Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped virus which binds its cellular receptor angiotensin-converting enzyme 2 (ACE2) and enters hosts cells through the action of its spike (S) glycoprotein displayed on the surface of the virion. Compared to the reference strain of SARS-CoV-2, the majority of currently circulating isolates possess an S protein variant characterized by an aspartic acid-to-glycine substitution at amino acid position 614 (D614G). Residue 614 lies outside the receptor binding domain (RBD) and the mutation does not alter the affinity of monomeric S protein for ACE2. However, S(G614), compared to S(D614), mediates more efficient ACE2-mediated transduction of cells by S-pseudotyped vectors and more efficient infection of cells and animals by live SARS-CoV-2. This review summarizes and synthesizes the epidemiological and functional observations of the D614G spike mutation, with focus on the biochemical and cell-biological impact of this mutation and its consequences for S protein function. We further discuss the significance of these recent findings in the context of the current global pandemic.
Keywords: SARS-CoV-2, COVID-19, Spike protein
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the COVID-19 pandemic. SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA virus, named for its genetic similarity to SARS-CoV (referred to as SARS-CoV-1 herein for clarity) which emerged in 2003. SARS-CoV-1 and SARS-CoV-2 are both members of the genus Betacoronavirus, a classification also shared with human coronavirus (HCoV)-OC43, HCoV-HKU1, and Middle East respiratory syndrome-related coronavirus (MERS-CoV). Among these human betacoronaviruses, only the sarbecoviruses SARS-CoV-1 and SARS-CoV-2, which share 79% nucleotide sequence identity [1], depend on angiotensin-converting enzyme 2 (ACE2) for target-cell entry [[2], [3], [4], [5], [6]], which we identified in 2003 as a receptor for SARS-CoV-1 [7]. Interestingly, the distantly related human alphacoronavirus NL-63 also utilizes ACE2 as its primary receptor [8]. The spike (S) glycoprotein, which is necessary for receptor binding and cellular entry, is displayed on the virion in the form of trimers. These S protein trimers give rise to the viruses’ coronated appearance by electron microscopy [9]. The S protein of SARS-CoV-1 and SARS-CoV-2 are both comprised of an N-terminal S1 subunit responsible for binding to the ACE2 receptor and a C-terminal membrane-spanning S2 subunit which is necessary for cellular fusion [10]. The receptor-binding domain (RBD) within S1 interacts with ACE2 when the virus engages a target cell [[11], [12], [13], [14]]. During SARS-CoV-1 entry, following interaction of S1 with ACE2, a cleavage site at the S1/S2 boundary is processed by proteases such as TMPRSS2 on the surface of the target cell or by lysosomal cathepsins to facilitate membrane fusion activity [[15], [16], [17], [18], [19], [20]]. In contrast to SARS-CoV-1 and all other described animal sarbecoviruses, SARS-CoV-2 S protein has acquired a multibasic cleavage site at the junction of the S1 and S2 subunits which is processed by furin-like proprotein convertases in the virus-producer cell [21]. Thus, like the entry proteins of HIV-1 and some influenza A viruses, the S protein is cleaved before encountering target cells. Therefore, intermolecular association between S1 and S2 is necessary to retain S1 on the virion. In both SARS-CoV-1 and SARS-CoV-2, a second cleavage at a site internal to S2, yielding a fragment known as S2’, is also necessary for membrane fusion with the target cell [[21], [22], [23]].
Due to the clear importance of S protein in the entry process, a number of tools have been developed to closely study its function. Pseudovirus (PV) systems based on lentiviral, gamma-retroviral, or vesicular stomatitis virus (VSV) vectors which are coated with the CoV S protein have facilitated rapid advances in understanding the entry of SARS-CoV-2. PVs may be pseudotyped with full-length S protein, although C-terminal truncation has often been observed to improve efficiency of incorporation of SARS-CoV-1 and SARS-CoV-2 S protein into the pseudovirion [[24], [25], [26]]. Substitution of non-native signal sequences to enhance pseudotyping has also been employed by some groups based on observations with other viral entry proteins, although a recent side-by-side comparison in SARS-CoV-2 S protein did not observe a difference between native and optimized signal peptide [27]. In general, experimental evidence reveals that SARS-CoV-2 S-mediated entry into ACE2-expressing cells is less efficient than that mediated by SARS-CoV-1 S and is more highly dependent on TMPRSS2 [28,29]. This is consistent with lower ACE2 binding affinity of SARS-CoV-2 compared to SARS-CoV-1 spike trimers, although the RBD of SARS-CoV-2 S in particular has a higher affinity for ACE2 [14].
Coronaviruses and other members of the order Nidovirales maintain the largest and most complex genomes known among RNA viruses and therefore coronavirus replication makes use of exonuclease proofreading activity of nsp14 to avoid accumulation of deleterious mutations [30]. Despite this uniquely high fidelity of replication, a number of mutations to the SARS-CoV-2 genome have been observed throughout the present course of the COVID-19 pandemic, including in the S gene. Indeed, analysis of 3090 isolates in June 2020 found that the number of unique alleles of S was greater than the other structural genes M, N, and E, suggesting that changes in S may provide an evolutionary benefit [31]. One non-synonymous mutation of S gene in particular, an aspartic acid-to-glycine substitution at amino acid position 614 (D614G), has attracted immense attention following reports of its enrichment discovered by epidemiological surveillance [32]. Initial responses called into question the functional relevance of this change and whether its predominance is due to a fitness advantage or founder effect [[32], [33], [34]]. Further work has demonstrated a clear phenotypic advantage conferred by D614G, including our own observation of enhanced infectivity by PVs pseudotyped with S(G614) versus S(D614) [35]. A consensus on the exact mechanism of this enhancement has not yet been attained. However, the picture has become increasingly clear with continued investigation in this area. This review aims to assemble the work to date on the D614G mutation, including published studies as well as unreviewed manuscripts available in pre-print.
2. S(G614) genotype of SARS-CoV-2 has supplanted S(D614) globally as the predominant form
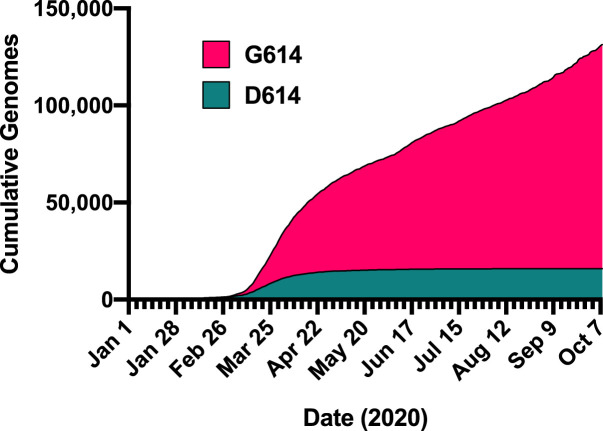
Initial attention to the D614G mutation was prompted by a preprint from Korber et al. using an “early warning” bioinformatic pipeline. SARS-CoV-2 bearing the S(G614) genotype was first detected in late January 2020 and began emerging in March 2020 as it steadily increased in global incidence to become the predominant form relative to the S(D614) virus (Fig. 1 ) [32]. The investigators also noted approximately 3-fold higher viral load in patients infected with S(G614) virus compared to those infected with S(D614) virus, a finding which was reproduced independently in a different patient population [36]. In the initial absence of clear functional evidence for a phenotype of S(G614), independent epidemiological analyses using other methodologies concluded that the D614G mutation did not confer a transmissibility advantage [33]. As attention to this mutation increased, however, a multitude of studies have been reported which supported the initial assertions of Korber and colleagues. Analysis of infection data from European countries, where the S(G614) genotype is dominant, estimated the doubling time at approximately 3 days [37], significantly shorter than the 6 day doubling time of the initial outbreak in China [38]. Further, a small but significant positive correlation was reported between the prevalence of S(G614) in a given geographic area and case-fatality rate in that region [39]. Another group observed a similar trend, although not statistically significant in their dataset [40]. Moreover, computational models of evolutionary dynamics suggest that D614G is under strong selective pressure [31]. Further, quantitative estimates of the basic reproductive number R0 from infection incidence data suggest that viruses bearing S(G614) are 31% more transmissible than those bearing S(D614), although this analysis did not consider the effect of concomitant changes at other loci in the circulating viral strains [41]. Various studies have identified co-segregation of changes in other viral genes along with the D614G mutation in spike, such as the P323L mutation in the viral polymerase RdRp which may attenuate replication to compensate for the increased transmissibility of D614G viruses [40,42,43].
Fig. 1.
Cumulative reported SARS-CoV-2 sequences harboring the D614 or G614 S gene. Quantities for the indicated dates were retrieved from the COVID-19 Viral Genome Analysis Pipeline hosted by the Los Alamos National Laboratory (https://cov.lanl.gov/content/sequence/TRACK_MUT/trackmut.html) which utilizes the data of the Global Initiative on Sharing All Influenza Data (GSAID).
3. The D614G mutation enhances the infectivity of pseudoviruses and live SARS-CoV-2
Enhanced entry by PVs displaying the S(G614) has now been demonstrated in a number of different model systems, ranging from roughly 2.5- to 45-fold over S(D614) depending on experimental conditions including target cell type, pseudotyping system, and infection procedure. At equal copy numbers, GFP reporter-expressing Maloney murine leukemia virus (MLV) pseudotyped with S(G614) versus S(D614) resulted in significantly higher transduction of ACE2-overexpressing HEK293T cells with or without TMPRSS2 overexpression, as well as ACE2-overexpressing NCI–H1975 human lung epithelial cells [35]. Enhanced transduction by lentiviral PVs carrying S(G614) has also been observed in ACE2-overexpressing or ACE2- and TMPRSS2-overexpressing HEK293T cells [25,28,32,39,40,44,45], ACE2-overexpressing A549 human lung epithelial cells [39], ACE2-overexpressing Huh7.5 human hepatocytes [39], Caco-2 human colon epithelial cells [39,44], and Calu-3 human lung epithelial cells [44]. Similarly, VSV PVs pseudotyped with S(G614) more efficiently infect Vero cells, ACE2-overexpressing HEK293T cells, and ACE2- and TMPRSS2-overexpressing HEK293T cells [32,45]. The effect of the D614G mutation in virus infectivity was durable in the presence or absence of TMPRSS2 overexpression and regardless of the modification of the S protein cytoplasmic domain, namely, full-length or C-terminally truncated S protein [35,40,45]. Work by Michaud and colleagues further augmented observations on D614G with quantitatively focused approaches. Using a doxycycline-inducible ACE2-expressing HEK293T cell line, investigators observed that S(G614) lentiviral PVs mediated more efficient transduction than S(D614) PVs over a wide range of ACE2 expression levels, which they argue are more representative of physiologically relevant cell types than stable cells that abundantly express the receptor [25]. The same group also employed a two-color reporter system to observe that GFP-expressing S(G614) PVs outcompete RFP-expressing S(D614) PVs in ACE2-overexpressing HEK293T cells. Cotransduction over a range of S(D614) to S(G614) PV ratios consistently resulted in a greater proportion of GFP-expressing cells relative to RFP expressing cells, compared to the control in which both RFP- and GFP- expressing vectors were pseudotyped with S(D614) [25].
Initial observations of the D614G effect from PV systems have now been borne out with live SARS-CoV-2 in cell culture and in animal models. Daniloski et al. developed a trans-complementation assay, wherein HEK293T cells were transfected with ACE2 and either S(D614) or S(G614), then infected with SARS-CoV-2 bearing S(D614). The investigators observed significantly increased viral replication in the cells transfected with S(G614), likely because progeny virus produced in those cells incorporated the exogenously provided S(G614) [39]. Mok et al. compared the infectivity in hamsters of three live SARS-CoV-2 isolates, two with S(D614) (HK-8 and HK-13) and one with S(G614) (HK-95). Significantly higher viral titers were observed in the lungs and nasal turbinates of naïve hamsters co-housed with HK-95-infected hamsters than in those co-housed with HK-8- or HK-13-infected hamsters [46]. The findings support the hypothesis that the D614G variant of S protein promotes transmissibility of the virus in mammals. However, one must consider that the viruses used in the study are not isogenic and thus other genetic differences may have contributed to the observed infectivity. Addressing this limitation, Plante et al. utilized an infectious cDNA clone to produce isogenic SARS-CoV-2 bearing either S(D614) or S(G614) [47]. The investigators observed increased replication of G614 virus upon infection of Calu-3 human lung epithelial cells [47]. This effect was also demonstrated in primary human airway cultures. Further, a competition experiment produced similar results to those seen in a PV system; G614 virus predominated in primary human airway cells even when co-inoculated with a 9-fold excess of D614 virus [47]. Infection of Syrian hamsters with the G614 virus also resulted in significantly increased replication in nasal tissues, as well as relatively more infectious (by PFU to RNA ratio) virus in nasal, tracheal, and lung tissues, although only a handful of conditions and timepoints attained statistical significance [47].
Hou et al. utilized an independently developed reverse genetics system to generate isogenic SARS-CoV-2 clones encoding a luciferase reporter and either S(D614) or S(G614). Consistent with the findings by Plante et al., enhancement of infection by S(G614) was observed in Vero-E6 cells, Huh7 cells, and ACE2-overexpressing A549 cells [48]. Likewise, in primary large airway epithelial cells infected with both D614 and G614 virus, the G614 virus became dominant after three passages even when the starting inoculum contained a ten-fold excess of D614 virus [48]. The same investigators also observed that infection of primary human nasal epithelial cells and large airway epithelial cells infected with G614 SARS-CoV-2 resulted in significantly higher titers than those infected with an equal amount of isogenic D614 virus [48]. This effect was not seen, however, in small airway epithelial cells. With the same isogenic viruses, investigators infected Syrian hamsters and observed a modestly greater weight loss in G614 virus-infected hamsters compared to D614, although viral titers in lung and nasal tissues were similar. Importantly, although direct inoculation did not reveal an appreciable effect of the D614G mutation, the G614 virus was remarkably more transmissible from infected hamsters to naïve hamsters. After 2 days of exposure, 5 of 8 hamsters exposed to G614 virus-infected hamsters had detectable infection and virus shedding, while 0 of 8 hamsters exposed to D614 virus-infected hamsters had detectable infection at the same timepoint [48].
4. Molecular mechanisms underlying the D614G effect are incompletely understood
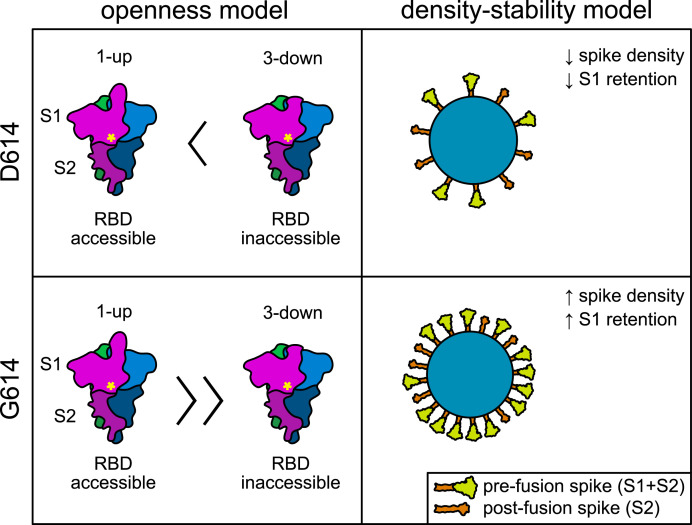
Extensive efforts have been undertaken to ascertain how the D614G mutation increases infectivity of SARS-CoV-2 and S-pseudotyped vectors. The hypotheses which have been discussed in the literature may be distilled into several central ideas (Fig. 2 ): the D614G mutation (a) modulates cleavage efficiency of S protein, (b) promotes a conformation favorable for RBD-ACE2 interaction (“openness” hypothesis), (c) facilitates more efficient S protein incorporation into the virion (“density” hypothesis), and (d) stabilizes the association of prefusion spike trimers (“stability” hypothesis). It is noteworthy that the D614G mutation does not modulate the S protein binding affinity for ACE2; independent studies have found that monomers of S(D614) and S(G614) have similar affinity for ACE2 as measured by surface plasmon resonance [35] or bio-layer interferometry [39]. While others reported affinity changes [44,49], caution should be taken to interpret the data, depending on the nature of the S protein used in the study. If soluble spike trimers are used, ACE2 binding is determined not only by affinity but also by S1 shedding. On the other hand, if soluble spike trimers containing a furin-null mutation is used, although this mutation addresses the S1-shedding problem, the difference between D614 and G614 may no longer be observed.
Fig. 2.
Schematic comparison of two independent models explaining increased infectivity by SARS-CoV-2 isolates possessing D614G mutant spike. On the left, the “openness” model explains increased infectivity by an increased propensity of S(G614) spike to assume the 1-up conformation thought to be necessary for ACE2 interaction with the RBD. Magenta, blue, and green colors represent individual S protein monomers. Yellow asterisk indicates position of residue 614 near the S1/S2 interface. On the right, the “density-stability” model explains increased infectivity by increased stability of S(G614) spike trimers, facilitating greater incorporation into virions and less shedding of the S1 subunit. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4.1. Modulating S protein cleavage
Using in silico predictions, Bhattacharyya et al. proposed that the D614G substitution introduces a cleavage site for elastase [50]. Another group used an independent software tool for prediction of protease cleavage sites and identified the same novel cleavage site introduced by the D614G substitution [51]. In that study, a cell lysate Western blot of HEK293T cells overexpressing S protein showed stronger S1 versus full-length S signal in cells transfected with S(G614) than in those transfected with S(D614), and further observed that the elastase inhibitor sivelestat significantly decreased S1 band intensity [51]. However, sivelestat also appeared to slightly decrease S1 intensity in the context of S(D614), and one must note that observations of spike in cell lysate my not be representative of that of mature virions. An independent group further observed that protease cleavage at the furin site is more efficient in S(G614) compared to S(D614) [52], which is supported by computational predictions of furin affinity for the two variants [53]. On the other hand, Daniloski et al. observed that S(G614) compared to S(D614) was more resistant to cleavage as determined by amount of uncleaved S in cell lysates and upon digestion with furin in vitro [39].
4.2. Promoting open RBD conformation
By performing molecular dynamics simulations, Mansbach and colleagues predicted that the D614G mutation effects changes in intra-protomer energetics which favor a “one-up” conformation of the spike trimer – one of three protomers in open conformation. This “one-up” conformation is considered the infection-capable state, because spike trimers in all-closed conformation do not expose RBD to ACE2, and conversely two or three simultaneously open protomers are unstable [54]. By applying global differential contact analysis, they further observe that contacts in CT1-CT2 (528-685) and FP-FPR (816-911) regions are major contributors to this effect, particularly the abolishment of the D614-T859 hydrogen bond [54]. These interactions are implicated by other groups as well [28,32,55,56]. Independent molecular dynamics simulations also support the hypothesis that the S(G614) variant favors an open conformation, specifically suggesting that the D614G mutation disrupts the formation of an inter-protomer D614-K835 salt bridge in the closed conformation [40]. Experimental data in support of these claims have been reported in the evaluation of purified spike trimers by cryo-electron microscopy (EM) [44] as well as negative-stain EM [57]. With both methodologies, investigators observed that the open states were more heavily populated in S(G614) spikes whereas significantly greater proportion of S(D614) spikes were observed in the “three-down” conformation thought to be unfavorable for RBD-ACE2 interaction. A significant difference between the two studies is that in negative-stain EM data from Weissman et al., 84% of G614 spikes were in “one-up” conformation and the remaining spikes were observed in “three-down” conformation [57], while in contrast, cryo-EM data on spike timers reported by Yurkovetskiy et al. exhibit “two-up” and “three-up” conformations of S(G614) spikes, exceeding the limited proportion of “one-up” spikes [44]. A limitation of both studies is that the S protein constructs had furin-null mutations and a di-proline mutation of residues 986-987 to stabilize the pre-fusion conformation [44,57]. Therefore, the analyzed macromolecular structures were uncleaved and effects of D614G on intermolecular association between S1 and S2 of processed S protein trimers could not be ascertained. Gobeil et al. studied the spike ectodomain structure with the native K986 and V987 residues while maintaining the furin-null mutation, and found that D614 spike trimers had similar melting temperature with or without the di-proline mutation [52]. Cryo-EM structures of S(D614) and S(G614) spike lacking the di-proline mutation did not identify any populations in “two-up” or “three-up” conformation, while they do show an enrichment of the “one-up” conformation in S(G614) [52].
4.3. Facilitating higher S protein incorporation into the virion
In our own study, we produced virus-like particles (VLPs) comprised of SARS-CoV-2 nucleoprotein (N), membrane protein (M), envelope protein (E), and S(D614) or S(G614). By Western blot, we found that VLPs with S(G614) had significantly higher density of incorporated S protein than S(D614) VLPs, when same number of VLP particles were analyzed and their quantities confirmed by N protein band intensity [35]. We also observed higher incorporation of S(G614) compared to S(D614) into MLV PVs. On the contrary, several studies observed no effect of the D614G mutation on spike density, using lentiviral PV or live SARS-CoV-2 [44,47,48]. Cytotoxicity can explain, at least partially, this apparent discrepancy. Significant cytotoxicity is associated with the production of lentiviral and VSV PVs, as well as live SARS-CoV-2 infection. In contrast, we found that MLV PV production exhibits no cytotoxicity at all, while SARS-CoV-2 VLP production, particularly expression of M protein, is toxic [35]. In the case of these cytotoxic systems, S protein released from lysed cells may have masked any difference in S protein density on the virion. One caveat is that visualization of isogenic D614 and G614 variant SARS-CoV-2 by scanning EM and transmission EM did not reveal significant differences in spike density [48], which cannot be explained by the masking effect of contaminating S protein. Because many studies have observed deletion of the furin cleavage site in live SARS-CoV-2 in as few as two to three passages in Vero cells [[58], [59], [60], [61]], care must be taken to ensure that studies of live viruses are not impacted by artifacts of this adaptation. In line with our own observations on VLPs and MLV PVs, Turonova et al. visualized live SARS-CoV-2 G614 virions by cryo-EM and observed much greater spike density than was reported in prior studies with D614 viruses [61,62]. Consistently, increased spike density associated with the D614G mutation was reproduced by Michaud et al. in a lentiviral PV system [25]. The investigators also observed that S(G614), when co-expressed with S(D614), is preferentially incorporated into pseudovirions, and that chimeric spike trimers on pseduovirions contain more S(G614) protomers than S(D614) protomers [25]. One explanation for increased spike density on the G614 virion is a greater propensity of the S(G614) monomer to form stable trimers, a requisite for virion incorporation.
4.4. Enhancing S protein stability
The stability of S(G614) likely underlies observations that the D614G mutation decreases S1 shedding. In our own study with both the PV and VLP experimental models, we observed stronger retention of the S1 subunit on the S(G614) compared to S(D614) virion [35]. Stronger intermolecular association of S1 with S2 is supported by biophysical analyses of Fernández, implicating stabilized S1–S2 interface in S(G614) [63]. Further, it is likely that S1 shedding is a limiting factor in the infectivity of S(D614) SARS-CoV-2 as an early cryo-EM study of live virus observed a major population of spikes in the post-fusion state which lacks S1 [62]. Several independent studies have resulted in experimental data supporting this hypothesis. Nguyen et al. expressed S protein alone in HEK293T cells and found significantly lower shedding of S1 into cell culture media by S(G614), while processing into S1 and S2 of S(G614) was only slightly higher than that of S(D614) [45]. Zhang et al. also made similar observations; full-length S protein was purified and analyzed by size exclusion chromatography (SEC), and the investigators found that while S(D614) eluted in three peaks representing the prefusion S trimer, postfusion S2 trimer, and dissociated S1 subunit, S(G614) eluted in only one major peak comprised 90% of prefusion S trimer [49]. Similar SEC profiles for S(D614) versus S(G614) were obtained by Gobeil et al., confirming greater S1 retention in S(G614) [52]. Additionally, Zhang et al. provide a mechanistic explanation for reduced shedding in their interpretation of cryo-EM structures of S protein trimers. They explain that in S(G614) trimers, a ∼20-residue segment in the CTD2 was ordered, whereas the same region was largely disordered in S(D614) trimers [49]. The authors posit that this structured loop in S(G614) stabilizes packing of the trimer by tucking between the NTD and CTD1, as well as sealing a hydrophobic surface of the CTD2, which in turn blocks S1 dissociation [49].
5. G614 virus is as sensitive to neutralization as D614 virus
One aspect of the D614G mutation which has been investigated more extensively than the mechanism of increased infectivity is its impact on vaccines and antibodies. Early discussions raised concern of possible antigenic drift, suggesting that vaccines targeting S(D614) may offer limited protection against viruses with S(G614) and that antibodies in exposed individuals would not offer cross-protection [64]. Indeed, though reinfections have been reported with two independent isolates harboring the same S protein variant [65], recent case reports identified first infection events with D614 virus followed by reinfection with G614 virus [66,67]. However, many studies have shown that humoral immunity is likely protective against both variants. Yuan and Li employed in silico prediction of 12 linear and 53 discontinuous B-cell epitopes from the S protein and found that the D614G mutation had a negligible impact on predicted epitopes [68]. Experimental data pertaining to neutralization of S(G614) and S(D614) pseudoviruses have been reported in abundance. The majority of experiments demonstrate similar neutralization of both S(D614) and S(G614) PVs by antibodies or patient antisera directed against one or the other S protein variant [28,35,51,[69], [70], [71], [72], [73], [74], [75], [76], [77], [78]], while in some cases S(G614) exhibited greater susceptibility to neutralization [51,57]. Similar neutralization or greater susceptibility of the S(G614) variant have been reproduced with live SARS-CoV-2 as well [47,48]. Several candidate vaccines have been evaluated in mouse, monkey, and ferret models and elicited antibodies with comparable neutralizing activity against S(D614) and S(G614) [[79], [80], [81]]. As underscored by these experimental data, single-residue mutations will not likely change viral sensitivity to neutralization unless they grossly alter S protein conformation.
6. Discussion
Current literature makes it clear that the D614G variant in SARS-CoV-2 S protein significantly enhances infectivity and transmissibility of the virus. In-depth functional virological studies confirm the effect of this change, and scrutiny of epidemiological data reveals that the proliferation of this genotype cannot be simply explained by founder effect. The D614 variant was well established in several regions before being supplanted by isolates harboring G614 [32], and data analyses support the assertion that predominance of G614 is due to selection [82]. However, the exact molecular mechanisms by which this mutation enhances infectivity are still incompletely understood. For example, data indicating S(G614) has equal or lower affinity for ACE2 than S(D614) are at odds with conclusions that S(G614) promotes a conformation favoring interaction with ACE2. Obtaining a clear conclusion on this issue suffers from technical difficulties to generate spike trimers without introducing artificial trimerization motifs. It also suffers from the complexity of data interpretation, owing to the furin-null mutation introduced to stabilize spike trimers, which prevents natural S1 shedding. On the other hand, stabilization of intermolecular interactions between S1 and S2, which leads to decreased S1 shedding and more efficient spike incorporation onto virions, can explain the enhanced infectivity of D614G viruses by an avidity effect. Ultimately, the field has not reached a working model which is consistent with the entirety of experimental data available at this time. However, the rapid proliferation of research on this topic which has resulted in an enormous amount of data is a testament to the crucial role of basic molecular virology in responding to this unprecedented pandemic. In addition to an incomplete understanding of the molecular mechanism by which the D614G mutation exerts its effect, the full health impacts of this mutation are also not yet clear. For example, Butowt et al. speculate that the concurrent emergence of S(G614) SARS-CoV-2 and clinical observations of anosmia are functionally linked [83]. The clinical and epidemiological data reviewed above suggest that although the D614G mutation confers an advantage in transmissibility, its effect on disease severity is uncertain, partially because other mutations frequently co-occur.
SARS-CoV-2 acquired a furin cleavage site not found in SARS-CoV-1 or any other known sarbecovirus. Studies have shown that acquisition of this furin cleavage site could be beneficial to the virus, because mutations at this site decrease viral infectivity [4,14,21,58], but in other contexts or other mutations of the furin cleavage site, infectivity is increased [4,35,58,59]. We currently do not know whether viruses without the furin cleavage site would replicate more efficiently in vivo. What we know is that none of the viruses isolated from the presumed SARS-CoV-2 reservoir species has the furin cleavage site and that its acquisition may have been recent, perhaps close to the timeframe of zoonosis. It is possible, then, that acquiring the furin cleavage site is critical to human infection or transmission, a possibility supported by the continued retention of this site during the pandemic. However, the furin cleavage site may have made the S protein less stable and therefore the virus may have acquired the D614G mutation to compensate for this instability. Regardless, the predominance of D614G indicates that it, like the furin cleavage site, is beneficial to the virus. During the current pandemic, there has so far been only one mutation in the S protein, D614G, which confers an apparent fitness advantage to the virus. It is not certain whether additional similar mutations will emerge in a prolonged pandemic, but there are reasons to be hopeful. First, there does not appear to be major selective pressure on the S protein from the antibody response, likely because the virus moves to a new host more quickly than a neutralizing antibody response can develop. Indeed, the single S protein mutation that did emerge, D614G, is buried within the S protein and does not have a significant impact on any critical antibody epitope. Second, SARS-CoV-2 currently occupies an excellent evolutionary niche, as demonstrated by its ability to sustain a global pandemic. The acute selective pressure of S protein instability, which has been an Achilles’ heel of the virus, was resolved by the D614G mutation. There may not be any further selective pressure similar to that posed by S protein instability.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by an administrative supplement to NIH R01 AI129868 for coronavirus research (to M.F. and H.C.).
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/JVI.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 14.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Hofmann-Winkler H., Pöhlmann S. In: Activation of Viruses by Host Proteases. Böttcher-Friebertshäuser E., Garten W., Klenk H.D., editors. Springer International Publishing; Cham: 2018. Priming time: how cellular proteases arm coronavirus spike proteins; pp. 71–98. [Google Scholar]
- 23.Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukushi S., Mizutani T., Saijo M., Matsuyama S., Miyajima N., Taguchi F., Itamura S., Kurane I., Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86:2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- 25.Michaud W.A., Boland G.M., Rabi S.A. The SARS-CoV-2 Spike mutation D614G increases entry fitness across a range of ACE2 levels, directly outcompetes the wild type, and is preferentially incorporated into trimers. bioRxiv. 2020 doi: 10.1101/2020.08.25.267500. 2020.2008.2025.267500. [DOI] [Google Scholar]
- 26.Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized pseudotyping conditions for the SARS-COV-2 spike glycoprotein. J. Virol. 2020;94 doi: 10.1128/jvi.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozono S., Zhang Y., Ode H., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. bioRxiv. 2020:2020. doi: 10.1101/2020.06.15.151779. 2006.2015.151779. [DOI] [Google Scholar]
- 29.Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. bioRxiv. 2020 doi: 10.1101/2020.07.22.216150. 2020.2007.2022.216150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan X.-Y., Zhang Y., Zhou X., Huang K., Qian Y., Leng Y., Yan L., Huang B., He Y. Molecular evolution of SARS-CoV-2 structural genes: evidence of positive selection in spike glycoprotein. bioRxiv. 2020:2020. doi: 10.1101/2020.06.25.170688. 2006.2025.170688. [DOI] [Google Scholar]
- 32.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. bioRxiv. 2020:2020. doi: 10.1101/2020.05.21.108506. 2005.2021.108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dearlove B., Lewitus E., Bai H., Li Y., Reeves D.B., Joyce M.G., Scott P.T., Amare M.F., Vasan S., Michael N.L., Modjarrad K., Rolland M. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. 2020.2006.2012.148726. Updated manuscript in press - Nat Commun. [DOI] [Google Scholar]
- 36.Wagner C., Roychoudhury P., Frazar C.D., Lee J., Müller N.F., Moncla L.H., Hadfield J., Hodcroft E.B., Pelle B., Richardson M., Behrens C., Huang M.-L., Mathias P., Pepper G., Shrestha L., Xie H., Addetia A., Nguyen T., Rachleff V.M., Gautom R., Melly G., Hiatt B., Dykema P., Adler A., Brandstetter E., Han P.D., Fay K., Ilcisin M., Lacombe K., Sibley T.R., Truong M., Wolf C.R., Cowgill K., Schrag S., Duchin J., Boeckh M., Englund J.A., Famulare M., Lutz B.R., Rieder M.J., Thompson M., Neher R.A., Baird G.S., Starita L.M., Chu H.Y., Shendure J., Lindquist S., Nickerson D.A., Greninger A.L., Jerome K.R., Bedford T. Comparing viral load and clinical outcomes in Washington State across D614G substitution in spike protein of SARS-CoV-2. 2020. https://github.com/blab/ncov-wa-d614g
- 37.Pellis L., Scarabel F., Stage H.B., Overton C.E., Chappell L.H.K., Lythgoe K.A., Fearon E., Bennett E., Curran-Sebastian J., Das R., Fyles M., Lewkowicz H., Pang X., Vekaria B., Webb L., House T.A., Hall I. Challenges in control of Covid-19: short doubling time and long delay to effect of interventions. medRxiv. 2020 doi: 10.1101/2020.04.12.20059972. 2020.2004.2012.20059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/s0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daniloski Z., Jordan T.X., Ilmain J.K., Guo X., Bhabha G., tenOever B.R., Sanjana N.E. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. bioRxiv. 2020:2020. doi: 10.1101/2020.06.14.151357. 2006.2014.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilmjärv S., Abdul F., Acosta-Gutiérrez S., Estarellas C., Galdadas I., Casimir M., Alessandrini M., Gervasio F.L., Krause K.-H. Epidemiologically most successful SARS-CoV-2 variant: concurrent mutations in RNA-dependent RNA polymerase and spike protein. medRxiv. 2020 doi: 10.1101/2020.08.23.20180281. 2020.2008.2023.20180281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung K., Pei Y., Leung G.M., Lam T.T.Y., Wu J.T. Empirical transmission advantage of the D614G mutant strain of SARS-CoV-2. medRxiv. 2020:2020. doi: 10.1101/2020.09.22.20199810. 2009.2022.20199810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corcoran D., Urban M.C., Wegrzyn J., Merow C. Virus evolution affected early COVID-19 spread. medRxiv. 2020:2020. doi: 10.1101/2020.09.29.20202416. 2009.2029.20202416. [DOI] [Google Scholar]
- 43.Kannan S.R., Spratt A.N., Quinn T.P., Heng X., Lorson C.L., Sönnerborg A., Byrareddy S.N., Singh K. Infectivity of SARS-CoV-2: there is something more than D614G? J. Neuroimmune Pharmacol. 2020:1–4. doi: 10.1007/s11481-020-09954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020 doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen H.T., Zhang S., Wang Q., Anang S., Wang J., Ding H., Kappes J.C., Sodroski J.G. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. bioRxiv. 2020:2020. doi: 10.1101/2020.10.22.351569. 2010.2022.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mok B.W.-Y., Cremin C.J., Lau S.-Y., Deng S., Chen P., Zhang A.J., Lee A.C.-Y., Liu H., Liu S., Ng T.T.-L., Lao H.-Y., Lee E.L.-K., Leung K.S.-S., Wang P., To K.K.-W., Chan J.F.-W., Chan K.-H., Yuen K.-Y., Siu G.K.-H., Chen H. SARS-CoV-2 spike D614G variant exhibits highly efficient replication and transmission in hamsters. bioRxiv. 2020 doi: 10.1101/2020.08.28.271635. 2020.2008.2028.271635. [DOI] [Google Scholar]
- 47.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv. 2020 doi: 10.1101/2020.09.01.278689. 2020.2009.2001.278689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Edwards C.E., Tse L.V., Boucher R.C., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G Variant Exhibits Enhanced Replication ex vivo and Earlier Transmission in vivo. bioRxiv. 2020 doi: 10.1101/2020.09.28.317685. 2020.2009.2028.317685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Cai Y., Xiao T., Lu J., Peng H., Sterling S.M., Walsh R.M., Rits-Volloch S., Sliz P., Chen B. Structural impact on SARS-CoV-2 spike protein by D614G substitution. bioRxiv. 2020 doi: 10.1101/2020.10.13.337980. 2020.2010.2013.337980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharyya C., Das C., Ghosh A., Singh A.K., Mukherjee S., Majumder P.P., Basu A., Biswas N.K. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. 2020 doi: 10.1101/2020.05.04.075911. 2020.2005.2004.075911. [DOI] [Google Scholar]
- 51.Hu J., He C.-L., Gao Q.-Z., Zhang G.-J., Cao X.-X., Long Q.-X., Deng H.-J., Huang L.-Y., Chen J., Wang K., Tang N., Huang A.-L. D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. bioRxiv. 2020:2020. doi: 10.1101/2020.06.20.161323. 2006.2020.161323. [DOI] [Google Scholar]
- 52.Gobeil S.M.C., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., Stalls V., Kopp M., Henderson R., Edwards R.J., Haynes B.F., Acharya P. D614G mutation alters SARS-CoV-2 spike conformational dynamics and protease cleavage susceptibility at the S1/S2 junction. bioRxiv. 2020:2020. doi: 10.1101/2020.10.11.335299. 2010.2011.335299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammad A., Alshawaf E., Marafie S.K., Abu-Farha M., Abubaker J., Al-Mulla F. Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D.C., Korber B., Gnanakaran S. The SARS-CoV-2 spike variant D614G favors an open conformational state. bioRxiv. 2020:2020. doi: 10.1101/2020.07.26.219741. 2007.2026.219741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., Wang Y., Yang Y., Chen K., Zheng W., Kong L., Wang F., Zuo Q., Huang Z., Cong Y. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. bioRxiv. 2020 doi: 10.1101/2020.06.30.177097. 2020.2006.2030.177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissman D., Alameh M.-G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., Mansouri K., Gobeil S., McDanal C., Pardi N., Hengartner N., Lin P.J.C., Tam Y., Shaw P.A., Lewis M.G., Boesler C., Sahin U., Acharya P., Haynes B.F., Korber B., Montefiori D.C. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. medRxiv. 2020 doi: 10.1101/2020.07.22.20159905. 2020.2007.2022.20159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pohl M.O., Busnadiego I., Kufner V., Schmutz S., Zaheri M., Abela I., Trkola A., Huber M., Stertz S., Hale B.G. Distinct phenotypes of SARS-CoV-2 isolates reveal viral traits critical for replication in primary human respiratory cells. bioRxiv. 2020 doi: 10.1101/2020.10.22.350207. 2020.2010.2022.350207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., Hartman A.L., McElroy A.K., Reed D.S., Rennick L.J., Duprex W.P. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. bioRxiv. 2020 doi: 10.1101/2020.04.20.049924. 2020.2004.2020.049924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turonova B., Sikora M., Schurmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bulow S., Gecht M., Bagola K., Horner C., van Zandbergen G., Landry J., de Azevedo N.T.D., Mosalaganti S., Schwarz A., Covino R., Muhlebach M.D., Hummer G., Krijnse Locker J., Beck M. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C., Mendonca L., Yang Y., Gao Y., Shen C., Liu J., Ni T., Ju B., Liu C., Tang X., Wei J., Ma X., Zhu Y., Liu W., Xu S., Liu Y., Yuan J., Wu J., Liu Z., Zhang Z., Liu L., Wang P., Zhang P. The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by cryo-EM and cryo-ET. Structure. 2020;28:1218–1224. doi: 10.1016/j.str.2020.10.001. e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández A. Structural impact of mutation D614G in SARS-CoV-2 spike protein: enhanced infectivity and therapeutic opportunity. ACS Med. Chem. Lett. 2020;11:1667–1670. doi: 10.1021/acsmedchemlett.0c00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9 doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., Farrell M.J., Van Hooser S., Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldman J.D., Wang K., Roltgen K., Nielsen S.C.A., Roach J.C., Naccache S.N., Yang F., Wirz O.F., Yost K.E., Lee J.-Y., Chun K., Wrin T., Petropoulos C.J., Lee I., Fallen S., Manner P.M., Wallick J.A., Algren H.A., Murray K.M., Su Y., Hadlock J., Jeharajah J., Berrington W.R., Pappas G.P., Nyatsatsang S.T., Greninger A.L., Satpathy A.T., Pauk J.S., Boyd S.D., Heath J.R. Reinfection with SARS-CoV-2 and failure of humoral immunity: a case report. medRxiv. 2020:2020. doi: 10.1101/2020.09.22.20192443. 2009.2022.20192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., Lee L.L., Wan P., Tso E., To W.K., Tsang D., Chan K.H., Huang J.D., Kok K.H., Cheng V.C., Yuen K.Y. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan X., li l. The influence of major S protein mutations of SARS-CoV-2 on the potential B cell epitopes. bioRxiv. 2020:2020. doi: 10.1101/2020.08.24.264895. 2008.2024.264895. [DOI] [Google Scholar]
- 69.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., Vacca F., Cardamone D., De Santi C., Benincasa L., Agrati C., Capobianchi M.R., Castilletti C., Emiliozzi A., Fabbiani M., Montagnani F., Depau L., Brunetti J., Bracci L., Montomoli E., Sala C., Ippolito G., Rappuoli R. Extremely potent human monoclonal antibodies from convalescent Covid-19 patients. bioRxiv. 2020 doi: 10.1101/2020.10.07.328302. 2020.2010.2007.328302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., Gokool L., Morrisseau C., Bégin P., Tremblay C., Martel-Laferrière V., Kaufmann D.E., Richard J., Bazin R., Finzi A. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11 doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao X., Maruyama J., Zhou H., Kerwin L., Sattler R., Manning J.T., Johnson S., Richards S., Li Y., Shen W., Blair B., Du N., Morais K., Lawrence K., Lu L., Pai C.-I., Li D., Brunswick M., Zhang Y., Ji H., Paessler S., Allen R.D. Discovery and development of human SARS-CoV-2 neutralizing antibodies using an unbiased phage display library approach. bioRxiv. 2020 doi: 10.1101/2020.09.27.316174. 2020.2009.2027.316174. [DOI] [Google Scholar]
- 72.Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., Murali M.R., Alter G., Charles R.C., Dighe A., Branda J.A., Lennerz J.K., Lingwood D., Schmidt A.G., Iafrate A.J., Balazs A.B. COVID-19 neutralizing antibodies predict disease severity and survival. medRxiv. 2020:2020. doi: 10.1101/2020.10.15.20213512. 2010.2015.20213512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., Zheng N.-Y., Utset H., Stamper C.T., Dugan H.L., Miller W.D., Huang M., Dai Y.-N., Nelson C.A., Hall P.D., Jansen M., Shanmugarajah K., Donington J.S., Krammer F., Fremont D.H., Joachimiak A., Kawaoka Y., Tesic V., Madariaga M.L., Wilson P.C. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. bioRxiv. 2020 doi: 10.1101/2020.09.12.294066. 2020.2009.2012.294066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee C.Y.-P., Amrun S.N., Chee R.S.-L., Goh Y.S., Mak T.-M., Octavia S., Yeo N.K.-W., Chang Z.W., Tay M.Z., Torres-Ruesta A., Carissimo G., Poh C.M., Fong S.-W., Bei W., Lee S., Young B.E., Tan S.-Y., Leo Y.-S., Lye D.C., Lin R.T.P., Maurer-Stroh S., Lee B., Cheng-I W., Renia L., Ng L.F.P. Neutralizing antibodies from early cases of SARS-CoV-2 infection offer cross-protection against the SARS-CoV-2 D614G variant. bioRxiv. 2020:2020. doi: 10.1101/2020.10.08.332544. 2010.2008.332544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Legros V., Denolly S., Vogrig M., Boson B., Rigaill J., Pillet S., Grattard F., Gonzalo S., Verhoeven P., Allatif O., Berthelot P., Pélissier C., Thierry G., Botelho-Nevers E., Paul S., Walzer T., Cosset F.-L., Bourlet T., Pozzetto B. A longitudinal study of SARS-CoV-2 infected patients shows high correlation between neutralizing antibodies and COVID-19 severity. medRxiv. 2020 doi: 10.1101/2020.08.27.20182493. 2020.2008.2027.20182493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li T., Han X., Wang Y., Gu C., Wang J., Hu C., Li S., Wang K., Luo F., Huang J., Long Y., Song S., Wang W., Hu J., Wu R., Mu S., Hao Y., Chen Q., Gao F., Shen M., Long S., Gong F., Li L., Wu Y., Xu W., Cai X., Qu D., Yuan Z., Gao Q., Zhang G., He C., Nai Y., Deng K., Du L., Tang N., Xie Y., Huang A., Jin A. A key linear epitope for a potent neutralizing antibody to SARS-CoV-2 S-RBD. bioRxiv. 2020;2020 doi: 10.1101/2020.09.11.292631. 2009.2011.292631. [DOI] [Google Scholar]
- 78.Klumpp-Thomas C., Kalish H., Hicks J., Mehalko J., Drew M., Memoli M.J., Hall M.D., Esposito D., Sadtler K. D614G spike variant does not alter IgG, IgM, or IgA spike seroassay performance. medRxiv. 2020 doi: 10.1101/2020.07.08.20147371. 2020.2007.2008.20147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.P., Schmidt S.D., Wang L., Zhang Y., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Stevens L.J., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Alvarado G.S., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McAuley A.J., Kuiper M.J., Durr P.A., Bruce M.P., Barr J., Todd S., Au G.G., Blasdell K., Tachedjian M., Lowther S., Marsh G.A., Edwards S., Poole T., Layton R., Riddell S.J., Drew T.W., Druce J.D., Smith T.R.F., Broderick K.E., Vasan S.S. Experimental and in silico evidence suggests vaccines are unlikely to be affected by D614G mutation in SARS-CoV-2 spike protein. NPJ Vaccines. 2020;5:96. doi: 10.1038/s41541-020-00246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel A., Walters J., Reuschel E.L., Schultheis K., Parzych E., Gary E.N., Maricic I., Purwar M., Eblimit Z., Walker S.N., Guimet D., Bhojnagarwala P., Doan A., Xu Z., Elwood D., Reeder S.M., Pessaint L., Kim K.Y., Cook A., Chokkalingam N., Finneyfrock B., Tello-Ruiz E., Dodson A., Choi J., Generotti A., Harrison J., Tursi N.J., Andrade V.M., Dia Y., Zaidi F.I., Andersen H., Lewis M.G., Muthumani K., Kim J.J., Kulp D.W., Humeau L.M., Ramos S., Smith T.R.F., Weiner D.B., Broderick K.E. Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv. 2020:2020. doi: 10.1101/2020.07.28.225649. 2007.2028.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Furuyama T.N., Antoneli F., Carvalho I.M.V.G., Briones M.R.S., Janini L.M.R. 2020. Temporal Data Series of COVID-19 Epidemics in the USA, Asia and Europe Suggests a Selective Sweep of SARS-CoV-2 Spike D614G Variant. arXiv.org. arXiv:2006.11609. [Google Scholar]
- 83.Butowt R., Bilinska K., Von Bartheld C.S. Chemosensory dysfunction in COVID-19: integration of genetic and epidemiological data points to D614G spike protein variant as a contributing factor. ACS Chem. Neurosci. 2020;11:3180–3184. doi: 10.1021/acschemneuro.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]