that ACE2-positive cells were in fact pericytes enveloping capillary endothelial cells of the vascular tube (Almaça et al., 2018). In addition, we observed ACE2-positive cytoplasmic processes extending along CD31-positive endothelial cells colocalized with the extracellular matrix marker collagen-IV within the vascular basement membrane (Figures 3D′, 3H′, S5D′, S5H′, S5L′, and S5P′), a pattern that is consistent with pericyte morphology (Almaça et al., 2018).
Figure 3.
ACE2 Protein Localization with Islet and Exocrine Capillaries in Adult and Juvenile Human Pancreas
(A–C') Representative images of endothelial cells (CD31, white) and ACE2-positive perivascular cells (red; antibody ab15348) in the adult islet microvasculature (n = 14 total images analyzed).
(D and D′) ACE2-positive perivascular cells (red; antibody ab15348) and the extracellular matrix marker collagen-IV (COL4, green) within the vascular basement membrane; DAPI counterstain (blue).
(E–G') Representative images of endothelial cells (CD31, white) and ACE2-positive perivascular cells (red; antibody ab15348) in the exocrine tissue microvasculature (n = 14 total images analyzed).
(H and H′) ACE2-positive perivascular cells (red; antibody ab15348) and the extracellular matrix marker collagen-IV (COL4, green) within the vascular basement membrane; DAPI counterstain (blue).
(I–L′) Representative images of endothelial cells (CD31, white) and ACE2-positive perivascular cells (red; antibody ab15348) in juvenile human islet microvasculature. Islet endocrine cells expressing insulin and glucagon (ENDO, green; L and L′) are shown with DAPI counterstain (blue) (n = 10 total images analyzed).
Yellow arrowheads point to CD31-positive endothelial cells while magenta arrowheads point to perivascular ACE2-positive cells. Inset areas (A′–L′) are marked by yellow boxes in (A)–(L). Human pancreatic donor information is available in Table S2 (A–H′, donor N2; I–L′, donor J4). See also Figure S5. Scale bars, 50 μm (A–L) and 10 μm (insets, A′–L′).
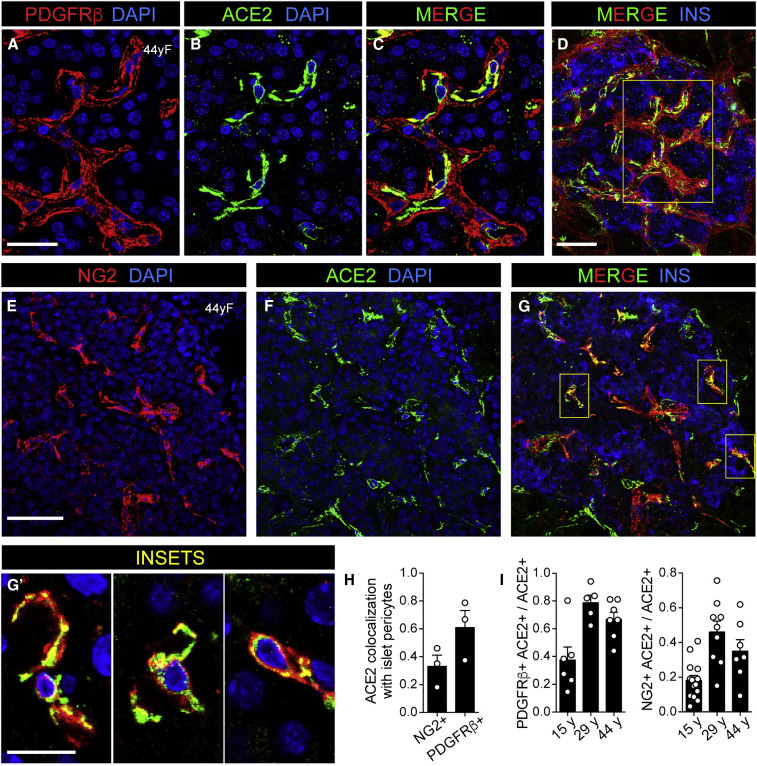
To address this observation more directly, we visualized ACE2 expression in relationship to known pericyte markers and found that ACE2 was enriched in pericyte populations expressing PDGFRβ (Figures 4A–4D). Indeed, colocalization analysis revealed that ∼60% of ACE2-positive cells were PDGFRβ positive (Figures 4H and 4I). Since PDGFRβ labels pericytes and perivascular fibroblasts (Almaça et al., 2020), we also stained tissues with a more specific pericyte marker, NG2, and found that ∼30% of ACE2-positive cells were NG2 positive (Figures 4E–4G′, 4H, and 4I). These data agree with our scRNA-seq analysis of the HPAP dataset, which identified a subset of pericytes, marked by PDGFRB, that expressed moderate levels of ACE2 (Figure 1B). In addition, our findings support recent reports of pericyte-specific vascular expression of ACE2 in brain and heart tissue (Chen et al., 2020; He et al., 2020). Emerging evidence from both pre-clinical models of SARS-CoV-2 infection (Aid et al., 2020) and patients with COVID-19 indicates that cells of the microvasculature (e.g., endothelial cells and pericytes) are requisite contributors to the initiation and propagation of severe disease (Teuwen et al., 2020). Although we found that TMPRSS2 expression was negligible in endothelial and stellate cells (which includes pericytes) of human islets, CTSL, ADAM17, FURIN, and ACE2 were moderately to highly expressed in these populations (Figure 1B), raising the intriguing possibility of direct infection of cells in the islet microvasculature by SARS-CoV-2. Such an occurrence could trigger β cell dysfunction and the metabolic sequelae of COVID-19. Future studies addressing this possibility in human islets and pancreatic tissue are urgently needed.
Figure 4.
ACE2 Colocalization with Islet Pericytes
(A–C) Representative images of an adult human islet labeled for platelet-derived growth factor receptor β-positive (PDGFRβ, red; A and C) and ACE2 (green; antibody ab15348; B and C) in perivascular cells within a normal donor. DAPI (blue) (n = 20 total images analyzed).
(D) Image of the same islet shown in (A)–(C) with insulin labeling (INS, blue). Inset areas in (A)–(C) are marked by the yellow box.
(E–G′) Representative images of an adult human islet labeled for neuron-glial antigen 2 (NG2, red; E and G), ACE2 (green; antibody ab15348; F and G), and insulin (INS, blue; G) in perivascular cells within a normal donor. DAPI (blue; E, F, and G′). Inset areas in (G′) are marked by yellow boxes in (G) and represent a subset of ACE2-expressing islet pericytes (n = 36 total images analyzed).
(H) Colocalization of ACE2 with NG2- or PDGFRβ-positive cells.
(I) Quantification of ACE2-positive cells expressing pericyte markers PDGFRβ and NG2. Shown are average values obtained for 3 confocal planes per islet for a minimum of 5 islets per each donor of a given age.
Human pancreatic donor information is available in Table S2 (donors HP1754, HP2041, and HP2091). See also Figure S5. Scale bars, 20 μm (A–C), 40 μm (D–G), and 10 μm (G′).
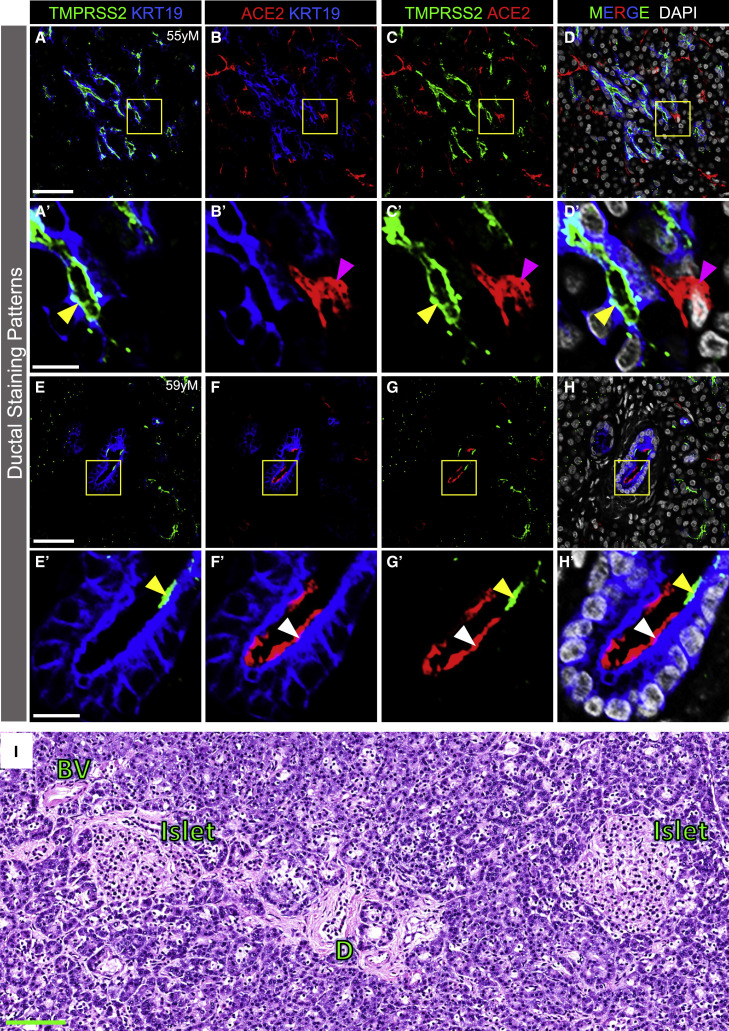
Both ACE2 and TMPRSS2 Proteins Are Present in Pancreatic Ducts but Rarely Are Co-expressed
Our work indicated that TMPRSS2 protein was localized to the apical surface of intercalated and larger ducts in ND (Figures 2B, 2C″, 2E, 2F″, and S3E–S3H), T2D (Figures 2H, 2I″, 2K, 2L″, S3M, and S3N), and T1D pancreatic tissues (Figures 2N, 2O″, 2Q, 2R″, and S3S–S3V). To further define ductal expression of TMPRSS2 and determine whether ACE2 was expressed beyond exocrine tissue capillaries, we visualized expression of these two proteins in relationship to cytokeratin-19 protein (KRT19), a ductal cell marker. TMPRSS2, but not ACE2, was expressed by KRT19-positive cells on the apical surface of intercalated and larger ducts throughout the exocrine compartment (Figures 5A–5D′). Rarely, both ACE2 and TMPRSS2 were found on the apical surface of ductal epithelial cells but were spatially distinct (Figures 5E–5H′). These findings agree with our scRNA-seq analysis in which TMPRSS2 was more highly expressed in ductal cells than ACE2, but cells positive for both ACE2 and TMPRSS2 were rare. In addition, although TMPRSS2 mRNA was detected in acinar cells by scRNA-seq (Figure 1B), TMPRSS2 protein was undetectable by immunofluorescence analysis (Figure 5), indicating that studies of mRNA and protein may be discordant.
Figure 5.
ACE2 and TMPRSS2 Protein in Pancreatic Ducts and Pancreas Histology from an Individual Infected with SARS-CoV-2
(A–D′) TMPRSS2 (green), ACE2 (red; antibody R&D AF933), and KRT19 (blue) immunostaining of the exocrine compartment. DAPI (white). Yellow arrowheads point to TMPRSS2 on the apical surface of ductal cells (KRT19-positive). Magenta arrowheads point to ACE2-positive non-ductal cells. Inset areas in (A′)–(D′) are marked by yellow boxes in (A)–(D) (n = 44 total images analyzed).
(E–H′) TMPRSS2 (green) and ACE2 (red; antibody R&D AF933) immunostaining of ductal epithelial cells (KRT19, blue). Yellow and white arrowheads point to TMPRSS2 and ACE2 labeling, respectively, localized to the apical surface of larger ducts (KRT19-positive cells). Inset areas in (E′)–(H′) are marked by yellow boxes in (E)–(H) (n = 44 total images analyzed).
(I) Representative H&E image of human pancreas upon autopsy after COVID-19 disease. BV, blood vessel; D, duct (n = 7 total images analyzed).
Human pancreatic donor information is available in Table S2 (A–D′, donor N8; E–H′, donor N9; I, COVID-19 donor 1). See also Figure S3. Scale bars, 50 μm (A–H), 10 μm (A′–H′), and 100 μm (I).
Because rare cells in the exocrine compartment co-express ACE2 and TMPRSS2, it is possible that SARS-CoV-2 could infect those cells, induce pancreatitis, and consequently impact islet function. While recent case reports of individuals with COVID-19 have noted mild elevations of amylase and lipase or frank pancreatitis (Karimzadeh et al., 2020; Meireles et al., 2020; Rabice et al., 2020; Wang et al., 2020a), the vast majority of individuals with COVID-19 have neither elevated pancreatic enzymes nor frank pancreatitis (Ashok et al., 2020; Bonney et al., 2020; Bruno et al., 2020). To investigate this possibility, we analyzed the histology of pancreatic tissue obtained at autopsy from seven individuals with COVID-19, three of whom had diabetes. We did not find signs of pancreatitis, interstitial edema, inflammatory infiltrate, hemorrhage, or necrosis (Figure 5I). It is important that exocrine inflammation and ACE2/TMPRSS2 expression be analyzed in a larger number of COVID-19 autopsy samples, particularly in those with new-onset diabetes. Such studies will be challenging since the pancreas rapidly undergoes autolysis upon death.
In summary, recent reviews, commentaries, and clinical guidelines (Apicella et al., 2020; Bornstein et al., 2020; Goldman et al., 2020; Gupta et al., 2020; Heaney et al., 2020) have highlighted an attractive hypothesis of direct cytotoxicity by SARS-CoV-2 to β cells. Here, we addressed the fundamental question of whether canonical SARS-CoV-2 cell entry machinery is present in β cells of human pancreatic tissue. By examining pancreatic ACE2 and TMPRSS2 expression in normal donors and in those with diabetes, we found that (1) β cells do not coexpress ACE2 and TMPRSS2 transcripts by scRNA-seq analysis and neither protein is detectable in α or β cells by immunofluorescence; (2) ACE2 is primarily expressed in the islet and exocrine tissue capillaries, including pericytes, as well as a subset of ductal cells; (3) TMPRSS2 is primarily expressed in ductal cells; (4) ACE2 and TMPRSS2 are infrequently co-expressed in pancreatic ducts; (5) ACE2 and TMPRSS2 protein expression and distribution appear similar in ND and T2D pancreata, but studies of additional pancreata are needed to definitively answer this question; and (6) alternative pathways facilitating SARS-CoV-2 viral infection in β cells cannot be excluded. These findings align considerably with independent observations from Kusmartseva et al. (2020) and do not support the hypothesis that SARS-CoV-2 binds and infects islet β cells via a canonical pathway mediated by ACE2 and TMPRSS2, eliciting a diabetogenic effect. As outlined below in the Limitations of Study section, additional studies are needed to investigate whether direct SARS-CoV-2 infection of β cells occurs or is detrimental to β cell health or function by other mechanisms. However, based on current data, it appears that the interaction of diabetes and SARS-CoV-2 is mediated by systemic inflammation and/or metabolic changes in other organs such as liver, muscle, or adipose tissue.
Limitations of Study
We did not directly measure SARS-CoV-2 binding or entry into human β cells but instead assessed expression of canonical co-receptors, ACE2 and TMPRSS2, in the human pancreas. While studies culturing SARS-CoV-2 and human islets in vitro will be of considerable interest, such experiments may be challenging to interpret as the islet isolation process itself may impact ACE2 or TMPRSS2 expression or susceptibility to viral infection. In addition, infection of islet cells within an intact, isolated islet with a variety of viruses has proven difficult, as is selection of an appropriate ratio of viral particles and islet cells reflective of the in vivo environment. Furthermore, it is not clear if infection of an isolated islet accurately models the physiology of a vascularized, innervated islet within the context of the pancreas.
While the preponderance of research suggests that ACE2 and TMPRSS2 are the primary means for SARS-CoV-2 entry into host cells, knowledge about additional pathways and/or mechanisms of SARS-CoV-2 cell entry and infection is rapidly evolving. We sought, but did not find, expression evidence, suggesting other proposed pathways (namely DPP4, CTSL, ADAM17, FURIN, or TMPRSS4) for β cell entry.
We did not assess the presence of SARS-CoV-2, its proteins, or RNA in the pancreas from individuals with COVID-19. It is possible that SARS-CoV-2 could infect pancreatic exocrine cells, ductal cells, endothelial cells, or microvasculature, leading to pancreatic inflammation that negatively impacts β cell health or function. We did not find evidence of inflammation in the pancreas of seven individuals with COVID-19 (three of whom had diabetes). It is also possible that COVID-19 could induce ectopic expression of SARS-CoV-2 cell entry factors in the endocrine pancreas and render islet cells susceptible to direct infection. Future efforts are needed to collect a large number of pancreata from individuals with COVID-19 with and without diabetes (long-standing and recent onset) to search for presence of SARS-CoV-2 or inflammation (islet or exocrine). Since the pancreas undergoes rapid autolysis after death, care must be taken to collect the pancreas very soon after death for this analysis.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Cytokeratin 19 (1:1000) | Abcam | Cat# ab194399 |
| Rabbit anti-TMPRSS2 (1:400) | Sigma | Cat# HPA035787; RRID: AB_2674782 |
| Mouse anti-Glucagon (1:500-1:1000) | Abcam | Cat# ab10988; RRID: AB_297642 |
| Guinea pig anti-Glucagon (1:500) | LSBio | Cat# LS-C202759-50 |
| Rat anti-C-peptide (1:1000) | DSHB | Cat# GN-ID4; RRID: AB_2255626 |
| Guinea pig anti-Insulin (1:500-1:1000) | Dako | Cat# A0564; RRID: AB_2617169 |
| Rabbit anti-ACE2 (1:500, Figure S2) | Atlas Antibodies | Cat# HPA000288; RRID: AB_1078160 |
| Rabbit anti-ACE2 (1:1000, Figures 1C, 3, 4, S2, S3, and S5) | Abcam | Cat# ab15348; RRID: AB_301861 |
| Goat anti-ACE2 Figures S1) (1:200, Figure S2; 1:400 in Figures 2 and 5) | R&D | Cat# AF933; RRID: AB_355722 |
| Mouse anti-ACE2 (1:33, Figure S2) | R&D | Cat# MAB933; RRID: AB_2223153 |
| Mouse anti-CD31 (1:50) | BD Biosciences | Cat# 550389; RRID: AB_2252087 |
| Goat anti-collagen IV alpha 1 (1:500) | Novus | Cat# NBP1-26549; RRID: AB_1853202 |
| Goat anti-DPP4 (1:200) | R&D | Cat# AF1180; RRID: AB_354651 |
| Mouse anti-NG2 [647] (1:50) | R&D | Cat# FAB2585R |
| Goat anti-PDGFRβ (1:100) | R&D | Cat# AF385; RRID: AB_355339 |
| Donkey anti-mouse-Cy2 (1:500) | Jackson ImmunoResearch | Cat# 715-225-150; RRID: AB_2340826 |
| Donkey anti-mouse-Cy3 (1:500) | Jackson ImmunoResearch | Cat# 715-165-150; RRID: AB_2340813 |
| Donkey anti-mouse-Cy5 (1:250 – 1:500) | Jackson ImmunoResearch | Cat# 715-175-151; RRID: AB_2340820 |
| Donkey anti-rabbit-Cy3 (1:500) | Jackson ImmunoResearch | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey anti-rat-Cy2 (1:500) | Jackson ImmunoResearch | Cat# 712-225-153; RRID: AB_2340674 |
| Donkey anti-guinea pig-Cy2 (1:500) | Jackson ImmunoResearch | Cat# 706-225-148; RRID: AB_2340467 |
| Donkey anti-goat-Cy2 (1:500) | Jackson ImmunoResearch | Cat# 705-225-147; RRID: AB_2307341 |
| Donkey anti-goat-Cy3 (1:500) | Jackson ImmunoResearch | Cat# 705-165-147; RRID: AB_2307351 |
| Donkey anti-guinea pig-Cy5 (1:250-1:500) | Jackson ImmunoResearch | Cat# 706-175-148; RRID: AB_2340462 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat# 017-000-121; RRID: AB_2337258 |
| Biological Samples | ||
| Human pancreatic islets | Integrated Islet Distribution Program (IIDP) | http://iidp.coh.org; RRID: SCR_014387 |
| Human pancreatic tissue | International Institute for the Advancement of Medicine (IIAM) | http://www.iiam.org; RRID: SCR_016172 |
| Human pancreatic tissue | National Disease Research Interchange (NDRI) | http://ndriresource.org; RRID: SCR_000550 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propidium Iodide | BD Biosciences | Cat# 556463 |
| Antigen Unmasking Solution, Citrate-Based | Vector Laboratories | Cat# H-3300-250 |
| 1X PBS | Corning | Cat# 46-013-CM |
| Triton X-100 | USB | Cat# 22686 |
| BSA | Sigma | Cat# A6003-25G |
| Human ACE2 Peptide | Abcam | Cat# ab15352 |
| Aqua-Poly/Mount | Polysciences | Cat# 18606-5 |
| 10% Neutral Buffered Formalin | Electron Microscopy Sciences | Cat# 15740-01 |
| 16% paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| O.C.T. compound | Fisher Scientific | Cat# 4585 |
| Critical Commercial Assays | ||
| RNAqueous-Micro Kit | Fisher Scientific (Invitrogen) | Cat# AM1931 |
| High Capacity cDNA Reverse Transcription Kit | Fisher Scientific (Invitrogen) | Cat# 4368814 |
| Deposited Data | ||
| RNA-seq data for FACS purified human alpha and beta cells (Blodgett et al., 2015) | NCBI Gene Expression Omnibus | GEO: GSE67543 |
| RNA-seq data for FACS purified human alpha and beta cells (Arda et al., 2016) | NCBI Gene Expression Omnibus | GEO: GSE57973 |
| Single-cell RNA-seq data for human islets (Baron et al., 2016) | NCBI Gene Expression Omnibus | GEO: GSE84133 |
| Single-cell RNA-seq data for human islets (Camunas-Soler et al., 2020) | NCBI Gene Expression Omnibus | GEO: GSE124742 |
| Single-cell RNA-seq data for human islets (Segerstolpe et al., 2016) | ArrayExpress | ArrayExpress: E-MTAB-5061 |
| Single-cell RNA-seq data for human islets (Human Pancreas Analysis Program [HPAP]) | HPAP Database | https://hpap.pmacs.upenn.edu; RRID: SCR_014393 |
| Software and Algorithms | ||
| FV31S-SW Viewer 2.4.1.198 | Olympus | https://www.olympus-lifescience.com/en/support/downloads/ |
| Seurat 3.1 R package | Butler et al., 2018; Stuart et al., 2019 | N/A |
| Prism for macOS v.8.4.3 | Graphpad Software | http://www.graphpad.com; RRID: SCR_002798 |
| JACoP: Just Another Co-localization Plugin | ImageJ | https://imagej.nih.gov/ij/plugins/track/jacop2.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alvin C. Powers (al.powers@vumc.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All data generated or analyzed during this study are included in this published article and in the data repositories listed in the Key Resources Table. Original/source data for Figure 1A (bulk RNA-seq of FACS sorted human islet α and β cells) is available under NCBI GEO accession numbers GSE67543 (Blodgett et al., 2015) and GSE57973 (Arda et al., 2016). Original/source data for Figures 1B and S1A (single cell RNA-seq of human islets) is available under NCBI GEO accession number GSE84133 (Baron et al., 2016), GSE124742 (Camunas-Soler et al., 2020), and ArrayExpress (EBI) accession number E-MTAB-5061 (Segerstolpe et al., 2016). This manuscript used data acquired from the Human Pancreas Analysis Program (HPAP) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network consortium.
Experimental Model and Subject Details
Human Subjects
Pancreata and islets from juvenile and adult human donors including individuals with T1D or T2D were obtained through partnerships with the International Institute for Advancement of Medicine (IIAM), National Disease Research Interchange (NDRI), Integrated Islet Distribution Program (IIDP), and local organ procurement organizations. Pancreata from normal donors were processed for islet isolation (Balamurugan et al., 2003) and/or histological analysis as described previously (described below and in Table S2) (Brissova et al., 2018; Dai et al., 2017). Pancreata from COVID-19 decedents after autopsy were obtained from the Translational Pathology Shared Resource (TPSR) at Vanderbilt University Medical Center (Nashville, TN) and processed for histological analysis as according to standard procedures for clinical diagnostics (VUMC Histology Peloris Processing Protocol). Human kidney samples were provided by Dr. Agnes B. Fogo, Vanderbilt University Medical Center. Subject samples were allocated by disease status. Samples from donors of both sexes were used in our analyses. Donor demographic information is summarized in Table S2. The Vanderbilt University Institutional Review Board has declared that studies on de-identified human pancreatic specimens do not qualify as human subjects research.
Mice
Adult NOD.Cg-Prkdc scid Il2rg tm1Wjl/ SzJ (NSG) male and female healthy mice ages 12 to 18 weeks (purchased as needed from Jackson Laboratory) were used in this study. Mice were maintained by Vanderbilt Division of Animal Care in group housing in sterile containers at ambient temperature within a pathogen-free barrier facility housed with a 12 h light/12 h dark cycle and access to free water and standard rodent chow. Mice were used directly after importation from Jackson Laboratory following acclimatization. All animal procedures were approved by the Vanderbilt Institutional Animal Care and Use Committee.
Method Details
Bulk RNA-seq Data Acquisition
Transcripts Per Million (TPM) and Reads Per Kilobase Per Million (RPKM) normalized counts were extracted from publicly available RNA-seq datasets (Arda et al., 2016; Blodgett et al., 2015). The sources of the datasets are summarized in the Key Resources Table. GraphPad Prism v8 was used to generate plots in Figure 1A.
Single Cell RNA-seq Data Acquisition
Raw gene count matrices were extracted from existing single cell RNA-seq datasets (Baron et al., 2016; Camunas-Soler et al., 2020; Segerstolpe et al., 2016) and from the Human Pancreas Analysis Program (HPAP) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network consortium. SMART-seq technology (Picelli et al., 2014) was used to generate datasets by Camunas-Soler et al. (2020) and Segerstolpe et al. (2016), while droplet-based scRNA-seq by InDrop (Klein et al., 2015) or 10x genomics was utilized for databases by Baron et al. (2016) and the HPAP (Kaestner et al., 2019), respectively. The sources of the datasets are summarized in the Key Resources Table. Gene count matrices were further analyzed using the R package Seurat version 3.1 (Butler et al., 2018; Stuart et al., 2019). Gene count measurements were normalized for each cell by library size and log-transformed using a size factor of 10,000 molecules per cell. The data was further scaled to unit variance and zero mean implemented in the “ScaleData” function. Cell types already annotated by the authors in the original study were used and thus, no clustering was performed. Seurat’s “DotPlot” function was used to generate plots shown in Figures 1B and S1A to visualize scaled expression.
Human Pancreas Preparation
Pancreata from juvenile, adult, T1D and T2D donors were obtained within 18 h from cross-clamp and maintained in cold preservation solution on ice until processing, as described previously (Balamurugan et al., 2003; Brissova et al., 2018). Briefly, connective tissue and fat were cleaned from the pancreas, 2- to 3-mm thick cross-sectional slices were collected from the head, body and distal tail, and each slice was cut into four quadrants. For cryosections, slices were fixed in 0.1 M PBS containing 4% paraformaldehyde for 3 h on ice with gentle agitation, washed four times over 2 h with 0.1 M PBS, equilibrated in 30% sucrose (prepared in 0.01 M PBS) overnight, and embedded in Tissue-Plus O.C.T. compound. For FFPE sections, slices were fixed in 10% neutral buffered formalin for 24-48 h at room temperature, washed four times in 0.01 M PBS at 4°C for 2-3 h with gentle agitation, transferred to 70% ethanol and kept at 4°C until paraffin embedding. Pancreata recovered from seven COVID-19 decedants were obtained from Vanderbilt University Medical Center (Nashville, TN) within 8-27 h of death. COVID-19 autopsy samples were fixed in 10% formalin for 5 days at room temperature prior to processing in the clinical pathology laboratory.
Mouse Kidney Preparation
Kidneys from adult NOD.Cg-Prkdc scid Il2rg tm1Wjl/ Sz (NSG) mice ages 12 to 18 weeks (Jackson Laboratory) were isolated. Tissue specimens were processed for OCT-embedded, 5-μm cryosections as described previously (Brissova et al., 2018).
Immunohistochemical Analysis
Immunohistochemical analysis was performed on 5-μm cryosections (Figures 1C, 3, 4, S2A–S2I, S3A–S3D, S3I–S3L, S3O–S3R, S4, and S5) or formalin-fixed paraffin embedded (FFPE) pancreatic sections (Figures 2, 5, S2J–S2W, S3E–S3H, S3M, S3N, and S3S–S3V) as indicated and described previously (Brissova et al., 2014, 2018; Wright et al., 2020). Briefly, cryosections were thawed at room temperature for 10-15 min, washed 3x for 5 min each in 1X PBS, permeabilized with 0.2% Triton X-100 in 1X PBS for 15 min at room temperature, and washed 3x for 5 min each in 1X PBS. FFPE sections were first deparaffinized and rehydrated in xylene and ethanol (3x for 3 min each, xylene; 2x for 2 min each, 100% ethanol; 1x for 2 min, 95% ethanol; 1x for 2 min, 90% ethanol; 1x for 2 min, 70% ethanol; 1x for 1 min, 50% ethanol; 1x for 5 min, distilled water) followed by heat-induced epitope retrieval in a citrate-based antigen unmasking solution (Key Resources Table), pH 6.0 (microwaved on high [1100W] for 14 min). Sections were cooled at room temperature for 20 min and then rinsed in distilled water for 10 min. Both cryosections and FFPE sections were blocked with 5% normal donkey serum at room temperature for 60-90 min in a humidified chamber. All primary and secondary antibodies were diluted in filter-sterilized antibody buffer (0.1% Triton X-100, 1% BSA, 1X PBS). Tissue sections in primary antibody were incubated in a humidified chamber overnight at 4°C, washed 3x for 10 min each in 1X PBS, incubated in secondary antibody in a humidified chamber for 60-90 min at room temperature (protected from light), counterstained with DAPI for 10 min at room temperature (protected from light), washed 3x for 15 min each in 1X PBS, and mounted with Aqua-Poly/Mount. Immunofluorescence analysis of isolated islets was performed on 8-μm cryosections of islets embedded in collagen gels as described previously and above (Brissova et al., 2005, 2018). Primary antibodies to all antigens and their working dilutions are listed in the Key Resources Table. The antigens were visualized using appropriate secondary antibodies listed in the Key Resources Table. Digital images were acquired with an Olympus FLUOVIEW FV3000 laser scanning confocal microscope (Olympus Corporation).
H&E Staining
FFPE pancreas tissue sections (5-μm sections, n = 7 donors) were processed according to standard procedures for clinical diagnostics (Vanderbilt University Medical Center Histology Peloris Processing Protocols). Slides were stained with hematoxylin & eosin and reviewed by a clinical pathologist (M.E.K.) at Vanderbilt University Medical Center.
Peptide Competition
We combined 1 μg of anti-ACE2 antibody (ab15348) with or without 10 μg of the immunizing human ACE2 peptide (ab15352) in antibody buffer solution (0.1% Triton X-100/1% BSA/1X PBS) and incubated overnight at 4°C with gentle agitation. Immunofluorescence staining of 5-μm serial cryosections from the same donor was performed as described above. One section was incubated with the neutralized antibody buffer while the other section was incubated with the non-neutralized antibody buffer. ACE2 staining patterns were visualized via confocal microscopy as described above.
Quantification and Statistical Analysis
Data Representation and Statistical Analysis
Details of statistical analyses, when applicable, can be found in the figure legends. Bulk RNA-seq (Figure 1A) and ACE2 colocalization (Figures 4H and 4I) data are expressed as mean ± standard error of mean. Sample size (n) is provided as the number of independent donor samples. Mander’s coefficients were determined to quantify colocalization between pericyte markers (NG2 and PDGFRβ) and ACE2 in confocal images of islets using the ImageJ plugin “Just Another Co-localization Plugin” (https://imagej.nih.gov/ij/plugins/track/jacop2.html). These data were all continuous and showed a normal distribution (Shapiro-Wilk normality test; p > 0.05). The remaining immunofluorescence data was not analyzed quantitatively. Blinding, power calculations and exclusion criteria were not utilized in this study. Sample sizes were determined based on tissue availability and disease status. There were no exclusion criteria in this study; donors were placed into categories based on disease status.
Acknowledgments
We are especially thankful to organ donors and their families. This research was supported by funding provided by the National Institute of Diabetes and Digestive and Kidney Diseases; the Human Islet Research Network (HIRN, RRID: SCR_014393, https://hirnetwork.org; DK112232, DK123716, U01DK123594, DK104211, DK108120); DK106755, DK117147, DK117147-01A1S1, DK111757, DK112217, and DK20593; and the Functional Genomics Core of DK19525. This manuscript used data acquired from and available at the Human Pancreas Analysis Program (HPAP, RRID: SCR_016202) Database (https://hpap.pmacs.upenn.edu), a HIRN consortium. This work was also supported by grants from the Doris Duke Charitable Foundation (DDCF 4043516256), Human Islet Research Network New Investigator Pilot Award (UC4 DK104162), JDRF, the Leona M. and Harry B. Helmsley Charitable Trust, and the Department of Veterans Affairs (BX000666). Human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program at the City of Hope (NIH grant # 2UC4 DK098085; RRID: SCR_014387; https://iidp.coh.org). Human kidney sections were provided by Dr. Agnes B. Fogo at Vanderbilt University Medical Center. We thank the Vanderbilt University Medical Center Translational Pathology Shared Resource (supported by NCI/NIH Cancer Center Support Grant 2P30 CA068485-14 and the Vanderbilt Mouse Metabolic Phenotyping Center 5U24DK059637-13). We also thank Amber M. Bradley for technical assistance. The graphical abstract was created with https://biorender.com/.
Author Contributions
K.C.C., J.C., M.B., and A.C.P. designed the experiments. K.C.C., J.C., M.B., and A.C.P. wrote the manuscript. K.C.C., J.C., S.S., W.W., L.M.G., J.A., M.E.K., M.F., A.M., C.D., D.C.S., R.B., R.J., R.S., K.H.K., G.V., M.B., and A.C.P. performed experiments or analyzed the data. All authors reviewed and edited the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: November 13, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2020.11.006.
Supplemental Information
References
- Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S. Vascular disease and thrombosis in SARS-CoV-2-infected Rhesus macaques. Cell. 2020 doi: 10.1016/j.cell.2020.10.005. Published online October 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaça J., Weitz J., Rodriguez-Diaz R., Pereira E., Caicedo A. The pericyte of the pancreatic islet regulates capillary diameter and local blood flow. Cell Metab. 2018;27:630–644.e4. doi: 10.1016/j.cmet.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaça J., Caicedo A., Landsman L. Beta cell dysfunction in diabetes: the islet microenvironment as an unusual suspect. Diabetologia. 2020;63:2076–2085. doi: 10.1007/s00125-020-05186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella M., Campopiano M.C., Mantuano M., Mazoni L., Coppelli A., Del Prato S. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. Lancet Diabetes Endocrinol. 2020;8:782–792. doi: 10.1016/S2213-8587(20)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda H.E., Li L., Tsai J., Torre E.A., Rosli Y., Peiris H., Spitale R.C., Dai C., Gu X., Qu K. Age-dependent pancreatic gene regulation reveals mechanisms governing human β cell function. Cell Metab. 2016;23:909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A., Faghih M., Singh V.K. Mild pancreatic enzyme elevations in COVID-19 pneumonia: synonymous with injury or noise? Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.086. Published online June 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan A.N., Gu Y., Miyamoto M., Wang W., Inoue K., Tabata Y. Isolation, culture, and functional characteristics of diabetic islets [corrected] Pancreas. 2003;26:102–103. doi: 10.1097/00006676-200301000-00021. [DOI] [PubMed] [Google Scholar]
- Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron E., Bakhai C., Kar P., Weaver A., Bradley D., Ismail H., Knighton P., Holman N., Khunti K., Sattar N. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: a whole-population study. Lancet Diabetes Endocrinol. 2020;8:813–822. doi: 10.1016/S2213-8587(20)30272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett D.M., Nowosielska A., Afik S., Pechhold S., Cura A.J., Kennedy N.J., Kim S., Kucukural A., Davis R.J., Kent S.C. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume C., Jackson C.L., Spalluto C., Legebeke J., Nazlamova L., Conforti F., Perotin-Collard J.-M., Frank M., Crispin M., Coles J. A novel isoform of ACE2 is expressed in human nasal and bronchial respiratory epithelia and is upregulated in response to RNA respiratory virus infection. bioRxiv. 2020 doi: 10.1101/2020.07.31.230870. [DOI] [PubMed] [Google Scholar]
- Bonney G.K., Gao Y., Chew C.A., Windsor J.A. SARS-COV-2 associated acute pancreatitis: cause, consequence or epiphenomenon? Pancreatology. 2020;20:1017–1018. doi: 10.1016/j.pan.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein S.R., Rubino F., Khunti K., Mingrone G., Hopkins D., Birkenfeld A.L., Boehm B., Amiel S., Holt R.I., Skyler J.S. Practical recommendations for the management of diabetes in patients with COVID-19. Lancet Diabetes Endocrinol. 2020;8:546–550. doi: 10.1016/S2213-8587(20)30152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach J.D., Dube P., Ghosh S., Abdullah B.N., Modyanov N.N., Malhotra D., Dworkin L.D., Haller S.T., Kennedy D.J. Impact of comorbidities on SARS-CoV-2 viral entry-related genes. J. Pers. Med. 2020;10:E146. doi: 10.3390/jpm10040146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M., Powers A.C. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005;53:1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- Brissova M., Aamodt K., Brahmachary P., Prasad N., Hong J.Y., Dai C., Mellati M., Shostak A., Poffenberger G., Aramandla R. Islet microenvironment, modulated by vascular endothelial growth factor-A signaling, promotes β cell regeneration. Cell Metab. 2014;19:498–511. doi: 10.1016/j.cmet.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M., Haliyur R., Saunders D., Shrestha S., Dai C., Blodgett D.M., Bottino R., Campbell-Thompson M., Aramandla R., Poffenberger G. α cell function and gene expression are compromised in type 1 diabetes. Cell Rep. 2018;22:2667–2676. doi: 10.1016/j.celrep.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno G., Fabrizio C., Santoro C.R., Buccoliero G.B. Pancreatic injury in the course of coronavirus disease 2019: a not-so-rare occurrence. J. Med. Virol. 2020 doi: 10.1002/jmv.26134. Published online June 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camunas-Soler J., Dai X.Q., Hang Y., Bautista A., Lyon J., Suzuki K., Kim S.K., Quake S.R., MacDonald P.E. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab. 2020;31:1017–1031.e4. doi: 10.1016/j.cmet.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou B., Hadjadj S., Wargny M., Pichelin M., Al-Salameh A., Allix I., Amadou C., Arnault G., Baudoux F., Bauduceau B., CORONADO investigators Phenotypic characteristics and prognosis of inpatients with COVID-19 and diabetes: the CORONADO study. Diabetologia. 2020;63:1500–1515. doi: 10.1007/s00125-020-05180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee Y.J., Ng S.J.H., Yeoh E. Diabetic ketoacidosis precipitated by Covid-19 in a patient with newly diagnosed diabetes mellitus. Diabetes Res. Clin. Pract. 2020;164:108166. doi: 10.1016/j.diabres.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.09.033. Published online September 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Hang Y., Shostak A., Poffenberger G., Hart N., Prasad N., Phillips N., Levy S.E., Greiner D.L., Shultz L.D. Age-dependent human β cell proliferation induced by glucagon-like peptide 1 and calcineurin signaling. J. Clin. Invest. 2017;127:3835–3844. doi: 10.1172/JCI91761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker D.J. Coronavirus infections and type 2 diabetes-shared pathways with therapeutic implications. Endocr. Rev. 2020;41:bnaa011. doi: 10.1210/endrev/bnaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fignani D., Licata G., Brusco N., Nigi L., Grieco G., Marselli L., Overbergh L., Gysemans C., Colli M., Marchetti P. SARS-CoV-2 receptor angiotensin I-converting enzyme type 2 (ACE2) is expressed in human pancreatic β-cells and in the human pancreas microvasculature. Front. Endocrinol. 2020 doi: 10.3389/fendo.2020.596898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi C.M., von Herrath M.G. Viral trigger for type 1 diabetes: pros and cons. Diabetes. 2008;57:2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N., Fink D., Cai J., Lee Y.N., Davies Z. High prevalence of COVID-19-associated diabetic ketoacidosis in UK secondary care. Diabetes Res. Clin. Pract. 2020;166:108291. doi: 10.1016/j.diabres.2020.108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Mae M.A., Sun Y., Muhl L., Nahar K., Vázquez Liébanas E., Fagerlund M.J., Oldner A., Liu J., Genove G. Pericyte-specific vascular expression of SARS-CoV-2 receptor ACE2 – implications for microvascular inflammation and hypercoagulopathy in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.05.11.088500. [DOI] [Google Scholar]
- Heaney A.I., Griffin G.D., Simon E.L. Newly diagnosed diabetes and diabetic ketoacidosis precipitated by COVID-19 infection. Am. J. Emerg. Med. 2020 doi: 10.1016/j.ajem.2020.05.114. Published online June 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16:e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein T., Schulte D.M., Schulz J., Glück A., Ziegler A.G., Bonifacio E., Wendorff M., Franke A., Schreiber S., Bornstein S.R., Laudes M. Autoantibody-negative insulin-dependent diabetes mellitus after SARS-CoV-2 infection: a case report. Nat Metab. 2020;2:1021–1024. doi: 10.1038/s42255-020-00281-8. [DOI] [PubMed] [Google Scholar]
- Holman N., Knighton P., Kar P., O’Keefe J., Curley M., Weaver A., Barron E., Bakhai C., Khunti K., Wareham N.J. Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: a population-based cohort study. Lancet Diabetes Endocrinol. 2020;8:823–833. doi: 10.1016/S2213-8587(20)30271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K.H., Powers A.C., Naji A., Atkinson M.A., HPAP Consortium NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: the Human Pancreas Analysis Program (HPAP) Diabetes. 2019;68:1394–1402. doi: 10.2337/db19-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimzadeh S., Manzuri A., Ebrahimi M., Huy N.T. COVID-19 presenting as acute pancreatitis: lessons from a patient in Iran. Pancreatology. 2020;20:1024–1025. doi: 10.1016/j.pan.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.Y., Ha E., Moon J.S., Lee Y.H., Choi E.Y. Acute hyperglycemic crises with coronavirus disease-19: case reports. Diabetes Metab. J. 2020;44:349–353. doi: 10.4093/dmj.2020.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M., Holmes D., Bennet N., The Lancet Diabetes Endocrinology COVID-19 and diabetes: a co-conspiracy? Lancet Diabetes Endocrinol. 2020;8:801. [Google Scholar]
- Kusmartseva I., Wu W., Syed F., Van Der Heide V., Jorgensen M., Joseph P., Tang X., Candelario-Jalil E., Yang C., Nick H. Expression of SARS-CoV-2 entry factors in the pancreas of normal organ donors and individuals with COVID-19. Cell Metab. 2020 doi: 10.1016/j.cmet.2020.11.005. Published online November 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee I.T., Nakayama T., Wu C.T., Goltsev Y., Jiang S., Gall P.A., Liao C.K., Shih L.C., Schurch C.M., McIlwain D.R. Robust ACE2 protein expression localizes to the motile cilia of the respiratory tract epithelia and is not increased by ACE inhibitors or angiotensin receptor blockers. medRxiv. 2020 doi: 10.1101/2020.05.08.20092866. [DOI] [Google Scholar]
- Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes. Metab. 2020 doi: 10.1111/dom.14057. Published online April 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Z., Yang L., Lian X., Xie Y., Li S., Xin S., Cao P., Lu J. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience. 2020;23:101160. doi: 10.1016/j.isci.2020.101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles P.A., Bessa F., Gaspar P., Parreira I., Silva V.D., Mota C., Alvoeiro L. Acalculous acute pancreatitis in a COVID-19 patient. Eur. J. Case Rep. Intern. Med. 2020;7:001710. doi: 10.12890/2020_001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G.G., Liu J.S., Russ H.A., Tran S., Saxton M.S., Chen R., Juang C., Li M.L., Nguyen V.Q., Giacometti S. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat. Cell Biol. 2019;21:263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Attig J., Bolland W., Young G.R., Major J., Wrobel A.G., Gamblin S., Wack A., Kassiotis G. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat. Genet. 2020 doi: 10.1038/s41588-020-00732-8. Published online October 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020 doi: 10.1038/s41588-020-00731-9. Published online October 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Rabice S.R., Altshuler P.C., Bovet C., Sullivan C., Gagnon A.J. COVID-19 infection presenting as pancreatitis in a pregnant woman: a case report. Case Rep. Womens Health. 2020;27:e00228. doi: 10.1016/j.crwh.2020.e00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique S., Ahmed F.W. A case of combined diabetic ketoacidosis and hyperosmolar hyperglycemic state in a patient with COVID-19. Cureus. 2020;12:e8965. doi: 10.7759/cureus.8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle M.C., Buse J.B., Franks P.W., Knowler W.C., Ratner R.E., Selvin E., Wexler D.J., Kahn S.E. COVID-19 in people with diabetes: urgently needed lessons from early reports. Diabetes Care. 2020;43:1378–1381. doi: 10.2337/dci20-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber B., Patel A., Verma A. Shedding light on COVID-19: ADAM17 the missing link? Am. J. Ther. 2020 doi: 10.1097/MJT.0000000000001226. Published online August 3, 2020. [DOI] [PubMed] [Google Scholar]
- Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.M., Andréasson A.C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedpour S., Khodaei B., Loghman A.H., Seyedpour N., Kisomi M.F., Balibegloo M., Nezamabadi S.S., Gholami B., Saghazadeh A., Rezaei N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: A systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2020 doi: 10.1002/jcp.30032. Published online September 9, 2020. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera J., El-Huneidi W., Hamad M., Mohammed A.K., Elaraby E., Hachim M.Y. Expression profile of SARS-CoV-2 host receptors in human pancreatic islets revealed upregulation of ACE2 in diabetic donors. Biology (Basel) 2020;9:215. doi: 10.3390/biology9080215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuwen L.A., Geldhof V., Pasut A., Carmeliet P. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 2020;20:389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth R., Wallace S., Oliver N.S., Yeung S., Kshirsagar A., Naidu H., Kwong R.M.W., Kumar P., Logan K.M. New-onset type 1 diabetes in children during COVID-19: multicenter regional findings in the U.K. Diabetes Care. 2020;43:e170–e171. doi: 10.2337/dc20-1551. [DOI] [PubMed] [Google Scholar]
- van den Brink S.C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C.S., Robin C., van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang H., Fan J., Zhang Y., Wang H., Zhao Q. Pancreatic injury patterns in patients with coronavirus disease 19 pneumonia. Gastroenterology. 2020;159:367–370. doi: 10.1053/j.gastro.2020.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ma P., Zhang S., Song S., Wang Z., Ma Y., Xu J., Wu F., Duan L., Yin Z. Fasting blood glucose at admission is an independent predictor for 28-day mortality in patients with COVID-19 without previous diagnosis of diabetes: a multi-centre retrospective study. Diabetologia. 2020;63:2102–2111. doi: 10.1007/s00125-020-05209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wright J.J., Saunders D.C., Dai C., Poffenberger G., Cairns B., Serreze D.V., Harlan D.M., Bottino R., Brissova M., Powers A.C. Decreased pancreatic acinar cell number in type 1 diabetes. Diabetologia. 2020;63:1418–1423. doi: 10.1007/s00125-020-05155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.K., Lin S.S., Ji X.J., Guo L.M. Binding of SARS coronavirus to its receptor damages islets and causes acute diabetes. Acta Diabetol. 2010;47:193–199. doi: 10.1007/s00592-009-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and in the data repositories listed in the Key Resources Table. Original/source data for Figure 1A (bulk RNA-seq of FACS sorted human islet α and β cells) is available under NCBI GEO accession numbers GSE67543 (Blodgett et al., 2015) and GSE57973 (Arda et al., 2016). Original/source data for Figures 1B and S1A (single cell RNA-seq of human islets) is available under NCBI GEO accession number GSE84133 (Baron et al., 2016), GSE124742 (Camunas-Soler et al., 2020), and ArrayExpress (EBI) accession number E-MTAB-5061 (Segerstolpe et al., 2016). This manuscript used data acquired from the Human Pancreas Analysis Program (HPAP) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network consortium.