Abstract
Understanding the hallmarks of the immune response to SARS-CoV-2 is critical for fighting the COVID-19 pandemic. We assessed antibody and T cell reactivity in convalescent COVID-19 patients and healthy donors sampled both prior to and during the pandemic. Healthy donors examined during the pandemic exhibited increased numbers of SARS-CoV-2-specific T cells, but no humoral response. Their probable exposure to the virus resulted in either asymptomatic infection without antibody secretion or activation of preexisting immunity. In convalescent patients, we observed a public and diverse T cell response to SARS-CoV-2 epitopes, revealing T cell receptor (TCR) motifs with germline-encoded features. Bulk CD4+ and CD8+ T cell responses to the spike protein were mediated by groups of homologous TCRs, some of them shared across multiple donors. Overall, our results demonstrate that the T cell response to SARS-CoV-2, including the identified set of TCRs, can serve as a useful biomarker for surveying antiviral immunity.
Keywords: SARS-CoV-2, COVID-19, Spike, T cell response, T cell epitopes, T cell receptor, TCR sequencing, clonal structure, MHC, HLA-A∗02:01
Graphical Abstract
Highlights
-
•
Healthy donors sampled in 2020 had an increased T cell response to SARS-CoV-2
-
•
SARS-CoV-2 glycoprotein S-specific TCR repertoire features public CDR3 motifs
-
•
Two epitopes are recognized by the majority of the HLA-A2+ COVID-19 convalescents
Shomuradova et al. assessed the immune response to SARS-CoV-2 in convalescent patients and healthy donors. Antigen-specific T cells were increased in convalescents and in donors sampled during the pandemic. The work identified two public epitopes from S-glycoprotein. T cell receptor repertoire profiling of S-glycoprotein-specific lymphocytes revealed public CDR3 motifs.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus responsible for the global coronavirus disease 2019 (COVID-19) pandemic (Lillie et al., 2020; Phelan et al., 2020; Wu et al., 2020b; Zhou et al., 2020; Zhu et al., 2020). Elucidating the mechanisms underlying the adaptive immune response to SARS-CoV-2 will be crucial for predicting vaccine efficacy and assessing the possibility of reinfection.
It is commonly assumed that antibody response is required for viral clearance (Huang et al., 2020). Multiple serological tests for detection of SARS-CoV-2-specific antibodies are being developed (Amanat et al., 2020; Guo et al., 2020; Krammer and Simon, 2020), and massive efforts are being undertaken in many countries to estimate the number of seropositive individuals in the population. Monoclonal antibodies (Pinto et al., 2020; Shanmugaraj et al., 2020; Wang et al., 2020) and plasma of convalescent patients (CPs) are also being developed as therapy for COVID-19 (Chen et al., 2020; Wong et al., 2003). For example, administration of monoclonal neutralizing antibodies against the spike (S) protein of SARS-CoV-2 protects experimental animals from high doses of virus (Rogers et al., 2020). However, about 30% of CPs have no or very low titers of SARS-CoV-2 neutralizing antibodies (Wu et al., 2020a), suggesting that other immune mechanisms are involved in viral elimination.
There is strong evidence for an important role of T cell immunity in the clearance of respiratory viruses, such as the SARS-CoV that caused an atypical pneumonia outbreak in 2003. Memory T cell responses to SARS-CoV epitopes were detectable in 50% of CPs at 12 months post-infection (Li et al., 2008). Moreover, CD8+ T cells specific to the immunodominant epitope of S protein of a mouse-adapted SARS-CoV strain protected aged mice from otherwise lethal infection (Channappanavar et al., 2014). In another study, the adoptive transfer of SARS-CoV-specific CD8+ or CD4+ T cells or immunization with a peptide-pulsed dendritic cell-based vaccine reduced viral titers in the lungs and enhanced survival of mice, showing that T cells are sufficient for virus clearance even in the absence of antibodies or activation of the innate immune system (Zhao et al., 2010). Other studies in mice likewise suggest a leading role for CD4+ T cells in SARS-CoV clearance. For example, depletion of CD4+ T cells at the time of SARS-CoV infection delayed viral clearance, whereas depletion of CD8+ T cells had no such effect (Chen et al., 2010). Furthermore, CD4+ T cell response was found to be correlated with a positive outcome of SARS-CoV infection (Zhao et al., 2010).
In humans, severe SARS-CoV infection was characterized by the delayed development of the adaptive immune response and delayed viral clearance (Cameron et al., 2008). Decreased numbers of T cells were strongly correlated with disease severity (Li et al., 2004). T cells not only contribute to the resolution of infection but also form a long-lasting memory response to SARS-CoV. For example, CD8+ and CD4+ cells isolated from CPs 4 years after recovery secreted interferon-γ (IFNγ) in response to stimulation by peptide pools derived from the S, N, and M proteins of SARS-CoV (Fan et al., 2009). SARS-CoV antigen-specific T cells have been shown to persist in CPs for up to 11 years post-infection (Ng et al., 2016). The S protein in particular has been shown to be the most immunogenic among all the SARS-CoV antigens (Fan et al., 2009; Li et al., 2008), and specific T cell epitopes have been identified for the SARS-CoV S protein. For example, two CD8+ epitopes were presented in HLA-A∗02:01 and elicited a specific response in SARS-CoV CPs but not in healthy donors (Wang et al., 2004b). Several CD8+ T cell epitopes have also been identified for the M (Yang et al., 2006, 2007) and N (Yang et al., 2006) proteins of SARS-CoV.
Growing evidence likewise supports an important role for the T cell response to SARS-CoV-2 in disease control. High CD8+ T cell counts in the lungs are correlated with better control of SARS-CoV-2 progression (Liao et al., 2020). The presence of T follicular helper cells and CD8+ T cells with activated phenotypes in the blood at the time of virus clearance suggests active involvement in the immune response in recovered patients (Thevarajan et al., 2020). On the other hand, an exhausted phenotype for CD8+ T cells in the peripheral blood may serve as an indicator of poor disease prognosis (Zheng et al., 2020).
In all likelihood, the immune memory is capable of protecting against reinfection by SARS-CoV-2. Studies in primates have demonstrated that repeated virus challenge failed to provoke reinfection once the initial infection was eliminated (Deng et al., 2020). The T cells of CPs are responsive to stimulation by peptide pools covering the SARS-CoV-2 proteome. In particular, S protein is a strong inducer of Th1-type response in CD4+ cells (Weiskopf et al., 2020). Interestingly, CD4+ cells reactive to SARS-CoV-2 antigens were found not only in COVID-19 CPs but also in healthy donors (Braun et al., 2020). A recent publication demonstrated that these T cells are cross-reactive to common cold coronaviruses (Mateus et al., 2020), although further studies are necessary to clarify whether this preexisting T cell response is protective. Other studies hint at a possible role of patient HLA genotype in response to the virus. According to bioinformatic predictions, some HLA alleles present more SARS-CoV-2 epitopes than others, possibly affecting the severity of disease (Nguyen et al., 2020). Some known immunogenic T cell epitopes of SARS-CoV are conserved in SARS-CoV-2 (Grifoni et al., 2020a), suggesting that they might also play a role in the immune response to SARS-CoV-2. However, no experimental data about the targets of T cell reactivity are currently available.
In order to investigate this question, we analyzed the adaptive immune response to SARS-COV-2 in COVID-19 CPs, with the aim of describing the underlying structure, clonality, and epitope specificity of the T cell immune response to the S protein. Here we examined the frequency of T cells specific to SARS-CoV-2 S protein in COVID-19 CPs and two cohorts of the healthy donors, revealing the higher magnitude of the immune response in individuals sampled during the COVID-19 pandemic compared with the biobanked samples. We were able to identify multiple similarity groups in the S protein-reactive TCR repertoires of CPs, some of which were statistically linked to particular HLA alleles and even tentative major histocompatibility complex (MHC) class I and II epitopes. The most prominent finding of our study was the discovery of two S protein epitopes, YLQ and RLQ, that elicit immune response in almost all HLA-A∗02:01+ CPs. The analysis of the epitope-specific TCR repertoire revealed that YLQ epitope is recognized by the public CDR3 motifs with germline-like features.
Results
Virus-Specific T Cells but Not Antibodies Were Present in Healthy Donors during the Pandemic
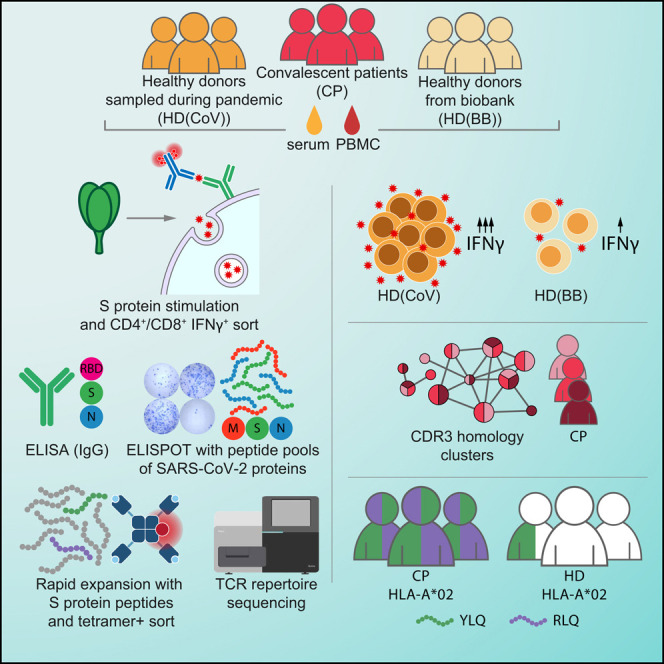
To study the adaptive immune response to SARS-CoV-2, we recruited 34 CPs. According to the classification developed by the U.S. National Institutes of Health, the patients were categorized as having asymptomatic (n = 2), mild (n = 20), or moderate to severe (n = 12) disease. None of the patients required treatment in the intensive care unit, oxygen supplementation, or invasive ventilation support. The cohort was gender balanced (17 male, 17 female), with ages ranging from 17 to 59 years and a median age of 35 years. Peripheral blood was collected between days 17 and 49 (median day 34) after the onset of symptoms or a positive PCR test result (Figure 1 A). The control group included 14 healthy volunteers recruited during the COVID-19 pandemic (HD(CoV)) with no symptoms and negative PCR test results. We also obtained 10 samples of peripheral blood mononuclear cells (PBMCs) from biobanked healthy hematopoietic stem cell donors (HD(BB)), which were cryopreserved no later than September 2019, and 10 serum samples from healthy blood donors that were cryopreserved no later than 2017 (HD(S)). We tested all serum samples for the presence of SARS-CoV-2-specific antibodies, and cell samples were used to study the T cell response to pools of membrane (M), nucleoprotein (N), and S protein-derived peptides and to recombinant S protein in order to determine the T cell receptor (TCR) repertoire of S protein-specific cells.
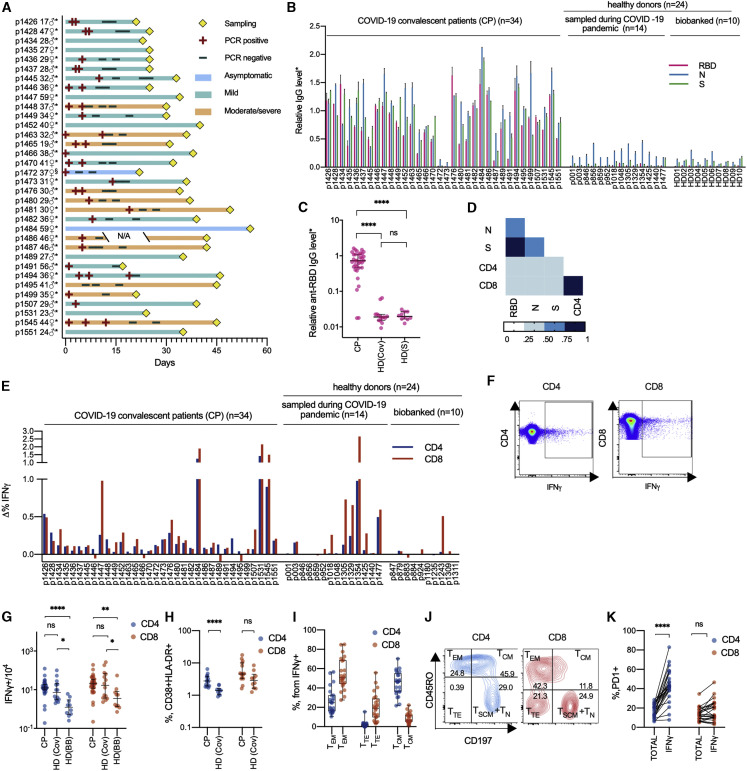
Figure 1.
Healthy Donors Sampled during the COVID-19 Pandemic Have Increased Numbers of SARS-CoV-2-Specific T Cells but Not Antibodies
(A) COVID-19 patient data. Age and gender are indicated at left. Time point of sampling, results of PCR tests, and severity of disease are provided in the swimmer’s plot. ∗Days are calculated since the onset of symptoms; §days are calculated since the positive PCR test result. N/A, not available.
(B and C) Relative levels of anti-RBD, anti-N, and anti-S IgG were measured by conventional ELISA in CPs (n = 34), HD(CoV) (n = 14), and HD(S) (n = 10). Plotted data are means of two independent measurements ± SD (B) and medians with bars representing interquartile range (C).
(D) Correlation between relative levels of IgG and T cell response in the CP group (n = 34). Heatmap of intervals of Spearman’s coefficient of correlation is shown.
(E–G) T cell response to S protein was measured by frequencies of IFNγ-producing CD4+ and CD8+ cells in CPs (n = 34), HD(CoV) (n = 14), and HD(BB) (n = 10). Plots show changes in frequencies of activated T cells in stimulated and unstimulated samples (E). Representative dot plots are shown for IFNγ-producing cells (F). Median frequencies with bars representing the interquartile range (G).
(H) Percentage of activated (CD38+ HLA-DR+) CD4+ and CD8+ cells in CP (n = 15) and HD(CoV) (n = 10) groups. Plots show medians with bars representing interquartile range.
(I and J) Phenotype of IFNγ-secreting CD4+ and CD8+ cells in CP (n = 24). TEM, T effector memory (CD45RO+, CD197−); TTE, T terminal effector (CD45RO−, CD197−); TCM, T central memory (CD45RO+, CD197+). Box represents interquartile range with the median line, and whiskers represent minimum and maximum (I). Representative dot plots are shown for distribution of phenotypes (p1481) (J).
(K) Comparison of frequencies of PD1+ cells in CD4+ and CD8+ non-naive T cells (TOTAL) and the non-naive activated subpopulation (IFNγ) in the CP group (n = 24).
For group comparisons, we used the Kruskal-Wallis test and Dunn’s multiple-comparison test (C, G, and H) and the Mann-Whitney test (H, I, and K). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. See also Figures S1–S3 and Table S1.
Analysis of the humoral immune response to SARS-CoV-2 demonstrated that the majority of CPs had IgG antibodies specific to all of the tested viral antigens. IgGs from the HD(CoV) and HD(BB) groups showed no reactivity to the S protein of SARS-CoV-2 or its receptor-binding domain (RBD) (Figures 1B and 1C). The presence of antibodies specific to the N protein in the various HD samples could be explained by either their cross-reactivity (N is among the most highly conserved SARS-CoV-2 genes) or by recognition of bacterial products co-purified with the antigen, which was expressed in E. coli (Figures 1B and S1A). Despite the variability of the antibody response, in most cases the levels of IgG specific to all three antigens distinguished CP from HD (Figures 1B, 1C, S1A, and S1B), and the response to RBD in particular exhibited the lowest background. Only two patients (p1472 and p1473) did not demonstrate IgG response to any of the tested viral antigens. In our cohort the level of humoral response did nor correlate with time since disease onset (Figures S1F–S1H), patient age (Figures S1I–S1K), or disease severity (Figures S1L–S1N). It should be noted that levels of IgG antibodies specific to different antigens positively correlated in CP (Figures 1D and S1C–S1E), but with the strongest correlation (r = 0.83, p < 0.0001) observed between RBD and S protein.
The T cell response as measured by IFNγ secretion assay was highly variable across donors, with some CPs lacking detectable virus-reactive T cells (Figures 1E–1G). We did not observe any clear association between the magnitude of T cell response and the time since disease onset, disease severity, or patient age (Figures S1O–S1T). We observed a significant increase in activated (CD38+, HLA-DR+) CD4+ cells in the CP group compared with HD(CoV) (Figure 1H). We also observed only mild correlation between the magnitude of the T cell and humoral response in our cohort (for anti-RBD IgG and CD8+ T cell response, r = 0.392 and p = 0.0219) whereas the magnitude of the CD8+ and CD4+ responses were interdependent (Figure 1D).
All tested HD(CoV) sera lacked antibodies against SARS-CoV-2 antigens. Surprisingly, some exhibited comparable frequencies of S protein-specific T cells to donors from the CP group (Figures 1E and 1F). In addition to the significant difference in T cell response between CP and HD(BB) (CD4+, p < 0.0001; CD8+, p = 0.0014), we also observed a significant increase in S protein-specific CD4+ and CD8+ T cells in HD(CoV) compared with HD(BB) (CD4+, p = 0.0108; CD8+, p = 0.045) (Figure 1G). This might indicate that some HD(CoV) patients were exposed to the virus but rapidly cleared it via T cells without developing a humoral response.
S protein-specific T cells in CPs exhibited a conventional phenotype distribution typical to CD4+ and CD8+ cells. S protein-reactive CD4+ T cells were represented predominantly by a central memory phenotype (CD45RO+, CD197+) and, to a lesser extent, an effector memory phenotype (CD45RO+, CD197−). Antigen-specific CD8+ cells mostly had an effector memory phenotype, with the terminal effector (CD45RO−, CD197−) phenotype second most abundant (Figures 1I and 1J). The level of PD-1 expression by CD4+, but not CD8+, cells was significantly higher in the IFNγ-secreting population (Figure 1K). The flow cytometry gating strategy for all populations is shown in Figure S2.
We also measured the T cell immune response to recombinant S protein using ELISPOT and to peptide pools covering the S, M, and N proteins. Some patients responded to recombinant S protein while demonstrating no response to S protein-derived peptide pools (Figure S3). This might be explained by incomplete coverage of the protein sequence (see Discussion for details). Activation of T cells upon stimulation with full-length S protein was equally effective in both CD4+ and CD8+ lymphocytes (Figures S3A and S3B). The M protein-directed immune response was significantly stronger compared with the response to S protein (p = 0.0125) (Figures S3C and S3D). All CPs exhibited either CD8+ or CD4+ T cell reactivity to at least one of the proteins of SARS CoV-2 (Figures 1E and S3).
Immune Response to Two HLA-A∗02:01-Restricted S Protein Epitopes Discriminates CP and HD Samples
The most common MHC I allele in the CP cohort was HLA-A∗02:01 (Table S1), present in 17 of the 34 patients. We selected 13 potential S protein epitopes that were predicted to be presented by HLA-A∗02:01; some of these shared 100% sequence homology with SARS-CoV and were previously shown to be immunogenic (Table 1 ). The magnitude of the S protein-directed response was less than 0.1% of the total CD8+ population in some patients, so we decided to perform rapid in vitro antigen-specific expansion of memory cells using a previously published protocol (Danilova et al., 2018). Epitope-specific cells were detected by flow cytometry using MHC-tetramers (Figures 2A and 2B). Strikingly, 16 of 17 CPs with the HLA-A∗02:01 allele reacted to a single epitope, S protein269–277 YLQPRTFLL (YLQ), while only one HD(CoV) sample had cells recognizing this epitope. Response to another epitope, S protein1000–1008 RLQSLQTYV (RLQ), was also characteristic for this group of patients, albeit to a lesser extent (Figure 2A). The proportion of positive wells that underwent expansion in response to both epitopes could be used to confirm previous COVID-19 (Figure 2C). The low level of homology of these two epitopes to common cold coronaviruses explains the almost complete absence of cross-reactive response in the HD group. The remaining 11 tested epitopes yielded only sporadic T cell responses (Figure 2A).
Table 1.
HLA-A∗02:01-Restricted Peptides of S Protein Used in This Study
| N | Start Position | End Position | Length | AA Sequence | Cleavage Probability | Binding Score | Binding Rank | SARS-CoV-1 Identity | Link |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 269 | 277 | 9 | YLQPRTFLL | 0.977383 | 0.973032 | 0.015 | 6/9 | Baruah and Bose (2020) |
| 2 | 417 | 425 | 9 | KIADYNYKL | 0.966616 | 0.908998 | 0.0583 | 8/9 | – |
| 3 | 424 | 433 | 10 | KLPDDFTGCV | 0.896237 | 0.591582 | 0.3676 | 8/10 | Grifoni et al. (2020a) |
| 4 | 691 | 699 | 9 | SIIAYTMSL | 0.951629 | 799,513 | 0.11 | 8/9 | Lv et al. (2009) |
| 5 | 821 | 829 | 9 | LLFNKVTLA | 0.859019 | 0.785743 | 0.1599 | 9/9 | Ishizuka et al. (2009) |
| 6 | 958 | 966 | 9 | ALNTLVKQL | 0.943025 | 0.450592 | 0.6159 | 9/9 | Grifoni et al. (2020a) |
| 7 | 976 | 984 | 9 | VLNDILSRL | 0.97061 | 0.950712 | 0.0356 | 9/9 | Lv et al. (2009) |
| 8 | 983 | 991 | 9 | RLDKVEAEV | 0.969105 | 0.860941 | 0.0962 | 9/9 | – |
| 9 | 996 | 1004 | 9 | LITGRLQSL | 0.891839 | 0.957880 | 1.98 | 9/9 | Wang et al. (2004b) |
| 10 | 1000 | 1008 | 9 | RLQSLQTYV | 0.748401 | 0.743108 | 0.199 | 9/9 | Wang et al. (2004a) |
| 11 | 1185 | 1193 | 9 | RLNEVAKNL | 0.9655 | 0.618928 | 0.3295 | 9/9 | Wang et al. (2004a) |
| 12 | 1192 | 1200 | 9 | NLNESLIDL | 0.945978 | 0.697243 | 0.2411 | 9/9 | Lv et al. (2009) |
| 13 | 1220 | 1228 | 9 | FIAGLIAIV | 0.179154 | 0.82072 | 0.1288 | 9/9 | Wang et al. (2004b) |
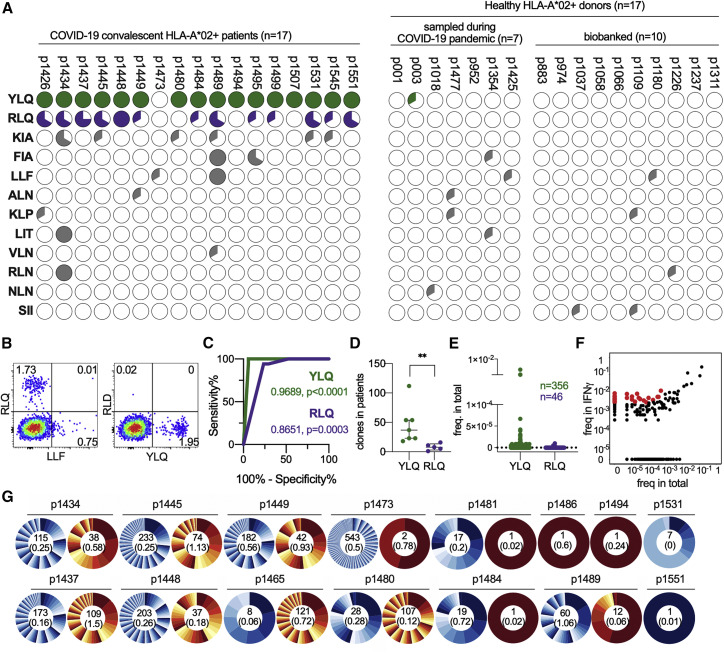
Figure 2.
Epitope Specificity of CD8+ T Cell Response to S Protein Differs Significantly between HLA-A∗02:01-Positive CPs and HD
(A and B) PBMCs of HLA-A∗02:01-positive CPs (n = 17), HD(CoV) (n = 7), and HD(BB) (n = 10) were stimulated with a mix of 13 predicted peptides and expanded for 8–12 days, followed by MHC-tetramer staining. (A) Pie charts represent fractions of wells containing tetramer-positive cells after expansion. (B) Representative MHC-tetramer staining at day 9 (patient p1445, well 3).
(C) Receiver operating characteristic (ROC) curve of the CP/HD classifier on the basis of the presence of YLQ or RLQ epitope-specific cells after expansion.
(D) MHC-tetramer-positive clones after rapid in vitro expansion were FACS-sorted, and their TCR repertoires were sequenced. Numbers of YLQ- and RLQ-specific clones in each patient are plotted (p = 0.0013 by Mann-Whitney test).
(E) YLQ- and RLQ-specific T cell clones occupy only a negligible fraction of the total repertoire. Frequencies of each antigen-specific T cell clone in the PBMCs are plotted.
(F) IFNγ-secreting CD4+ and CD8+ cells were FACS-sorted after stimulation with S protein, and their TCRβ repertoire was sequenced. A representative enrichment plot for patient p1448, showing IFNγ-secreting CD8+ versus total PBMCs. Red dots represent clones that are both strongly (>10×) and significantly (p < 10−8, Fisher’s exact test) enriched.
(G) Clonal structure of the CD4+ (blue) and CD8+ (orange) antigen-specific T cell populations. Numbers inside the pie charts indicate the total number of antigen-specific clones, and numbers below in parentheses indicate their combined share in the total T cell repertoire.
To describe the structure and clonality of the SARS-CoV-2 T cell response, we analyzed the TCR repertoires of fluorescence-activated cell sorting (FACS)-sorted IFNγ-secreting CD8+/CD4+ cells and MHC-tetramer-positive populations as well as the total fraction of PBMCs by Illumina high-throughput sequencing. We observed only negligible intersection between the MHC-tetramer-positive and IFNγ-secreting populations (a single YLQ-specific clone was enriched in the IFNγ+ population of p1445), indicating only a minimal presence of RLQ- and YLQ-specific clones in the peripheral blood. The YLQ-specific response was significantly more diverse than the RLQ-specific clones, with medians of 37 and 8 clones per individual, respectively (Figure 2D). T cell clones specific for both antigens were either undetectable or observed at a very low frequency in the total TCR repertoire of the peripheral blood (Figure 2E).
We designated antigen and epitope-specific TCRs as those sequences that were strongly (≥10-fold) and significantly (p < 10−8, Fisher’s exact test) enriched (Figure 2F). The clonality of IFNγ-secreting cells was higher in CD8+ T cells (Figure 2G); the number of IFNγ-secreting CD4+ clones ranged between 0 and 545 (median 18), whereas the CD8+ IFNγ-secreting cell population ranged from 0 to 121 (median 2) clones. Sequences of YLQ- and RLQ-specific clonotypes were deposited into the VDJdb database (http://vdjdb.cdr3.net) according to the recommendations of the Adaptive Immune Receptor Repertoire Community (Rubelt et al., 2017). Clonotypes significantly enriched in the IFNγ-secreting CD4+ and CD8+ populations are listed in Table S2.
TCRs Specific to Two SARS-CoV-2 Epitopes Display Prominent Motifs Shared across Individuals
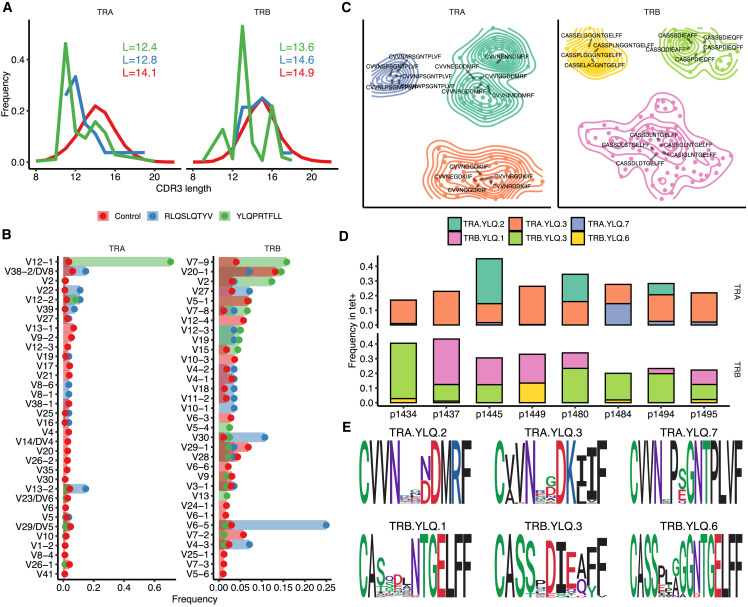
Statistical analysis of V(D)J rearrangements in RLQ- and YLQ-specific TCRs revealed biases in the complementarity-determining region 3 (CDR3) length distribution (Figure 3 A) and V gene usage (Figure 3B) for both TCRα (TRA) and β (TRB) chains. The CDR3 regions of these TCRs, with the exception of RLQ-specific TRB, appear to be substantially shorter than in a control PBMC repertoire. This is a feature previously shown to be associated with public TCRs that have higher V(D)J rearrangement probability and incidence rate across individuals (Pogorelyy et al., 2018). TRB CDR3 length difference can also explain why YLQ-specific T cells are more frequent than RLQ-specific ones. We also observed some notable differences in the frequency of certain V genes. For example, TRAV12-1 and TRBV7-9 were used by 71% and 16% of YLQ-specific TCRs, and TRAV13-2 and TRBV6-5 were used by 15% and 25% of RLQ-specific TCRs, compared with just 3%–4% gene usage in control TCRs. Strong biases in V-gene usage in YLQ-specific α and β chains and RLQ-specific β chain (Figures S4A and S4B) might suggest the importance of germline-encoded features in TCR recognition of these peptides, as previously seen in other antiviral immune responses (Bovay et al., 2018; Minervina et al., 2020b; Chen et al., 2017; Culshaw et al., 2017; Miles et al., 2011; Price et al., 2004).
Figure 3.
YLQ-Specific Clones Display Prominent CDR3 Motifs that Are Shared across Individuals
(A) Length distribution of CDR3 amino acid sequences of tetramer-positive T cell repertoires and a control dataset. Blue, TCRs specific to RLQ; green, TCRs specific to YLQ; red, control TCR repertoire. Inset shows mean CDR3 lengths.
(B) A histogram of V gene usage across datasets. Only V genes with frequency ≥ 1% in any of the datasets are shown.
(C) CDR3 sequence similarity maps of TCR motifs discovered in TCRα (TRA) and β (TRB) chain tetramer-positive T cell repertoires. Sets of highly homologous CDR3 sequences with at least ten members are shown. Contour plots show connected components of the corresponding sequence similarity graph, and labels highlight five representative CDR3 sequences for each cluster.
(D) Distribution of TCRs corresponding to each CDR3 motif across donor samples. The color of each bar corresponds to the specified CDR3 motif, and the height of each bar corresponds to the total fraction of T cells having a given CDR3 in each donor.
(E) Position-weight matrices for CDR3 sequences of TRA and TRB chain motifs found in tetramer-positive T cell repertoires.
See also Figure S4.
We performed TCR sequence similarity analysis to extract the set of motifs that governs the recognition of RLQ and YLQ epitopes (see STAR Methods). Our analysis revealed a set of three distinct CDR3α and three distinct CDR3β motifs, each containing more than 10 highly similar sequences, from YLQ-specific T cells (Figure 3C). Interestingly, all of these motifs were encountered in the majority of donors surveyed (Figure 3D), suggesting the public nature of the response and little difference between motifs in terms of publicity. Moreover, some YLQ-specific TCRs were shared between multiple individuals, while others exhibited a high degree of global similarity (Figures S4C–S4F). Position-weight matrices of the motifs demonstrated a set of highly dissimilar consensuses (Figure 3E), suggesting that while RLQ- and YLQ-specific TCR repertoires are highly public, they are also diverse.
Repertoire Analysis of CD4+ and CD8+ T Cell Responses to S Protein Reveals Public TCR Motifs
We analyzed the repertoire sequencing data for bulk T cell responses to full-length S protein using the TCRNET algorithm described previously (Ritvo et al., 2018). This algorithm detects groups of homologous TCR sequences that are unlikely to arise because of convergent V(D)J recombination and thus indicate an antigen-specific response. We scored each TCR sequence in a pooled dataset of IFNγ-producing CD4+ and CD8+ T cells by quantifying the number of similar TCR sequences observed in the same dataset and in the dataset of pooled corresponding PBMCs (Figure 4 A). This analysis revealed 1,388 and 1,017 unique TRB clonotypes that are part of homologous TCR clusters for CD4+ and CD8+ subsets, respectively (Table S3). Of these, 227 CD4+ and 109 CD8+ TRB clonotypes overlapped with those determined above on the basis of the increased clonotype abundance in the stimulated fraction compared with control. Only two CDR3 amino acid sequences of tetramer-positive T cells were matched to these clusters when allowing a single amino acid substitution, both coming from YLQ-specific T cells.
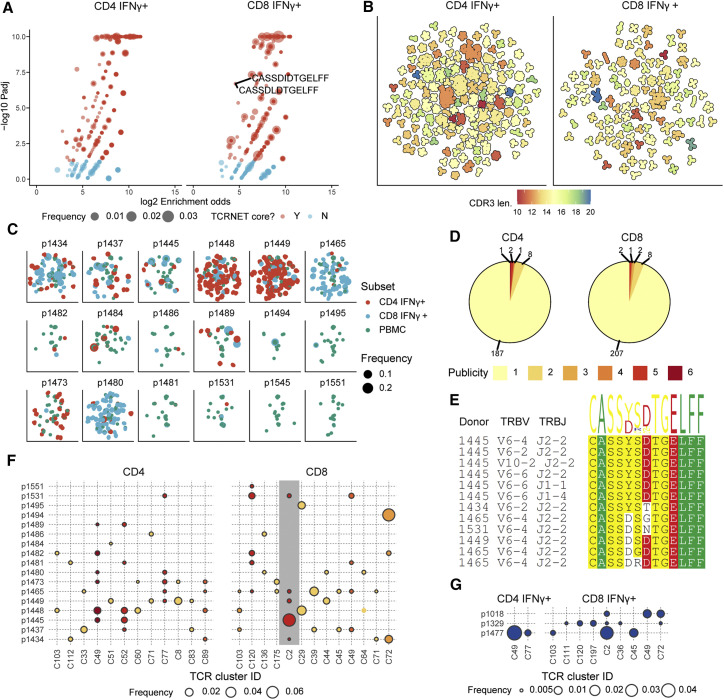
Figure 4.
IFNγ-Producing T Cells from Stimulated Pools Feature Public TCR Motifs
(A) T cell neighborhood enrichment (TCRNET) analysis results showing TCRs related to expanded T cell families. The plot shows the number of homologous TCRs expanded upon stimulation compared with the control dataset (pooled corresponding PBMCs). Each point represents a TCR sequence, and point size represents total frequency in the pooled sample. The x axis corresponds to the logarithm of the ratio of the observed to expected number of homologous TCRs (neighbors) for a given sequence, and the y axis shows the logarithm of the adjusted p value for TCRNET test, capped at 10−10. Red points specify selected TCRs (enrichment odds greater than 4×, padj < 0.05). Labels indicate two shared TCRs found in tetramer-positive T cell datasets that are YLQ specific. Only TCRs with padj < 1 are shown in order to avoid overplotting.
(B) CDR3 similarity map showing the structure of motifs discovered for CD4+ and CD8+ T cells. Points representing individual CDR3 sequences are placed according to the layout of a TCR similarity graph, built on the basis of CDR3 sequences identified by TCRNET. Black outline shows groups of homologous CDR3 sequences, color indicates the CDR3 length for a given cluster. Only clusters with three or more members are shown.
(C) Distribution of TCRs coming from TCRNET-identified clusters across donors. Each point represents a unique TCR; points are scaled according to TCR frequency in a given donor and subset and colored by T cell subset.
(D) Publicity of CDR3 motifs discovered in IFNγ-producing CD8+ and CD4+ T cell datasets. Labels indicate the number of distinct CDR3 motifs found in a given number of donors.
(E) Multiple sequence alignment of the CDR3 region and the V and J genes of distinct TCR variants corresponding to cluster C2 found in IFNγ-producing CD8+ T cell populations from different donors.
(F) Distribution and frequency of TCRs associated with public CDR3 motifs in IFNγ-producing CD8+ and CD4+ T cell populations from different donors. Point size and color correspond to cluster frequency in a given donor and publicity, respectively (see D). Cluster C2, which contains TCRs found in tetramer-positive T cells (see A), is highlighted with a gray background. Only clusters present in at least two donors are shown.
(G) Intersection of S protein-reactive TCR repertoires of CPs and HD(CoV). Distribution and frequency of TCRs associated with public CDR3 motifs in IFNγ-producing CD8+ and CD4+ T cell populations from CPs were intersected with IFNγ-producing populations of HD(CoV) donors. Point size corresponds to cluster frequency in a given donor. Donors p1354 and p1305 had no similar TCRs in their IFNγ-producing fractions. See also Figure S5 and Tables S3 and S4.
Plotting the CDR3 similarity graph revealed many large (in terms of the number of members) homologous clusters of various CDR3 lengths, with the CD4+ subset displaying higher cluster density than CD8+ (Figure 4B). The total number of clusters was higher for the CD8+ subset than CD4+ (220 versus 199 clusters), and the average CD4+ cluster size was larger than CD8+ in terms of the number of unique CDR3 amino acid sequences (5.8 versus 3.9). However, the average frequency of a given cluster in the corresponding datasets in terms of number of cells was larger for CD8+ than CD4+ (0.24% versus 0.11%), highlighting greater diversity of CD4+ response, in line with previous reports (Qi et al., 2014).
We then mapped the set of detected TCR clusters back to the original donor samples (Figure 4C) and found that all donors exhibited some level of homologous TCR response. Yet there were also some prominent inter-donor differences. For example, donors p1448 and p1449 displayed many CD4+ clusters; donors p1465 and p1480 displayed almost exclusively CD8+ clusters; and donor p1484 displayed few clusters, although some exhibited high frequency in terms of the number of cells. Analysis of cluster sharing across donors (Figures 4D and 4E) revealed that most clusters are private to donors. That is expected, as donors have multiple unmatched HLAs, and cells are stimulated with whole protein. However, there were 12 CD4+ and 13 CD8+ clusters shared between stimulated cells of multiple donors (Figure 4F). One of these public clusters (C2) contained sequences matching those found in YLQ-specific T cells. We explored this cluster further by mapping it to IFNγ-producing CD8+ T cell populations from all donors (Figure 4E); this revealed a set of TCR sequences in multiple donors matching cluster C2, all sharing the highly-conserved CDR3 motif CASS[YD][SGR][DTGN]TGELFF.
To investigate the origins of the increased levels of S protein-specific CD4+ and CD8+ cells in the HD(CoV) cohort, we additionally analyzed TCR repertoires of FACS-sorted IFNγ-secreting CD8+/CD4+ cells from the top five responders in this group. In three donors, we found TCRs homologous to six and two public CD8+ and CD4+ clusters, respectively (Figure 4G), whereas we found no public clusters within the HD(CoV) group. However, two clusters that were private in CP became public when we included HD(CoV) samples. This suggests that T cell response in healthy donors sampled during the pandemic is largely focused on the same set of epitopes.
Next, we analyzed co-occurrence of public TCR clusters with HLA alleles in our CP cohort. Four CD8+ clusters and two CD4+ clusters were associated with HLA class I and II alleles with p value < 0.05 by Fisher’s exact test (Figures S5A and S5B). In particular, CD8+ cluster 72 and CD4+ cluster 112 were strongly associated with HLA-B∗15:01 (p = 0.0059) and HLA-DRB1∗04:01 (p = 0.0074), respectively. This suggested that TCRs belonging to those clusters might recognize one of the S protein-derived peptides presented by these alleles. As some peptides might be presented by more than one HLA allele, we predicted all S protein-derived epitopes presented by class I and class II HLA alleles of the donors in our cohort, and analyzed the association of TCR cluster occurrence with the ability of the donor cells to present a particular peptide. This allowed us to pinpoint HLA-associated CD8+ and CD4+ clusters to a limited set of HLA class I- and class II-presented peptides (Figures S5C and S5D). Furthermore, some clusters lacking any association with HLA alleles were strongly correlated with the potential presence of particular peptides. For example, CD8+ cluster 103 was associated with peptide DIADTTDAV, which can be presented by HLA-A∗25:01, HLA-A∗26:01 and HLA-B∗35:02. For a complete list of predicted peptides and associated TCR clusters, refer to Table S4.
Discussion
We analyzed T cell and humoral immune response to SARS-COV-2 in 34 donors who had recently recovered from COVID-19 and control cohorts of healthy donors sampled before or during the pandemic. Two patients (p1472 and p1473) had no detectable antibodies to any of the tested SARS-CoV-2 antigens and no T cell response to any of the peptide pools, but both had CD4+ and CD8+ T cells reactive to recombinant S protein. This finding is in line with other recent studies, where certain CPs had a T cell response in the absence of virus-specific antibodies (Gallais et al., 2020; Long et al., 2020; Sekine et al., 2020). This suggests that the T cell-mediated immune response has the potential to achieve virus clearance without the participation of the humoral immune response. We also cannot exclude reactivity to other viral proteins, such as ORF1ab, for which a strong T cell response has been demonstrated (Gangaev et al., 2020; Grifoni et al., 2020b; Nelde, 2020).
Like Phan et al. (2020), we found that detection of anti-RBD IgG yielded more reliable results than other tested antigens. On the other hand, although it was previously demonstrated that titers of anti-SARS-CoV-2 antibodies positively correlate with patient age (Wu et al., 2020a), we did not observe this pattern in our cohort.
Accumulating data indicate that some healthy COVID-19-naive donors have T cells specific to SARS-CoV-2 antigens, in particular S protein (Braun et al., 2020; Grifoni et al., 2020b; Ni et al., 2020). Our findings support this, with the caveat that the significant increase in S protein-reactive T cells that we observed in healthy donors sampled during the pandemic was accompanied by a complete lack of SARS-CoV-2-specific antibodies in that group. This suggests that some donors may have had contact with SARS-CoV-2 before blood sampling and were either protected by the preexisting cross-reactive T cell response induced by other coronaviruses or developed an asymptomatic infection that was cleared without the help of the humoral response. This is illustrated by donor p1477, who is known to have cohabited with a COVID-19 patient but was negative in multiple PCR tests, did not have any COVID-19-typical or flu-like symptoms and had no detectable antibodies to any SARS-CoV-2 antigens. However, this hypothesis needs to be validated on a larger cohort of donors.
As others have shown before (Braun et al., 2020; Thevarajan et al., 2020), we observed that significantly more CD4+ T cells in convalescent donors express HLA-DR and CD38. We have also shown the same tendency for CD8+ T cells, although the difference here was not significant. As shown before (Weiskopf et al., 2020), the majority of SARS-CoV-2-specific CD4+ cells belonged to the T central memory (TCM) cell subpopulation, whereas CD8+ cells predominantly exhibited T terminal effector (TTE) or T effector memory (TEM) phenotypes. However, it should be noted that the expression of surface markers might have been affected by the antigen stimulation used in our assay.
We aimed to describe the structure of the antigen-specific repertoire, as was done previously for other viruses (Chen et al., 2017) and for SARS-CoV-2 (Schultheiß et al., 2020). In this work, we used IFNγ secretion upon antigen stimulation as a criterion for identifying antigen-specific cells. This approach might miss some relevant T cells, as there are other modes of T cell reactivity, particularly for CD4+ T cells. Nevertheless, previous studies have shown that CD4+ T cells react to stimulation by SARS-CoV-2 antigens (and S protein in particular) primarily by Th1-type response (Weiskopf et al., 2020). In another study, IFNγ was the predominant cytokine produced by memory T cells after stimulation with SARS-CoV-2 peptides (Fan et al., 2009). The results we obtained for T cells stimulated by full-length S protein reveal a highly specific response in terms of the TCR repertoire structure of both CD8+ and CD4+ T cells. The IFNγ-producing T cell repertoires of CPs featured multiple groups of homologous TCR sequences that are in a good agreement with TCR variants found to be enriched on the basis of corresponding clonal abundances. Thus, we were able to identify hundreds of TCR motifs, 25 of which were shared across multiple donors. Moreover, in three of the five HD(CoV) donors with the strongest T cell response to SARS-CoV-2 S protein, the IFNγ-secreting fraction contained TCRs belonging to three or more of the public clusters. Combined with the absence of TCR clusters characteristic for HD(CoV), this suggests that the T cell response in that group is by a large extent is focused on the same epitopes. Furthermore, for 19 of the public TCR clusters, we were able to predict a potential cognate epitope or restricting HLA allele using a statistical approach. These predicted S protein-derived peptides represent attractive targets for further studies.
In this work, we tested 13 epitopes; for 11 of these, presentation by HLA-A∗02 was previously confirmed and immunogenicity demonstrated in SARS-CoV CPs (Zhou et al., 2006) and healthy donors (Lv et al., 2009). Eight were also proposed as potential immune targets in a recent study (Ahmed et al., 2020). Despite using antigen-specific expansion for all peptides to detect even minor T cell clones, we observed a consistent epitope-specific response to only 2 of the 13 peptides. Notably, we did not see a strong response to the RLN peptide despite its previously reported immunodominance in SARS-CoV-2 CPs (Wang et al., 2004a). KLP-specific T cells were detected only in one CP, contrary to findings from SARS-CoV, which showed this epitope to be highly immunogenic in CPs but not healthy donors (Zhou et al., 2006). The KLP peptide of SARS-CoV-2 differs from SARS-CoV by a single amino acid substitution (M-T), whereas the RLN peptide is 100% homologous. The lack of an immune response to these epitopes in SARS-CoV-2 patients may be attributable to the existence of more immunodominant epitopes, which elicit T cell expansion that outcompetes RLN- and KLP-specific T cells. It was previously shown that the T cell immune response in healthy donors is focused predominantly on epitopes in the C-terminal part of the SARS-CoV-2 S protein, which may be explained by higher homology of this region with common cold coronaviruses (Braun et al., 2020). In our study, only one epitope (YLQ) was derived from the N-terminal part of the S protein. We described an immune response to YLQ and RLQ in 16 and 13 of 17 convalescent donors, respectively. YLQ-specific TCRs corresponded to one of the detected motifs in IFNγ-secreting repertoires. This motif was extensively shared between donors and was also found in an unstimulated PBMC fraction. In contrast, we saw almost no response to this epitope in healthy donors. This is unsurprising, as these epitopes have very limited homology to common cold coronaviruses. Nevertheless, T cells specific to both epitopes occupied only a negligible fraction of the total TCR repertoire, explaining the almost complete lack of intersection between tetramer-positive and IFNγ-secreting repertoires. It is possible that YLQ- and RLQ-specific clones are localized in the peripheral tissues, and that only a limited number of cells are present in the circulation. Indeed, it was previously shown that clonal CD8+ T cell expansions in SARS-CoV-2 are tissue-resident (Liao et al., 2020).
The YLQ epitope was previously identified as potentially immunogenic in SARS-CoV-2 (Baruah and Bose, 2020; Romero-López et al., 2020) and was predicted to bind to HLA-A∗01:01 and HLA-C∗07:02 as well as HLA-A∗02:01 (Lee and Koohy, 2020). In this study, we showed that it is not only highly immunogenic but is also recognized by T cells sharing the same TRAV segment (TRAV12-1), suggesting an important role for the TCRα chain in recognizing this epitope. Notably, this epitope was not present in the utilized S protein-derived peptide pool, possibly explaining the discrepancy between reactivity to the recombinant S protein and peptide pools seen in Figure S3. Our analysis of T cells specific to RLQ and YLQ epitopes using tetramer-based enrichment revealed a set of highly conserved TCR sequences shared across multiple donors. These sequences feature highly restricted V segment usage and relatively short CDR3 length, suggesting that RLQ and YLQ are targeted by public TCRs with germline-based motifs. YLQ is recognized by several unrelated motifs that are shared across several donors, suggesting that the response is both public and diverse.
Limitations of Study
Natural limitations of our study arise from the size of the studied cohort (n = 34) and the fact that it lacked any patients who required treatment in the intensive care unit. Because of the limitations of the methods used (IFNγ cytokine secretion assay and IFNγ ELISPOT), we were not able to detect T cells responding by expressing other cytokines, so the magnitude of the virus-specific T cell response was potentially underestimated in this study. Predicted S protein-derived epitopes are limited to the set of MHC class I and II alleles common in the population of the European decent and require further experimental validation.
Alongside the work described by Minervina et al., 2020a, this study provides a glimpse into the structure of T cell response to SARS-CoV-2. Further studies on the specificity of the SARS-CoV-2 targeted response and deconvolution of SARS-CoV-2 epitopes will provide crucial information for vaccine design and disease diagnosis.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AF700 anti-human CD3 (OKT3) | Sony Biotechnology | Cat# 2186700; RRID: AB_2861432 |
| FITC anti-human CD4 (RPA-T4) | Sony Biotechnology | Cat# 2102690; RRID: AB_2861437 |
| PerCP/Cy5.5 anti-human CD8 (RPA-T8) | Sony Biotechnology | Cat# 2105160; RRID: AB_2861438 |
| PE/Dazzle594 anti-human CCR7(CD197) (G043H7) | Sony Biotechnology | Cat# 2366180; RRID: AB_2861439 |
| BV421 anti-human PD1(CD279) (EH12.2H7) | Sony Biotechnology | Cat# 2249600; RRID: AB_2861440 |
| BV711 anti-human CD27 (O323) | Sony Biotechnology | Cat# 2114170; RRID: AB_2861442 |
| BV785 anti-human CD28 (CD28.2) | Sony Biotechnology | Cat# 2114750; RRID: AB_2861443 |
| PE/Cy7 anti-human CD45RO (UCHL1) | Sony Biotechnology | Cat# 2121150; RRID: AB_2861444 |
| Bacterial and Virus Strains | ||
| Escherichia coli strain BL21(DE3) pLysS | Novagen | Cat# 69451 |
| Biological Samples | ||
| Blood sample from healthy donors sampled during the pandemic | National Research Center for Hematology | N/A |
| Blood sample from biobanked healthy donors | National Research Center for Hematology | N/A |
| Blood sample from COVID-19 convalescent patients | National Research Center for Hematology | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human IL-2 | Miltenyi Biotec | Cat# 130-097-743 |
| Human IL-7 | Miltenyi Biotec | Cat# 130-095-367 |
| Human IL-15 | Miltenyi Biotec | Cat# 130-095-760 |
| SARS-CoV-2 S-glycoprotein-His (truncated recombinant) | In house | N/A |
| SARS-CoV-2 S-glycoprotein receptor-binding domain (RBD-His; recombinant) | In house | N/A |
| SARS-CoV-2 N protein (full-length recombinant) | In house | N/A |
| SARS-CoV-2 S protein peptide pool | Miltenyi | Cat# 130-126-701 |
| SARS-CoV-2 M protein peptide pool | Miltenyi | Cat# 130-126-702 |
| SARS-CoV-2 N protein peptide pool | Miltenyi | Cat# 130-126-698 |
| SARS-CoV-2 S protein A02-restricted peptides | Shemyakin-Ovchinnikov IBCH RAS | custom-made |
| UV-cleavable peptide KILGFVFJV | Thermo Fisher Scientific custom peptide synthesis | custom-made |
| UV-cleavable peptide AARGJTLAM | Thermo Fisher Scientific custom peptide synthesis | custom-made |
| MES buffer | Sigma Aldrich | Cat# M3671 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#11360070 |
| DMSO | Sigma Aldrich | Cat# 472301 |
| Expi293 expression medium | Thermo | Cat# A1435101 |
| ExpiFectamine 293 transfection kit | Thermo | Cat# A14524 |
| Opti-MEM I medium | Thermo | Cat# 31985070 |
| Filter cartridge UFP-10-C-4X2MA | Cytiva | Cat# 56-4102-11 |
| Ni-NTA agarose | QIAGEN | Cat# 30210 |
| Slide-A-Lyzer dialysis cassette, 20K MWCO, 3.0 ml | Thermo Fisher Scientific | Cat# 87735 |
| Superdex 75 pg | Cytiva | Cat# 17104401 |
| Imidazole | Sigma Aldrich | Cat# 56748 |
| Reduced glutathione | Sigma Aldrich | Cat# G6529 |
| Oxidized glutathione | Sigma Aldrich | Cat# G4626 |
| Protease inhibitor cocktail, tablets | Thermo | Cat# A32963 |
| Protease inhibitor cocktail, EDTA-free, tablets | Thermo | Cat# A32965 |
| GeneJET endo-free plasmid maxiprep kit | Thermo | Cat# K0861 |
| Critical Commercial Assays | ||
| ELISPOT interferon gamma | CTL | Cat# HIFNGP-2M/5 |
| Human CD8 microbeads | Miltenyi | Cat# 130-097-057 |
| Human CD4 microbeads | Miltenyi | Cat# 130-091-893 |
| SARS-CoV-2-IgG-ELISA kit | National Research Center for Hematology | N/A |
| IFNγ secretion assay - detection kit (PE) | Miltenyi | Cat#130-054-2020 |
| IFNγ secretion assay - detection kit (APC) | Miltenyi | Cat#130-090-762 |
| Deposited Data | ||
| Tetramer-positive TCR sequencing data | vdjdb.cdr3.net | N/A |
| Experimental Models: Cell Lines | ||
| Expi293FTM | ThermoFisher Scientific | Cat# A14527 |
| Software and Algorithms | ||
| FLOWJO X 10.0.7r2 | https://www.flowjo.com/solutions/flowjo/ | RRID: SCR_008520 |
| GraphPad Prism 8 | https://www.graphpad.com/scientific-software/prism/ | RRID: SCR_002798 |
Resource Availability
Lead contact
Further information and requests for resources and reagent should be directed to and will be fulfilled by the lead contact, Grigory Efimov (efimov.g@blood.ru)
Materials availability
This study did not generate new unique reagents
Data and code availability
Tetramer-positive TCR sequencing data was formatted and deposited to the VDJdb database (vdjdb.cdr3.net) - issue#315.
R markdown notebooks used for data analysis are available at https://github.com/antigenomics/covid19-tcr-analysis.
Experimental Model and Subject Details
Human subjects
34 COVID-19 convalescent patients from Moscow, Russia were recruited voluntarily. COVID-19 was confirmed either by positive SARS-CoV-2 RT-PCR test, or retrospectively by the detection of anti-RBD antibodies. All donors signed the informed consent form approved by the National Research Center for Hematology ethical committee before enrollment. The severity of disease was defined from the patient’s case history according to the classification scheme used by the US National Institutes of Health: asymptomatic (lack of symptoms), mild severity (fever, cough, muscle pain, but without respiratory difficulty or abnormal chest imaging) and moderate/severe (lower respiratory disease at CT scan or clinical assessment, oxygen saturation (SaO2) > 93% on room air, but lung infiltrates less than 50%). We also recruited 7 healthy volunteers who were sampled during COVID-19 pandemic but without known contact with COVID-19 patients (except p1477, who was cohabiting with a COVID-19 patient) and confirmed negative infection. Additionally, 10 healthy hematopoietic stem cell donor samples and 10 healthy donor serum samples were obtained from the blood bank with the approval of the local ethical committee. Cell and serum samples were cryopreserved no later than September 2019 and 2017, respectively.
Cell lines
Expi293F cells were maintained in Expi293 expression medium at 37°C in 8% CO2.
Method Details
Peripheral blood mononuclear cell (PBMC) isolation
30 mL of venous blood from healthy donors and recovered COVID-19 patients was collected into EDTA tubes (Sarstedt) and subjected to Ficoll (Paneco) density gradient centrifugation (400 x g, 30 min). Isolated PBMCs were washed with ice-cold PBS containing 2 mM EDTA and used for multiple assays.
HLA genotyping
For most donors HLA genotyping was performed using the One Lambda ALLType kit, which uses multiplex PCR to amplify full HLA-A/B/C gene sequences, and from exon 2 to the 3′ UTR of the HLA-DRB1/3/4/5/DQB1 genes. Prepared libraries were run on an Illumina MiSeq sequencer using standard flow-cell with 2 × 150 paired-end sequencing. Reads were analyzed using the One Lambda HLA TypeStream Visual Software (TSV), version 2.0.0.27232 and the IPD-IMGT/HLA database 3.39.0.0. Other donors were HLA genotyped by Sanger sequencing for loci HLA-A, B, C, DRB1 and DQB1, using Protrans S4 and Protrans S3 reagents, respectively. The PCR products were prepared for sequencing with BigDye Terminator v1.1. Capillary electrophoresis was performed on a Genetic Analyzer Nanophore05. One donor’s HLA genotyping was determined based on existing exome sequencing data.
SARS-CoV-2 S protein peptides
Putative HLA-A∗02:01 epitopes of S protein were included in the analysis if they meet the following criteria: 1) weak binders (0.5 < rank < 2) or strong binders (rank < 0.5) according to NetMHCpan 4.0, and 2) full or partial homologs of existing SARS-CoV S protein epitopes (identity > 60%). Detailed information about selected peptides is listed in Table 1. Predicted proteasomal cleavage score of the C-terminal amino acid was estimated using NetChop 3.1 (Nielsen et al., 2005). HLA-A∗02:01 binding affinity score and rank were estimated by NetMHCpan 4.0 (Reynisson et al., 2020). SARS-CoV identity was measured as the count of identical positions in an amino acid alignment of SARS-CoV and SARS-CoV-2 (MN908947.3) S protein performed by QIAGEN CLC Genomic Workbench software. Peptides were synthesized by solid-phase synthesis and purified by high-performance liquid chromatography (HPLC) (greater than 95% purity). All peptides were dissolved in DMSO, except cysteine-containing peptides, which were dissolved in MES buffer, pH 6.5 / isopropanol mixture (1:1 vol.).
Flow cytometry
Surface staining and phenotype analysis of PBMCs was performed with the following antibodies: CD3-AF700 (OKT3; Sony), CD4-FITC (RPA-T4; Sony), CD8-PerCP/Cy5.5 (RPA-T8; Sony), CCR7 (CD197)-PE/Dazzle594 (G043H7; Sony), PD1 (CD279)-BV421 (EH12.2H7; Sony), CD27-BV711 (0323; Sony), CD28-BV785 (CD28.2; Sony) and CD45RO-PE/Cy7 (UCHL1; Sony). Cells were analyzed on the FACS Aria III cell sorter (BD Biosciences). FlowJo Software (version 10.6.1) was used for analysis.
IFNγ ELISPOT
Measurement of antigen-specific IFNγ production by T cells was performed with the ImmunoSpot human IFNγ single-color ELISPOT kit (CTL) with a 96-well nitrocellulose plate precoated with human IFNγ capture antibody. CD8+ and CD4+ T cells isolated from thawed PBMCs were plated at a density of 105 cells/well in duplicate in CTL-test medium. Unlabeled cells obtained after the selection of CD8+ and CD4+ T cells were used as antigen-presenting cells at a density of 2 × 105/well. SARS-CoV-2 S protein was added at a final concentration of 10 μg/mL in serum-free testing medium (CTL) containing 1 mM GlutaMAX (GIBCO) at a final volume 200 μL/well. Total PBMCs were seeded at a concentration of 5 × 105/well in duplicate and pulsed with M, N or S protein-derived peptide pools (Miltenyi Biotec) at a final concentration of 1 μM. Plates were incubated for 18h at 37°C in 5% CO2. Assays were then performed according to manufacturer’s instructions. Briefly, plates were washed twice with PBS and then twice with PBS + 0.05% Tween-20, followed by incubation with biotinylated anti-human IFNγ detection antibody for 2h at room temperature (RT). Wells were washed three times with PBS + 0.05% Tween-20, and streptavidin-AP was added for 30 min at RT. After a few washes, the colorimetric reaction was started by adding substrate components for 15 min at RT. The reaction was stopped by gently rinsing the plate with tap water. Spots were counted by CTL ImmunoSpot Analyzer using ImmunoSpot software. For group comparison, the data were log(2)-transformed, normality was assessed by Shapiro-Wilk test, and two-way ANOVA with Tukey’s multiple comparisons test was performed. The negative control was subtracted from each value as background.
Expression and purification of recombinant proteins
Recombinant SARS-Co-2 S protein-His6 was expressed in Expi293F cells (Thermo Fisher Scientific) maintained in Expi293 expression medium (Thermo Fisher Scientific), which were transfected with the ExpiFectamine 293 transfection kit (ThermoFisher Scientific) for five days according to manufacturer`s instructions. After harvesting, the medium was centrifuged at 15,600 x g, and the supernatant was concentrated 10 times and diafiltered into buffer A (10 mM phosphate buffer, 2.7 mM KCl, 500 mM NaCl, pH 8.0) using the ÄKTATM flux tangential flow filtration system (Cytiva, filter cartridge UFP-10-C-4X2MA). 50 volumes of concentrate were mixed with 3 volumes of Ni-NTA agarose resin (QIAGEN), which had been previously equilibrated with buffer A and incubated 2h at 22°C with agitation. The resin mix was packed into a Tricorn 10/150 column (Cytiva) and washed with 10 volumes of buffer A with 30 mM imidazole and eluted with buffer A with 200 mM imidazole. The eluate was dialyzed against 100 volumes of PBS (10 mM phosphate buffer, 2.7 mM KCl, 137 mM NaCl, pH 7.5) using Slide-A-Lyzer Dialysis cassettes (20K MWCO, Thermo Fisher Scientific).
Biotinylated MHC class I/UV-cleavable peptide complexes for UV-mediated ligand exchange were produced as described (Bakker et al., 2008; Rodenko et al., 2006). Briefly, heavy (HLA-A∗02:01 with biotinylation tag) and light (β-2 microglobulin) chains were expressed in E. coli strain BL21(DE3) pLysS as inclusion bodies. Proteins were dissolved in denaturation buffer (50 mM Tris-HCl, pH 8.0, and 8 M urea) to a final concentration of 10–20 mg/ml. In vitro folding was set up in 50 mL of folding buffer (100 mM Tris-HCl, 400 mM arginine, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 2 mM EDTA, protease inhibitors, 1 mM PMSF, рН 8.0). UV-cleavable peptide (KILGFVFJV for HLA-A∗02:01 and AARGJTLAM for HLA-B∗07:02, Thermo Fisher Scientific custom peptide synthesis), light and heavy chains were mixed in folding buffer at a 30:2:3 molar ratio. Correctly-folded complexes were purified on a Superdex 75 pg 16/600 column (Cytiva) using Tris-buffered saline (20 mM Tris-HCl, 150 mM NaCl, pH 8.0) as mobile phase. Complexes were biotinylated with in-house made biotin ligase and purified on a Superdex 75 pg 10/300 column. Concentrations were determined using specific absorbance (A280) = 1.03, 2.36, and 1.68 for S protein-His6, HLA-A, and hB2M, respectively (calculated based on amino acid sequence).
Recombinant SARS-CoV-2 N protein was a generous gift from Vasily Lazarev. The plasmid encoding S protein-His6 of SARS-CoV-2 was kindly provided by Prof. Florian Krammer (Amanat et al., 2020). The plasmids encoding HLA and b2-microglobulin were kindly provided by Ton Schumacher (the Netherlands Cancer Institute, Amsterdam, the Netherlands).
ELISA
The IVD ELISA kit developed by the National Research Center for Hematology for the detection of anti-RBD IgG was used according to the manufacturers’ instructions. For N protein and full-length S protein, we used an in-house ELISA assay according to a protocol adapted from Krammer et al. (Amanat et al., 2020). In brief, 96-well plates (Thermo Fisher Scientific) were coated with 50 μL per well of 1.4 μg/mL N protein or 0.4 μg/mL S protein in coating buffer (100 mM bicarbonate/carbonate, pH 9.6). After 14h, the plates were washed three times with 250 μL PBS + 0.1% Tween 20 (TPBS) and blocked with 200 μl of 3% non-fat dry milk (Thermo Fisher Scientific) in PBS for 1.5h. Then the plates were washed thrice, and 100 μl of serum samples diluted 1:100 in TPBS + 1% non-fat dry milk were added in duplicate and incubated for 2h. The wells were washed three times and then incubated for one more hour with 100 μL of anti-human IgG monoclonal HRP-conjugated antibodies (supplied with the RBD ELISA kit). Finally, the plates were washed three times, and 100 μL of 3′,5,5′-tetramethylbenzidine (TMB) substrate was added to each well. 10 min later, 50 μl of 1 M H3PO4 was added as a stop solution, and the optical density (OD) was measured at 450 nm with a reference of 650 nm on a MultiScan FC (Thermo Fisher Scientific) instrument. To compare samples within one plate and normalize values across different plates for comparison, we performed serial dilutions of p1484 serum (from 1:200 to 1:51,200) in each plate. A sigmoid four-parameter logistic (4PL) fitting curve model was used to fit the calibration curve based on the serial dilution. For each plate, an EC50 value (the half OD between the top and bottom segment of a curve) was used as a coefficient of normalization. The mean of two OD values for each sample was divided by the coefficient of normalization for the given plate. For group comparison, the Kruskal–Wallis test and Dunn’s multiple comparison test were used.
Antigen-specific T cell expansion
PBMCs of HLA-A∗02:01 positive donors were used for rapid in vitro expansion. Briefly, 3 × 106 cells were incubated for 8–12 days in RPMI 1640 culture medium supplemented with 10% normal human A/B serum, 1 mM sodium pyruvate, IL-7 (25 ng/mL), IL-15 (40 ng/mL), and IL-2 (50 ng/mL) (Miltenyi) at a final volume of 2 ml/well. Half of the medium was replaced on days 3, 5, and 7. A mix of HLA-A∗02:01-restricted peptides (see Table 1) of S protein in DMSO or MES buffer (Sigma-Aldrich) (final concentration of each peptide in medium = 10 ng/mL) was added at day 0.
Tetramer staining
Antigen-specific cells were detected by staining with CD3-AF700, CD8-FITC, and 7AAD (Sony), along with seven combinations of two different peptide-tetramer complexes conjugated with streptavidin-allophycocyanin and streptavidin-R-phycoerythrin (Thermo Fisher Scientific). A FACS Canto II cell analyzer and Aria III cell sorter (BD Biosciences) were used. Data were analyzed using FlowJo Software.
IFNγ-secretion assay
Measurement of IFNγ secretion in CD4+ and CD8+ T cells was performed using the IFN-γ secretion assay - detection kit (Miltenyi) according to the manufacturer’s protocol. Briefly, fresh PBMCs of CPs and thawed PBMCs of HD(BB) and HD(CoV) were resuspended in RPMI 1640 culture medium (GIBCO) supplemented with 5% normal human A/B serum (obtained from pooled, inactivated human AB Rh- male serum) and 1 mM sodium pyruvate (GIBCO) and plated at a density of 1–10 × 106 cells/ml. Cells were treated for 16h with 10 μg/mL of SARS-CoV-2 S protein, followed by incubation for 5 min at 4°C with IFNγ Catchmatrix reagent (Miltenyi). Cells were then transferred into warm medium (37°C) for 45 min to re-initiate secretion of IFNγ, washed with ice-cold PBS containing 2 mM EDTA and 0.5% BSA, and stained with surface and phenotype markers together with IFNγ detection antibody-APC (Miltenyi) for 10 min at 4°C. CD4+IFNγ+ and CD8+IFNγ+ populations were sorted directly into TRIzol reagent (Thermo Fisher Scientific) using a FACS Aria III cell sorter. Data were analyzed using FlowJo software. For group comparison, we used the Kruskal–Wallis test, Dunn’s multiple comparison test, and the Mann-Whitney test.
Immunomagnetic isolation of CD4+ and CD8+ T cells
CD4+ and CD8+ T cells were isolated using human CD4+ and human CD8+ microbeads (Miltenyi), respectively, according to the manufacturer’s protocol. Briefly, PBMCs isolated from CP donors were incubated for 15 min at 4°C with lyophilized CD8+ microbeads, washed with ice-cold PBS containing 2 mM EDTA and 0.5% BSA, and loaded onto an MS MACS column (Miltenyi), and placed in the MACS Separator. After the columns were removed and magnetically-labeled CD8+ cells were eluted, the flow-through fraction was collected and used for isolation of CD4+ cells by lyophilized CD4+ microbeads according to manufacturer’s instructions. Isolated CD8+ and CD4+ T cells and unlabeled cells were counted using the LUNA Automated Cell Counter and used for the IFNγ ELISPOT assay.
TCR repertoire sequencing
TCR libraries were processed as described previously (Zvyagin et al., 2017). The cDNA synthesis reaction for TCRα and β chains was carried out with a primer to the C-terminal region and SMART-Mk, providing a 5′ template switch effect and containing a sample barcode for contamination control and unique molecular identifier. TCR repertoire data were analyzed using MIXCR software with default settings.
TCR repertoire motif discovery and analysis
Motif discovery for TCR repertoires corresponding to tetramer-positive TCRs specific to SARS-CoV-2 epitopes was performed as described previously Bagaev et al., 2019 using TCRNET method (Pogorelyy and Shugay, 2019; Ritvo et al., 2018). Briefly, the TCR similarity network was constructed, allowing a single amino acid substitution in the CDR3 sequence (Hamming distance of 1). The number of similar sequences (“neighbors”) for each CDR3 was counted and compared to the expected number of neighbors predicted using a reference dataset containing ∼107 amino acid sequences of TCRα or β chains. CDR3 sequences having more neighbors than would be expected at random were considered to be representative (“core”) sequences, TCR motifs were defined as connected components of the TCR similarity network containing at least one core sequence.
A similar analysis was performed to detect TCR motifs in pooled IFNγ+ fractions of stimulated CD4+ and CD8+ T cells, using a control made from the pooled PBMC repertoire of the convalescent patients cohort and counting neighbors based on CDR3 nucleotide sequences as described in Pogorelyy et al. (2018).
Quantification and Statistical Analysis
Shannon diversity of V-gene usage was calculated using python3. All data comparisons were performed using GraphPad Prizm 8 software. Association between the presence of cluster-related TCRs and HLA alleles was calculated by Fisher exact test using the SciPy python3 library. The patient-specific set of peptides predicted to bind to HLA class I (8-11 amino acids) and class II (15 amino acids) was calculated by NetMHCpan 4.1, and NetMHCIIpan 4.0, respectively. Associations between the presence of cluster-related TCRs and S protein-derived peptides in the donor immunopeptidome were calculated by Fisher’s exact test using the SciPy python3 library. Only public clusters and the instances where TCR was present in the relevant fraction (i.e., a TCR belonging to a CD4+ cluster was detected in a CD4+ population) were taken into account. A pseudocount of 0.5 was added to each value.
Acknowledgments
We would like to express our gratitude to all donors who volunteered for our study; to the nurses who performed the venipuncture (Yulia Fadeeva, Anastasya Nisanova, Valentyna Mirponova, and Lubov Piskunova); and to our colleagues Anastasia Minervina, Mikhail Pogorelyy, Vasily Lazarev, Maria Lagarkova, Ivan Zvyagin, Igor Fabrichny, Anastassia Semikhina, Alexey Panov, Ekaterina Avilova, Artem Demidenko, Alexander Veretennikov, Fedor Rozov, Elena Osipova, Nikolay Mugue, Igor Fabrichny, Yuri Lebedin, and Ekaterina Morozova, for their kind help with experiments and reagents and their most valuable advice. We thank Michael Eisenstein for careful and thoughtful manuscript editing. Wet lab experiments (Figures 1, 2, and S1–S4) were supported by Russian Science Foundation grant 20-15-00395 (G.A.E.). The bioinformatic work of D.V.B., A. Pivnyuk, D.S.S., and M.S. (Figures 3, 4, and S5) was supported by a grant from the Ministry of Science and Higher Education of the Russian Federation (075-15-2019-1789) and by Russian Foundation for Basic Research (RFBR) grant 19-34-70011 (M.S.). A.I. was supported by the Ministry of Science and Higher Education of the Russian Federation (agreement 075-15-2019-1660).
Author Contributions
Conceptualization, G.A.E.; Methodology, G.A.E., A.Shomuradova, M.S.V., and M.S.; Software, M.S., D.S.S., D.V.B., A. Pivnyuk, A.K., and D.B.M.; Formal Analysis, M.S.V., A.T., A.K., and M.S.; Investigation, A.Shomuradova., M.S.V., K.V.Z., S.A.S., I.P., Y.S., A.T., A.K., A.V.M., N.T.S., B.B., and E.G.K.; Resources, D.K, M.M., D.V.D., A.Shmelev., A. Pilunov, and A.I.; Writing – Original Draft, G.A.E., A.Shomuradova., and M.S.; Writing – Review & Editing, G.A.E., A.Shomuradova, I.P., A.Shmelev., S.A.S., A.T., and M.S.; Project Administration, G.A.E.; Funding Acquisition, G.A.E., A.I., and M.S.
Declaration of Interests
The authors declare no competing interests.
Published: December 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.11.004.
Supplemental Information
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaev D.V., Vroomans R.M.A., Samir J., Stervbo U., Rius C., Dolton G., Greenshields-Watson A., Attaf M., Egorov E.S., Zvyagin I.V. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Research. 2019;48(D1) doi: 10.1093/nar/gkz874. In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A.H., Hoppes R., Linnemann C., Toebes M., Rodenko B., Berkers C.R., Hadrup S.R., van Esch W.J.E., Heemskerk M.H.M., Ovaa H., Schumacher T.N.M. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl. Acad. Sci. U S A. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovay A., Zoete V., Dolton G., Bulek A.M., Cole D.K., Rizkallah P.J., Fuller A., Beck K., Michielin O., Speiser D.E. T cell receptor alpha variable 12-2 bias in the immunodominant response to Yellow fever virus. Eur. J. Immunol. 2018;48:258–272. doi: 10.1002/eji.201747082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Yang X., Ko A., Sun X., Gao M., Zhang Y., Shi A., Mariuzza R.A., Weng N.P. Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep. 2017;19:569–583. doi: 10.1016/j.celrep.2017.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culshaw A., Ladell K., Gras S., McLaren J.E., Miners K.L., Farenc C., van den Heuvel H., Gostick E., Dejnirattisai W., Wangteeraprasert A. Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8+ T cell response. Nat. Immunol. 2017;18:1228–1237. doi: 10.1038/ni.3850. [DOI] [PubMed] [Google Scholar]
- Danilova L., Anagnostou V., Caushi J.X., Sidhom J.W., Guo H., Chan H.Y., Suri P., Tam A., Zhang J., Asmar M.E. The mutation-associated neoantigen functional expansion of specific T cells (MANAFEST) assay: a sensitive platform for monitoring antitumor immunity. Cancer Immunol. Res. 2018;6:888–899. doi: 10.1158/2326-6066.CIR-18-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.-Y., Huang Z.-T., Li L., Wu M.-H., Yu T., Koup R.A., Bailer R.T., Wu C.-Y. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais F., Velay A., Wendling M.-J., Nazon C., Partisani M., Sibilia J., Candon S., Fafi-Kremer S. Intrafamilial exposure to SARS-CoV-2 induces cellular immune response without seroconversion. medRxiv. 2020 doi: 10.1101/2020.06.21.20132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaev A., Ketelaars S.L.C., Patiwael S., Dopler A., Isaeva O.I., Hoefakker K., Biasi S.D., Mussini C., Guaraldi G., Girardis M. Profound CD8 T cell responses towards the SARS-CoV-2 ORF1ab in COVID-19 patients. Research Square. 2020 https://www.researchsquare.com/article/rs-33197/v1 [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka J., Grebe K., Shenderov E., Peters B., Chen Q., Peng Y., Wang L., Dong T., Pasquetto V., Oseroff C. Quantitating T cell cross-reactivity for unrelated peptide antigens. J. Immunol. 2009;183:4337–4345. doi: 10.4049/jimmunol.0901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Res. 2020;9:145. doi: 10.12688/f1000research.22507.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Qiu Z., Zhang L., Han Y., He W., Liu Z., Ma X., Fan H., Lu W., Xie J. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J. Infect. Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lillie P.J., Samson A., Li A., Adams K., Capstick R., Barlow G.D., Easom N., Hamilton E., Moss P.J., Evans A., Phe Incident Team. The Airborne HCID Network Novel coronavirus disease (COVID-19): the first two patients in the UK with person to person transmission. J. Infect. 2020;80:578–606. doi: 10.1016/j.jinf.2020.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lv Y., Ruan Z., Wang L., Ni B., Wu Y. Identification of a novel conserved HLA-A∗0201-restricted epitope from the spike protein of SARS-CoV. BMC Immunol. 2009;10:61. doi: 10.1186/1471-2172-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J.J., Thammanichanond D., Moneer S., Nivarthi U.K., Kjer-Nielsen L., Tracy S.L., Aitken C.K., Brennan R.M., Zeng W., Marquart L. Antigen-driven patterns of TCR bias are shared across diverse outcomes of human hepatitis C virus infection. J. Immunol. 2011;186:901–912. doi: 10.4049/jimmunol.1003167. [DOI] [PubMed] [Google Scholar]
- Minervina A.A., Komech E.A., Titov A., Bensouda Koraichi M., Rosati E., Mamedov I.Z., Franke A., Efimov G.A., Chudakov D.M., Mora T. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T cell memory formation after mild COVID-19 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minervina A.A., Pogorelyy M.V., Komech E.A., Karnaukhov V.K., Bacher P., Rosati E., Franke A., Chudakov D.M., Mamedov I.Z., Lebedev Y.B. Primary and secondary anti-viral response captured by the dynamics and phenotype of individual T cell clones. eLife. 2020;9:e53704. doi: 10.7554/eLife.53704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelde A. SARS-CoV-2 T-cell epitopes define heterologous and COVID-19-induced T-cell recognition. Research Square. 2020 https://www.researchsquare.com/article/rs-35331/v1 [Google Scholar]
- Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020 doi: 10.1128/JVI.00510-20. https://jvi.asm.org/content/94/13/e00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O., Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- Phan I.Q., Subramanian S., Kim D., Carter L., King N., Anishchenko I., Barrett L.K., Craig J.K., Tillery L., Shek R. In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19. bioRxiv. 2020 doi: 10.1101/2020.05.22.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323:709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Pogorelyy M.V., Fedorova A.D., McLaren J.E., Ladell K., Bagaev D.V., Eliseev A.V., Mikelov A.I., Koneva A.E., Zvyagin I.V., Price D.A. Exploring the pre-immune landscape of antigen-specific T cells. Genome Med. 2018;10:68. doi: 10.1186/s13073-018-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelyy M.V., Shugay M. A framework for annotation of antigen specificities in high-throughput T-Cell repertoire sequencing studies. Front. Immunol. 2019;10:2159. doi: 10.3389/fimmu.2019.02159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.A., West S.M., Betts M.R., Ruff L.E., Brenchley J.M., Ambrozak D.R., Edghill-Smith Y., Kuroda M.J., Bogdan D., Kunstman K. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Qi Q., Liu Y., Cheng Y., Glanville J., Zhang D., Lee J.-Y., Olshen R.A., Weyand C.M., Boyd S.D., Goronzy J.J. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo P.-G., Saadawi A., Barennes P., Quiniou V., Chaara W., El Soufi K., Bonnet B., Six A., Shugay M., Mariotti-Ferrandiz E., Klatzmann D. High-resolution repertoire analysis reveals a major bystander activation of Tfh and Tfr cells. Proc. Natl. Acad. Sci. U S A. 2018;115:9604–9609. doi: 10.1073/pnas.1808594115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenko B., Toebes M., Hadrup S.R., van Esch W.J.E., Molenaar A.M., Schumacher T.N.M., Ovaa H. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López J.P., Carnalla-Cortés M., Pacheco-Olvera D.L., Ocampo M., Oliva-Ramirez J., Moreno-Manjón J., Bernal-Alferes B., García-Latorre E., Domínguez-López M.L., Reyes-Sandoval A., Jiménez-Zamudio L. Prediction of SARS-CoV2 spike protein epitopes reveals HLA-associated susceptibility. Research Square. 2020 https://www.researchsquare.com/article/rs-25844/v1 [Google Scholar]
- Rubelt F., Busse C.E., Bukhari S.A.C., Bürckert J.-P., Mariotti-Ferrandiz E., Cowell L.G., Watson C.T., Marthandan N., Faison W.J., Hershberg U., AIRR Community Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat. Immunol. 2017;18:1274–1278. doi: 10.1038/ni.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53:442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Karolinska COVID-19 Study Group Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N., Zhou X., Wu Y., Yang F., Yu Y. Identification of an HLA-A∗0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.D., Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W., He X.Y., Zhang H.G., Ng J.N., Cheng C.S. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V.W., Dai D., Wu A.K., Sung J.J. Treatment of severe acute respiratory syndrome with convalescent plasma. Hong Kong Med. J. 2003;9:199–201. [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Peng H., Zhu Z., Li G., Huang Z., Zhao Z., Koup R.A., Bailer R.T., Wu C. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J. Gen. Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.-T., Peng H., Zhu Z.-L., Li G., Huang Z.-T., Zhao Z.-X., Koup R.A., Bailer R.T., Wu C.-Y. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin. Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Xu D., Li X., Li H., Shan M., Tang J., Wang M., Wang F.-S., Zhu X., Tao H. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J. Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvyagin I.V., Mamedov I.Z., Tatarinova O.V., Komech E.A., Kurnikova E.E., Boyakova E.V., Brilliantova V., Shelikhova L.N., Balashov D.N., Shugay M. Tracking T-cell immune reconstitution after TCRαβ/CD19-depleted hematopoietic cells transplantation in children. Leukemia. 2017;31:1145–1153. doi: 10.1038/leu.2016.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tetramer-positive TCR sequencing data was formatted and deposited to the VDJdb database (vdjdb.cdr3.net) - issue#315.
R markdown notebooks used for data analysis are available at https://github.com/antigenomics/covid19-tcr-analysis.