Abstract
The novel coronavirus disease 2019 (COVID-19) pandemic has imposed significant public health problems for the human populations worldwide after the 1918 influenza A virus (IVA) (H1N1) pandemic. Although numerous efforts have been made to unravel the mechanisms underlying the coronavirus, a notable gap remains in our perception of the COVID-19 pathogenesis. The innate and adaptive immune systems have a pivotal role in the fate of viral infections, such as COVID-19 pandemic. MicroRNAs (miRNAs) are known as short noncoding RNA molecules and appear as indispensable governors of almost any cellular means. Several lines of evidence demonstrate that miRNAs participate in essential mechanisms of cell biology, regulation of the immune system, and the onset and progression of numerous types of disorders. The immune responses to viral respiratory infections (VRIs), including influenza virus (IV), respiratory syncytial virus (RSV), and rhinovirus (RV), are correlated with the ectopic expression of miRNAs. Alterations of the miRNA expression in epithelial cells may contribute to the pathogenesis of chronic and acute airway infections. Hence, analyzing the role of these types of nucleotides in antiviral immune responses and the characterization of miRNA target genes might contribute to understanding the mechanisms of the interplay between the host and viruses, and in the future, potentially result in discovering therapeutic strategies for the prevention and treatment of acute COVID-19 infection. In this article, we present a general review of current studies concerning the function of miRNAs in different VRIs, particularly in coronavirus infection, and address all available therapeutic prospects to mitigate the burden of viral infections.
Keywords: COVID-19, MicroRNAs, SARS-CoV-2, Respiratory viruses, Immune response
1. Introduction
For centuries, infectious diseases have caused a significant damage to human health and devastated the entire organisms [1]. Currently, there is no particular medication or prophylactic vaccine for the eradication of all types of coronaviruses, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), and Middle East respiratory syndrome coronavirus (MERS-CoV) [2], [3], [4], [5], [6]. The novel human coronavirus, named SARS-CoV-2, is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, initially identified in Wuhan, Hubei province, China, which has now affected millions of individuals over the world [7], [8]. The SARS-CoV-2 is a beta coronavirus (a positive-sense, enveloped, and single-stranded RNA) belonging to the Coronaviridae family [9], [10]. Human coronaviruses (HcoVs) cause respiratory tract infections (RTIs). Some of these infectious agents, such as human coronavirus NL63 (HCoV-NL63), human coronavirus OC43 (HCoV-OC43), and human coronavirus HKU1 (HCoV-HKU1), may be benign and cause the common cold. Nonetheless, the science of virology has characterized a group of highly pathogenic human coronaviruses, including SARS-CoV, over the past two decades, which caused infectious diseases in about 8000 people in the world with a fatality rate of ~10% or MERS-CoV in 2012 that infected 2500 cases with a mortality rate of 37% [9]. Numerous studies indicated that these pathogenic coronaviruses led to the emergence of acute respiratory distress syndrome (ARDS), leading to extensive pulmonary inflammation, dyspnea, and even death. Compared with MERS and SARS-CoV-1, SARS-CoV-2 tends to spread more quickly than other types of coronaviruses; nevertheless, it has a lower mortality rate of 2–3% [11]. By November 3, 2020, more than 46, 591, 622 million cases have been diagnosed in 219 countries, and more than 1, 201, 200 deaths have been reported [12].
Micro-RNAs (miRNAs) are classified as small molecules with a length of 17–24 nucleotides [13], [14]. They are noncoding single-stranded RNAs that can suppress the translation of mature messenger RNAs (mRNAs) [13]. MicroRNAs are significant regulators of the gene expression process and play a decisive function in different biological phenomena, including cell propagation, differentiation, growth, and apoptosis [15]. Recently, it has been known that miRNAs contribute to the regulation of both innate and adaptive immune systems by controlling the expansion and activation of immune cells [15]. As mentioned earlier, there is no definite therapy or vaccine to treat and prevent highly pathogenic coronaviruses. Therefore, seeking new biological strategies for the therapy of viral diseases is highly recommended. Data imply that miRNAs have important functions in modulating the immune system toward respiratory viruses, such as HcoVs that include SARS-CoV-1, MERS, SARS-CoV-2, and other types of HcoVs, human metapneumovirus (hMPV), human rhinovirus (hRV), IV, and RSV. Also, changes in the expression of miRNAs in epithelial cells may participate in the pathogenesis of both severe and chronic respiratory infections [16]. This review will summarize the recent discoveries associated with miRNAs in various respiratory infections caused by viruses, especially coronavirus, and address all feasible therapeutic options to mitigate the burden of VRIs.
2. Immune reactions to the SARS-CoV-2
The humoral immunity is immunologically categorized as an acquired immune response in which T helper cells collaborate with B cells to differentiate these types of cells to plasma cells [17], [18], [19]. Plasma cells can synthesize specific antibodies (Abs) against a particular viral antigen (Ag) [17]. Secreted Abs are capable of blocking viruses to prevent them from entering the host cells. Henceforth, they play an influential defensive role in preventing infection and its recurrence [17]. The capability of humoral immune responses as well as B and T cell epitopes against SARS-CoV have been broadly investigated and mapped to analyze the envelope as well as spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins (structural proteins) [20]. Angiotensin-Converting Enzyme 2 (ACE2) acts as a receptor for the entrance of SARS-CoV-2 when infecting the epithelial cells of the lungs. It has been discovered that the S proteins of 2019-nCoV and SARS-CoV-1 have similar structural dominant T cell epitopes [21]. For the generation of compensating Abs, two proteins are needed to support B cell epitopes in the form of receptor binding domains (RBD) [17]. A potent T cell response is a prerequisite for generating a higher concentration of neutralizing Abs [17]. It has been demonstrated that a specific site is not required for T cell epitopes compared with B cell epitopes; thus, they are abundantly expressed in viral infections [17]. T helper cells participate in the switching of isotypes, stimulating B cells to secrete Immunoglobulin M (IgM) and Immunoglobulin G (IgG) Abs against SARS-CoV-1. Also, seroconversion may occur, which could be mediated by the production of T helper cells [22]. Following this phase, at the end of week 12, IgM starts to disappear. Simultaneously, IgG can persist for a more extended period, implying that IgG Abs may be potent protectors during infection [23]. Current evidence strongly suggests that the T helper 1 (Th1) response is a crucial factor for successful controlling of MERS-CoV and SARS-CoV-1, and it might be the same for SARS-CoV-2 [24].
Cellular immunity is immunologically named as acquired immunity [17]. In contradiction of the humoral immune responses, cellular immunity can kill infected cells mediated by T cells [17]. T helper cells are responsible for the overall direction of adaptive immune responses. In contrast, cytotoxic T cells are implicated in killing and eliminating cells infected with viral agents [17]. For the construction of potent vaccines, cellular immunity granted by T cells would be indispensable, as shown by murine SARS-CoV and MERS-CoV models [24]. It has been demonstrated that T cell deficiency results in failure of viral clearance in infected mice, suggesting the significance of these cells in viral clearance [25]. Pointing back to MERS-CoV and SARS-CoV-1, it has been indicated that CD8+ (Interferon (IFN)-γ and Tumor necrosis factor (TNF)-α) and CD4+ (TNF-α, IFN-β, and Interleukin 2 (IL-2)) memory T cells can be persevered in patients recovered from SARS-CoV for up to four years while having a functionality by the propagation of T cells, generation of IFN-γ, and delayed-type hypersensitivity (DTH) responses [26]. Currently, the study results show a distinct memory T cell response to the S library of peptides belonging to SARS-CoV years after the onset of infection [20]. Different CD8+ T cells were detected through the clearance of MERS-CoV in a murine model [27]. These data could be analyzed in the case of SARS-CoV-2 [17]. Nevertheless, the current study demonstrated that the numbers of CD8+ and CD4+ T cells are reduced in individuals infected with SARS-CoV-2, resulting in the compromised generation of memory T cells in patients recovered from SARS-CoV-2 [17]. Several reports have studied the diverse lymphocyte populations in subjects with SARS-CoV-2 infection.
In a study by Qin et al. [28] lymphocytes were evaluated in 44 subjects and according to their results, the total amount of B, T, and NK cells was significantly diminished in the infected group as NK and T cells were below the usual rate, while B lymphocytes were within the lower range of the reference rates [28], [29]. T lymphocytes are usually the most changed by the viral infection, nevertheless, the numbers of CD8+ T, CD4+ T and NK cells were within the normal range in Qi’s study [28]. Moreover, studying several subsets of T lymphocytes showed that both cytotoxic T (CD3+ CD8+) and helper (CD3+ CD4+) lymphocytes were below the normal rate in subjects with COVID-19, and the T helper/suppressor ratio within the normal rates [28]. Additionally, COVID-19 subjects showed lower numbers of regulatory T lymphocytes (Treg) (CD3+ CD4+ CD25+ CD127low+), and this decrease was particularly documented in critical COVID-19 cases [28]. Also, a reduction in stimulated (CD45RO+ CD3+ CD4+ CD25+ CD127low+) regulatory T and naïve (CD45RA+ CD3+ CD4+ CD25+ CD127low+) lymphocytes was documented in critical COVID-19 subjects [28].
Cytokine storm syndrome (CSS) occurs; meanwhile, massive amounts of proinflammatory cytokines are secreted by immune cells due to the overreaction of the human immune system to the external signals [17], [30]. One study showed CSS as one of the complications of COVID-19 [27]. This study was in line with the data on death-related risk factors of SARS-CoV-2 infection [17]. ARDS is highly correlated with CSS as the immune effector cells secrete high amounts of proinflammatory cytokines and chemokines during ARDS, leading to the uncontrolled systemic inflammatory responses [31]. In former pandemics prompted by other coronaviruses, including MERS-CoV and SARS-CoV-1, the analysis of cytokines and chemokines showed excessive amounts of chemokines, including C-X-C Motif Chemokine Ligand 8 (CXCL8), C-X-C Motif Chemokine Ligand 9 (CXCL9), C-X-C Motif Chemokine Ligand 10 (CXCL10), C-C Motif Chemokine Ligand 2 (CCL2), C-C Motif Chemokine Ligand 3 (CCL3), C-C Motif Chemokine Ligand 5 (CCL5), along with cytokines, such as Interleukin-1β (IL-1β), Interleukin 6 (IL-6), Interleukin 12 (IL-12), Interleukin 18 (IL-18), Interleukin 23 (IL-23), IFN-α, IFN-γ, TNF-α, and transforming growth factor beta (TGF-β) [17]. In the case of MERS-CoV, increased expression of proinflammatory cytokines, such as IFN-α, IL-6, as well as chemokines, including CCL-5, CXCL-8, and CXCL-10, have been reported [32]. In a study, Xu et al. found that peripheral blood of COVID-19 subjects had an extremely high rate of CCR6 T-helper 17 (Th17) lymphocytes, sustaining a Th17-type CSS in this infection [29], [33].
Cytokine storm-induced ARDS has destructive effects on the immune system, resulting in multiple organ failure and subsequent death [34]. This deleterious effect has caused similar mortality in the SARS-CoV-2 outbreak as in SARS-CoV-1 and MERS-CoV infections [17]. It is now known that drugs that target IL-1, IL-6, IL-18, and IFN-γ are useful for attenuating cytokine storm syndrome in other viral infections. This casts a doubt on the effectiveness of these therapeutic agents for mitigating the severity of COVID-19 [35]. Recently, one of the specific inhibitors of IL-6 was demonstrated to be effective in reducing COVID-19 severity in a few cases of patients in China [36]. CSS is a crucial factor that determines the intensity of signs, the mortality rate, and the onset of extrapulmonary signs during COVID-19 [29]. Severe infections show features suggestive of a series of pathologies associated with CSS, in which multi-organ failure and hyper inflammation are demonstrated following excessive cytokines produced from an enhanced immune stimulation [29]. Hence, studies with CSS are promising and could urge further pathogenetic studies on identification of effective therapeutic biomarkers.
2.1. Role of microRNAs in host immune responses to VRIs
The innate immunity is regarded as the primary defense line against most infectious agents, such as bacteria, viruses, and other pathogens, posing a hazard to the host [16]. It has been reported that miRNAs contribute to regulating the function of dendritic cells, epithelial cells, monocytes, granulocytes, NK cells, and macrophages that participate in innate immune responses [16]. Activation of the intracellular signaling pathway triggers the innate immune response and stimulates the coupling of pathogen-associated molecular patterns (PAMPs) with pattern recognition receptors (PRPs), which are specialized transmembrane proteins. This family of proteins includes NOD-like receptors, toll-like receptors (TLRs), and RIG-I-like receptors (RLRs), which have a fundamental function in antimicrobial protection [37]. Since the primary target of VRIs is the epithelial cells, these types of cells are involved in viral-induced immune responses. For instance, stimulating the epithelial cells by TLR agonists incites the expression and synthesis of chemokines, IFNs, and cytokines [16]. The most effective stimulating response is achieved by treating epithelial cells with TLR3 ligands, which detect double-strand RNAs (dsRNAs) produced as intermediate products during the replication of viral genomes [38]. Recently, it has been shown that miRNAs effectively participate in signal transduction of TLRs [16]. Studies have indicated the enhanced expression of miR-146a and miR-146b in response to lipopolysaccharide (LPS) when exposed to the THP-1 cell line. This finding was in agreement with other related studies, indicating that similar responses occur upon the activation of the TLR signaling pathway in myeloid cells in response to fungal, viral, and bacterial agonists of TLRs [39], [40]. Interestingly, no significant rise was seen in the expression of miR-146a after the stimulation of TLR-3, TLR-7, and TLR-9 in response to bacterial and viral nucleic acids [16]. These results were in contrast to findings obtained from the expression of miR-155, which showed increased levels following the activation of TLR-3 and TLR-9 in response to poly I: C and CpG, respectively [16]. Several lines of evidence showed that miR-155 is a definite regulator of the pro-inflammatory cytokines, as demonstrated by research carried out on the transgenic mouse model [40], [41]. Also, reports indicated that miR-146a could modulate the TLR signaling pathway by targeting TRAF6 and IRAK1, which are introduced as protein complexes involved in the activation of IL-1βR, TLR2, TLR4, and TLR5 signal transduction [42], [43]. Studies showed that miR-9 is overexpressed upon TLR4 activation in human monocytes and neutrophils [44], [45]. Remarkably, it has been reported that the stimulation of TLR3, a signal transduction activator dependent on MyD88, did not influence the expression of miR-9, denoting that miR-9 induction is linked with signal transduction dependent on MyD88 activation in monocytes. Current evidence indicated that the activation of TLR2, TLR4, and TLR7/8 promotes the activation of the MyD88 signaling pathway, leading to an improvement in the expression of miR-9 in human monocytes and neutrophils [16]. Following the exposure of murine macrophages to TLR2, TLR3, and TLR4 agonists and lipopolysaccharide (LPS), the expression of miR-147 is activated [38], [46]. A recent study showed that miR-147 has anti-inflammatory properties and attenuates excessive inflammatory responses [16]. MiRNAs control the expression of some cytokines involved in the activation of innate immune responses [16]. The generation of some cytokines, such as TNF-α, IL-6, and IL-12 are lowered after the induction of miR-152 and miR-148, associated with TLR3, TLR4, and TLR9 activation [42]. Except for IFN-γ, proinflammatory cytokines, such as IL-1β, induce miR-9 expression [38].
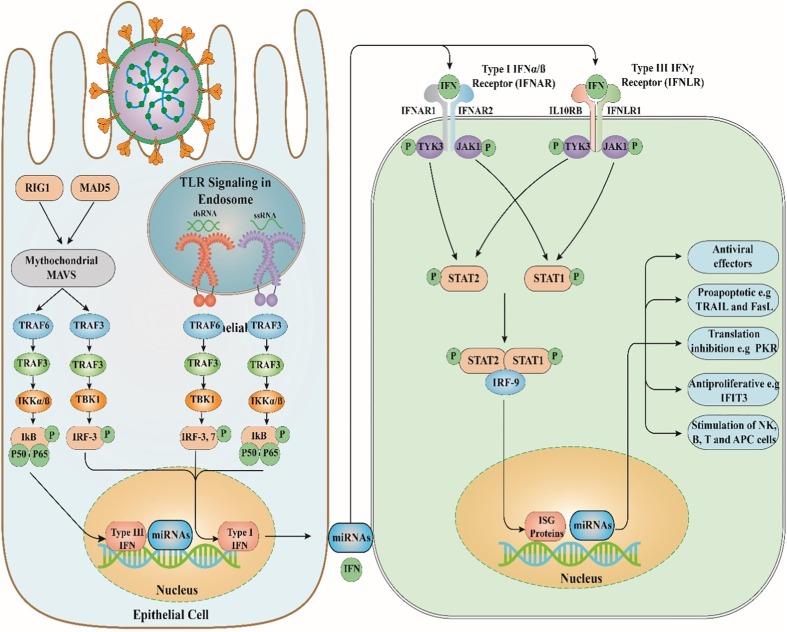
Other receptors belonging to the PRPs family, such as RLRs, consist of melanoma differentiation-associated gene 5 (MDA5) and retinoic acid-inducible gene (RIG-I) [16]. RLRs are surrounded in the cytoplasm and contribute to the identification of viral components [16]. Viral infections caused by negative- and positive-sense RNA viruses, such as IV and RSV, can lead to RIG-1 activation, while MDA5 can identify Picornaviruses, such as RVs [47]. RV can induce antiviral responses by triggering RIG-I, TLR3, and MDA5 activation [16]. Furthermore, the stimulation of HeLa cells infected with RV led to the enhanced levels of miR-23b, implying that the expression of this miRNA is regulated by IFN- and RIG-mediated signaling pathways (Fig. 1 ) [48].
Fig. 1.
The antiviral immune response and role of miRNAs. TLR, Toll-like receptor; RIG1, Retinoic acid-inducible gene I; MAD5, melanoma differentiation-associated protein 5; Mitochondrial antiviral-signaling protein; TRAF3, TNF receptor-associated factor3; TRAF6, TNF receptor-associated factor6; IKK, IκB kinase; TANK-binding kinase 1; IRF3, Interferon regulatory factor 3; IFN, Interferon; IFNAR1, interferon-α/β receptor 1; STAT1, Signal transducer and activator of transcription 1; STAT2, Signal transducer and activator of transcription 2; IRF9, Interferon regulatory factor 9; ISG, interferon-stimulated gene; miRNAs, microRNAs.
Adaptive immunity is a member of the antigen-specific defense system, mediated by T and B lymphocytes [16]. Recently, it has been indicated that the miR-17–92 cluster is the primary regulator of T cell proliferation involved in T cell expansion, leading to the increased endurance of these cells by the impairment of the expression of pro-apoptotic protein-encoding mRNAs, such as phosphatase, tensin homolog, and Bcl-2 [49]. The expression of some sets of miRNAs contributes to the polarization of T helper cells. For instance, miR-155 enhanced the differentiation of Th-2 cells, whereas miR-326 promotes the development of Th-17 cells [16]. Besides, miRNAs are responsible for the regulation of mechanisms underlying the central-adaptive immune responses, such as presenting Ags, engaging miR-155, and promoting the development of B and T cells in the presence of miR-181a [49], [50]. Li and colleagues [51] observed the expression of miR-181a in immature T cells resident in the thymus. This miRNA can also modulate T-cell receptor signal transduction by suppressing the activation of various phosphatase enzymes [51].
In contrast to human naive T cells, miR-146a is highly expressed in human TCM [50], [52]. Studies indicated that miR-146a interferes with apoptosis initiation and targets the Fas-associated death domain [16]. Correspondingly, the engagement of miR-146a in the acquired immunity is more pronounced when the overproduction of miR-146a leads to the disturbed production of IL-2 and action of activator protein-1 [53]. Several studies showed that the memory and effector CD8+ T cell-mediated antiviral responses recruit miR-155 for their activation [16]. CD8+ T cells lacking miR-155 exhibit decreased effector responses, and they are unable to be differentiated into memory cells [54]. Another study conducted on mice knock-out for miR-155 and infected with IV demonstrated the impaired clearance of viral infection in these genetically-manipulated animals [55].
Besides, miR-155-deficient CD8+ T cells are more resistant to the anti-proliferative properties of IFNs and show increased IFN-I signaling [16]. A study conducted by Lind et al. [55] showed that immunity to the lymphocytic choriomeningitis virus is compromised in miR-155-deficient mice. Remarkably, miR-155 is overexpressed in resting primary human B cells infected with EBV that cause the increased survival of virus within the lymphoblastic cells. This implies the importance of miR-155 for EBV latency. Therefore, miR-155 seems to be a vital component of antiviral responses.
The immune responses against VRIs, such as IV, hRV, human coronavirus (HcoV), hMPV, and RSV, are correlated with the aberrant expression of several miRNAs in epithelial cells and participate in the pathogenesis of chronic and acute forms of respiratory disorders (Table 1 ) [16]. In this section, the role of miRNAs in VRIs is comprehensively reviewed.
Table 1.
Role of microRNAs in viral respiratory infections.
| MicroRNA | Viral infection | Effect | Reference |
|---|---|---|---|
| hs-miR-a, miR-b | Human rhinovirus | The levels of hs-miR-a and hs-miR-b were reliant on HRV replication. | [61] |
| miR-128, miR-155 | human rhinovirus | The bioinformatics investigation indicated that the miR-128 and miR-155 contribute to the innate immune response toward HRV-1B. | [62] |
| miR-23b | human rhinovirus | The RIG-I- like receptor-inducible miRNAs, miR-23b, hinders infections HRV-1B by decreasing the very low-density lipoprotein receptor. | [48] |
| miR-323, miR-491, miR-654 | influenza virus | The mutational examination determined that miR-323, miR-491, and miR-654 were increased following H1N1 infection. | [83] |
| miR-17-3p, miR-221 | influenza virus | The expression levels of miR-17-3p and miR-221 decreased in human alveolar basal epithelial cells (HBEpC) through the initial step of IAV infection. | [133] |
| let-7c | influenza virus | The miR-let-7c expression was remarkably raised in IV-infected A549 cells. | [134] |
| let-7f | human metapneumovirus | Following RSV infection, the let-7f expression was significantly increased. | [135] |
| miR-185–5p | human metapneumovirus | The hMPV provokes miR-185–5p expression. | [136] |
| miR-16 | human metapneumovirus | During hMPV infection, has-miR-185–5p expression was increased. hMPV (Wild type) infection does not provoke miR-30a and miR-16 expression, but the virus deficient the M2-2 gene enhancing miR-16 and miR-30a. | [137] |
| miR-374a, miR-192 | human metapneumovirus | The miR-374a* was diminished by hMPV infection. | [79] |
| miR-221 | respiratory syncytial virus | The RSV infection significantly diminished the expression of miR-221. | [138] |
| miR-30, let-7i | respiratory syncytial virus | The expression of let-7i was observed in healthy HBEpC cultures infected with an RSV that deficient NS1 and NS2 proteins. | [71], [136] |
| let-7 | respiratory syncytial virus | The increase in the expression of let-7b was observed in HBEpC cultures infected with an RSV. | [56], [71] |
| miR-339-5p | respiratory syncytial virus | Following the RSV infection, miR-339-5p targets the TrKB and APAF1 genes and result in TrKB- Brain-derived Neurotrophic Factor binding as well as apoptosis. | [138], [139] |
| miR-453 | respiratory syncytial virus | During the RSV disease, the low-affinity p75 NTR receptor gene was declined via miR-453. | [138], [139] |
| miR-34 | respiratory syncytial virus | In patients infected with RSV, the expression levels of miR-34b and miR-34c were reduced. | [56] |
| miR-125 | respiratory syncytial virus | Among the miR-125 family, in patients infected with RSV, the expression of miR-125a and miR-125c were reduced. | [56], [140] |
| miR-29 | respiratory syncytial virus | Among the human miR-29 family, the expression levels of miR-29c were declined in RSV infected patients contrasted to the control group. | [56], [141] |
| miR-429 | respiratory syncytial virus | According to the severity of the mild group's illness in patients infected with RSV, miR-429 levels were downregulated. | [56], [70], [142] |
| miR-27 | respiratory syncytial virus | The miR-27 family was deregulated during RSV infection. For instance, the miR-27b is downregulated in patients infected with RSV. | [56], [143], [144], [70], [138], [145] |
| miR-155 | respiratory syncytial virus | miR-155 overexpressed in patients infected with RSV. This miR represses IFN pathways for reducing anti-viral status. | [56], [146] |
| miR-31 | respiratory syncytial virus | The miR-31 overexpressed in patients infected with RSV. | [56], [147] |
RV causes upper respiratory tract infections (URTIs) in adults and children. The epithelial cells resident in the respiratory tract is the main target of RV [56]. RV is a single-stranded-RNA (ssRNA) virus that possesses icosahedral capsids and belongs to the Picornaviridae family [57], [58]. The dsRNA is generated during viral replication and is identified by RIG-I and TLR3 [47], [59]. Bioinformatics software have recently enabled us to efficiently predict whether the natural or designed miRNAs can target some viruses to act as therapeutic agents [60]. Several studies showed that miR-155 and miR-128 could regulate innate immune responses against RV-1B due to targeting the genetic components of RV [61]. It has been shown that the knock-down of these two miRNAs can stimulate RV replication [62]. MiR-23b contributes to the immune response against RV, leading to the decreased expression of the transmembrane region of VLDLR and LPR5 [16]. These two receptors are hijacked by twelve RV types, including RV23, RV25, RV29, RV30, RV31, RV1A, RV1B, RV2, RV44, RV47, RV49, and RV62 [48].
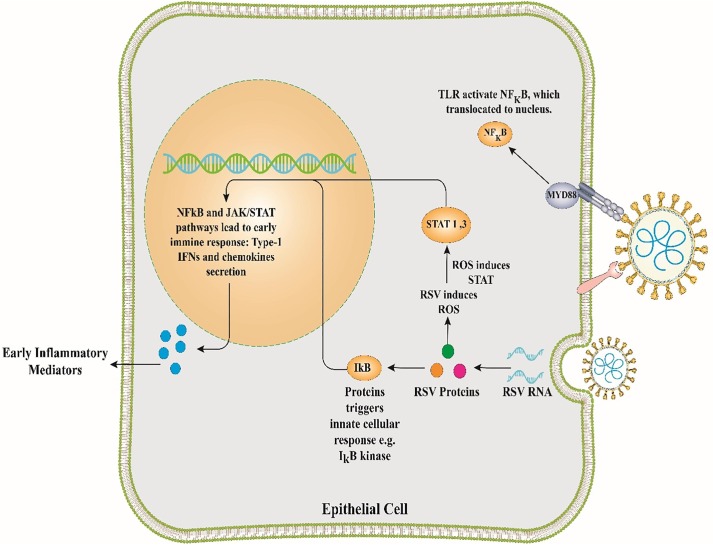
The genome of RSV is a negative-sense ssRNA, encoding 11 proteins (NS1, NS2, M, P, N, SH, M2-1, M2-2, G, F, and L), which is related to the Paramyxoviridae family [63], [64], [65]. RSV is a prevalent human pathogenic respiratory virus that attacks the lower respiratory tract (LRT) and causes similar clinical symptoms to those of the common cold in children and adults. The virus is classified as a respiratory virus regularly isolated from hospitalized children with bronchitis [56]. Primary infection typically causes acute illness, whereas secondary infection can provoke obstructive bronchitis [66], [67], [68]. In HBEpC, RSV can reduce the production of miR-221, while the degree of let-7i and miR-30b production is increased after 48 h of infection [56]. Increased expression of let-7i and miR-30b has been reported in RSV-infected HBEpC lacking NS1 and NS2 proteins, implying that these proteins have antagonistic properties against miR-30b and let-7i; thus, suppressing the generation of type I IFN [56]. Among miRNAs aberrantly expressed during the infection period of RSV-A2, miR-221, miR-27a, miR-339-5p, miR-574, miR-453, and miR-744 are more affected compared with others. Except for the miR-744, the expression levels of all the aforementioned miRNAs are increased [69]. A case-control study explained that patients infected with RSV had a marked decrease in the expression of miR-34c, miR-34b, miR-29c, miR-125b, miR-125a, miR-27b, and miR-429 when compared with healthy individuals. In contrast, the expression levels of miR-31, miR-155, let-7d, miR-203a, and miR-16 were elevated [56]. In this case-control study, subjects were allocated to three groups based on the severity of the disease: mild, moderate, and severe. The results demonstrated that miR-125a and miR-429 expression decreased in the moderate group [70]. Some investigations indicated that RSV could provoke different expression levels of miRNAs in at least two various ways. First, the induction of let-7b and let-7i is dependent on the expression of IFN-β in HBEpC and monocyte-derived dendritic cells (MDDCs) [71]. Second, miR-30b is not induced by IFN-β while being reliant on the expression of NF-κB in HBEpC (Fig. 2 ) [71]. Evidence also demonstrates that RSV can diminish miR-221 expression in HBEpC [71]. RSV infection can lower the expression of some miRNAs, such as miR-337-3p, let-7f, miR-24, miR-26b, miR-520a-5p, miR-595, and miR-198 [72]. All of these miRNAs have related victims, such as genes encoding the cell cycle proteins (CCND1, DYRK2, and ELF4), suppressor of cytokine signaling 3 gene (SOCS3), and chemokines (CCL7) [72]. Moreover, the RSV G-protein can increase the expression of let-7f, having an antagonistic activity against DYRK2 and CCND1. These proteins contribute to cell cycle check in the G1 phase and enhance viral propagation [72]. It has been demonstrated that miR-let-7 is vital for inducing the host genes during viral infections [72].
Fig. 2.
Cellular activation following respiratory syncytial virus infection. JAK/STAT, Janus kinase/Signal transducer and activator of transcription; NF-KB, nuclear factor-kappa B; IFN, Interferon; RSV, respiratory syncytial virus.
hMPV is a recently identified prominent member of the Paramyxoviridae family [46], including the human parainfluenza virus [73], [74]. NS1 and NS2 are non-structural genes, which are not expressed in the genome of hMPV with eight open-reading frames: 3′-N-P-M-F-M2-SH-G-L-5′ [75]. Some clinical trials demonstrated that hMPV is responsible for developing pediatric LRTI [76], [77], [78]. Previous works have indicated that hMPV causes some alterations in the expression of a collection of miRNAs, such as miR-4552, let-7f, miR-16, miR-30a, miR-192, and miR-374a* which are expressed in epithelial cells resident in the respiratory tract [76], [77], [78]. It has been reported that hMPV is responsible for regulating the expression of 174 miRNAs in A549 cells. Among these miRNAs highly expressed in A549 cells is let-7f, which can target the RNA polymerase enzyme of hMPV to control the viral replication [79].
The influenza virus (IV) is an ssRNA virus related to the Orthomyxoviridae family, consisting of three influenza viruses: A, B, and C [56]. Type A influenza virus (IVA) is sub-categorized based on two proteins expressed on the exterior surface of the virus: neuraminidase and hemagglutinin (N and H proteins, respectively) [56]. To date, 16 types of H proteins, along with 9 types of N enzymes, have been characterized [80]. Influenza A (H1N1 or H3N2) and B have currently the highest incidence rate in the united states [81]. IV is a highly contagious respiratory disorder, characterized by the emergence of featured signs, including cephalea, fever, coryza, myalgia, sore throat, and coughing [81]. The IV preferably infects the upper respiratory tract (URT); nevertheless, in severe cases, the lower respiratory tract (LRT) (bronchioles) might also be infected with the virus [82]. The miRNAs expression during IV infection might be dysregulated [56]. Many miRNAs, including miR-491, miR-323, and miR-654, can hinder the replication of IVA H1N1, resulting in the reduced IVA gene expression in infected cells [83]. The mRNA degeneration of the PB1 protein (contributing to the viral propagation of IVA) by the host miR-491, miR-323, and miR-654 is one of the exemplary mechanistic roles of miRNAs [84]. Suppression of the M1 protein expression by let-7c regulates the replication of the IVA in A549 cells [84], [85]. Studies indicated a substantial reduction in the production of miR-221 and miR-17-3p in human alveolar basal epithelial cells during the IV infection [86].
2.2. MicroRNAs in the infection period of coronaviruses
Coronaviruses are responsible for a broad range of respiratory infectious diseases, from mild URTIs to critical and life-threatening forms of the disease [87]. Up to now, the role of miRNAs in the infectivity of coronaviruses has not been examined in in-vivo studies. A limited number of in-vitro studies were carried out on the OC43 virus, and some in-silico analyses have been conducted on SARS-CoV and MERS-CoV [88]. The coronavirus OC43 causes the common cold [89]. The N protein of coronavirus is requisite for viral replication and binding the genomic RNA to produce helical capsids. The OC43 N protein triggers NF-kB activation by attaching to its negative regulator called miR-9 [88]. Whether the activation of NF-kB contributes to viral replication directly or acts as a bystander to limit the viral replication has not been so far revealed [88].
Compared with other pathogenic coronaviruses, a lower rate of OC43 virulence, along with minor symptoms, may affect the interaction between infected and non-infected subjects and consequently increase the distribution of the virus within a community [90]. This innovative mechanism to boost gene expression may explain how RNA viruses suppress the host immune scheme or induce pathological alterations in humans [88]. SARS-CoV-1 is a newly-identified coronavirus that caused a global pandemic in 2002–2003. In-silico examination of miRNAs interacting with SARS-CoV-1 mRNAs recommended that the virus may inhibit its replication throughout infection by upregulating miR-214, miR-574-5p, and miR-17 [88]. The virulent proteins, including N, S, M, and E proteins, serve as targets for host miRNAs. [91]. The abolition of viral replication may support the viral evasion from the immune system surveillance until the infection of other cells [88]. These outcomes describe how SARS-CoV-1 can cause some changes in the host miRNA expression pattern to its authority.
Regarding the SARS-CoV-1, it is thought that this virus cannot encode its miRNAs [91]. However, in due progress of evolution, the virus may evolve some highly sophisticated mechanisms to take advantage of the host cellular biosynthetic system and evade from the cellular defense mechanisms [91]. Under specific conditions, SARS-CoV-1 employs cellular miRNA transcripts to promote its own progress [91]. In a pioneer work, Mallick et al. [91] assessed miRNA–interfered host–SARS-CoV-1 interplays in bronchoalveolar stem cells (BASCs) at the early stage of the disease. They found that BASCs could be categorized as a subset of Oct-4+ ACE2+ cells and major targets for SARS-CoV infection [91]. Recently, it has been discovered that SARS-CoV-1 could enter the host cells, contaminate pulmonary BASCs, and regulate its developmental steps through molecular controls-miRNAs [91]. The interaction between the host miRNAs and viruses exhibits a planned scenario to control the viral replication to prevent immune elimination until the infection of other cells [91]. Afterwards, the virus undergoes rapid mutations to optimize target-miRNA mismatches and promote its propagation until cells reach an entirely differentiated status [91]. Mallick et al. [91] indicated that BASCs are the primary target cells for SARS-CoV. They revealed that the miRNAs-17*, -574-5p, and -214 are upregulated following SARS-CoV infection in BASCs to suppress the viral replication and escape from the immune system [91]. It has also been noted that N and S proteins downregulate miR-223 and miR-98 in BASCs to manage several steps of the differentiation process in these cells and activate inflammatory chemokines to downregulate the activity of the ACE2 enzyme [91]. These processes effectively account for a fruitful viral transference and reproduction within BASCs, leading to the ongoing destruction of lung cells and loss of repair capacity [91]. Generally, this study revealed the different modes of exploiting cellular miRNA tools by a virus to its benefit.
MERS is introduced as a new human coronavirus named MERS-CoV [92]. An in-silico study recognized some miRNAs (Table 2 ) that possess remarkable sequence identity to hairpin constructions in the MERS-CoV genome. These miRNAs may accordingly downregulate the viral gene expression to hinder its replication [93]. In a recent study, Zhanga et al. [94], investigated the impact of host ceRNA (competitive endogenous RNA) alternations along with the biological circRNAs (circular RNAs) on Calu-3 (human lung adenocarcinoma epithelial) infected with MERS-CoV. This study indicated that infection with MERS-CoV in Calu-3 causes an alternation in miRNA expression (Table 2) [94]. These phenomena pave the way to better understand host-virus interactions and design novel antiviral agents against MERS-CoV, a highly fatal respiratory disorder.
Table 2.
MicroRNAs in coronaviruses infection.
| MicroRNA | Coronavirus | Description | Reference |
|---|---|---|---|
| miR-9 | OC43 | The OC43 nucleocapsid protein, via direct interaction with mir-9, can modulate NF-kB expression. | [90], [88] |
| miR-17, miR-223, miR-574-5p, miR-214, miR-223, miR-98 | SARS | Viral Nucleocapsid and Spike protein reduced the expression of miR-223 and miR-98 within the BASC. | [91] |
| miR-16-1-3p, miR-26a-1-3p, miR-425-5p, miR-500b-5p, miR-627-5p, miR-1257, miR-1275, miR-2277-5p, miR-2392, miR-4448, miR-4455, miR-4521, miR-6807-5p, and miR-6847-3p, miR-329-5p, miR-539-5p, miR-619-5p, miR-762, and miR-6836-5p | MERS | The host ceRNA and circRNAs analysis in human lung adenocarcinoma epithelial cells contaminated with the highly pathogenic MERS-CoV indicated that MERS-CoV infection could impact on host gene expression. | [94] |
| miR628-5p, miR-6804-3p, miR-4289, miR-208a-3p, miR-510-3p, miR-18a-3p, miR-329-3p, miR-548ax, miR-3934-5p, miR-4474-5p, miR-7974, miR-6865- 5p, and miR-342-3p | MERS | The computational approach provided an exciting hypothesis that those miRNAs involved in MERS-CoV pathogenesis, and this approach may help to understand host-pathogen interplay better and promote new antiviral treatment toward MERS-CoV. | [93] |
| miR-146a-5p, -21-5p, and -126-3p | SARS-CoV-2 | In patients with COVID-19 that not respond to the tocilizumab treatment, the levels of miR-146a-5p were decreased in the serum. Also, this study's data suggest miRs in the blood, such as miR-146a-5p, can act as biomarkers and provide a molecular link between inflammation and the COVID-19 clinical course. | [116] |
| miR-200c | SARS-CoV-2 | Aa study investigated several identified miRNAs that could regulate ACE2 which may be exploited to regulate the SARS-CoV-2 receptor. Their data reveal that both ACE2 mRNA and ACE2 protein levels are inhibited by miR-200c in rat primary cardiomyocytes and, importantly, in human-derived cardiomyocytes | [120] |
| miR-98-5p | SARS-CoV-2 | In a study evaluated the microRNAs that specifically target TMPRSS2. Through a bioinformatic approach, they identified miR-98-5p as a suitable candidate and they mechanistically validated miR-98-5p as a regulator of TMPRSS2 transcription in two different human endothelial cell types, derived from the lung and from the umbilical vein | [121] |
3. MicroRNAs in SARS-CoV-2
The host miRNA expression has a fundamental effect on viral pathogenesis control through interfering with T cells and immune reactions to viral infections [95]. The miR-32 is the primary described example of a cellular miRNA that targets the viral RNA genome that decreases viral replication within the cells [96]. Besides, it has been demonstrated that two proteins (L and P) of VSV are targets of miR-24 and miR-93, while miR-29a targets the Nef protein of HIV to inhibit its replication. Moreover, some miRNAs, including miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431, and miR-448, target the HCV proteins such as C and NS5A proteins to suppress the translation/replication by provoking IFN signaling [97], [98], [99]. Hence, miRNA can provide a featured concept for elucidating the pathogenesis and infectivity of the new pandemic SARS-CoV-2. While the SARS-CoV-1 is far related to SARS- CoV-2, some relationships have been found in their manifestations. However, there are marked discrepancies between the two disorders [100].
Currently, numerous countries deal with SARS-CoV-2, and such a prevalence makes the virus prone to undergoing some mutations and developing variations in different populations [101]. Several lines of evidence showed that viral pathogens could evade the immune system surveillance by utilizing the host miRNAs [102], [103]. Khan et al. [101] analyzed the host miRNAs profile and the epigenetic regulators of the different expression levels of miRNAs among SARS-CoV and SARS-CoV-2. They applied computational procedures to prognosticate possible host and viral miRNAs and their potential tasks in various operative pathways [101]. They also distinguished multiple presumed host miRNAs (with antiviral function) capable of targeting SARS viruses and their miRNAs to target the host genes [101]. The contrast among the host miRNA profiles associated with distinct SARS-CoV-2 genomes among 24 distinct nations with corresponding normalized deaths elucidated some miRNA clusters correlated with raised mortality rates [101]. In this context, Khan and colleagues [80] detected that the provoked cellular miRNAs could act as a double-edged sword for host protection, as they have significant functions in compensating the viral infection.
Additionally, miRNAs can function as proviral determinants. Khan et al. [101] indicated that both SARS-CoV-1 and SARS-CoV-2 viral miRNAs can target a wide range of signaling pathways associated with the immune system, and some SARS-CoV-2 miRNAs can target a set of immune-associated signaling pathways, including autophagy and IFN-I signaling, suggesting their immune-evasion tools for continued latency inside hosts [101]. Besides, various crucial cellular pathways can be modulated by SARS-CoV-2, which may result in extended aberrations in cases with comorbidities such as cardiovascular disorders, diabetes, and respiratory difficulties [101]. This event suggests that miRNAs can be considered as critical epigenetic modulators in developing complications in cases with COVID-19. The study of Khan et al. [101] supported the fact that miRNAs from both the host and virus (SARS-CoV-2) can contribute to pathological events. These results will further be beneficial in developing RNA-based therapeutics to mitigate the complexities of COVID-19.
In a research conducted by Wyler et al. [104], in three Caco-2, Calu-3, and H1299 cells (human cell lines), gene expression of both SARS-CoV-1 and SARS-CoV-2 were examined with mass and single-cell transcriptomics. RNA profiling exhibited the robust induction of the protection and inflammation-linked microRNAs, such as miRNA-155, following infection with these viruses [104]. Infection of the Calu-3 with SARS-CoV-2 approximately causes a 2-fold increase in the expression of the IFN-stimulated response element genes and the expression of cytokines, such as CXCL10 or IL6 compared with SARS-CoV-1 [104]. They showed the significant increased expression of miR-155 in cells infected with SARS-CoV-2 [104]. This miRNA has been correlated with numerous viral diseases [105], [106], [107], and has been recognized as a regulator of immune cell differentiation, especially T-cells [108], [109]. The involvement of this miRNA in directing the innate immune responses has further been addressed elsewhere [110]. Recently, miR-155-5p induction was determined to be provoked in a murine model infected with the IVA [111]. Significantly, in the study of Wyler and colleagues, the pulmonary damage caused by ARDS was reduced by the elimination of miR-155, denoting that this miRNA could be a possible curative target for the treatment of COVID-19 [104].
Additionally, in bioinformatics studies by another team, some miRNAs, such as miR-218-5p, let-7g-5p, miR-506-3p, miR-133b, miR-124-3p, miR-22-3p, and miR-133a-3p have been identified in SARS-CoV-2 disease [112]. In another bioinformatics work by Sardar et al. [113], a unified sequence-based examination of SARS-CoV-2 genome from diverse geographic places was carried out to recognize notable genomic characteristics of SARS-CoV-2 by integrated analysis. Their analysis revealed nine host miRNAs that can conceivably target SARS-CoV-2 genes (Table 2) [113]. Interestingly, they found that nine miRNAs did not have targets in another related viral genomes, including those of SARS-CoV-1 and MERS [113].
Liu et al. [114] used computational methods to investigate the SARS-CoV-2 genome for alleged miRNAs and detected the viral miRNA targets on the virus, and human genome simultaneously and also detected the human miRNAs that can target the viral genome. Besides, they examined miRNAs implicated in dysregulation prompted by a viral disease that involved the immunity and cytoskeleton structure, which are the common critical biological means modulated by the miRNAs, induced by infection [114]. Notably, they observed that hsa-miR-4661-3p could target the S gene of SARS-CoV-2, and that miR147-3p (virus-encoded miRNA) enhanced the induction of TMPRSS2 by increasing the virulence of SARS-CoV-2 in the intestine.
Ivashchenko et al. [5] studied how miRNAs can protect humans from COVID-19 using bioinformatics tools. They found that among 2565 miRNAs, three miRNAs, including miR-6864-5p, miR-5197-3p, and miR 4778-3p, were recognized as the common significant miRNAs communicating with the guide RNA of SARS-CoV-1, MERS-CoV, and SARS-CoV-2 [5]. Using the genome sequences of several miRNAs, including miR-6864-5p, miR-4778-3p, and miR-5197-3p, they identified the cc-miR (complete complementary miRNA) in the guide RNA of coronaviruses [5]. The indicated that the binding sites of these cc-miRs did not create intramolecular networks in the 2D construction of the guide RNA of SARS-CoV-1, MERS-CoV, and SARS-CoV-2 [5]. Hence, the cc-miRs can join guide RNA via these sites out of the race [5]. It has been revealed that in these coronaviruses, cc-miRs are devoid of target genes among 17,508 subjects encoding genes with a ΔG/ΔGm of higher than 85%, implying that these cc-miRs do not affect the translation of the mRNA in humans [5]. It has been shown that cc-miRs could be employed as remedial molecules by being fused with exosomes or additional biological vesicles to be transferred within the lung through inhalation [5]. The transfer of cc-miR within the blood can hinder the propagation of the virus in the bloodstream and the entire organs it can penetrate [5]. The recommended approach of the hindrance of coronaviruses replication can further be employed for other viruses.
In another study, Tak-Sum Chow and colleagues predicted and examined the microRNAs in the human lung epithelium (HLE) that could target SARS-CoV-2 [115]. They have recognized 128 microRNAs (from human) with the ability of targeting the SARS-CoV-2 genome [115]. This study indicated that six of these microRNAs are differentially expressed simultaneously in in-vitro SARS-CoV-2 infection. Besides, 28 microRNA targets (for SARS-CoV-1) and 23 microRNA targets (for MERS-CoV) have been found [115]. Furthermore, 48 and 32 microRNAs have been typically distinguished in two other investigations. More investigations into distinguishing authentic microRNAs that target the coronavirus will help understand the importance of microRNAs as a cellular security mechanism toward pathogenic coronavirus diseases [115]. Sabbatinelli et al. [116] investigated the levels of miR-21-5p, miR-146a-5p, and miR-126-3p in the serum of 29 subjects with COVID-19 receiving tocilizumab along with sex and age-matched healthy control. The study results indicated that the serum levels of miR-146a-5p were significantly lowered in cases who did not respond to tocilizumab [116]. They also demonstrated that among non-responders, those with the lowest serum levels of miR-146a-5p experienced the most adverse outcomes [116]. Finally, they proposed that the biomarkers in blood, such as miR-146a-5p, can present a molecular connection between inflammation and the COVID-19 clinical course, thus expanding the drug arsenal toward this global health threat [116]. The authors also demonstrated that between non-responders, those with the lowermost levels of miR-146a-5p in serum experienced a majority of unfavorable consequences.
It has been found that ACE2 helps in maintaining the blood pressure as well as electrolyte balance and it also decreases the level of angiotensin II in the circulation by suppressing the renin-angiotensin-aldosterone system (RAAS) conferring anti-hypertensive influences [117]. Currently, it has also been found that ACE2 acts as a receptor for the S protein of SARS-CoV-2 that plays a pivotal activity in COVID-19 [118]. Of note, in severe COVID-19 patients, various health complications, such as hypertension, cardio-vascular disorders, and diabetes have been reported [119]. In particular, it has been found that ACE2 is produced in cardiomyocytes and raised in individuals with cardiac disorders [120]. In an outstanding study by Lu et al. [120], several miRNAs with the ability of modulating ACE2 which can be exploited to control the SARS-CoV-2 receptor were investigated. Their findings showed that both ACE2 mRNA and ACE2 protein rates are suppressed by miR-200c in animal cardiomyocytes and more importantly, in cardiomyocytes derived from humans [120]. Lu et al. [120] found the first miRNA candidate that could target ACE2 in cardiomyocytes that could be used as a preventive approach for cardiovascular complications in COVID-19 patients [120].
ACE2 is one of the crucial factors by which SARS-CoV-2 accesses the human cells, and it has been noted that internalization of the SARS-CoV-2 requires not only binding to ACE2 but also the TMPRSS2 (transmembrane protease serine 2) [121]. Of note, the potential involvement of TMPRSS2 and targeting it in COVID-19 as a novel therapeutic method has not been fully evaluated. In a study by Matarese et al. [121] the microRNAs that specifically target TMPRSS2 were evaluated by an in-silico approach. They found miR-98-5p as a proper candidate and mechanistically validated it as a modulator of TMPRSS2 transcription in two different umbilical vein endothelial cells derived from the human lung [121]. Overall, these data show that TMPRSS2 can be a promising target by specific non-coding-RNAs in the treatment of COVID-19 patients.
4. MicroRNAs: As therapeutic and diagnostic biomarkers in viral infections
To date, there is no available drug to increase/inhibit any miRNAs in VRIs; nevertheless, progress has been made in other disorders. The primary inhibitory medication designed for miR-122 was constructed in 2010, and it is currently in a phase II trial for hepatitis C therapy [56]. MRX34 is the primary artificial miRNA (2013) used to treat advanced hepatocellular carcinoma [122]. In further investigations, artificial miRNAs have been produced to transfect mononuclear cells of peripheral blood with liposomal carriers [56]. These procedures boost particular proinflammatory cytokines, such as TNF-α, and promote innate immunity [123]. The most current purpose of these miRNAs was to establish modern vaccines by attenuated viruses filled with an expression cassette (EC) encoding an artificial miRNA to target necessary viral proteins [56]. The viral PR8-amiR-93NP was produced by inserting an EC for miR-93 in attenuated IV within viral genes (genes encoding non-structural proteins). The target of this miRNA is the nucleoproteins of IV [56]. This vaccine is given by intranasal injection, protecting against various dissimilar viral strains [124]. Plants also generate miRNAs that can control the replication of viruses [56].
For example, the expression of PB2 and NS1 proteins in IVAs (H1N1 and H7N9) was repressed by miR-2911 (a miRNA in honeysuckle) [125]. The absence of an in-vivo delivery system is a fundamental challenge for the advancement of miRNA-based therapeutics. Up to now, aerosolization is the most traditional and valuable tool for passing small RNAs in the respiratory tract with a micro sprayer [126], [127]. This method presents the possibility of miRNA transmission for potential applications in RIs.
Concerning COVID-19, the viral glycoprotein called the S protein mediates viral entry to the target cells via binding to the ACE2 expressed on cell surface [128]. The S glycoprotein is activated through proteolytic cleavage by a host protease called furin [128]. ADAM17 (a disintegrin and metalloprotease 17 controls the ACE2 expression on the plasma membrane, promoting the protein shedding [128]. The Notch signaling elevates furin expression, while repressing ADAM17 via miRNA-145. Hence, blocking the initiation of Notch signaling pathway may serve as an approach to hinder the entry of virus into the cells by diminishing furin and increasing ADAM17 shedding (such as γ-secretase inhibitor) [128]. Besides, approaches that can increase the expression of ADAM17, such as the application of antagomir to miRNA-145, must be considered [128].
Commercializing a novel medicine is time-consuming and expensive owing to the administrative manners correlated to pre-clinical and clinical analyses. Notwithstanding, we believe that various molecules previously investigated and expanded after the former SARS-CoV-1 disease in 2002 can be used as a platform for developing active molecules for the current SARS-CoV-2 disease. One of the exciting facets of these molecules is the feasibility of changing the functionality of the viral replication machinery and disrupting the secondary construction of the central 5′UTR genomic area [129]. Major molecules reported in recent works are natural RNA molecules which have exhibited no side effects in in-vivo and can penetrate the cells without the need for transmission vehicles or transfecting tools. This is a beneficial characteristic as, usually, RNA molecules are not much alive per se. They demand a means to enter the cells; for example, mRNA-1273 is transported via lipid nanoparticles to COVID-19 patients (phase-I clinical trial) [130].
Additionally, antagomir has been characterized to act on miRNAs turnover [131]. Antagomir is a new type of engineered oligonucleotide, which can specifically silence the endogenous miRNAs [131]. In this regard, such approaches can act with a good efficacy on COVID-19 patients with severe conditions due to the CSS [132]. Antagomirs - easy to be produced in the laboratory, and cheaper than traditional biological approaches, especially when produced in a large scale - could be customized upon the immunological hallmarks of a given condition to block the activation of a range of compounds, effectively tailoring the whole cytokine storm to prevent the patient from having serious, life-threatening complications associated with the COVID-19. Taken together, antagomir can be used to specifically tailor the action of key miRNAs that contribute to the inflammatory process in COVID-19 patients, and can improve the clinical symptoms of these patients.
Using the administration of miRNAs, the armory that can operate to combat SARS-CoV-2 is encouraging and conceivably very efficacious. As postulated, a few of these curative possibilities could mitigate COVID-19 signs or diminish viral replication. We firmly propose that none of these opportunities should be dropped unexamined, as identifying an efficacious vaccine could be an extraordinarily long and unpredictable way.
5. Conclusion
COVID-19 pandemic is a newly discovered viral disease with a high mortality rate by which the human has been afflicted in the 21st century after the pandemic influenza outbreak of 1918. To date, there are no vaccines or the Food and Drug Administration (FDA)-approved drugs for the prevention or treatment of COVID-19. Nevertheless, much effort has been made for unraveling the immunopathogenesis of COVID-19 to find better therapeutic options to overcome this virus. However, given the paucity of effective therapeutic strategies, innovative approaches are warranted for prevention, diagnosis, and treatment of this infection. Host antiviral miRNAs contribute to the regulation of immune responses during viral infection. Countless numbers of human miRNAs appear to be capable of targeting viral genes and their functions, including interfering with replication, translation, and expression. The dual function of host cellular miRNAs cannot be underestimated in the viral replication or inhibition. Host miRNAs may have an antiviral role, which is useful by the host, or might promote viral replication and infection by miRNA-viral genome interaction. Finally, future investigations on the role of miRNAs in respiratory viral infections can open up a new horizon for the application of miRNAs to prevent, diagnose, and cure viral infections.
Funding
This work did not receive any particular grant from funding agencies in the commercial and public sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Hamadan University of Medical Sciences, Hamadan, Iran.
References
- 1.Srivastava N., Baxi P., Ratho R., Saxena S.K. Springer; 2020. Global Trends in Epidemiology of Coronavirus Disease 2019 (COVID-19), Coronavirus Disease 2019 (COVID-19) pp. 9–21. [Google Scholar]
- 2.T.M. Nguyen, Y. Zhang, P.P. Pandolfi, Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses, Nature Publishing Group, 2020. [DOI] [PMC free article] [PubMed]
- 3.Sardar T., Ghosh I., Rodó X., Chattopadhyay J. A realistic two-strain model for MERS-CoV infection uncovers the high risk for epidemic propagation. PLoS Neglected Trop. Dis. 2020;14(2):e0008065. doi: 10.1371/journal.pntd.0008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. Shirato, N. Nao, H. Katano, I. Takayama, S. Saito, F. Kato, H. Katoh, M. Sakata, Y. Nakatsu, Y. Mori, Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan, Japanese J. Infect. Dis. (2020) JJID. 2020.061. [DOI] [PubMed]
- 5.A. Ivashchenko, A. Rakhmetullina, D. Aisina, How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV, 2020.
- 6.Mirzaei R., Mohammadzadeh R., Mahdavi F., Badrzadeh F., Kazemi S., Ebrahimi M., Soltani F., Kazemi S., Jeda A.S., Darvishmotevalli M. Overview of the current promising approaches for the development of an effective severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine. Int. Immunopharmacol. 2020:106928. doi: 10.1016/j.intimp.2020.106928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodarzi P., Mahdavi F., Mirzaei R., Hasanvand H., Sholeh M., Zamani F., Sohrabi M., Tabibzadeh A., Jeda A.S., Niya M.H.K. Coronavirus disease 2019 (COVID-19): Immunological approaches and emerging pharmacologic treatments. Int. Immunopharmacol. 2020:106885. doi: 10.1016/j.intimp.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., Dashtbin S., Jalalifar S., Mohammadzadeh R., Teimoori A. Bacterial co-infections with SARS-CoV-2. IUBMB Life. 2020;72(10):2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.T.W.H. Organization, Coronavirus disease (COVID-19) pandemic, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019?gclid=CjwKCAiAnIT9BRAmEiwANaoE1VHPRA-7ZPlmOBOUFRDpjbqDqatunz5v4pFnSJ_j3F2khKTQtpMpQhoCriUQAvD_BwE.
- 13.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom., Proteom. Bioinform. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirzaei R., Mohammadzadeh R., Mirzaei H., Sholeh M., Karampoor S., Abdi M., Alikhani M.Y., Kazemi S., Ahmadyousefi Y., Jalalifar S. Role of microRNAs in Staphylococcus aureus infection: potential biomarkers and mechanism. IUBMB Life. 2020 doi: 10.1002/iub.2325. [DOI] [PubMed] [Google Scholar]
- 15.Raisch J., Darfeuille-Michaud A., Nguyen H.T.T. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol.: WJG. 2013;19(20):2985. doi: 10.3748/wjg.v19.i20.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Głobińska A., Pawełczyk M., Kowalski M.L. MicroRNAs and the immune response to respiratory virus infections. Exp. Rev. Clin. Immunol. 2014;10(7):963–971. doi: 10.1586/1744666X.2014.913482. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Host immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus Dis. 2019 (COVID-19) 2020:43–53. [Google Scholar]
- 18.Rasoul M., Rokhsareh M., Mohammad S.M., Sajad K., Ahmadreza M. The human immune system against Staphylococcus epidermidis. Critical Rev.™ Immunol. 2019;39(3) doi: 10.1615/CritRevImmunol.2019031282. [DOI] [PubMed] [Google Scholar]
- 19.Mirzaei R., Sadeghi J., Talebi M., Irajian G. Prevalence of atlE, ica, mecA, and mupA Genes in Staphylococcus epidermidis isolates. Infect. Dis. Clin. Practice. 2017;25(1):37–40. [Google Scholar]
- 20.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59(1–3):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L., Saxena S.K. Structural, glycosylation and antigenic variation between, 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020:1–9. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C.K.-f. Li, H. Wu, H. Yan, S. Ma, L. Wang, M. Zhang, X. Tang, N.J. Temperton, R.A. Weiss, J.M. Brenchley, T cell responses to whole SARS coronavirus in humans, J. Immunol. 181(8) (2008) 5490–5500. [DOI] [PMC free article] [PubMed]
- 23.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. New Engl. J. Med. 2003;349(5):508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 24.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S., Stokes K.L., Currier M.G., Sakamoto K., Lukacs N.W., Celis E., Moore M.L. Vaccine-elicited CD8+ T cells protect against respiratory syncytial virus strain A2-line19F-induced pathogenesis in BALB/c mice. J. Virol. 2012;86(23):13016–13024. doi: 10.1128/JVI.01770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuri T., Weber F. Interferon interplay helps tissue cells to cope with SARS-coronavirus infection. Virulence. 2010;1(4):273–275. doi: 10.4161/viru.1.4.11465. [DOI] [PubMed] [Google Scholar]
- 27.Coleman C.M., Sisk J.M., Halasz G., Zhong J., Beck S.E., Matthews K.L., Venkataraman T., Rajagopalan S., Kyratsous C.A., Frieman M.B. CD8+ T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J. Virol. 2017;91(1):e01825–e1916. doi: 10.1128/JVI.01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allegra A., Di Gioacchino M., Tonacci A., Musolino C., Gangemi S. Immunopathology of SARS-CoV-2 infection: immune cells and mediators, prognostic factors, and immune-therapeutic implications. Int. J. Mol. Sci. 2020;21(13):4782. doi: 10.3390/ijms21134782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirzaei R., Karampoor S., Sholeh M., Moradi P., Ranjbar R., Ghasemi F. A contemporary review on pathogenesis and immunity of COVID-19 infection. Mol. Biol. Rep. 2020;47(7):5365–5376. doi: 10.1007/s11033-020-05621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respirat. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song P., Li W., Xie J., Hou Y., You C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81(16):8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 38.Sha Q., Truong-Tran A.Q., Plitt J.R., Beck L.A., Schleimer R.P. Activation of airway epithelial cells by toll-like receptor agonists. Am. J. Respir. Cell Mol. Biol. 2004;31(3):358–364. doi: 10.1165/rcmb.2003-0388OC. [DOI] [PubMed] [Google Scholar]
- 39.Taganov K.D., Boldin M.P., Chang K.-J., Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonkoly E., Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J. Cell Mol. Med. 2009;13(1):24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tili E., Michaille J.-J., Calin G.A. Expression and function of micro RNAs in immune cells during normal or disease state. Int. J. Med. Sci. 2008;5(2):73. doi: 10.7150/ijms.5.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T., Garcia J.G., Zhang W. Integrating microRNAs into a system biology approach to acute lung injury. Translat. Res. 2011;157(4):180–190. doi: 10.1016/j.trsl.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nahid M.A., Pauley K.M., Satoh M., Chan E.K. miR-146a is critical for endotoxin-induced tolerance implication in innate immunity. J. Biol. Chem. 2009;284(50):34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oglesby I.K., McElvaney N.G., Greene C.M. MicroRNAs in inflammatory lung disease-master regulators or target practice? Respir. Res. 2010;11(1):148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu G., Friggeri A., Yang Y., Park Y.-J., Tsuruta Y., Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. 2009;106(37):15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slater L., Bartlett N.W., Haas J.J., Zhu J., Message S.D., Walton R.P., Sykes A., Dahdaleh S., Clarke D.L., Belvisi M.G. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouda R., Onomoto K., Takahasi K., Edwards M.R., Kato H., Yoneyama M., Fujita T. Retinoic acid-inducible gene I-inducible miR-23b inhibits infections by minor group rhinoviruses through down-regulation of the very low density lipoprotein receptor. J. Biol. Chem. 2011;286(29):26210–26219. doi: 10.1074/jbc.M111.229856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anglicheau D., Muthukumar T., Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation. 2010;90(2):105. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Sonkoly, M. Ståhle, A. Pivarcsi, MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation, in: Seminars in cancer biology, Elsevier, 2008, pp. 131–140. [DOI] [PubMed]
- 51.Li Q.-J., Chau J., Ebert P.J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Allantaz F., Cheng D.T., Bergauer T., Ravindran P., Rossier M.F., Ebeling M., Badi L., Reis B., Bitter H., D'Asaro M. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtale G., Citarella F., Carissimi C., Goldoni M., Carucci N., Fulci V., Franceschini D., Meloni F., Barnaba V., Macino G. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115(2):265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 54.Tsai C.-Y., Allie S.R., Zhang W., Usherwood E.J. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J. Virol. 2013;87(4):2348–2351. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lind E.F., Elford A.R., Ohashi P.S. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J. Immunol. 2013;190(3):1210–1216. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- 56.Leon-Icaza S.A., Zeng M., Rosas-Taraco A.G. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1(1):1. doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renwick N., Schweiger B., Kapoor V., Liu Z., Villari J., Bullmann R., Miething R., Briese T., Lipkin W.I. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 2007;196(12):1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossmann M.G., Arnold E., Erickson J.W., Frankenberger E.A., Griffith J.P., Hecht H.-J., Johnson J.E., Kamer G., Luo M., Mosser A.G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 59.Calvén J., Yudina Y., Uller L. Rhinovirus and dsRNA induce RIG-I-like receptors and expression of interferon β and λ1 in human bronchial smooth muscle cells. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0062718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dweep H., Sticht C., Gretz N. In-silico algorithms for the screening of possible microRNA binding sites and their interactions. Curr. Genom. 2013;14(2):127–136. doi: 10.2174/1389202911314020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Megremis S., Taka S., Oulas A., Kotoulas G., Iliopoulos I., Papadopoulos N.G. O20-Human rhinovirus replication-dependent induction of micro-RNAs in human bronchial epithelial cells. Clin. Translat. Allergy. 2014;4(1):O20. [Google Scholar]
- 62.Bondanese V.P., Francisco-Garcia A., Bedke N., Davies D.E., Sanchez-Elsner T. Identification of host miRNAs that may limit human rhinovirus replication. World J. Biol. Chem. 2014;5(4):437. doi: 10.4331/wjbc.v5.i4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bossert B., Conzelmann K.-K. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 2002;76(9):4287–4293. doi: 10.1128/JVI.76.9.4287-4293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P.L. Collins, G.W. Wertz, Human respiratory syncytial virus genome and gene products, Concepts in Viral Pathogenesis II, Springer, 1986, pp. 40–46.
- 65.Fuentes S., Tran K.C., Luthra P., Teng M.N., He B. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 2007;81(15):8361–8366. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005;352(17):1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 67.Glezen W.P., Taber L.H., Frank A.L., Kasel J.A. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 1986;140(6):543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 68.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89(4):422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 69.Rossi G.A., Silvestri M., Colin A.A. Respiratory syncytial virus infection of airway cells: role of microRNAs. Pediatr. Pulmonol. 2015;50(7):727–732. doi: 10.1002/ppul.23193. [DOI] [PubMed] [Google Scholar]
- 70.Inchley C.S., Sonerud T., Fjærli H.O., Nakstad B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect. Dis. 2015;15(1):150. doi: 10.1186/s12879-015-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thornburg N.J., Hayward S.L., Crowe J.E. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio. 2012;3(6):e00220–e312. doi: 10.1128/mBio.00220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakre A., Mitchell P., Coleman J.K., Jones L.P., Saavedra G., Teng M., Tompkins S.M., Tripp R.A. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 2012;93(Pt 11):2346. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prins J., Wolthers K., Kamphuisen P., Rosendaal F., Loffeld R., van der Putten A., Janssen M., Laheij R., Jansen J., de Boer W. Human metapneumovirus: a new pathogen in children and adults 177. Medicine (Baltimore) 2004;2(1):1.6. [PubMed] [Google Scholar]
- 74.Bhella D., Ralph A., Murphy L.B., Yeo R.P. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 2002;83(8):1831–1839. doi: 10.1099/0022-1317-83-8-1831. [DOI] [PubMed] [Google Scholar]
- 75.van den Hoogen B.G., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295(1):119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- 76.Gern J.E., Rosenthal L.A., Sorkness R.L., Lemanske R.F., Jr Effects of viral respiratory infections on lung development and childhood asthma. J. Allergy Clin. Immunol. 2005;115(4):668–674. doi: 10.1016/j.jaci.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez F.D. Heterogeneity of the association between lower respiratory illness in infancy and subsequent asthma. Proc. Am. Thoracic Soc. 2005;2(2):157–161. doi: 10.1513/pats.200504-044AW. [DOI] [PubMed] [Google Scholar]
- 78.Williams J.V., Harris P.A., Tollefson S.J., Halburnt-Rush L.L., Pingsterhaus J.M., Edwards K.M., Wright P.F., Crowe J.E., Jr Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 2004;350(5):443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng J., Ptashkin R.N., Wang Q., Liu G., Zhang G., Lee I., Lee Y.S., Bao X. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Mol. Therapy-Nucl. Acids. 2014;3 doi: 10.1038/mtna.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.W.C.o.t.S.W.H.O.C.o.C.A.o.H.I.w.A.I.A. Virus, Update on avian influenza A (H5N1) virus infection in humans, New Engl. J. Med. 358(3) (2008) 261–273. [DOI] [PubMed]
- 81.Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3(8):591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 82.García-García J., Ramos C. Influenza, an existing public health problem. Salud Publica Mex. 2006;48(3):244–267. doi: 10.1590/s0036-36342006000300009. [DOI] [PubMed] [Google Scholar]
- 83.Song L., Liu H., Gao S., Jiang W., Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010;84(17):8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Izzard L., Stambas J. Harnessing the power of miRNAs in influenza A virus research. Br. J. Virol. 2015;2:28. [Google Scholar]
- 85.Ma Y.J., Yang J., Fan X.L., Zhao H.B., Hu W., Li Z.P., Yu G.C., Ding X.R., Wang J.Z., Bo X.C. Cellular micro RNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell Mol. Med. 2012;16(10):2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakamura S., Horie M., Daidoji T., Honda T., Yasugi M., Kuno A., Komori T., Okuzaki D., Narimatsu H., Nakaya T. Influenza A virus-induced expression of a GalNAc transferase, GALNT3, via MicroRNAs is required for enhanced viral replication. J. Virol. 2016;90(4):1788–1801. doi: 10.1128/JVI.02246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gralinski L.E., Baric R.S. Molecular pathology of emerging coronavirus infections. J. Pathol. 2015;235(2):185–195. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tahamtan A., Inchley C.S., Marzban M., Tavakoli-Yaraki M., Teymoori-Rad M., Nakstad B., Salimi V. The role of microRNAs in respiratory viral infection: friend or foe? Rev. Med. Virol. 2016;26(6):389–407. doi: 10.1002/rmv.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren L., Zhang Y., Li J., Xiao Y., Zhang J., Wang Y., Chen L., Paranhos-Baccalà G., Wang J. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015;5:11451. doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lai F.W., Stephenson K.B., Mahony J., Lichty B.D. Human coronavirus OC43 nucleocapsid protein binds MicroRNA 9 and potentiates NF-κB activation. J. Virol. 2014;88(1):54–65. doi: 10.1128/JVI.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mallick B., Ghosh Z., Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE. 2009;4(11) doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banik G., Khandaker G., Rashid H. Middle East respiratory syndrome coronavirus “MERS-CoV”: current knowledge gaps. Paediatr. Respir. Rev. 2015;16(3):197–202. doi: 10.1016/j.prrv.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hasan M.M., Akter R., Ullah M., Abedin M., Ullah G., Hossain M. A computational approach for predicting role of human microRNAs in MERS-CoV genome. Adv. Bioinform. 2014;2014 doi: 10.1155/2014/967946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X., Chu H., Wen L., Shuai H., Yang D., Wang Y., Hou Y., Zhu Z., Yuan S., Yin F. Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation. Emerg. Microbes Infect. 2020;9(1):733–746. doi: 10.1080/22221751.2020.1738277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dickey L.L., Worne C.L., Glover J.L., Lane T.E., O’Connell R.M. MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J. Neuroinflam. 2016;13(1):240. doi: 10.1186/s12974-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lecellier C.-H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saïb A., Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308(5721):557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 97.Otsuka M., Jing Q., Georgel P., New L., Chen J., Mols J., Kang Y.J., Jiang Z., Du X., Cook R. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27(1):123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 98.Ahluwalia J.K., Khan S.Z., Soni K., Rawat P., Gupta A., Hariharan M., Scaria V., Lalwani M., Pillai B., Mitra D. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5(1):117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V., David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2):244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.M.A.-A.-K. Khan, M.R.U. Sany, M.S. Islam, M.S. Mehebub, A.B. Islam, Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2 and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19, bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 102.Islam M.S., Khan M.A.A.K., Murad M.W., Karim M., Islam A.B.M.M.K. In silico analysis revealed Zika virus miRNAs associated with viral pathogenesis through alteration of host genes involved in immune response and neurological functions. J. Med. Virol. 2019;91(9):1584–1594. doi: 10.1002/jmv.25505. [DOI] [PubMed] [Google Scholar]
- 103.Mishra R., Kumar A., Ingle H., Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.03079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.E. Wyler, K. Mösbauer, V. Franke, A. Diag, L.T. Gottula, R. Arsie, F. Klironomos, D. Koppstein, S. Ayoub, C. Buccitelli, Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention, bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 105.Badry A., Jaspers V.L., Waugh C.A. Environmental pollutants modulate RNA and DNA virus-activated miRNA-155 expression and innate immune system responses: insights into new immunomodulative mechanisms. J. Immunotoxicol. 2020;17(1):86–93. doi: 10.1080/1547691X.2020.1740838. [DOI] [PubMed] [Google Scholar]
- 106.Dickey L.L., Hanley T.M., Huffaker T.B., Ramstead A.G., O'Connell R.M., Lane T.E. MicroRNA 155 and viral-induced neuroinflammation. J. Neuroimmunol. 2017;308:17–24. doi: 10.1016/j.jneuroim.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gottwein E. Springer; 2013. Roles of microRNAs in the Life Cycles of Mammalian Viruses, Intrinsic Immunity; pp. 201–227. [DOI] [PubMed] [Google Scholar]
- 108.Mehta A., Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016;16(5):279. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 109.Thai T.-H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 110.Zhou H., Huang X., Cui H., Luo X., Tang Y., Chen S., Wu L., Shen N. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood, J. Am. Soc. Hematol. 2010;116(26):5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 111.Woods P.S., Doolittle L.M., Rosas L.E., Nana-Sinkam S.P., Tili E., Davis I.C. Increased expression of microRNA-155-5p by alveolar type II cells contributes to development of lethal ARDS in H1N1 influenza A virus-infected mice. Virology. 2020 doi: 10.1016/j.virol.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.A. Shiek, P. Paramasivam, K. Raj, V. Kumar, R. Murugesan, V. RamaKrishanan, Interplay of host regulatory network on SARS-CoV-2 binding and replication machinery, bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 113.R. Sardar, D. Satish, S. Birla, D. Gupta, Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis, bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 114.Z. Liu, J. Wang, Y. Xu, M. Guo, K. Mi, R. Xu, Y. Pei, Q. Zhang, X. Luan, Z. Hu, Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2, arXiv preprint arXiv:2004.04874 (2020).
- 115.Chow J.T.-S., Salmena L. Prediction and analysis of SARS-CoV-2-targeting microRNA in human lung epithelium. Genes. 2020;11(9):1002. doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.J. Sabbatinelli, A. Giuliani, G. Matacchione, S. Latini, N. Laprovitera, G. Pomponio, A. Ferrarini, S.S. Baroni, M. Pavani, M. Moretti, Decreased serum levels of inflammaging marker miR-146a are associated with clinical response to tocilizumab in COVID-19 patients, medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 117.Ames M.K., Atkins C.E., Pitt B. The renin-angiotensin-aldosterone system and its suppression. J. Vet. Intern. Med. 2019;33(2):363–382. doi: 10.1111/jvim.15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Novel C.P.E.R.E. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi. 2020;41(2):145. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 120.Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., Bär C., Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matarese A., Gambardella J., Sardu C., Santulli G. miR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. 2020;8(11):462. doi: 10.3390/biomedicines8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.A. Bouchie, First microRNA mimic enters clinic, Nature Publishing Group, 2013. [DOI] [PubMed]
- 123.Mobergslien A., Sioud M. Exosome-derived miRNAs and cellular miRNAs activate innate immunity. J. Innate Immun. 2014;6(1):105–110. doi: 10.1159/000351460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li J., Arévalo M.T., Diaz-Arévalo D., Chen Y., Choi J.-G., Zeng M. Generation of a safe and effective live viral vaccine by virus self-attenuation using species-specific artificial microRNA. J. Control. Release. 2015;207:70–76. doi: 10.1016/j.jconrel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou Z., Li X., Liu J., Dong L., Chen Q., Liu J., Kong H., Zhang Q., Qi X., Hou D. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25(1):39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rosas-Taraco A.G., Higgins D.M., Sánchez-Campillo J., Lee E.J., Orme I.M., González-Juarrero M. Intrapulmonary delivery of XCL1-targeting small interfering RNA in mice chronically infected with Mycobacterium tuberculosis. Am. J. Respir. Cell Mol. Biol. 2009;41(2):136–145. doi: 10.1165/rcmb.2008-0363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rosas-Taraco A.G., Higgins D.M., Sánchez-Campillo J., Lee E.J., Orme I.M., González-Juarrero M. Local pulmonary immunotherapy with siRNA targeting TGFβ1 enhances antimicrobial capacity in Mycobacterium tuberculosis infected mice. Tuberculosis. 2011;91(1):98–106. doi: 10.1016/j.tube.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rizzo P., Dalla Sega F.V., Fortini F., Marracino L., Rapezzi C., Ferrari R. COVID-19 in the heart and the lungs: could we “Notch” the inflammatory storm? Basic Res. Cardiol. 2020;115(3) doi: 10.1007/s00395-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.A. Baldassarre, A. Paolini, S.P. Bruno, C. Felli, A.E. Tozzi, A. Masotti, Non-Coding RNAs and Innovative Therapeutic Strategies to Target the 5’UTR of SARS-CoV-2 (2020). [DOI] [PMC free article] [PubMed]
- 130.ClinicalTrials.gov, Oxygen-Ozone as Adjuvant Treatment in Early Control of COVID-19 Progression and Modulation of the Gut Microbial Flora (PROBIOZOVID), 2020. https://clinicaltrials.gov/ct2/show/NCT04366089.
- 131.Murdaca G., Tonacci A., Negrini S., Greco M., Borro M., Puppo F., Gangemi S. Effects of antagomiRs on different lung diseases in human, cellular, and animal models. Int. J. Mol. Sci. 2019;20(16):3938. doi: 10.3390/ijms20163938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.S. Gangemi, A. Tonacci, AntagomiRs: A novel therapeutic strategy for challenging COVID-19 cytokine storm, Cytokine & growth factor reviews (2020). [DOI] [PMC free article] [PubMed]
- 133.Nakamura S., Horie M., Daidoji T., Honda T., Yasugi M., Kuno A., Komori T., Okuzaki D., Narimatsu H., Nakaya T., Tomonaga K. Influenza A virus-induced expression of a GalNAc transferase, GALNT3, via MicroRNAs is required for enhanced viral replication. J. Virol. 2016;90(4):1788–1801. doi: 10.1128/JVI.02246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ma Y.J., Yang J., Fan X.L., Zhao H.B., Hu W., Li Z.P., Yu G.C., Ding X.R., Wang J.Z., Bo X.C., Zheng X.F., Zhou Z., Wang S.Q. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell Mol. Med. 2012;16(10):2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng J., Ptashkin R.N., Wang Q., Liu G., Zhang G., Lee I., Lee Y.S., Bao X. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Mol. Ther. Nucleic Acids. 2014;3(5) doi: 10.1038/mtna.2014.18. e163–e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baños-Lara M.D.R., Zabaleta J., Garai J., Baddoo M., Guerrero-Plata A. Comparative analysis of miRNA profile in human dendritic cells infected with respiratory syncytial virus and human metapneumovirus. BMC Res. Notes. 2018;11(1):432. doi: 10.1186/s13104-018-3541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu W., Choi E.-J., Lee I., Lee Y.S., Bao X. Non-coding RNAs and their role in Respiratory Syncytial Virus (RSV) and Human Metapneumovirus (hMPV) infections. Viruses. 2020;12(3):345. doi: 10.3390/v12030345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Othumpangat S., Walton C., Piedimonte G. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0030030. e30030–e30030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lánczky A., Nagy Á., Bottai G., Munkácsy G., Szabó A., Santarpia L., Győrffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016;160(3):439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 140.Sun Y.-M., Lin K.-Y., Chen Y.-Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013;6(1):6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kriegel A.J., Liu Y., Fang Y., Ding X., Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genom. 2012;44(4):237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang H.F., Wang W.H., Zhuang H.W., Xu M. MiR-429 regulates the proliferation and apoptosis of nephroblastoma cells through targeting c-myc. Eur. Rev. Med. Pharmacol. Sci. 2018;22(16):5172–5179. doi: 10.26355/eurrev_201808_15713. [DOI] [PubMed] [Google Scholar]
- 143.Fernández-Hernando C., Suárez Y., Rayner K.J., Moore K.J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 2011;22(2):86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lin Q., Gao Z., Alarcon R.M., Ye J., Yun Z. A role of miR-27 in the regulation of adipogenesis. The FEBS J. 2009;276(8):2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou R., Gong A.-Y., Eischeid A.N., Chen X.-M. miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PLoS Pathog. 2012;8(5) doi: 10.1371/journal.ppat.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Forster S.C., Tate M.D., Hertzog P.J. MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Front. Immunol. 2015;6:334. doi: 10.3389/fimmu.2015.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.O'Day E., Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12(2) doi: 10.1186/bcr2484. 201–201. [DOI] [PMC free article] [PubMed] [Google Scholar]