Abstract
Considering our previous findings on the remarkable activity exhibited by cobalt(III) with 2-acetylpyridine-N(4)-R-thiosemicarbazone (Hatc-R) compounds against Mycobacterium tuberculosis, the present study aimed to explored new structure features of the complexes of the type [Co(atc--R)2]Cl, where R = methyl (Me, 1) or phenyl (Ph, 2) (13C NMR, high-resolution mass spectrometry, LC–MS/MS, fragmentation study) together with its antibacterial and antiviral biological activities. The minimal inhibitory and minimal bactericidal concentrations (MIC and MBC) were determined, as well as the antiviral potential of the complexes on chikungunya virus (CHIKV) infection in vitro and cell viability. [Co(atc-Ph)2]Cl revealed promising MIC and MBC values which ranged from 0.39 to 0.78 µg/mL in two strains tested and presented high potential against CHIKV by reducing viral replication by up to 80%. The results showed that the biological activity is strongly influenced by the peripheral substituent groups at the N(4) position of the atc-R1− ligands. In addition, molecular docking analysis was performed. The relative binding energy of the docked compound with five bacteria strains was found in the range of −3.45 and −9.55 kcal/mol. Thus, this work highlights the good potential of cobalt(III) complexes and provide support for future studies on this molecule aiming at its antibacterial and antiviral therapeutic application.
Keywords: cobalt(III) complexes, biological application, mass spectrometry, molecular docking
1. Introduction
The clinical achievement of inorganic drugs, such as cisplatin and its derivates chemotherapeutics agents, has significantly improved the study of metal ions on the biological system for the treatment of several diseases [1,2,3]. The ability to form complexes with different coordination numbers, the geometries, the varied oxidation number and the possibility to bind to biomolecules are some features that prove the potential of metal ions as interesting means to explore innovation in drug discovery. Over the years, many transition metal complexes of Cu, Ni, Zn, Co, and Pd have been reported to present remarkable biological activity in a considerable number of illnesses [4,5,6,7,8].
In the bioinorganic chemistry field, besides the strong research on active drugs containing essential metals including Fe [9], Zn [8,10], Cu [11], Co [12], and Mg [13], among others, metallodrugs based on those metal ions have not successfully advanced in clinical trials so far. Moreover, a great advantage of potential drugs containing an essential metal would be the lower levels of toxicity on healthy cells. As vital metal ions, they play essential roles, being responsible for transporters, carriers, binding proteins and cofactors of a wide range of metalloproteins [14]. Cobalt, an essential trace element present in human body, is the main component of cobalamin, vitamin B12 [15]. The Co(III) ion in cobalamin is stabilized by a chelating tetradentate macrocycle in which four nitrogen donor atoms are located in the equatorial plane, and the axial positions are filled by adenosyl residue in an octahedral geometry [16]. This important metal ion is even involved in essential biological roles such as amino acid and fatty acid metabolism and hematopoiesis [17].
Interestingly, to the best of our knowledge, only one cobalt complex containing Schiff base as a ligand, named bis(2-methylimidazole)[(bis(acetylacetone)(ethylenediimine)]-cobalt(III), known as CTC-96 (Doxovir) [18], has recently been tested in a phase II clinical trial for anti-viral treatment [6]. Doxovir presents antiviral biological proprieties for herpes simplex virus (HSV) and for the treatment of two viral eye infections (adenoviral conjunctivitis and ophthalmic herpetic keratitis) [19]. In vitro and in vivo studies have explained the activity of CTC-96 against HSV by interactions of the complex with the viral envelope membrane, which consequently prevent the entry of the virus into the cells, but the exact mechanism is not fully understood [20].
Despite the extensive investigation on metal complexes as chemotherapeutic candidates, only a small number of cobalt(III) complexes are known as inorganic pharmaceuticals when compared to other metals (platinum and gold, for example). In view of the constant evolution of microorganism resistance, virus mutations and the limited existence of potential drugs with antibacterial and antiviral activity, it is necessary to develop new therapeutic alternatives. In this context, thiosemicarbazones Schiff bases are a recognized class of bioactive molecules with antimicrobial and antiviral purposes [21,22]. It is well established that the complexation of thiosemicarbazone ligands to metal ions can show considerable efficacy to improve biological activity [3,7,23,24]. Furthermore, this class of ligands has shown reduced toxic effects when chelated to metal ions [23]. In addition, the number of reports on the coordination chemistry modifications of thiosemicarbazone ligands has appeared to be associated with a rich variety, which is directly correlated to their biological activity [25].
Researchers have described cobalt complexes as promising anti-bacterial agents [26], and few studies show its antiviral potential [21]. Additionally, the activity of Co(III) with Schiff base 2-acetyl-piridine-N(4)-R-thiosemicarbazone (R = hydrogen, methyl, ethyl, cyclohexyl and phenyl) has been described in the literature. In addition to the satisfactory antibacterial activity of this type of complex on Mycobacterium tuberculosis H37Rv (ATCC 27294) bacteria with a minimal inhibitory concentration (MIC) of less than 10 µg/mL [27], the toxicity evaluated on normal cells (VERO and J774A.1) discovered the phenyl derivative complex as the highest selective agent (selective index of 17). The authors noted that the selectivity was highly dependent on the bulkiness of the R group, which probably increases the relative lipophilicity of the molecule. However, there are still few studies on the activity of Co(III) thiosemicarbazonate-containing compounds. Thus, the exploration of the potential of octahedral cationic cobalt(III) complexes with Schiff bases as inorganic pharmaceuticals seemed to be worthwhile.
Based on this, the aim of this study was to synthesize a known Schiff base Co(III) complex to design new structural experimental studies together with the antibacterial and antiviral activity, as well as further investigation of the interactions between the complex and bacteria through molecular docking analysis. Thus, this study is expected to provide new structural information and biological insights of cobalt(III) thiosemicarbazone-based compounds, encouraging the investigation of this substance as an alternative treatment for dental bacteria and chikungunya virus.
2. Results and Discussion
2.1. Structural Characterization
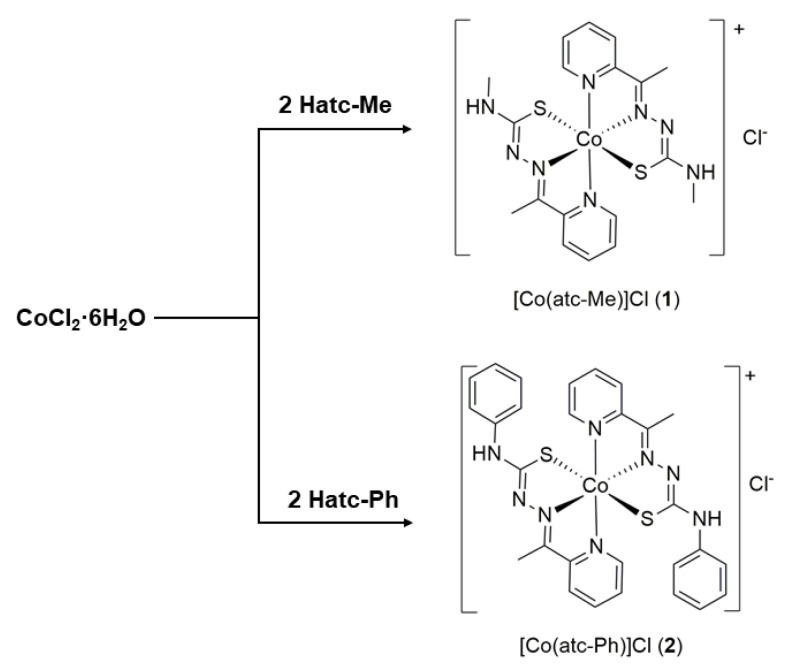
Reaction of thiosemicarbazone ligands Hatc-R (R = methyl or phenyl) with cobalt(II) chloride hexahydrate precursor in ethanolic solution resulted in the products [Co(atc-Me)2]Cl (1) and [Co(atc-Ph)2]Cl (2) (Scheme 1). The product was isolated as a brown precipitate and separated by vacuum filtration after reduction the solution volume. The identity of the complexes was confirmed predominant by 1H, 13C NMR, LC–MS/MS and conductivity measurements, which confirmed the electrolytes 1:1. The results are in accordance with the oxidation of cobalt(II) center to cobalt(III) upon the coordination due to the greater stabilization of the oxidation state III caused by chelating binders [16]. Both complexes are very soluble in methanol and ethanol solutions, while complex 1 is water soluble. Additional and unprecedented characterization techniques, such as 13C NMR, LC–MS, and MS/MS, were employed for the Co(III) complexes prepared here to better understand the complexes’ behavior in solution.
Scheme 1.
Synthesis of the cobalt(III) complexes.
The 13C NMR spectroscopic data for complexes 1 and 2 recorded in DMSO-d6 presented the expected signals, and the results are showed in Table S1. Upon coordination, a downfield shift around 4 ppm is observed for the C=N signals, while the signal for carbon in C=S fragment appears at 178.2 for complex 1 (Figure S1) and 175.5 ppm for 2 (Figure S2), which is in agreement with the N,N,S coordination and thioenolization of the C=S units of the thiourea and thiosemicarbazone moieties. The observed shifts are consistent with the N,N,S coordination of the (atc-R)− ligands.
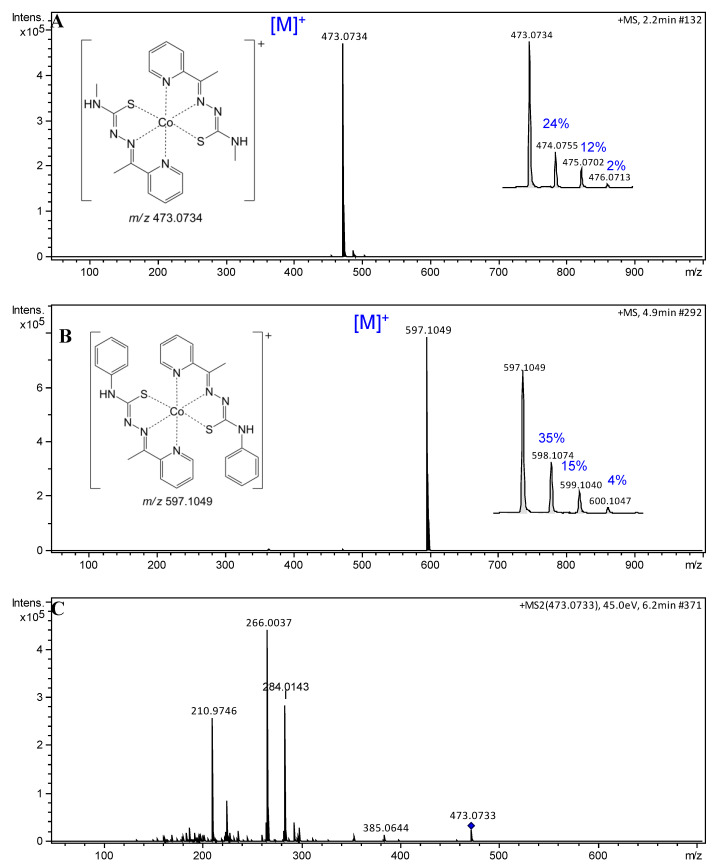
Information of interest is the structure–property relationship studies regarding the lipophilicity [28]. This factor represents the extent to which a drug and/or a substance prefers a hydrophilic or a lipophilic medium. A second important aspect is the different polarities that can suggest the behavior of a compound in biologic environments and even affect the cellular uptake [29,30]. A simple method of assessing the lipophilicity of two compound is to analyze their retention times (tR) in a chromatographic analysis [31]. Thus, compounds 1 and 2 were analyzed by LC–MS, and structural identifications were carried out through mass spectra analysis. Compound 1 (methyl derived) was identified as a majority signal at tR 4.8 min and showed the charged complex ion peak [M]+ at m/z 473. Due to the presence of a phenyl group, complex 2 presented a higher retention time at tR 7.7 min and showed a charged complex ion peak [M]+ at m/z 597. Thus, among them, complex 2 was more lipophilic. Both compounds showed high purity grade, determined to be 91 and 96% for 1 and 2, respectively, according to LC–MS measurement (Figures S3 and S4, Supplementary Materials). Additionally, mass spectra of cobalt complexes are characterized by the presence of a clear isotopic pattern due to the fact that cobalt is a monoisotopic element 59Co (100%), with the nuclide abundance in parentheses [32]. High-resolution mass spectra of compounds 1 and 2 were recorded in the positive mode, and the obtained data confirm the established patterns (Figure 1A,B).
Figure 1.
Electrospray ionization (ESI) mass spectra of cobalt(III) complexes 1 and 2. (A) (+)-high-resolution electrospray ionization mass spectra (HRESIMS) spectrum of 1, m/z 473.0734 [M]+ (calcd. for C18H22CoN8S2, 473.0735, Δ –0.21 ppm); (B) (+)-HRESIMS spectrum of 2, m/z 597.1049 [M]+ (calcd. for C28H26CoN8S2, 597.1048, Δ 0.17 ppm); (C) ESI–MS/MS spectrum of the molecular ion m/z 473.0733; and (D) ESI–MS/MS spectrum of the molecular ion m/z 597.1053.
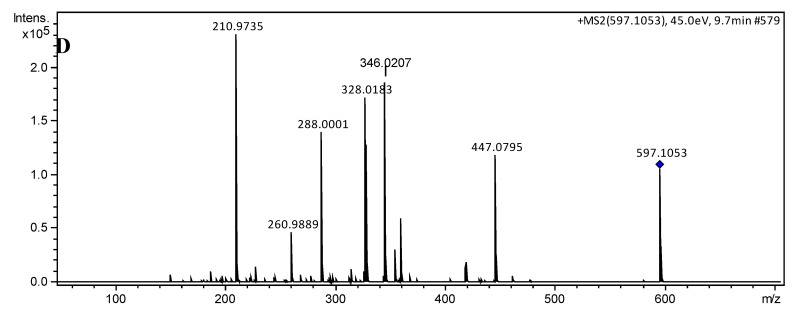
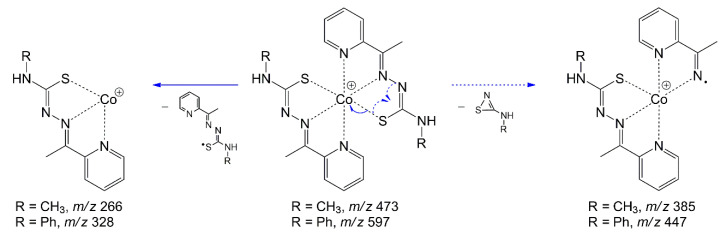
The charged complex ions were observed at m/z 473.0734 [M]+ (1) and 597.1049 [M]+ (2), in agreement with calculated values for C18H22CoN8S2 (473.0735; Δ −0.21 ppm) and C28H26CoN8S2 ( 597.1048; Δ 0.17 ppm), respectively. Collision-induced dissociation (CID) experiments (MS/MS) with an increasing collisional energy (5 to 50 eV) using N2 as collision gas under the selected ions at m/z 473 [M]+ (1) and 597 [M]+ (2) showed a similar fragmentation pathway for both complexes (Figure 2), which can be seen in the mass spectra with collisional energy of 45 eV (Figure 1C,D). When these signals were subjected to MS/MS analysis (Figure 1C,D), the fragment ions at m/z 266 and 328 arose, respectively, and were attributed to homolytic cleavage between the sulfur and the metal, reducing it to Co(II). Neutral losses of 88 or 150 amu produced fragment ions at m/z 385 and 447 for 1 and 2, respectively, and were proposed for neutral elimination of a strained heterocyclic thiozone ring through homolytic cleavages between S–Co and N–N chemical bonds, according to the mechanism pathway proposed. Furthermore, MS/MS analysis suggested that the complexes have high stability after observing the fragments at high energies only.
Figure 2.
Fragmentation pathway proposed for 1 (m/z 473) and 2 (m/z 597) analyzed by (+)-ESI–MS/MS.
2.2. Stability Studies
The stability of complexes 1 and 2 was monitored by 1H NMR in DMSO-d6 solution for a 24 h period. The spectra remained unchanged during the assay time, showing that the compound does not hydrolyze under such conditions (Figure S5).
2.3. Biological Activity Studies
Many efforts in bioinorganic chemistry field have been dedicated to the development of new agents based on a variety of metal ions compounds [33]. These metal-based compounds normally show promising activity against viruses [34], bacteria [35] and on cancer cell lines [36]. Due to the remarkable biological proprieties of thiosemicarbazone and the lower number of inorganic drugs on clinical trials, we believed that it was worth evaluating the antibacterial and antiviral activity of the cobalt(III) compounds developed in this work. As the adequate purity and stability in solution were certified, the compounds were submitted to the biological tests. Thus, the activity of the free ligands HLR and their cobalt complexes was evaluated against the dental caries pathogens and on chikungunya virus (CHIKV).
2.3.1. Antibacterial Activity
The compounds were evaluated in relation to their antibacterial activity. Table 1 shows the MIC and minimal bactericidal concentration (MBC) values determined for the free ligands Hatc-Me and Hatc-Ph, the complexes [Co(atc-Me)2]Cl (1) and [Co(atc-Ph)2]Cl (2), and the starting material for the synthesis CoCl2ꞏ6H2O (3) against seven cariogenic bacteria. The samples concentration evaluated against oral bacteria were between 0.195 µg/mL to 400 µg/mL. The positive control concentrations used (CHD: chlorhexidine) ranged from 0.115 µg/mL to 400 µg/mL. DMSO at 5% concentration was used as untreated control and had no effect on the tested bacteria.
Table 1.
Minimum inhibitory concentration (MIC, µg/mL) and minimum bactericidal concentration (MBC, µg/mL) in µg/mL against buccal bacteria strains.
| Bacterial Strains | Hatc-Me | Hatc-Ph | [Co(atc-Me)2]+ | [Co(atc-Ph)2]+ | CoCl2·6H2O | CHD * | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| Streptococcus mutans (ATCC 251755) | 12.5 | 12.5 | 100 | 100 | 400 | 400 | 0.78 | 0.78 | 100 | 100 | 0.46 | 0.46 |
| Streptococcus mitis (ATCC 49456) | 12.5 | 12.5 | 100 | 100 | 400 | 400 | 1.56 | 1.56 | 100 | 100 | 1.84 | 1.84 |
| Streptococcus sanguinis (ATCC 10556) | 6.25 | 6.25 | 100 | 100 | 400 | 400 | 0.39 | 0.39 | 100 | 100 | 0.92 | 0.92 |
| Streptococcus sobrinus (ATCC 33478) | 25 | 25 | 400 | 400 | 400 | 400 | 12.5 | 25 | >400 | >400 | 0.46 | 0.46 |
| Lactobacillus paracasei (ATCC 11578) | 50 | 50 | 400 | 400 | 400 | 400 | 0.78 | 0.78 | >400 | >400 | 0.92 | 0.92 |
| Streptococcus salivarius (ATCC 25975) | 25 | 25 | 100 | 100 | 400 | 400 | 3.12 | 3.12 | 100 | 100 | 0.92 | 0.92 |
| Enterococcus faecalis (ATCC 4082) | >400 | >400 | >400 | >400 | 400 | 400 | 6.25 | 6.25 | >400 | >400 | 3.68 | 3.68 |
* CHD: Chlorhexidine, positive control.
The free ligand Hatc-Ph and compounds 1 and 3 showed MIC and CBM values over 100 or 400 µg/mL for all assayed microorganisms, classifying them as not active against those bacteria. Hatc-Me presented considerable MIC and MBC values (<12.5 µg/mL) in three tested strains, while its cobalt(III) complex (1) derivate has not shown relevant biological activity (MIC and MCB = 400 µg/mL). Surprisingly, the phenyl containing ligand Hatc-Ph did not present activity, but complex 2 showed promising activity against all the investigated bacteria, especially for S. sanguinis strain in which the MIC and MCB are equal to 0.39 µg/mL, showing bactericidal and bacteriostatic effects, and, in this case improving the activity by complexation, as previously observed [3,13,27]. Neutral nickel [37] and manganese [13] complexes of the type [M(atc-R)2] (R = Me, Ph) reported previously presented similar MIC values for Mycobacterium tuberculosis, (MIC of [M(atc-Ph)2] was 0.78 µg/mL for M = Mn and 0.95 µg/mL for M = Ni) as the analogous cobalt complex [Co(atc-Ph)2]+ (2) presented in this work, but not showing the same trend in the biological activity with the change in the substituting R group.
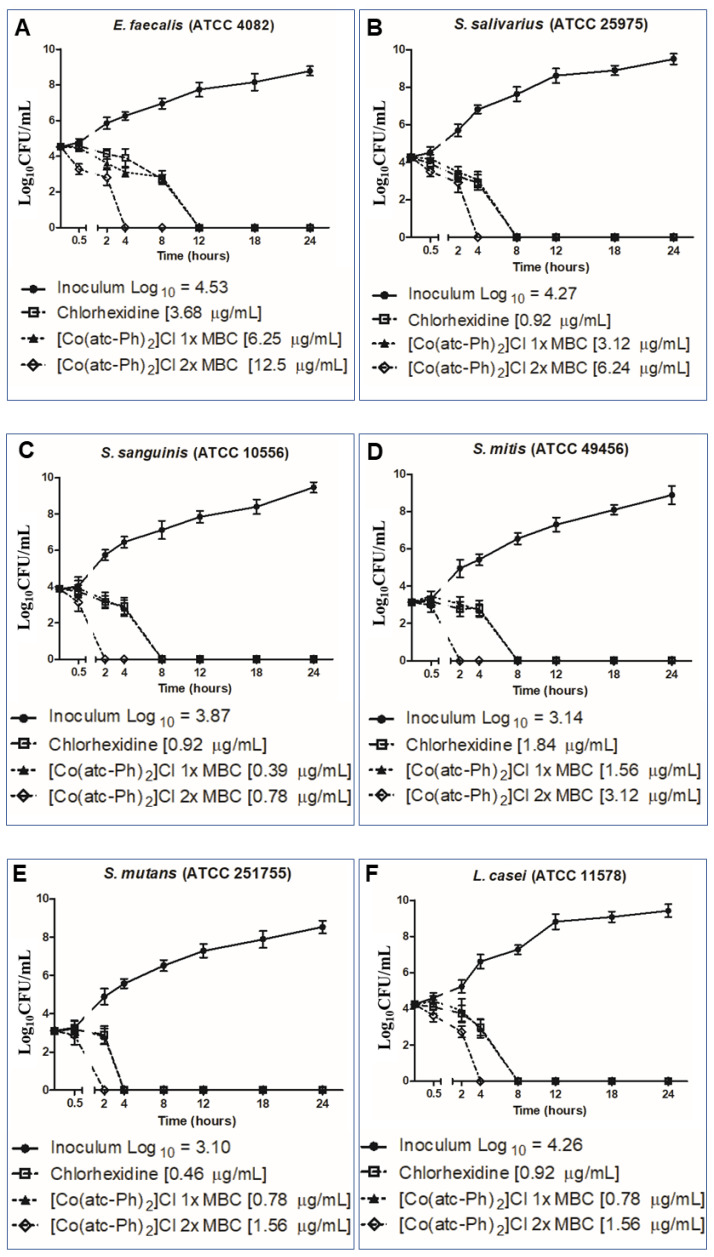
To investigate even further the antimicrobial potential of the complexes, time–kill curve assays were performed against six bacteria strains of complex 2, the most active compound. The curves were drawn using minimal bactericidal concentration (MBC) results obtained for complex 2 and are shown in Figure 3. Complex 2 had bactericidal action for all strains evaluated in less than 24 h, in the two concentrations evaluated. The compound at one time the MBC killed Streptococcus mutans in 4 h; S. salivarius, S. sanguinis, S. mitis and Lactobacillus paracasei in 8 h; and Enterococcus faecalis in 12 h. The compound at two times the MBC concentration killed all colonies of S. sanguinis and S. mutans in 2 h; E. faecalis, S. salivarius, S. sanguinis and S. mitis in 4 h; and L. paracasei in 8 h. The chlorhexidine used as positive control also presented the same bactericidal effected in less than 24 h, but the only strain that the antimicrobial eliminated faster than the compound was L. paracasei, which suggests that complex 2 has antibacterial action that is as fast as chlorhexidine.
Figure 3.
Time–kill curve profiles for complex 2 for caries bacteria as an antibacterial agent in different concentrations of MBC results, (A) E. faecalis; (B) S. salivarius; (C) S. sanguinis; (D) S. mitis; (E) S. mutans; (F) L. paracasei.
The structure–activity relation between the complexes and their antimicrobial activity is clearly affected by the peripheral group change in the thiosemicarbazone, since the antibacterial activity is dependent on the compound permeation through the cell walls of microorganisms [38] and their membrane–cell interaction. The presence of hydrophilic groups in the organic ligands, for instance, considerably reduces the biological activity as shown previously [39]. In our case, changing the CH3 group to the phenyl group led to an improvement in the antibacterial activity potency, which corroborates with the higher lipophilicity predicted for the phenyl-derived group.
2.3.2. Antiviral Activity
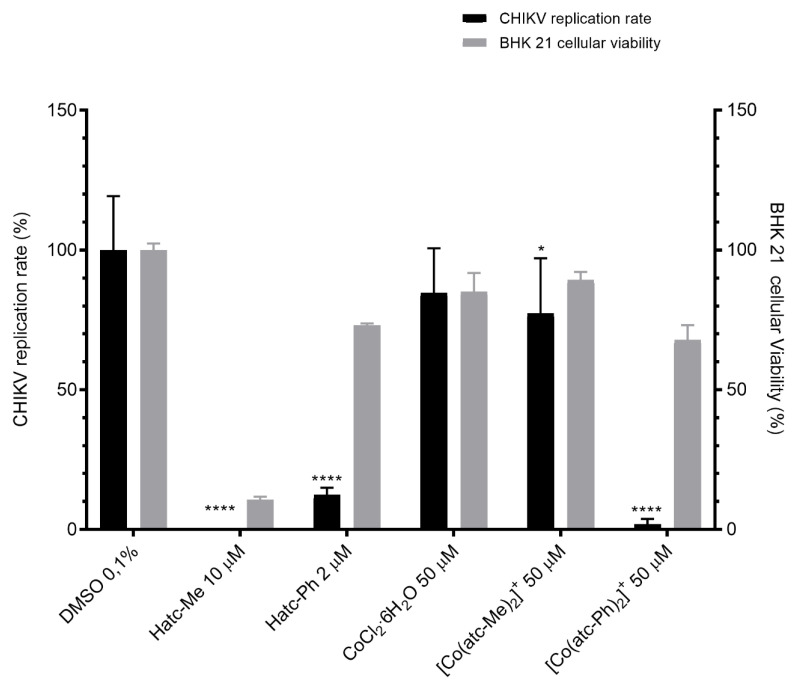
The effects of the free ligands and complexes on CHIKV infection were also studied, and the cellular viability was evaluated on baby hamster kidney (BHK) 21 cells, which were used for infection in the antiviral assays. The antiviral activity of the compounds against CHIKV was determined by using a recombinant CHIKV replicon that expresses the nanoluciferase reporter (CHIKV nanoluciferase) [40]. First, BHK 21 cells were treated with each compound at 50, 10 and 2 µM (0.1% of DMSO) diluted in cell culture media for 16 h, and cell viability was measured through a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay to access cytotoxicity. DMSO was used as non-treated control during the experiment. Cells tolerated each compound at different concentrations, and data of the maximum tolerated concentration are shown for each compound (Figure 4). Then, BHK 21 cells were infected with CHIKV nanoluc at a multiplicity of infection (MOI) of 0.01 and simultaneously treated with Hatc-Me 10 µM, Hatc-Ph 2 µM, CoCl2ꞏ6H2O 50 µM, [Co(atc-Me)2]Cl 50 µM (1) and [Co(atc-Ph)2]Cl 50 µM (2). The inhibition of the CHIKV infection in the presence of the free ligands Hatc-Me at 10 µM and Hatc-Ph 2 µM was significantly reduced to 0 and 12% (p < 0.01). Otherwise, the methyl-containing ligand presented low cell viability on BHK 21 cells (11%), demonstrating that the anti-CHIKV effect observed for this free ligand was a consequence of cell cytotoxicity. Alternatively, the phenyl ligand was well tolerated (73%) to BHK 21 cells, suggesting the potential antiviral property of this compound (Figure 4).
Figure 4.
Activity of compounds on chikungunya virus (CHIKV) infection and viability of baby hamster kidney (BHK) 21 cells. BHK 21 cells were infected with CHIKV nanoluc and simultaneously treated with the compound. After 16 h post-infection (p.i.), cells were lysed, and a Renilla luciferase assay was performed to assess CHIKV replication rates. In parallel, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was performed. Mean values of a minimum of three independent experiments each measured in triplicate are represented. **** p < 0.001/* p < 0.05 were considered significant.
In order to better understand the structure–activity relationship, the metal precursor was also evaluated in regard to its activity on CHIKV replication. It was observed that the cobalt starting material salt at 50 µM presented low inhibition of the virus replication and had no toxicity on BHK 21 cells (Figure 4). As expected, the effect of the complexes on virus replication showed highly significant inhibition only for complex 2 compared to the untreated control. Treatment of CHIKV-infected cells with [Co(atc-Ph)2]Cl 50 µM reduced up to 85% of virus replication, and cell viability was not affected compared to the treatment with the Hatc-Ph ligand. Surprisingly, the results confirmed that the complexation significantly affected the activity on virus replication without increasing the toxicity. This was mainly observed when comparing the concentrations of treatment of the free ligand (2 µM) or complexed molecule (50 µM). It seems that cells tolerated higher concentrations of the Hatc-Ph when complexed to the metal. This fact suggests that the metal ions play a role in the mechanism of action of the compounds.
Taken together, these biological results suggested that the higher volume and increased lipophilicity of the substituent group on the N(4) position of the thiosemicarbazone moiety play a vital role in determining both antibacterial and antiviral properties, and in the present investigation, the phenyl group is most optimal. From our results, it appears that, in general, the best activity is found in compounds of higher lipophilicity. Overall, the present study identified cobalt(III) complex 2 as a potential antiviral against CHIKV replication.
2.4. Molecular Docking
Antibacterial activity of the most active complex was also theoretically investigated and compared with that of CHD (positive control) to identify the interactions by binding sites on the structure of S. mutans, S. mitis, S. sanguinis, L. paracasei and E. faecalis bacteria. For the validation of the protocols performed in molecular docking, redocking of the cocrystal of ligands was performed using AutoDock4 program to validate structure–activity relationships.
The comparison between antibacterial activity and those calculated theoretically for the [Co(LPh)2]+ complex and CHD are in agreement for all bacteria, except for S. sanguinis. In the docking, the complex showed better results (high binding affinity) with S. mutans and L. paracasei bacteria when compared to CHD, while CHD presented better interaction with S. mitis, S. sanguinis and E. faecalis bacteria strains. Thus, the observed trend in the binding free energy was found to be consistent with the experimental inhibition trend for the [Co(atc-Ph)2]+ complex and CHD. The obtained ΔG values and estimated inhibition constant (Ki) of the [Co(atc-Ph)2]+ and CHD with S. mutans, S. mitis, S. sanguinis, L. paracasei and E. faecalis bacteria are shown in Table S3.
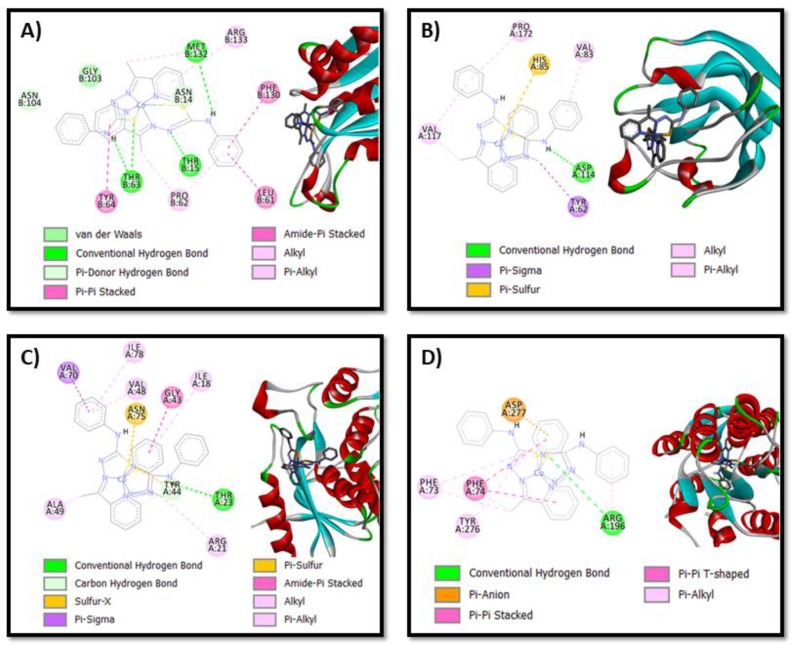
In Figure 5 and Figures S6–S11, the images of the interaction conformations of the [Co(atc-Ph)2]+ complex and CHD are shown. It is evident from the results obtained that both the [Co(atc-Ph)2]+ complex and CHD can successfully bind to one or more amino acids of all bacteria. The results of ΔG obtained were highly exothermic, which reveals good orientation and proximity, demonstrating the increment of the binding affinity [41]. The free binding energy between the [Co(atc-Ph)2]+ complex and bacteria was found to be −9.55 kcal/mol for L. paracasei, followed by −8.66 kcal/mol found for S. sanguinis. The greatest energy value was found for the S. mitis protein structure (−3.45 kcal/mol). On the other hand, the best free binding energy between CHD and bacteria was found for E. faecalis (−11.24 kcal/mol), followed by S. sanguinis (−9.49 kcal/mol), and the highest value was found for S. mitis (−3.03 kcal/mol). Energy derived from the cobalt complex interaction with the L. paracasei structure was lower compared to the energy derived from CHD interaction with the same bacteria structure, suggesting that the complex couple is more stable, a fact observed for other metal complexes in the literature [41,42]. This observation is also in accordance with the antibacterial assay results which show a higher biological activity for [Co(atc-Ph)2]+ (MIC/MBC = 0.39/0.39 µg/mL) than for CHD (MIC/MBC = 0.92/0.92 µg/mL). In all the investigated systems, predominantly hydrogen bonds were observed, and in almost all cases, aromatic–aromatic, aromatic–alkyl or aromatic-sulfur interactions were found.
Figure 5.
Interaction network between the [Co(atc-Ph)2]+ complex and (A) S. mutans; (B) S. mitis; (C) S. sanguinis; and (D) L. paracasei protein structures. For clarity, the non-polar hydrogen atoms of the metal complex are omitted.
The docked pose between the [Co(LPh)2]+ complex and L. paracasei (Figure 5D) shows a considerably strong H bond (3.18 Å in length) involving the N-H group from thiosemicarbazone ligand and a sulfur atom of amino acid Arg 196. Additionally, two pi–pi interactions between the pyridinic rings of both ligands in the complex and the Ph group of amino acid Phe 74, which are 4.74 Å and 4.78 Å in length, respectively, were observed. Similarly, the [Co(atc-Ph)2]+ complex and S. sanguinis also have a hydrogen bond between the N-H group of the complex and sulfur atom of amino acid Thr 63 with a length of 3.76 Å. Furthermore, two shorts H bonds between the N-H group of the complex and an oxygen atom of amino acids Thr 63 (1.81 Å) and Met 132 (2.35 Å) are observed. These short H bonds may justify the fact that the antibacterial activity of S. sanguinis had the lowest MIC and CBM value among all the tested bacteria. More details about the calculated H bond and pi–pi and pi–S distances of intermolecular interactions of the cobalt complex with amino acid residues of all the bacteria are shown in Table S4. Finally, according to all these presented interactions, the [Co(atc-Ph)2]+ complex can effectively bind to all bacteria and, subsequently, displays great potential as antibacterial agents.
3. Experiment
3.1. Materials
All chemicals were reagent grade and used without purification. The reagents and solvents 4-methyl-3-thiosemicarbazide, 4-phenyl-3-thiosemicarbazide, 2-acetylpyridine and analytical were obtained commercially. The compounds Hatc-Me and Hatc-Ph were synthesized as previously reported [43].
3.2. Preparation of the Complexes
The complexes [Co(atc-Me)2]Cl (1) and [Co(atc-Ph)2]Cl (2) were synthesized by slight modifications of the reported procedures described by Oliveira et al. [27] An amount of 0.25 mmol of CoCl2·6H2O was added to 0.5 mmol of Hatc-R in 10 mL ethanol. A brown solid was formed, separated by filtration and washed with n-hexane. The purity of the solids was confirmed by 1H, 13C NMR, LC–MS/MS and conductivity measurements.
R = Me (1). Yield 97.0 mg (60%). 1H NMR (400 MHz, DMSO-d6, δ in ppm): 2.85 (s, 9H, 2CH3C=N and 1CH3NH), 2.95 (s, 3H, CH3NH), 7.44 (s, 2H, Py-H), 7.85 (d, J = 4 Hz, 2H, Py-H), 8.03–8.09 (m, 4H, Py-H), 8.15 (s, 1H, NHCH3), 8.79 (s, 1H, NHCH3). 13C NMR (101 MHz, DMSO-d6, δ in ppm): 15.0 (CH3), 32.1 (CH3), 124.6 (CH), 126.6 (CH), 139.9 (CH), 148.7 (CH), 155.4 (CH), 157.2 (C), 178.2 (C). (+)-HRESIMS m/z 473.0734 [M]+ (calcd for C18H22ClCoN8S2, 473.0735 (Δ −0.21 ppm)). HPLC: tR = 4.8 min.
R = Ph (2). Yield 107.0 mg (68%). 1H RMN (400, DMSO-d6, δ in ppm): 3.00 (s, CH3, 6H), 7.08 (t, J = 8 Hz, Ph, 2H), 7.38 (t, J = 8 Hz, Ph, 4H), 7.52 (dd, J = 7 Hz, J = 2 Hz, Py, 2H), 7.76 (dd, J = 8 Hz, Ph, 4H), 8.02 (d, J = 8 Hz, Py, 2H), 8.14–8.19 (m, Py, 4H), 10.42 (s, NH, 2H). 13C NMR (101 MHz, DMSO-d6, δ in ppm): 15.8 (CH3), 120.4 (CH), 123.3 (CH), 125.5 (CH), 127.4 (CH), 128.7 (CH), 139.9 (CH), 140.1 (C), 149.2 (CH), 156.7 (CH), 158.7 (C), 175. (C). (+)-HRESIMS m/z 597.1049 [M]+ (calcd for C28H26ClCoN8S2, 597.1048 (Δ 0.17 ppm)). HPLC: tR = 7.7 min.
3.3. Instruments
Nuclear magnetic resonance (NMR) experiments were accomplished with a Bruker AscendTM 400 Avance III HD spectrometer (9.2 T) at 400 MHz (1H) and 100 MHz (13C) at 25 °C and referenced using Tetramethylsilane (TMS)(δTMS = 0.00) as internal standard or residual solvent signals of dimethylsulfoxide-d6 (DMSO-d6) at δ 2.50 and 39.5 ppm, respectively, for 1H and 13C. The molar conductivities of the complexes were measured using an Orion Star Series conductometer (1 × 10−3 mol L–1 MeOH solutions). High-resolution electrospray ionization mass spectra (HRESIMS) were measured on a Bruker Daltonics (microTOF-Q II) spectrometer, operating in the positive mode. Methanol–water (7:3, v/v) was used as solvent system, and the samples were infused into the Electrospray Ionization (ESI) source at a flow rate of 180 µL/h. N2 was employed as drying gas at a flow rate of 4 L/min and as nebulizer gas at 130 psi. The temperature of the nebulizer was set at 180 °C, and a potential of 3.5 kV was applied in the capillary. The collision energies were adjusted according to the precursor ion (charged complexes [M]+). The calculated values for the charged complex ions were made using ChemDraw Ultra 14.0. LC–MS data were recorded on Agilent mode Infinity 1260 coupled with a Q-TOF spectrometer of the Agilent® model 6520 B with an electrospray ionization source (IES) at the Nanobiotechnology Laboratory of the Federal University of Uberlândia (IBTC-UFU). The HPLC column was an Agilent model Poroshell, 100 × 3 mm, with a pore size of 2.7 µm. The mobile phase consisted of (A) deionized water 0.1% formic acid and (B) methanol. The following solvent gradient was 10% of B (0 min), 98% of B (0–10 min), and 100% of B (10–17 min). The ionization parameters were nebulizer pressure of 20 psi, drying gas at a flow rate of 8 L/min at a temperature of 220 °C and a capillary voltage of 4.5 kV.
3.4. Antibacterial Assays
3.4.1. Determination of MICs and MBC
The minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC) of the compounds were determined in triplicate. For the MIC, the microdilution broth method in 96-well microplates was utilized. Standard strains acquired from the American Type Culture Collection were used: Streptococcus mutans (ATCC 251755), S. mitis (ATCC 49456), S. sanguinis (ATCC 10556), S. sobrinus (ATCC 33478), Lactobacillus paracasei (ATCC 11578), S. salivarius (ATCC 25975), and Enterococcus faecalis (ATCC 4082). The methodology employed was in accordance with the guidelines of the Clinical and Laboratory Standards Institute, with some modifications that have been previously described in other studies by our research group [44], using brain heart infusion broth (BHI, Difco), and interpretation of the results was made using resazurin.
Solutions of the samples in concentration of 1.0 mg mL−1 were prepared in DMSO (Sigma-Aldrich, St. Louis, MO, USA), followed by dilution in BHI until obtaining concentrations ranging from 0.195 to 400 µg mL−1 with a final DMSO content of 5% (v/v), and this solution was used as a negative control. The inoculum was adjusted for each organism to yield a cell concentration of 5 × 106 colony-forming unit (CFU) per mL. Chlorhexidine was added as positive control, and the concentrations ranged from 0.115 to 50 µg/mL. The microplates were incubated at 37 °C for 24 h in microaerophilic conditions. Afterwards, resazurin (30 uL) in aqueous solution (0.02% v/v) was added to the microplates to indicate bacteria viability [44]. The MIC was interpreted as the minimum concentration of the sample that can inhibit the growth of bacteria.
To determine the MBC, an aliquot (10 µL) of the inoculum from each well of the MIC’s microplates was removed prior of resazurin and seeded in brain heart infusion agar (BHIa). Afterwards, the plates were incubated at 37 °C for 24 h. At the end of the incubation, it was defined as the lowest concentration of the sample at which no bacterial growth occurred.
3.4.2. Time–Kill Curves
Time–kill assays against the strains Streptococcus mutans (ATCC 251755), S. mitis (ATCC 49456), S. sanguinis (ATCC 10556), Lactobacillus paracasei (ATCC 11578), S. salivarius (ATCC 25975), and Enterococcus faecalis (ATCC 4082) were performed in triplicate, as described by D’arrigo et al. (2010) [45], with some modifications. The 96-well microplates containing the most promising compound at concentrations of one and two times the MBC value for the respective strains were inoculated with the target microorganism at an initial bacterial density of 5 × 106 CFU/mL and incubated in microaerophilic conditions. To count viable colonies, aliquots were removed at 0 min and 30 min and at 2, 4, 8, 12, 18 and 24 h. The diluted samples (50 µL) were spread onto BHI agar plates, incubated at 37 °C under appropriate atmosphere, and counted after the growth period. Time–kill curves were constructed by plotting log10 CFU/mL vs. time on the GraphPad Prism (version 8.0) software. Chlorhexidine and bacteria suspension without added compounds were used as the positive and negative control, respectively.
3.5. Antiviral Assays
3.5.1. Cell Culture
Baby hamster kidney cells (BHK 21, ATCC® CCL-10™), which are susceptible to CHIKV infection [46], were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich) supplemented with 100 U/mL penicillin (Hyclone Laboratories, Logan, UT, USA), 100 mg/mL streptomycin (Hyclone Laboratories, USA), 1% non-essential amino acids (Hyclone Laboratories, USA) and 1% fetal bovine serum (FBS, Hyclone Laboratories, USA) in a humidified 5% CO2 incubator at 37 °C.
3.5.2. In Vitro Cytotoxic Activity Evaluation by MTT Assays
Stock solutions of the free ligands and cobalt complexes were firstly prepared in DMSO at 50, 10 and 2 mM, following dilutions in cell culture media to achieve a concentration in micromolar and DMSO at 0.1%. Dimethyl sulfoxide (DMSO) was used as a non-treated control. Cell viability was measured through MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (Sigma-Aldrich) assay. BHK 21 cells were cultured in 96-well plates and treated with 50, 10 and 2 µM of each compound for 16 h at 37 °C with 5% of CO2. After treatment, compound-containing media was removed from 96-well plates, and MTT at 1 mg/mL solution was added to each well, incubated for 1 h and replaced with 100 μL of DMSO to solubilize the formazan crystals. The absorbance was measured at 560 nm on a Glomax microplate reader (Promega). Cell viability was calculated according to the equation (T/C) × 100%, in which T and C represent the optical density of the treated well and control groups, respectively. The highest non-cytotoxic concentration, ≥80%, was selected for further analyses in the antiviral assay.
3.5.3. Assessment of CHIKV Replication
To analyze the antiviral activity of each compound, BHK 21 cells were seeded at a density of 5 × 104 cells per well into 48-well plates 24 h prior to infection. Then, CHIKV nanoluciferase at a multiplicity of infection (MOI) of 0.01 and compounds, at the highest non-cytotoxic concentration selected in the MTT assay, were simultaneously added to cells in cell culture media. Samples were harvested in Renilla luciferase lysis buffer (Promega) at 16 h post-infection (h.p.i.), and virus replication was quantified by measuring nanoluciferase activity using the Renilla luciferase Assay System (Promega). DMSO 0.1% was used as non-treated control.
3.6. Statistical Analysis
Individual experiments were performed in triplicate, and all assays were performed a minimum of three times in order to confirm the reproducibility of the results. Differences between means of readings were compared using analysis of variance (one-way or two-way ANOVA) or Student’s t-test using Graph Pad Prism 8.0 software (Graph Pad Software). p values ≤ than 0.01 were considered statistically significant.
3.7. Molecular Docking Study
Molecular docking studies were performed using autodock4.2 (AD4) [47] with the [Co(atc-Ph)2]+ complex and CHD (positive control). The crystal structures of the complexes [Co(atc-Ph)2]Cl (CCDC 1416048) and CHD (CCDC 988972) were obtained from The Cambridge Crystallographic Data Centre, and the crystal structures of S. mutans (PDB 5C2O), S. mitis (PDB 3LE0), S. sanguinis (PDB 4N82), L. paracasei (PDB 6CHK) and E. faecalis (PDB 4LRL) bacteria were obtained from Protein Data Bank. No structures were found for S. sobrinus and S. salivarius bacteria. The re-dock procedures were conducted using the AutoDockTools 1.5.6 graphical interface along with autogrid4 and AD4. To prepare the cobalt complex and bacteria structures for molecular docking, AutoDockTools 1.5.6 was added to Gasteiger charges [48] and converted to the crystal structures required for AD4. The Lamarckian genetic algorithm in AutoDock was as follows: population (150 individuals); maximum energy evaluations (25,000,000); maximum generations (27,000); one individual surviving into next generation; genetic algorithm docking runs (150). A ranked cluster analysis was performed using a root mean square (RMS) tolerance of 2.0 Å on each docking calculation. The grid map and centers used can be seen in Table S2. The interaction pictures were generated using the Discovery Studio Visualizer program.
4. Conclusions
Screening of two cobalt(III) complexes in seven bacteria strains and on the chikungunya virus revealed a remarkably high potency for the phenyl group (2) compared to the methyl-derived group (1). The coordination of Hatc-Ph to the Co(III) was able to improve the antibacterial and antiviral activities when compared with the free ligand without an increase in the cytotoxicity, as verified by the test on healthy cells. Furthermore, the biological studies showed that small structural changes in the peripheral group of the thiosemicarbazone can substantially affect the antibacterial and antiviral activities. The results of biological activity have shown to be dependent on the metal ion and the phenyl group. The antiviral assays revealed a remarkable level of activity of free ligands Hatc-Me and Hatc-Ph. The coordination of the ligands to the metal ion Co(III) led to a substantial increment in the antiviral activity and low cytotoxicity on healthy cells, especially for complex 2. Additionally, the data of both antibacterial and antiviral assays successfully established a structure–activity relationship, in which the lipophilic characteristic was important for both biological assays. Based on this study, complex 2 was identified as a potential antibacterial and antiviral agent. A molecular docking study was also carried out to explain the binding pattern of most active cobalt complex (2) toward S. mutans, S. mitis, S. sanguinis, L. paracasei, and E. faecalis bacteria strains. This study revealed that complex 2 shows the best affinity towards two bacteria, predominantly L. paracasei and S. sanguinis. This may be explained by the enhanced lipophilic property of the central metal ion. The results presented here could provide useful insights for future practical applications of cobalt(III) complexes as antiviral and antibacterial agents. Furthermore, studies are underway in our laboratories to evaluate the promising compound in order to investigate the real participation of cobalt in the biological activity.
Acknowledgments
We thank Andres Merits (Institute of Technology, University of Tartu, Tartu, Estonia) for the provision of the CHIKV-expressing nanoluciferase reporter. The authors thank FAPESP (Grants 2009/54011-8, 2011/16380-1 and 2013/14957-5) and the Multi-user Laboratory of Institute of Chemistry of Federal University of Uberlândia for the NMR analysis. The authors also thank Norberto Peporine Lopes (USP, Ribeirão Preto-SP, Brazil) for the mass spectrometry analyses, and Mario Machado Martins and Luiz Ricardo Goulart (UFU, Laboratory of Nanobiotechnology (NANOS), Institute of Biotechnology) for the support in LC–MS/MS experiments. Work in the ACGJ laboratory is funded by the Royal Society—Newton Advanced Fellowship (grant reference NA 150195), CNPq (grant 445021/2014-4), FAPEMIG (Minas Gerais Research Foundation-APQ-00587-14, SICONV 793988/2013 and APQ-03385-18) and CAPES (Finance Code 001).
Abbreviations
| MIC | Minimum inhibitory concentration |
| CBM | Minimum bactericidal concentration |
| PDB | Protein Data Bank |
| CCDC | Cambridge Crystallographic Data Centre |
| AD4 | Autodock4.2 |
| RMS | Root mean square |
| ESI | Electrospray Ionization |
| TMS | Tetamethylsilane |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/21/8355/s1.
Author Contributions
L.d.P.F. synthesized and characterized the complexes and assisted in drafting the paper. J.M.B.S., M.B.S. and C.H.G.M. performed the in vitro antimicrobial assays together with the time–kill curve experiment and assisted in drafting the paper. D.O.S.M. and A.C.G.J. performed the cytotoxicity assays on CHIKV and assisted in drafting the paper. V.M.D. synthesized and characterized the complexes and assisted in drafting the paper. G.S.O. and A.E.H.M. assisted in drafting the paper. M.P. performed the proposed fragmentation pathway and assisted in drafting the paper. E.d.F.F. and R.A.C.S. performed the molecular docking analyses and assisted in drafting the paper. C.G.O. provided guidance to the primary author (L.d.P.F.), conceived of the project, and assisted in drafting the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Konkankit C.C., Marker S.C., Knopf K.M., Wilson J.J. Anticancer activity of complexes of the third row transition metals, rhenium, osmium, and iridium. Dalton Trans. 2018;47:9934–9974. doi: 10.1039/C8DT01858H. [DOI] [PubMed] [Google Scholar]
- 2.Budimir A. Metal ions, Alzheimer’s disease and chelation therapy. Acta Pharm. 2011;61:1–14. doi: 10.2478/v10007-011-0006-6. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira C.G., Romero-Canelón I., Silva M.M., Coverdale J.P.C., Maia P.I.S., Batista A.A., Castelli S., Desideri A., Sadler P.J., Deflon V.M. Palladium(II) complexes with thiosemicarbazones derived from pyrene as topoisomerase IB inhibitors. Dalton Trans. 2019;48:16509–16517. doi: 10.1039/C9DT02570G. [DOI] [PubMed] [Google Scholar]
- 4.Kostelidou A., Kalogiannis S., Begou O.-A., Perdih F., Turel I., Psomas G. Synthesis, structure and biological activity of copper(II) complexes with gatifloxacin. Polyhedron. 2016;119:359–370. doi: 10.1016/j.poly.2016.09.012. [DOI] [Google Scholar]
- 5.Carneiro Z.A., Lima J.C., Lopes C.D., Gaspari A.P.S., de Albuquerque S., Dinelli L.R., Veloso-Silva L.L.W., Paganelli M.O., Libardi S.H., Oliveira C.G., et al. Heterobimetallic nickel(II) and palladium(II) complexes derived from S-benzyl-N- (ferrocenyl)methylenedithiocarbazate: Trypanocidal activity and interaction with Trypanosoma cruzi Old Yellow Enzyme (TcOYE) Eur. J. Med. Chem. 2019;180:213–223. doi: 10.1016/j.ejmech.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Alghamdi N.J., Balaraman L., Emhoff K.A., Salem A.M.H., Wei R., Zhou A., Boyd W.C. Cobalt(II) Diphenylazodioxide Complexes Induce Apoptosis in SK-HEP-1 Cells. ACS Omega. 2019;4:14503–14510. doi: 10.1021/acsomega.9b01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira C.G., Romero-Canelón I., Coverdale J.P.C., Maia P.I.S., Clarkson G.J., Deflon V.M., Sadler P.J. Novel tetranuclear PdII and PtII anticancer complexes derived from pyrene thiosemicarbazones. Dalton Trans. 2020;49:9595–9604. doi: 10.1039/D0DT01133A. [DOI] [PubMed] [Google Scholar]
- 8.Lopes E.O., Oliveira C.G., Silva P.B., Eismann C.E., Suárez C.A., Menegário A.A., Leite C.Q.F., Deflon V.M., Pavan F.R. Novel Zinc(II) Complexes [Zn(atc-Et)2] and [Zn(atc-Ph)2]: In Vitro and in Vivo Antiproliferative Studies. Int. J. Mol. Sci. 2016;17:781. doi: 10.3390/ijms17050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitorino H.A., Mantovanelli L., Zanotto F.P., Espósito B.P. Iron Metallodrugs: Stability, Redox Activity and Toxicity against Artemia salina. PLoS ONE. 2015;10:e0121997. doi: 10.1371/journal.pone.0121997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez M.R., Plá J.D., Balsa L.M., León I.E., Piro O.E., Echeverría G.A., García-Tojal J., Pis-Diez R., Parajón-Costa B.S., González-Baró A.C. Cu(II) and Zn(II) complexes with a poly-functional ligand derived from o-vanillin and thiophene. Crystal structure, physicochemical properties, theoretical studies and cytotoxicity assays against human breast cancer cells. New J. Chem. 2019;43:7120–7129. doi: 10.1039/C8NJ06274A. [DOI] [Google Scholar]
- 11.Slator C., Molphy Z., McKee V., Long C., Brown T., Kellett A. Di-copper metallodrugs promote NCI-60 chemotherapy via singlet oxygen and superoxide production with tandem TA/TA and AT/AT oligonucleotide discrimination. Nucleic Acids Res. 2018;46:2733–2750. doi: 10.1093/nar/gky105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambika S., Manojkumar Y., Arunachalam S., Gowdhami B., Meenakshi Sundaram K.K., Solomon R.V., Venuvanalingam P., Akbarsha M.A., Sundararaman M. Biomolecular Interaction, Anti-Cancer and Anti-Angiogenic Properties of Cobalt(III) Schiff Base Complexes. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-39179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira C.G., da S Maia P.I., Souza P.C., Pavan F.R., Leite C.Q.F., Viana R.B., Batista A.A., Nascimento O.R., Deflon V.M. Manganese(II) complexes with thiosemicarbazones as potential anti-Mycobacterium tuberculosis agents. J. Inorg. Biochem. 2014;132:21–29. doi: 10.1016/j.jinorgbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Zoroddu M.A., Aaseth J., Crisponi G., Medici S., Peana M., Nurchi V.M. The essential metals for humans: A brief overview. J. Inorg. Biochem. 2019;195:120–129. doi: 10.1016/j.jinorgbio.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Rodionov D.A., Gelfand M.S., Gladyshev V.N. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:1–26. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang E.L., Simmers C., Knight D.A. Cobalt Complexes as Antiviral and Antibacterial Agents. Pharmaceuticals. 2010;3:1711–1728. doi: 10.3390/ph3061711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czarnek K., Terpiłowska S., Siwicki A.K. Selected aspects of the action of cobalt ions in the human body. Cent. Eur. J. Immunol. 2015;40:236–242. doi: 10.5114/ceji.2015.52837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heffern M.C., Yamamoto N., Holbrook R.J., Eckermann A.L., Meade T.J. Cobalt Derivatives as Promising Therapeutic Agents. Curr. Opin. Chem. Biol. 2013;17:189–196. doi: 10.1016/j.cbpa.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall M.D., Failes T.W., Yamamoto N., Hambley T.W. Bioreductive activation and drug chaperoning in cobalt pharmaceuticals. Dalton Trans. 2007;36:3983–3990. doi: 10.1039/b707121c. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz J.A., Lium E.K., Silverstein S.J. Herpes simplex virus type 1 entry is inhibited by the cobalt chelate complex CTC-96. J. Virol. 2001;75:4117–4128. doi: 10.1128/JVI.75.9.4117-4128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelosi G., Bisceglie F., Bignami F., Ronzi P., Schiavone P., Re M.C., Casoli C., Pilotti E. Antiretroviral Activity of Thiosemicarbazone Metal Complexes. J. Med. Chem. 2010;53:8765–8769. doi: 10.1021/jm1007616. [DOI] [PubMed] [Google Scholar]
- 22.Pahontu E., Julea F., Rosu T., Purcarea V., Chumakov Y., Petrenco P., Gulea A. Antibacterial, antifungal and in vitro antileukaemia activity of metal complexes with thiosemicarbazones. J. Cell Mol. Med. 2015;19:865–878. doi: 10.1111/jcmm.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonçalves A.C.R., Carneiro Z.A., Oliveira C.G., Danuello A., Guerra W., Oliveira R.J., Ferreira F.B., Veloso-Silva L.L.W., Batista F.A.H., Borges J.C., et al. PtII, PdII and AuIII complexes with a thiosemicarbazone derived from diacethylmonooxime: Structural analysis, trypanocidal activity, cytotoxicity and first insight into the antiparasitic mechanism of action. Eur. J. Med. Chem. 2017;141:615–631. doi: 10.1016/j.ejmech.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Andres S.A., Bajaj K., Vishnosky N.S., Peterson M.A., Mashuta M.S., Buchanan R.M., Bates P.J., Grapperhaus C.A. Synthesis, Characterization, and Biological Activity of Hybrid Thiosemicarbazone–Alkylthiocarbamate Metal Complexes. Inorg. Chem. 2020;59:4924–4935. doi: 10.1021/acs.inorgchem.0c00182. [DOI] [PubMed] [Google Scholar]
- 25.Pavan F.R., da S Maia P.I., Leite S.R.A., Deflon V.M., Batista A.A., Sato D.N., Franzblau S.G., Leite C.Q.F. Thiosemicarbazones, semicarbazones, dithiocarbazates and hydrazide/hydrazones: Anti—Mycobacterium tuberculosis activity and cytotoxicity. Eur. J. Med. Chem. 2010;45:1898–1905. doi: 10.1016/j.ejmech.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Rosu T., Gulea A., Nicolae A., Georgescu R. Complexes of 3dn Metal Ions with Thiosemicarbazones: Synthesis and Antimicrobial Activity. Molecules. 2007;12:782–796. doi: 10.3390/12040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira C.G., da S Maia P.I., Miyata M., Pavan F.R., Leite C.Q.F., de Almeida E.T., Deflon V.M. Cobalt(III) complexes with thiosemicarbazones as potential anti-Mycobacterium tuberculosis agents. J. Braz. Chem. Soc. 2014;25:1848–1856. doi: 10.5935/0103-5053.20140149. [DOI] [PubMed] [Google Scholar]
- 28.Di L., Kerns E.H. Lipophilicity Methods. In: Di L., Kerns E.H., editors. Drug-Like Properties. 2nd ed. Academic Press; Boston, MA, USA: 2016. pp. 299–306. [Google Scholar]
- 29.Zhang R., Qin X., Kong F., Chen P., Pan G. Improving cellular uptake of therapeutic entities through interaction with components of cell membrane. Drug Deliv. 2019;26:328–342. doi: 10.1080/10717544.2019.1582730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Echeverría J., Opazo J., Mendoza L., Urzúa A., Wilkens M. Structure-Activity and Lipophilicity Relationships of Selected Antibacterial Natural Flavones and Flavanones of Chilean Flora. Molecules. 2017;22:608. doi: 10.3390/molecules22040608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen S.M. A Bioinorganic Approach to Fragment-Based Drug Discovery Targeting Metalloenzymes. Acc. Chem. Res. 2017;50:1–21. doi: 10.1021/acs.accounts.7b00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meija J., Coplen T.B., Berglund M., Brand W.A., Bièvre P.D., Gröning M., Holden N.E., Irrgeher J., Loss R.D., Walczyk T., et al. Atomic weights of the elements 2013 (IUPAC Technical Report) Pure Appl. Chem. 2016;88:265–291. doi: 10.1515/pac-2015-0305. [DOI] [Google Scholar]
- 33.Mjos K.D., Orvig C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014;114:4540–4563. doi: 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- 34.Khan R.A., Tăbăcaru A., Ali F., Koo B.H. Anticancer and Antimicrobial Properties of Inorganic Compounds/Nanomaterials. Bioinorg. Chem. Appl. 2019;2019:1–2. doi: 10.1155/2019/6019632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rijt S.H., Sadler P.J. Current applications and future potential for bioinorganic chemistry in the development of anticancer drugs. Drug Discov. Today. 2009;14:1089–1097. doi: 10.1016/j.drudis.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P., Sadler P.J. Redox-Active Metal Complexes for Anticancer Therapy. Eur. J. Inorg. Chem. 2017;2017:1541–1548. doi: 10.1002/ejic.201600908. [DOI] [Google Scholar]
- 37.Maia P.I.S., Pavan F.R., Leite C.Q.F., Abram U., Lang E.S., Batista A.A., Deflon V.M. Thiosemicarbazone complexes of group 10 metals. Preparation, structural analysis and anti-Mycobacterium tuberculosis activity. In: Pele L., Powell J.J., Kinrade S., Jugdaohsingh R., Collery P., Maymard I., Badawi A., editors. Metal Ions in Biology and Medicine. Volume 11. John Libbey Eurotext; Paris, France: 2011. pp. 164–171. [Google Scholar]
- 38.O’Shea R., Moser H.E. Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 39.da S Maia P.I., Graminha A., Pavan F.R., Leite C.Q.F., Batista A.A., Back D.F., Lang E.S., Ellena J., Lemos S.D.S., Salistre-de-Araujo H.S., et al. Palladium(II) complexes with thiosemicarbazones: Syntheses, characterization and cytotoxicity against breast cancer cells and Anti-Mycobacterium tuberculosis activity. J. Braz. Chem. Soc. 2010;21:1177–1186. doi: 10.1590/S0103-50532010000700004. [DOI] [Google Scholar]
- 40.Pohjala L., Utt A., Varjak M., Lulla A., Merits A., Ahola T., Tammela P. Inhibitors of Alphavirus Entry and Replication Identified with a Stable Chikungunya Replicon Cell Line and Virus-Based Assays. PLoS ONE. 2011;6:e28923. doi: 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pahonțu E., Proks M., Shova S., Lupașcu G., Ilieș D.-C., Bărbuceanu Ș.-F., Socea L.-I., Badea M., Păunescu V., Istrati D., et al. Synthesis, characterization, molecular docking studies and in vitro screening of new metal complexes with Schiff base as antimicrobial and antiproliferative agents. Appl. Organomet. Chem. 2019;33:e5185. doi: 10.1002/aoc.5185. [DOI] [Google Scholar]
- 42.Gorgulu G., Cicek M.B., Dede B. Novel Aminoketooxime Ligand and Its Cu(II) and Mn(II) Complexes: Synthesis, Characterization and Molecular Docking Studies. Acta Phys. Pol. A. 2018;133:250–255. doi: 10.12693/APhysPolA.133.250. [DOI] [Google Scholar]
- 43.Richardson D.R., Kalinowski D.S., Richardson V., Sharpe P.C., Lovejoy D.B., Islam M., Bernhardt P.V. 2-Acetylpyridine thiosemicarbazones are potent iron chelators and antiproliferative agents: Redox activity, iron complexation and characterization of their antitumor activity. J. Med. Chem. 2009;52:1459–1470. doi: 10.1021/jm801585u. [DOI] [PubMed] [Google Scholar]
- 44.Santiago P.H.O., Santiago M.B., Martins C.H.G., Gatto C.C. Copper(II) and zinc(II) complexes with Hydrazone: Synthesis, crystal structure, Hirshfeld surface and antibacterial activity. Inorg. Chim. Acta. 2020;508:119632. doi: 10.1016/j.ica.2020.119632. [DOI] [Google Scholar]
- 45.D’Arrigo M., Ginestra G., Mandalari G., Furneri P.M., Bisignano G. Synergism and postantibiotic effect of tobramycin and Melaleuca alternifolia (tea tree) oil against Staphylococcus aureus and Escherichia coli. Phytomedicine. 2010;17:317–322. doi: 10.1016/j.phymed.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Davis J.L., Hodge H.M., Campbell W.E. Growth of Chikungunya Virus in Baby Hamster Kidney Cell (BHK-21-Clone 13) Suspension Cultures. Appl. Microbiol. 1971;21:338–341. doi: 10.1128/AEM.21.2.338-341.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mf S. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.