Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) mediates viral entry into cells and is critical for vaccine development against coronavirus disease 2019 (COVID-19). Structural studies have revealed distinct conformations of S, but real-time information that connects these structures is lacking. Here we apply single-molecule fluorescence (Förster) resonance energy transfer (smFRET) imaging to observe conformational dynamics of S on virus particles. Virus-associated S dynamically samples at least four distinct conformational states. In response to human receptor angiotensin-converting enzyme 2 (hACE2), S opens sequentially into the hACE2-bound S conformation through at least one on-path intermediate. Conformational preferences observed upon exposure to convalescent plasma or antibodies suggest mechanisms of neutralization involving either competition with hACE2 for binding to the receptor-binding domain (RBD) or allosteric interference with conformational changes required for entry. Our findings inform on mechanisms of S recognition and conformations for immunogen design.
Keywords: SARS-CoV-2 spike protein, single-molecule FRET, real-time conformational dynamics, receptor ACE2, antibody neutralization
Graphical Abstract
Highlights
-
•
SARS-CoV-2 S protein dynamically samples at least 4 distinct conformational states
-
•
hACE2 activates S from the ground state to the activated state via an intermediate
-
•
Proteolytic processing of S accelerates hACE2-dependent activation
-
•
Antibodies can antagonize S by two different mechanisms of neutralization
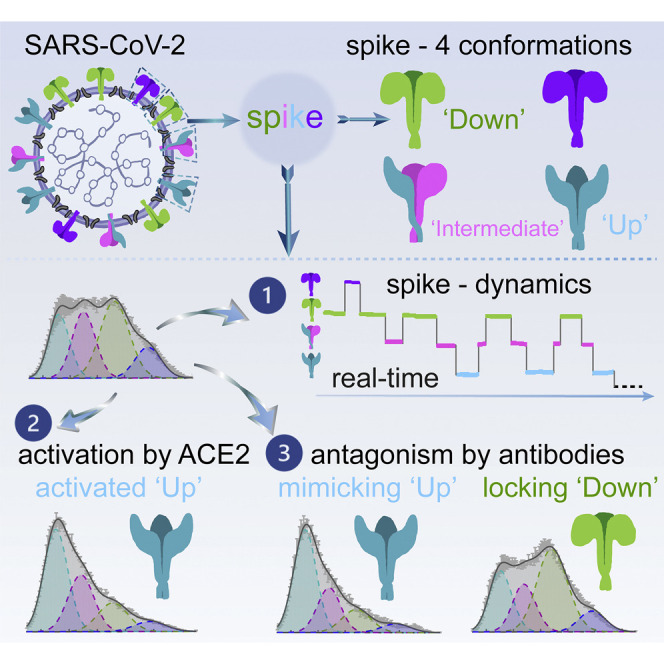
The SARS-CoV-2 spike protein has been observed to adopt different structural states. Lu et al. directly visualize the conformational dynamics of spike protein on the surface of virus particles and describe how the conformational landscape changes upon activation by the host receptor or antagonism by antibodies.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) mediates viral entry into cells and is a main target for antibody responses against the virus (Gao et al., 2020; Jackson et al., 2020; Mercado et al., 2020; Yu et al., 2020). S is synthesized as a precursor, processed into S1 and S2 by furin proteases, and activated for fusion when human angiotensin-converting enzyme 2 (hACE2) engages the receptor-binding domain (RBD) and when the N terminus of S2 is proteolytically processed (Hoffmann et al., 2020b; Walls et al., 2020; Wrobel et al., 2020). Structures of soluble ectodomains and native virus particles have revealed distinct conformations of S, including a closed trimer with all RBDs oriented downward, trimers with one or two RBDs up, and hACE2-stabilized conformations with up to three RBDs oriented up (Benton et al., 2020; Cai et al., 2020; Henderson et al., 2020; Hsieh et al., 2020; Ke et al., 2020; Lan et al., 2020; McCallum et al., 2020; Shang et al., 2020; Walls et al., 2020; Wang et al., 2020; Wrapp et al., 2020b; Wrobel et al., 2020; Yao et al., 2020; Zhou et al., 2020). Real-time information that connects these structures, however, has been lacking. Single-molecule fluorescence (Förster) resonance energy transfer (smFRET) is well suited to inform on conformational dynamics of proteins reporting domain movements in the millisecond to second range and has previously been applied to study HIV-1, influenza A, and Ebola spike glycoproteins via measurements of the distance-dependent energy transfer from an excited donor to a nearby acceptor fluorophores in real time (Das et al., 2020; Das et al., 2018; Lu et al., 2019a; Ma et al., 2018; Munro et al., 2014).
Results
Establishing Real-Time Observations of SARS-Cov-2 Spikes on Virus Particles
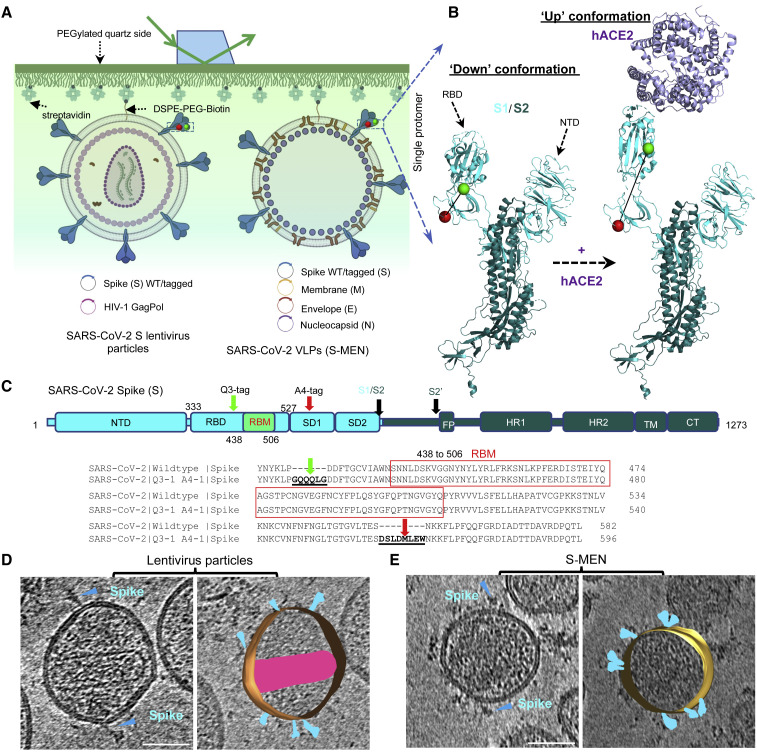
To probe dynamics of SARS-CoV-2 spikes, we used available high-resolution structures of the SARS-CoV-2 S trimer to identify sites of fluorophore pair labeling that have the potential to inform on distance changes expected to accompany conformational changes between the RBD-down and receptor hACE2-induced RBD-up trimer structures (Lan et al., 2020; Wrapp et al., 2020b) (Figures 1A and 1B). Accordingly, we engineered A4 and Q3 labeling peptides before and after the receptor-binding motif (RBM) to allow site-specific introduction of donor and acceptor fluorophores at these positions (Figures 1B and 1C). We optimized retroviral and lentiviral pseudoviral particles carrying the SARS-CoV-2 S protein (Figure S1A) to test the impact of these peptides on infectivity and found that they were well tolerated, both individually and in combination (Figures S1B and S1C). To measure conformational dynamics of the SARS-CoV-2 S trimer on the surface of virus particles, we applied the S protein carrying the Q3 and A4 labeling peptides at positions 427 and 556, respectively (Figure 1C). We established two types of particles, lentiviral particles carrying S, as well as coronavirus-like particles generated by expression of S, membrane (M), envelope (E), and nucleocapsid (N) protein (S-MEN) (Gordon et al., 2020; Siu et al., 2008) (Figure 1A). S-MEN particles co-express coronavirus surface proteins M and E. Particle quality and the presence of the corona-like S proteins on both particle surfaces were confirmed by cryo-electron microscopy (Figures 1C and 1D).
Figure 1.
Experimental Design for Single-Molecule FRET Imaging of Real-Time Conformational Dynamics of the SARS-CoV-2 Spike Protein on Virus Particles
(A and B) Experimental set-up.
(A) Virus particles carrying a two-dye-labeled SARS-CoV-2 spike protein protomer among wildtype spikes were immobilized on a quartz slide and imaged on a customized prism-based total internal reflection fluorescence (TIRF) microscope. The quartz slide was passivated with PEG/PEG-biotin to allow coating with streptavidin and subsequent immobilization of viruses carrying a biotin-lipid (DSPE-PEG-biotin). Two virus particle systems were developed to incorporate SARS-CoV-2 spike proteins on their surfaces. HIV-1 lentivirus particles comprise HIV-1 cores and SARS-CoV-2 spike proteins on the surface. S-MEN virus-like particles consist of four structural proteins, in which S represents spike, M the membrane protein, E the envelope protein, and N the nucleocapsid protein of SARS-CoV-2.
(B) Placement of labeling tags for the site-directed introduction of fluorophores (Cy3B, green; LD650, red) was guided by conformational changes in S1 induced by binding of the cellular receptor hACE2 from the “RBD-down” to the “RBD-up” conformation. NTD, N-terminal domain. Structures were adapted from RCSB PDB: 6VSB (“Down” S1/S2 protomer: S1, light cyan; S2, dark blue) and 6VYB/6M0J (“Up” protomer S1/S2 engaged with hACE2: hACE2, magenta).
(C) Domain organization of full-length (1,273 amino acids) wild-type SARS-CoV-2 spike protein (S1+S2) colored by domain and sequence for the region carrying the peptide-labeling tags. The insertion sites of labeling tags Q3 and A4 for the donor and acceptor fluorophores are indicated. S1/S2, S2′, protease cleavage sites; SD1, subunit domain 1; SD2, subunit domain 2; FP, fusion peptide; HR1/HR2, heptad repeat 1/heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Peptide insertion sites into RBD and SD1 before and after RBM for the dually tagged S protein applied in these studies.
(D and E) A representative tomographic slice (left) and the segmented 3D representation (right) of HIV-1 lentivirus particles carrying S (D) and S-MEN corona viral-like particles (E). Spikes in cyan; HIV-1 capsid in pink. The scale bar represents 50 nm.
For smFRET, lentivirus particles and S-MEN particles were generated (see STAR Methods) by transfecting HEK293T cells with an excess of plasmid-encoding wild type, doped with trace amounts of plasmid expressing labeling-peptide-carrying S (427-Q3/556-A4) to ensure the production of virus particles that contain, on average, only a single engineered S protein. As for analogous investigations of HIV-1 E protein (Lu et al., 2019a; Munro et al., 2014), donor (Cy3B[3S]) and acceptor fluorophore (LD650) were enzymatically conjugated to the engineered S proteins presented on the virus particle surface in situ (see STAR Methods). A biotinylated lipid was then incorporated into the virus particle membrane to allow their immobilization within passivated microfluidic devices coated with streptavidin to enable imaging by total internal reflection microscopy (Figure 1A). Donor fluorophores on single, immobilized virus particles were excited by a single-frequency 532-nm laser, and fluorescence emissions from both donor and acceptor fluorophores were recorded at 25 Hz (Figure 2 A). From the recorded movies, we computationally extracted hundreds of smFRET traces exhibiting anti-correlated donor and acceptor fluorescence intensities, the telltale signature of conformational changes within the S protein on individual virus particles.
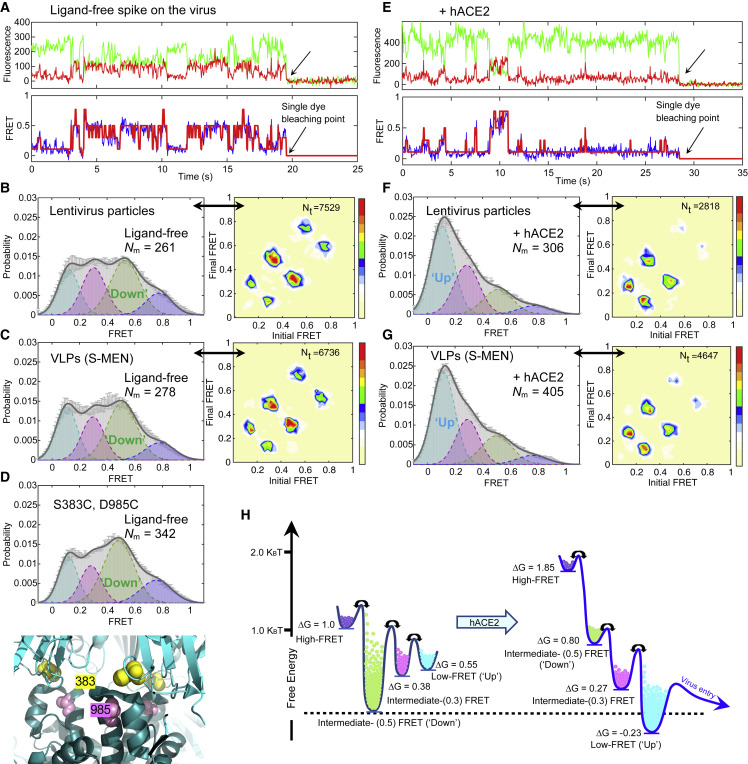
Figure 2.
SARS-CoV-2 Spike Protein Is Dynamic, and hACE2 Shifts Conformational Landscape from the Ground State to the Receptor-Bound State through One Necessary Intermediate
(A–D) The ligand-free S on virus particles primarily resides in “RBD-down” conformation (ground state).
(A) Example fluorescence trace (Cy3B, green; LD650, red) and resulting quantified FRET traces (FRET efficiency, blue; hidden Markov model initialization, red) of a dually labeled ligand-free spike protein on the surface of HIV-1 lentivirus particle. Arrows point to the single-step photobleaching steps of dyes at the single-molecule level and define the baseline.
(B and C) FRET histograms (left) and TDPs (right) of ligand-free spikes on lentivirus particles (B) and S-MEN viral-like particles (C). Also shown is the number (Nm) of individual dynamic molecules/traces compiled into a conformation-population FRET histogram (gray lines) and fitted into a 4-state Gaussian distribution (solid black) centered at 0.1-FRET (dashed cyan), 0.3-FRET (dashed red), 0.5-FRET (dashed green), and 0.8-FRET (dashed magenta). TDPs, displayed as initial FRET versus final FRET with relative frequencies, trace the locations of state-to-state transitions and their relative frequencies (max red scale = 0.01 transitions/second), originated from the idealization of individual FRET traces in FRET histograms.
(D) A modified spike (S383C and D985C) (Henderson et al., 2020; McCallum et al., 2020) stabilized in RBD-down conformation, observed from the FRET histogram (upper panel). The small increase in the population of the ground state (~0.5 FRET) likely reflects the partial nature of the formation of the disulfide in this mutant, which has 40% the infectivity of wild-type (Figure S3C). Modified S383C and D985C depicted in the high-resolution structure of S 6ZOY (lower panel).
(E–G) Experiments as in (A)–(C), respectively, conducted in the presence of 200 μg/mL monomeric hACE2. The soluble hACE2 activates spike proteins on the virus by shaping the conformational landscape toward stabilizing the RBD-up conformation (activated state). FRET histograms represent mean ± SEM, determined from three randomly assigned populations of all FRET traces under corresponding experimental conditions. N, number of individual FRET traces. Evaluated state occupancies see Table S1.
(H) Relative free-energy model of conformational landscapes of SARS-CoV-2 spikes in response to the hACE2 binding. The differences in free energies between states were roughly scaled based upon relative state occupancies of each state.
S Is Dynamic in Real Time, Sampling at Least Four Distinct Conformational States on the Virus Surface
Analyses of smFRET data from ligand-free S protein on lentiviral particles revealed that the SARS-CoV-2 S protein is dynamic, sampling at least four distinct conformational states characterized by low (∼0.1), intermediate (∼0.3 and ∼0.5), and high (∼0.8) FRET efficiency (FRET). Population FRET histograms, comprised of hundreds of smFRET traces, revealed that the conformation exhibiting intermediate FRET (∼0.5) to be the most abundantly occupied (Figure 2B; Table S1). Comparable findings were obtained for S protein incorporated into S-MEN coronavirus-like particles (Figure 2C). Notably, the intermediate-FRET (∼0.5) conformation of the S protein was stabilized by a disulfide bridge between amino acids 383 and 985 (Figures 2D and S2A–S2C). Because electron microscopy (EM) methods have identified the disulfide-bridge stabilized state as the three-RBD-down conformation (Henderson et al., 2020; McCallum et al., 2020), these experiments suggest that the intermediate-FRET (∼0.5) S protein conformation represents the closed trimer with all three RBDs oriented downward (RBD-down conformation).
S Exhibits a Defined Sequence of Conformational Transitions, and Receptor hACE2 Activates S from the Ground State to the Receptor-Bound State through One Necessary Intermediate
To identify the receptor-bound conformation of the SARS-CoV-2 S protein by smFRET, we measured the conformational consequences of soluble, monomeric hACE2 binding. The addition of the monomeric hACE2 receptor to surface-immobilized virus particles leads to increased occupancy of the low-FRET (∼0.1) S protein conformation from ∼22% to 48% (Figure 2E; Table S1), which was observed at the single-molecule level (Figure 2F). Similar hACE2 receptor impacts on the SARS-CoV-2 S protein were observed in both lentiviral particle and S-MEN coronavirus-like particle contexts (Figures 2E–2G). Dimeric hACE2, a more potent ligand (Figure S2D) (Lui et al., 2020), stabilized the low-FRET (∼0.1) S protein conformation more efficiently from ∼22% to 58% (Figures S2E and S2F), suggesting that the observed low-FRET state (demonstrated in Figures 2E, S2G, and S2H) likely represents the receptor-bound conformation in which all three RBD domains are oriented upward (RBD-up conformation).
A unique strength of single-molecule imaging is its capacity to directly reveal both the structural and kinetic features that define biological function (Lu et al., 2019b; Roy et al., 2008). To extract such information for the SARS-CoV-2 S protein, we employed hidden Markov modeling (HMM) (McKinney et al., 2006) to idealize individual smFRET traces. These data allowed quantitative analyses of the thousands of discrete FRET transitions observed for the S protein on the surface of lentiviral and S-MEN particles in the absence and presence of ligands to gain insights into the order and timing of conformational changes across conditions (Figures 2B, 2C, 2F, 2G, and S3). Transition density plots (TDPs), which conveniently display the nature, order, and frequency of the transitions observed in the ensemble of molecules examined (Roy et al., 2008), immediately revealed two salient features that informed on the nature of S protein dynamics. First, the symmetry of the evidenced transitions in each TDP with respect to the diagonal axis indicated that the unliganded S protein is in equilibrium exchange between distinct conformations under ambient conditions—i.e., physiological buffer at room temperature. Second, the transitions evidenced in the TDP appeared to exhibit a defined transition order, from low- to intermediate-FRET and from intermediate- to high-FRET conformations. These findings suggest that the SARS-CoV-2 S protein predominantly exhibits a defined sequence of activating structural transitions wherein the ground-state RBD-down conformation (intermediate- FRET; ∼0.5) converts to the receptor-activated RBD-up conformation (low-FRET; ∼0.1) via at least one intermediate-FRET (∼0.3) conformation. They further reveal that the RBD-down conformation of the S protein can reversibly access at least one additional high-FRET (∼0.8) conformation. The most frequent transitions evidenced were those between RBD-down (∼0.5 FRET) and the on-path intermediate-FRET (∼0.3) states, from which relatively infrequent transitions to the low-FRET (∼0.1), RBD-up conformation—akin to those observed upon hACE2 binding—could be achieved spontaneously.
As expected, the binding of the hACE2 receptor modified the dynamic S protein conformational landscape toward the RBD-up conformation (∼0.1 FRET), rendering it the most populated (Figures 2B, 2C, 2F, and 2G). This change resulted from an increased transition rate from the RBD-down conformation (∼0.5 FRET) toward the intermediate-FRET (∼0.3) state and subsequently the RBD-up (∼0.1 FRET) conformation, which was also modestly stabilized. The energy barriers for reverse transitions toward the RBD-down conformation (∼0.5 FRET) were also elevated, explaining receptor-bound conformation accumulation over time (Figure S3). These analyses lead to a qualitative model for hACE2 activation of the SARS-CoV-2 S protein from the ground state to the receptor-bound state through at least one intermediate conformation (Figure 2H). The summary of relative state occupancies and transition rates among conformations, as well as errors, are listed in Tables S1 and S2, respectively.
Proteolytic Processing by Serine Protease Trypsin Stimulates hACE2-Dependent Activation of S
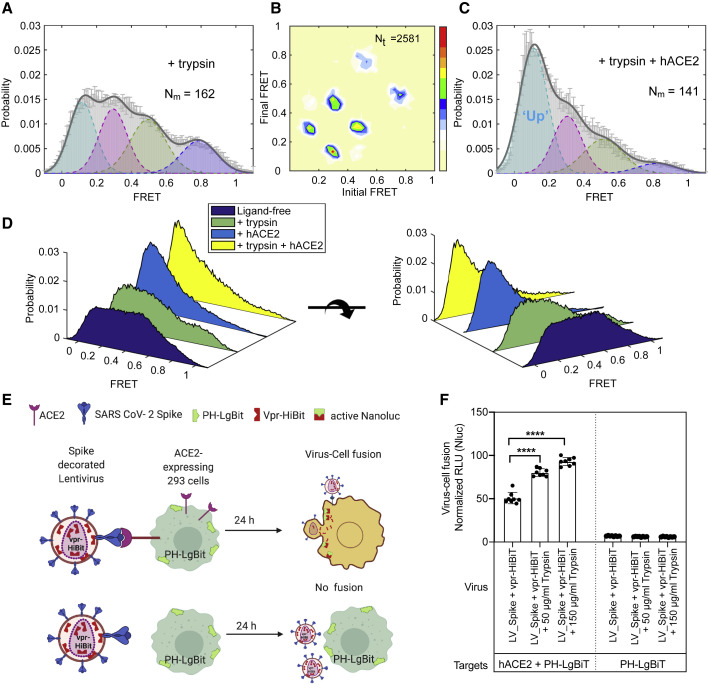
In most cell types, the serine protease TMPRSS2 is required for pH-independent SARS-CoV-2 entry (Bestle et al., 2020; Hoffmann et al., 2020a; Hoffmann et al., 2020b). In vitro, the actions of TMPRSS2 are mimicked by the serine protease trypsin, which has similar cleavage specificity (Bestle et al., 2020; Hoffmann et al., 2020b). smFRET analysis of trypsin-treated S protein on lentiviral particles in the absence of receptor revealed a clear shift toward activation (Figures 3A and 3B). After trypsin treatment, the addition of hACE2 receptor was more effective at stabilizing the S protein in the RBD-up (∼0.1 FRET) conformation (Figures 3C, 3D, and S4A).
Figure 3.
Conformational Effects of Trypsin Treatment of Spikes Follow the hACE2-Dependent Activation Pathway
(A–C) The serine protease trypsin remodels conformational landscape of spike proteins toward down-stream conformations on the path of hACE2-dependent activation.
(A and B) The FRET histogram (A) and TDP (B) of spike proteins on HIV-1 lentivirus particles in the presence of 50 μg/mL trypsin.
(C) An experiment as in (A), for spikes in the presence of both 50 μg/mL trypsin and 200 μg/mL hACE2.
(D) Three-dimensional presentations of FRET histograms of spike proteins on the virus in the presence and the absence of hACE2 and trypsin. FRET histograms represent mean ± SEM, determined from three randomly assigned populations of FRET traces. For evaluated state occupancies, see Table S1.
(E and F) Trypsin enhances SARS-CoV-2 spike-mediated hACE2-dependent virus-cell fusion.
(E) Assay design to monitor virus-cell fusion using the HiBit and LgBiT split NanoLuc system (Yamamoto et al., 2019). Vpr-HiBit was packaged into lentiviral particles carrying SARS-CoV-2 spike (LV_Spike). HEK293 target cells transiently expressing LgBiT tagged to PH domain of human phospholipase Cδ at the N terminus alone or together with hACE2. hACE2-dependent virus-cell fusion was determined by monitoring reconstituted NanoLuc activity in target cells 24 h after infection.
(F) Normalized relative luciferase units (RLU; mean ± SD, two replicates with quadruplicates) measured 24 h post-infection to quantify virus-cell fusion in stated target cells after treating viruses with or with indicated amounts of trypsin for 15–20 min at 37°C. NanoLuc activities were normalized to luciferase activity detected in uninfected target cells. p values derived from unpaired t test; ∗∗∗∗ corresponds to p < 0.0001.
To further validate this finding, we measured the effects of trypsin pre-treatment in virus-cell and cell-cell fusion assays by using split NanoLuc system consisting of LgBiT and HiBiT (Figures 3E, 3F, S4B, and S4C). Here, membrane fusion restored luciferase activity between lentiviral particles carrying the S protein as well as a Vpr-HiBiT fusion protein with cells expressing the LgBiT counterpart fused to a PH domain. This assay revealed fusion to be strictly dependent on the hACE2 receptor and to be stimulated by trypsin treatment (Figures 3E and 3F). Nearly identical results were observed for cell-cell fusion between donor cells expressing S and target cells expressing hACE2 (Figures S4B and S4C), confirming the activating role of trypsin treatment.
Spike-Conformational Preferences of RBD-Directed Monoclonal Antibodies and Convalescent Patient Plasma
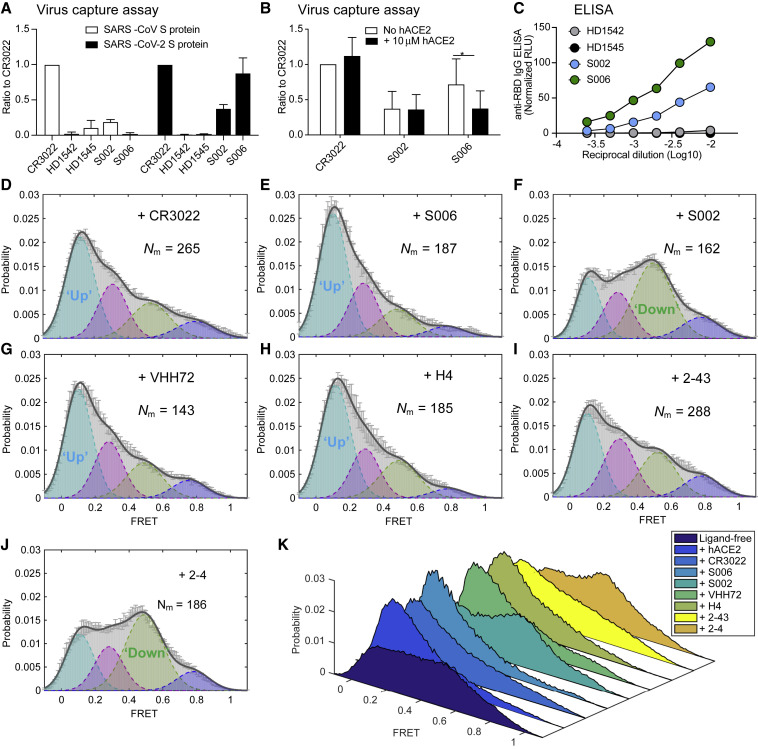
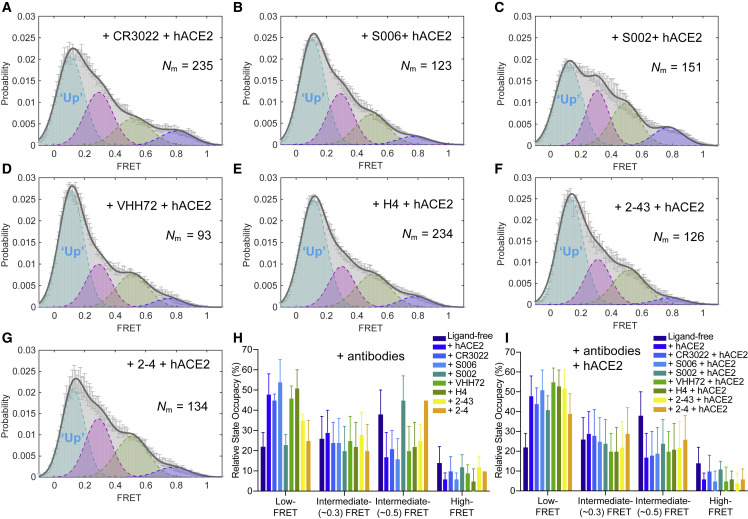
We next explored the suitability of the smFRET assay to characterize the conformational consequences of antibody binding to the SARS-CoV-2 S protein. Multiple studies on antibodies generated from coronavirus disease 2019 (COVID-19) patients have shown that one type of antibody often dominates immune responses (Barnes et al., 2020; Brouwer et al., 2020; Ju et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Wu et al., 2020a). This prompted us to screen plasma from convalescent patients with neutralizing activity that can bind to the S protein on lentiviral particles (Beaudoin-Bussières et al., 2020) by using a modified virus-capture assay (VCA) (Ding et al., 2019). Cross-reactive CR3022 (Yuan et al., 2020), one of the very first reported antibodies from SARS-CoV-1 that also binds to SARS-CoV-2 S RBD domain, served as a good indicator of RBD binding (Figure 4 A). We identified two plasma samples (S002 and S006) able to specifically bind the RBD, recognize S expressed at the cell surface and to neutralize viral particles (Figures 4A–4C and S5). smFRET analysis of antibody-bound S revealed that both CR3022 and plasma from patient S006 stabilized S in the RBD-up (∼0.1 FRET) conformation, as analogous to receptor hACE2 (Figures 4D and 4E). These data point to the presence of RBD-directed antibodies in patient S006. By contrast, smFRET indicated that plasma from patient S002 contained an activity that stabilized the RBD-down (∼0.5 FRET) conformation (Figure 4F). Plasma S002 antagonized hACE2 binding, but RBD competition did not affect its recognition of S, suggesting that its neutralization activity does not solely rely on blocking the receptor interface. We then assessed the conformational preference of four RBD-directed antibodies: the neutralization nanobody VHH72, and the potently neutralizing antibodies H4, 2-43, and 2-4, each of which binds RBD in a different way (Liu et al., 2020; Wrapp et al., 2020a; Wu et al., 2020b). Nanobody VHH72 and antibody H4 stabilized the S protein in an RBD-up (∼0.1 FRET) conformation similar to hACE2, CR3022, and S006 (Figures 4G and 4H), whereas the very potent neutralizing antibody 2-43 (Liu et al., 2020) showed a partial shift to the RBD-up (∼0.1 FRET) conformation (Figure 4I). Meanwhile, antibody 2-4 shifted the conformational landscape toward RBD-down (∼0.5 FRET) conformation (Figure 4J), similar to S002 (Figure 4K). The absence or presence of hACE2 did not appear to affect the RBD-up stabilization evidenced for antibodies CR3022, S006, VHH72, or H4 (Figure 5 ). However, plasma S002, and to a lesser extent antibody 2-4, reduced the hACE2-dependent stabilization of the RBD-up (∼0.1 FRET) conformation (Figure 5), suggesting that they could interfere with hACE2 receptor binding via an allosteric mechanism. These findings indicate that SARS-CoV-2 neutralization can be achieved in two ways: (1) antibodies that conformationally mimic hACE2 and directly compete with hACE2 receptor binding or (2) by allosterically stabilizing the S protein in its RBD-down conformation.
Figure 4.
Spike-Conformational Preferences of RBD-Directed Monoclonal Antibodies and Convalescent Patient Plasma
(A and B) Bar graphs of a VCA showing that convalescent plasma from SARS-CoV-2-positive patients S006 and S002 can bind to SARS-CoV-2 S on viral particles but not to SARS-CoV-1 S in reference to a cross-reactive SARS-CoV-1 monoclonal antibody CR3022.
(B) The competition VCA depicts sensitivity of S006 binding to S to soluble ACE2. p values derived from unpaired t test; ∗ corresponds to p < 0.05. Plasma from healthy donors HD1542 and HD1545 as controls. S006, S002, HD1542, and HD1542 represent plasma. Virus capture experiments were repeated three times and represented as mean ± SD.
(C) The binding affinity of both S006 and S002 toward SARS-CoV-2 RBD in comparison to plasma from heathy donors by anti-RBD IgG ELISA.
(D–J) FRET histograms of CR3022 (D) at 200 μg/mL, convalescent patient plasma from S006 (E) and S002 (F) at 1:100 dilution, and RBD-directed monoclonal antibodies VHH72 (G), H4 (H), 2-43 (I), and 2-4 (J) at 200 μg/mL.
(K) Three-dimensional presentations of FRET histograms of spikes on the retrovirus under different conditions (D)–(I) in reference to ligand-free (Figure 2B). FRET histograms represent mean ± SEM, determined from three randomly assigned populations of FRET traces. For state occupancies see Table S1.
Figure 5.
hACE2 Mediate the Antibodies-Equilibrated Conformational Landscape toward Down-Stream Conformations
(A–G) FRET histograms of SARS-CoV-2 S on the lentivirus in the presence of both 200 μg/mL of indicated antibody and 200 μg/mL hACE2, or 1:100 dilution of indicated plasma and 200 μg/mL hACE2. FRET histograms represent mean ± SEM, determined from three randomly assigned populations of all FRET traces.
(H and I) Evaluated relative state occupancies of S on the lentivirus in the presence of indicated individual antibody (H) as in Figures 4D–4G and both indicated antibody and hACE2 (I) as in (A)–(G). Relative state occupancies are presented as mean ± SEM, derived from histograms. Determining parameters of state occupancies see Table S1.
Discussion
The strength of the presented smFRET approach is revealed by the capacity to examine the dynamic properties of the S protein in real time, including the following: (1) the distinct conformational states that it spontaneously transits under physiological conditions, (2) the impact of sequence alterations on S protein dynamics, and (3) the responses of the S protein to cognate hACE2 receptor and antibody recognition.
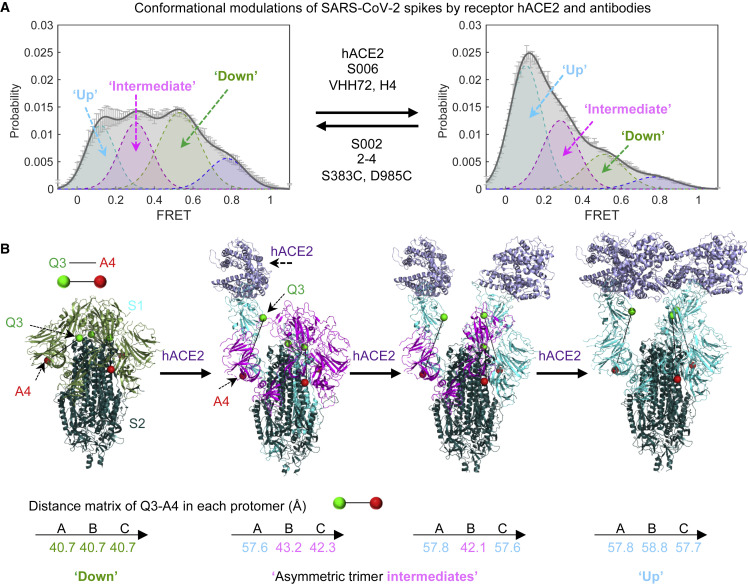
The present analyses of dynamic S protein molecules provide three lines of evidence that indicate that the intermediate-FRET (∼0.5) state observed represents the RBD-down, ground-state conformation of the S protein, in which all three RBD domains are oriented toward the viral particle membrane (Figure 6 ). First, in line with previous EM investigations (Cai et al., 2020; Henderson et al., 2020; Hsieh et al., 2020; Ke et al., 2020; Lan et al., 2020; McCallum et al., 2020; Shang et al., 2020; Walls et al., 2020; Wang et al., 2020; Wrobel et al., 2020; Yao et al., 2020), the RBD-down state is the most populated. In further agreement with recent EM studies, both a disulfide bridge (S383C, D985C) (Henderson et al., 2020; McCallum et al., 2020) and antibody 2-4 stabilized the S protein in a conformation with all three RBDs oriented down (Liu et al., 2020). Although our smFRET observations highlight considerable conformational flexibility in these contexts in comparison with that seen in EM of soluble trimers, these distinctions could be attributed to a tendency of our analysis approach to over-emphasize dynamic features, while EM could over-emphasize static conformations rigidified by cryogenic temperatures that could be more readily identified and classified (Li et al., 2020). Multiple lines of evidence also facilitated assignment of the RBD-up (∼0.1 FRET) conformation of the S protein with all three RBD domains oriented away from the virus particle membrane (Figure 6). For instance, this conformation was stabilized by soluble monomeric hACE2 receptor and even further stabilized in the presence of soluble, dimeric hACE2 receptor as well as RBD-targeting antibodies, such as CR3022, that are known to access their epitopes when the S protein is in an activated, RBD-up conformation (Wrapp et al., 2020a; Wu et al., 2020b; Yuan et al., 2020). The structure of the on-path (∼0.3 FRET) intermediate observed during S opening could represent an initial receptor-binding conformation that is globally similar in its overall architecture to the all-RBD down ground state. Cryo-EM structures of soluble SARS-CoV-2 S trimers (Wrapp et al., 2020b), which are asymmetrically engaged with only one or two hACE2 molecules receptors—rather than one for each S protomer (Zhou et al., 2020), reveal that the distance between the two labeling sites increases in the ligand-free protomers adjacent to a protomer engaged to hACE2 (Figure 6). The additional, highly compacted S conformation (∼0.8 FRET) evidenced, which is also depopulated by activating ligands, remains unknown.
Figure 6.
Conceptualization of the Observed FRET States within Current High-Resolution Structures
(A) Modulation of the conformational landscape observed by smFRET upon binding of receptor hACE2 or antibodies. The four conformational states are 0.1-FRET (dashed cyan), 0.3-FRET (dashed red), 0.5-FRET (dashed green), and 0.8-FRET (dashed magenta). smFRET histograms for ligand-free S and hACE2-bound are from Figures 2B and 2F, respectively.
(B) Interpretation of three of the observed smFRET states within the framework of cryo-EM structures of a soluble trimeric SARS-CoV-2 S, with GSAS and PP mutations and the T4 phage fibritin trimerization domain (Wrapp et al., 2020b). Ligand-free (PDB: 6VXX) and recognizing single, double, and triple ACE2 receptors molecules (Zhou et al., 2020), showing the distances between Q3-1 and A4-1 insertion sites on S1 in each protomer. The same color code is applied for conformational states within the smFRET histogram, individual S1 protomer in each cryo-EM structure and the distances between the fluorophores in each protomer of the spike trimer. See Discussion for details.
These smFRET analyses are thus in global agreement with the conformational states observed at the single-particle EM and cryoET level (Cai et al., 2020; Henderson et al., 2020; Ke et al., 2020; Klein et al., 2020; Lan et al., 2020; McCallum et al., 2020; Shang et al., 2020; Song et al., 2018; Walls et al., 2020; Wang et al., 2020; Wrapp et al., 2020a; Wrapp et al., 2020b; Wu et al., 2020b; Yao et al., 2020; Yuan et al., 2020), while also revealing dynamic information relevant to S protein activation and ligand recognition. The observed FRET states, interpreted within the framework provided by extant high-resolution structures, results in a working model for S activation (Figure 6). The model agrees with the expected increase in the distance between the labeling-peptide insertion sites that carry the fluorophores in the RBD-down and RBD-up conformations of the S trimer upon sequentially engaging with receptor hACE2 molecules. We note in this context that the FRET-defined conformations evidenced using the present labeling strategies are in line with measured distances in both symmetric S configurations (three-RBD-down or three-RBD-up) and asymmetric S configurations (one-RBD-up or two-RBD-up).
The capacity to examine the conformational preferences of RBD-directed antibodies to the S protein enabled us to identify conformational signatures of antibodies in patient plasma. This approach identified patients with antibody activities that either mimicked ACE2 (indicating anti-RBD activity) or stabilized the ground state of S, thereby interfering with hACE2 receptor-mediated activation of the SARS-CoV-2 S protein. We anticipate that ground-state stabilization by antibodies, small molecules, or the rational engineering of trimers could represent an effective avenue of antagonism as well as immunogen design.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cross-reactive SARS-CoV-1 monoclonal antibody CR3022 | ter Meulen et al., 2006 | RRID: AB_2848080 |
| Anti-SARS-CoV-2 monoclonal antibody VHH72 | Wrapp et al., 2020a | N/A |
| Anti-SARS-CoV-2 monoclonal antibody H4 | Wu et al., 2020b | N/A |
| Anti-SARS-CoV-2 monoclonal antibody 2-4 | Liu et al., 2020 | N/A |
| Anti-SARS-CoV-2 monoclonal antibody 2-43 | Liu et al., 2020 | N/A |
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody, HRP | Invitrogen | Cat # A18823; RRID: AB_2535600 |
| Biological Samples | ||
| Human Plasma from SARS-CoV-2-infected or uninfected donors | Beaudoin-Bussières et al., 2020; Prévost et al., 2020 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| GIBCO™ RPMI 1640 medium | Thermo Fisher Scientific | Cat # 11875093 |
| GIBCO™ Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | Cat # 11965118 |
| GIBCO™ MEM Non-essential amino acid (NEAA) solution | Thermo Fisher Scientific | Cat # 11140050 |
| GIBCO™ Penicillin-streptomycin solution (10,000 U/mL) | Thermo Fisher Scientific | Cat # 15140122 |
| GIBCO™ Dulbecco’s Phosphate Buffered Saline (DPBS) | Thermo Fisher Scientific | Cat # 14190144 |
| GIBCO™ L-Glutamine (200mM) | Thermo Fisher Scientific | Cat # 25030081 |
| GIBCO™ 0.05% Trypsin-EDTA, phenol red | Thermo Fisher Scientific | Cat # 25300054 |
| GIBCO™ Trypsin-EDTA (0.5%), no phenol red | Thermo Fisher Scientific | Cat # 15400054 |
| Fetal bovine serum | Atlanta Biologicals | Cat # S11550 |
| Tris-buffered saline (TBS) | Thermo Fisher Scientific | Cat # BP24711 |
| Bovine Serum Albumin (BSA) | BioShop | Cat # ALB001.100 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate |
Perkin Elmer Life Sciences | Cat # NEL105001EA |
| Tween20 | Thermo Fisher Scientific | Cat # BP337-500 |
| Passive lysis buffer | Promega | Cat # E1941 |
| D-Luciferin potassium salt | Thermo Fisher Scientific | Cat # L2916 |
| Transglutaminase | Sigma | Cat # 80146-85-6 |
| AcpS (acyl-carrier-protein synthase) | Mothes Lab | N/A |
| DSPE-PEG2000-biotin | Avanti Polar Lipids | Cat # 880129P |
| Streptavidin | Invitrogen | Cat # S888 |
| Opti-prep | Sigma | Cat # D1556 |
| PEG-passivated, streptavidin-coated quartz slides | This Study | N/A |
| Trolox | Sigma | Cat # 238813 |
| FuGENE 6 | Promega | Cat # E2311 |
| Cyclooctatetraene | Sigma | Cat # 138924 |
| Nitrobenzyl alcohol | Sigma | Cat # N12821 |
| Protocatechuic acid | Sigma | Cat # 37580 |
| Protoatechuate 3,4-deoxygenase | Sigma | Cat # P8279 |
| Acetone, EM-Grade, Glass-Distilled | Electron Microscopy Sciences | Cat #10015 |
| Cy3B(3S)-cadaverine or Cy3B or Cy3B(3S) | Lumidyne Technologies | N/A |
| LD650-CoA | Lumidyne Technologies | N/A |
| Monomeric hACE2 | Zhou et al., 2020a | N/A |
| Dimeric hACE2 | Zhou et al., 2020a | N/A |
| Turbo293 transfection reagent | SPEED BioSystems | Cat # PXX1002 |
| rProtein A Sepharose Fast Flow | GE Healthcare | Cat # 17–1279-03 |
| Pierce™ IgG Elution Buffer | ThermoFisher Scientific | Cat # 21004 |
| Critical Commercial Assays | ||
| KAPA SYBR FAST qPCR Master Mix (2X) Kit | KAPA Biosystems | Cat # KK4600 and KK4601 |
| Nano-Glo Luciferase Assay System | Promega | Cat # N1120 |
| Pierce™ Gaussia Luciferase Glow Assay Kit | ThermoFisher Scientific | Cat # 16158 |
| Experimental Models: Cell Lines | ||
| HEK293 | ATCC | Cat # CRL-1573 |
| HEK293T | ATCC | Ca t# CRL-3216 |
| Huh7.5 | Dr. Brett Lindenbach | N/A |
| Expi293F cells | ThermoFisher Scientific In | Cat # A14527 |
| 293T-ACE2 | Prévost et al., 2020 | N/A |
| Experimental Models: Organisms/Strains | ||
| HIV-1 lentiviral particles carrying spikes | This Study | N/A |
| MLV lentiviral particles carrying spikes | This Study | N/A |
| VSV(G)-Pseudoviruses carrying spikes | This Study | N/A |
| SARS-CoV-2 viral-like particles (VLPs) | This Study | N/A |
| Recombinant DNA | ||
| pCMV-S: pCMV3-SARS-CoV-2 Spike (S1+S2)-long | Sino Biological | Cat # VG40589-UT |
| pCMV-S Q3-1 | This Study | N/A |
| pCMV-S Q3-2 | This Study | N/A |
| pCMV-S A4-1 | This Study | N/A |
| pCMV-S A4-2 | This Study | N/A |
| pCMV-S Q3-1 A4-1 | This Study | N/A |
| pCMV-S Q3-1 A4-2 | This Study | N/A |
| pCMV-S Q3-2 A4-1 | This Study | N/A |
| pCMV-S Q3-2 A4-2 | This Study | N/A |
| pCMV-S S383C D985C | This Study | N/A |
| pCMV-S Q3-1 A4-1 S383C D985C | This Study | N/A |
| pCMV-S Δ19 | This Study | N/A |
| pCMV delta R8.2 | Addgene | Cat #12263 |
| HIV-1 GagPol-InGluc | Johnson et al., 2020 | N/A |
| S Δ19 | Johnson et al., 2020 | N/A |
| HIV-1-inGluc | Mothes Lab | N/A |
| MLV-inGluc | Mothes Lab | N/A |
| pLVX-M | Dr. Nevan Krogan | N/A |
| pLVX-E | Dr. Nevan Krogan | N/A |
| pLVX-N | Dr. Nevan Krogan | N/A |
| psPAX2 | Addgene | Cat # 12260 |
| pLL3.7 | Addgene | Cat # 11795 |
| pLL3.7-NanoLuc | Ventura et al., 2019 | N/A |
| pCMV-Vpr-HiBiT | Yamamoto et al., 2019 | N/A |
| PH-PLCd-LgBiT | Yamamoto et al., 2019 | N/A |
| pcDNA3.1 | ThermoFisher Scientific | Cat # V79020 |
| pVRC8400 vector | Addgene | Cat # 63160 |
| antibody expression constructs | Gene Universal Inc | N/A |
| pCG1-SARS-CoV-2 Spike | Hoffmann et al., 2020b | N/A |
| pCG1-SARS-CoV Spike | Hoffmann et al., 2013 | N/A |
| pCAGGS-229E Spike | Hofmann et al., 2005 | N/A |
| pCAGGS-NL63 Spike | Hofmann et al., 2005 | N/A |
| pCAGGS-OC43 Spike | Prévost et al., 2020 | N/A |
| pCDNA3.1(+)-SARS-CoV-2 RBD | Beaudoin-Bussières et al., 2020 | N/A |
| pNL4.3 R-E- Luc | NIH AIDS Reagent Program | Cat # 3418 |
| pSVCMV-IN-VSV-G | Lodge et al., 1997 | N/A |
| pIRES-GFP vector | Alsahafi et al., 2015 | N/A |
| Software and Algorithms | ||
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| MATLAB | Mathworks | https://www.mathworks.com |
| Spartan | St. Jude Children’s Research Hospital | https://www.scottcblanchardlab.com/software |
| PyMOL | Schrödinger | https://pymol.org/2/ |
| Chimera | University of California, San Francisco | http://plato.cgl.ucsf.edu/chimera; RRID: SCR_004097 |
| SerialEM software package | David N. Mastronarde, University of Colorado Boulder | https://bio3d.colorado.edu/SerialEM/ |
| IMOD software package | David N. Mastronarde, University of Colorado Boulder | https://bio3d.colorado.edu/imod/ |
| Other | ||
| Prism-based TIRF Microscope | Mothes Lab | N/A |
| Luminometer | Berthold Technologies | N/A |
| Nanodrop Spectrophotometer ND-1000 | ThermoFisher Scientific | N/A |
| C1000 Touch thermal cycler | Bio-Rad | N/A |
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| TriStar LB 942 Microplate Reader | Berthold Technologies | N/A |
| FEI Titan Krios G2 300kV Transmission Electron Microscope | ThermoFisher Scientific | https://cryoem.yale.edu/equipment |
| Gravity-driven plunger apparatus | Mothes Lab | N/A |
| QUANTIFOIL® holey carbon grids | Electron Microscopy Sciences | Cat # Q250-CR1 |
| 96-well white plates for luciferase assays | Costar | Cat # 3917 |
| Individual PCR tubes 8-tube Strip, clear | Bio-Rad | Cat # TLS0801 |
| ThermalGrid Rigid Strip PCR tubes | Denville Scientific Inc | Cat # C18064 |
| Acrodisc 25 mm Syringe Filter w/0.45 μm HT Tuffryn Membrane | PALL Life Sciences | Cat # 4184 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Walther Mothes (walther.mothes@yale.edu).
Materials Availability
All other unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. The full source code of SPARTAN, which was used for analysis of smFRET data, is publicly available (http://www.scottcblanchardlab.com/software). Custom MATLAB code are available upon request from the Lead Contact.
Experimental Model and Subject Details
Cell Lines
293 (or HEK293) cells, 293T (or HEK293T) cells, 293T-ACE2, Expi293F, and Huh7.5 cells were cultured in DMEM media, supplemented with 10% FBS, 100 U/mL penicillin/ streptomycin, 2 mM L-glutamine, and in the presence of 5% CO2. Cell culture media was exchanged before transfection. 293 cells are human embryonic kidney cells obtained from ATCC (cat # CRL-1573). CF2Th cells used in the virus capture assay were from ATCC (cat # CRL-1430). Expi293F cells are derived from the 293 cell line and were from ThermoFisher Scientific Inc (cat # A14528; RRID: CVCL_D615). 293T cells were derived from 293 cells, into which the simian virus 40 T-antigen was inserted. Huh7.5 is an immortal cell line of epithelial-like adult hepatocellular carcinoma cells expressing hACE2 on cell surfaces. 293T-ACE2 cells stably expressing human ACE2 is derived from 293T cells (Prévost et al., 2020).
Ethics Statement
All SARS-CoV-2-positive patient plasma work was conducted in accordance with the Declaration of Helsinki in terms of informed consent and approval by an appropriate institutional board (CHUM, 19.381). The donors met all donor eligibility criteria: previous confirmed COVID-19 infection by PCR and complete resolution of symptoms for at least 14 days.
Convalescent Plasma
Donor S002: Male, 65 years old, sample recovered 25 days after symptoms onset.
Donor S006: Male, 30 years old, sample recovered 41 days after symptoms onset.
Method Details
Construction of Full-Length Tagged SARS-CoV-2 Spike (S)
A full-length wild-type pCMV3-SARS-CoV-2 Spike (S1+S2)-long (termed as pCMV-S, codon-optimized, Sino Biological, cat # VG40589-UT) plasmid was used as a template to generate tagged pCMV-S. The translated amino acid sequence of pCMV-S is identical to QHD43416.1 (GenBank). Labeling tags Q3 (GQQQLG) and A4 (DSLDMLEM) (Lin and Ting, 2006; Yin et al., 2006) were placed before and after the receptor-binding motif (RBM) where the cellular receptor ACE binds (Figure S1). The short peptide labeling tag Q3 was inserted at either of two positions in the receptor-binding domain (RBD) to generate the pCMV-S Q3-1 and Q3-2 constructs. The labeling tag A4 was inserted at either of two sites in subunit domain 1 (SD1) to generate the constructs pCMV-S A4-1 and A4-2, respectively. Insertion of single tags into S proteins did not affect viral infectivity. Four different combinations of double-tagged S (designated as pCMV-S Q3-1 A4-1, Q3-1 A4-2, Q3-2 A4-1, Q3-2 A4-2) were constructed to permit smFRET imaging. Each pair of inserted tags did not compromise S-dependent lentivirus infectivity.
Infectivity Measurements
The infectivity of lentivirus particles carrying SARS-CoV-2 spike proteins was determined using a vector containing an HIV-1 or MLV long terminal repeat (LTR) that expresses a Gaussia luciferase reporter (Gluc) (Jin et al., 2009; Zhong et al., 2013) 293T cells were cultured in DMEM media, supplemented with 10% FBS, 100 U/mL penicillin/streptomycin, 2 mM L-glutamine, and in the presence of 5% CO2. Cell culture media was exchanged before transfection. Cells were transfected at 60%–80% confluency with the plasmid encoding respective spike protein, the plasmid encoding an intron-regulated Gluc (HIV-1-inGluc), and a plasmid pCMV delta R8.2 encoding HIV-1 GagPol (Addgene, plasmid # 12263) using FuGENE 6 (Promega, # E2311). Variants of plasmids encoding spike included the wild-type pCMV-S, single or dually tagged pCMV-S, pCMV-S Δ19 (S lacking the last 19 amino acids of the cytoplasmic tail), and an additional S Δ19 expressing plasmid (a gift from Marc Johnson, University of Missouri). To generate retroviral particles with an MLV core carrying full-length S, an MLV GagPol construct, and an intron-regulated Gaussia luciferase reporter construct (MLV-inGluc) were used (Jin et al., 2009). An HIV-1 GagPol-InGluc construct (Johnson et al., 2020) (a gift from Marc Johnson, University of Missouri) in which HIV-1 GagPol and HIV-1-inGluc genes are expressed from a single plasmid were also tested. Viruses were harvested 40 h post-transfection, filtered, and concentrated 7-fold by ultracentrifugation at 25,000 rpm in an SW28 rotor for 2 h at 4°C. Pelleted viruses were resuspended in above DMEM media and titered on Huh7.5 cells that endogenously express hACE2. Gluc activity was measured in the cell supernatant at 48 h using a Pierce™ Gaussia luciferase flash assay kits (ThermoFisher Scientific, # 16158).
Antibody and hACE2 Production
Antibody CR3022, VHH74, H4, 2-43, 2-4, and human ACE2 expression constructs were synthesized (Gene Universal Inc, Newark DE) and subcloned into corresponding pVRC8400 vectors. To express antibodies (Kong et al., 2019), 0.15 mL of Turbo293 transfection reagent (Speed BioSystems) were added to 2.5 mL Opti-MEM medium (Life Technology) and incubated for 5 min at room temperature (RT). Meanwhile, 25 μg of heavy chain and 25 μg of light chain plasmid DNA were added to 2.5 mL of Opti-MEM medium in another tube. The Opti-MEM medium containing Turbo293 were then mixed with the medium containing plasmid DNAs, incubated for 15 min at RT, and added to 40 mL of Expi293 cells (Life Technology) at 2.5 million cells/mL. The transfected cells were cultured in shaker incubator at 120 rpm, 37°C, 9% CO2 for 5 days. Culture supernatants were harvested and purified over 0.5 mL Protein A (GE Healthcare) resin in columns. Each antibody was eluted with IgG elution buffer (Pierce), immediately neutralized with one tenth volume of 1M Tris-HCL pH 8.0. The antibodies were then buffer exchanged in PBS by dialysis. Monomeric (residues 1-615) and dimeric (residues 1-732) human ACE2 proteins were produced as described previously (Zhou et al., 2020a). In brief, DNA sequence encoding monomeric or dimeric ACE2 followed by an HRV3C cleavage site, monomeric Fc tag and 8xHisTag at the 3¢ end were synthesized and cloned into the pVRC8400 vector. The proteins were expressed by transient transfection of 293F cells and purified by Protein A affinity columns. The Fc and 8xHis tags were removed by overnight HRV3C digestion at 4°C. hACE2 proteins were further purified with a Superdex 200 16/60 column in 5 mM HEPES, pH7.5 and 150 mM NaCl.
Soluble Human ACE2 (hACE2) Inhibition Assay
NanoLuc-expressing SARS-CoV-2 decorated lentiviruses were first generated by transfecting a plasmid mixture containing 5 μg of pCMV-S, 5 μg of pCMV delta R8.2 (expresses HIV-1 GagPol), 2 μg of pLL3.7 NanoLuc (packaging reporter to express NanoLuc) into 60%–70% HEK293 cells using FuGENE 6 (3ul of FuGENE 6 for every μg of DNA) in a 10 cm TC dish. The culture supernatants were harvested at 48 h after transfection, clarified using 0.45 μm filter, and viruses partially purified by sedimentation through a layer of 15% sucrose solution made in 1X PBS. The virus pellet was resuspended in OptiMEM to achieve a 20-fold concentration over original culture volume. Infectivity inhibition assays were performed by incubating NanoLuc expressing lentiviruses either alone or with soluble hACE2 (monomeric and dimeric) at indicated concentrations in triplicates for 90 min at room temperature prior to infection of the 293T cells stably expressing hACE2 seeded in a tissue-culture treated 96-well solid white assay plate (Costar Inc, catalog #3917). The culture supernatants were completely removed 24 h p.i. and cells lysed using 50 μl NanoLuc assay buffer. Luciferase activity was then measured using Tristar multiwell luminometer (Berthold Technology, Bad Wildbad, Germany) for 2.5 s by adding 20 μl of Nano-Glo® substrate (Promega Inc, WI, USA; diluted 1:40 in NanoLuc assay buffer). HEK293T cells (not expressing hACE2) infected similarly served as controls to determine basal luciferase activity for obtaining normalized relative light units in infected samples. Infectivity obtained in mock-treated samples were set to 100%. The data were processed and plotted using GraphPad Prism 8 v8.4.3.
Antibody Neutralization Assays
In the case of VSV-G, SARS-CoV-1, and SARS-CoV-2 parallel neutralization assay (Figure S5B), experiments were performed as follows. Target cells were infected with single-round luciferase-expressing lentiviral particles. Briefly, 293T cells were transfected by the calcium phosphate method with the pNL4.3 R-E- Luc plasmid (NIH AIDS Reagent Program) and a plasmid encoding for SARS-CoV-2 Spike, SARS-CoV-1 Spike or VSV-G at a ratio of 5:4. Two days post-transfection, cell supernatants were harvested and stored at –80°C until use. 293T-ACE2 (Prévost et al., 2020) target cells were seeded at a density of 1 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24 h before infection. Recombinant viruses in a final volume of 100 μL were incubated with the indicated plasma dilutions (1/50; 1/250; 1/1250; 1/6250; 1/31250) for 1 h at 37°C and were then added to the target cells followed by incubation for 48 h at 37°C; cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15 mM MgSO4, 15 mM KH2PO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM d-luciferin potassium salt (Prolume). The neutralization half-maximal inhibitory dilution (ID50) represents the plasma dilution to inhibit 50% of the infection of 293T-ACE2 cells by recombinant viruses bearing the indicated surface glycoproteins. The plasmids expressing the human coronavirus Spikes of SARS-CoV-2 and SARS-CoV-1 were previously described (Hoffmann et al., 2020b; Hoffmann et al., 2013; Hofmann et al., 2005; Prévost et al., 2020). The pNL4.3 R-E- Luc was obtained from NIH AIDS Reagent Program. The vesicular stomatitis virus G (VSV-G)-encoding plasmid (pSVCMV-IN-VSV-G) was previously described (Lodge et al., 1997).
Virus-Cell Fusion Assays
We used the split nanoluc assay to monitor virus-cell fusion (Yamamoto et al., 2019). For the preparation of Vpr-HiBiT–containing SARS CoV-2 decorated lentiviruses, a plasmid mixture containing 4 μg of pCMV-S, 4 μg of psPAX2 (Gag-pol, Rev, and Tat expression vector; does not express Vpr), 2 μg of pLL3.7 (packaging reporter to express GFP), and 2 μg of pCMV-Vpr-HiBiT (Vpr-HiBiT expression vector)) was transfected into 60%–70% HEK293 cells using FuGENE 6 (3ul of FuGENE 6 for 1μg of DNA). The culture supernatants were harvested at 48 h after transfection, clarified using 0.45 μm filter, and viruses partially purified by sedimentation through a layer of 15% sucrose solution made in 1X PBS. The virus pellet was resuspended in Opti-MEM to achieve a 20-fold concentration over original culture volume. Virus preparations were mock-treated or treated with trypsin (50 and 150 μg/mL) at 37°C for 15 min. Neat serum at a final concentration of 10% was added to terminate both trypsin and mock-treated samples. 2.5 × 105 HEK293 cells in 24-wells were transfected with 400 ng of hACE2-expressing plasmid or pcDNA3.1 vector along with 100 ng PH-LgBiT (LgBiT-tagged to pleckstrin homology domain of human phospholipase Cδ at the N terminus) expressing plasmid using FuGENE 6 for use as target cells. 24 h post-transfection, 2 × 105 cells were seeded in tissue-culture treated 96-well solid white assay plate (Costar Inc, # 3917) in 100 μl RPMI complete medium. They were infected with 25 μL of prepared Vpr-HiBiT-containing viruses in quadruplicates for 24 h at 37°C. Light signals derived from reconstituted HiBiT/LgBiT association were measured using Tristar multiwell luminometer (Berthold Technology, Bad Wildbad, Germany) for 2.5 s by adding 20 μl of Nano-Glo® substrate (Promega Inc, WI, USA; diluted 1:40 in PBS). Mock-infected cells served as controls to determine basal luciferase activity for obtaining normalized relative light units in infected samples. The data were processed and plotted using GraphPad Prism 8 v8.4.3.
Cell-Cell Fusion Assays
2.5 × 105 HEK293 cells seeded per well of 24-well plate were transfected with 400 ng of hACE2 and 100 ng PH-LgBiT expressing plasmid (Yamamoto et al., 2019) using FuGENE 6 (3 ul of FuGENE 6 for 1 μg of DNA). The second set of HEK293 cells for co-culture were transfected with 400 ng of SARS CoV-2 spike-expressing plasmid or pcDNA 3.1 vector along with 100 ng of Vpr-HiBiT (HiBiT tagged to HIV-1 Vpr at the N- terminus) expressing plasmid. 24 h later, HEK293 cells expressing Vpr-HiBiT alone or co-expressing spike and Vpr-HiBiT were either untreated or treated with 50 μg/mL trypsin solution for 10 min at 37°C. Trypsin was neutralized by adding serum-containing medium and cells washed once before resuspending in complete RPMI medium. 1 × 105 cells from various Vpr-HiBiT and PH-LgBiT expressing populations were mixed (1:1 ratio, 50 μl each population) in tissue culture treated 96-well solid white assay plate (Costar Inc, catalog #3917) in quadruplicates. Membrane fusion was initiated by spinning down the mixed cells in a 96-well plate (1600 rpm for 5 min, swing-out buckets, Heraeus, Sorvall centrifuge). The cells were then incubated at 37°C in tissue culture incubator for 4 h or 24 h before measuring reconstituted HiBiT/LgBiT activity using a Tristar multiwell Luminometer (Berthold Technology, Bad Wildbad, Germany) for 2.5 s by adding 20 μl of Nano-Glo® substrate (Promega Inc, WI, USA; diluted 1:40 in PBS). Individual unmixed populations of cells were treated identically and served as controls to determine basal luciferase activity to obtain normalized relative light units. The data were processed and plotted using GraphPad Prism 8 v8.4.3.
Preparation of Lentivirus Particles Carrying SARS-CoV-2 Spikes for smFRET Imaging
Lentiviruses carrying SARS-CoV-2 spikes were prepared similarly as previously described for HIV-1 (Alsahafi et al., 2019; Lu et al., 2019a; Ma et al., 2018; Munro et al., 2014). Two short peptides labeling tags (Q3: GQQQLG; A4: DSLDMLEM), either single or in pairwise combinations were introduced into indicated positions in the S1 subunit on the plasmid pCMV-S (see Figure S1). Viruses carrying 100% single-tagged or double-tagged pCMV-S showed no noticeable defect in virus infectivity. The spike carrying one pairwise combination of peptide tags (designated as pCMV-S Q3-1 A4-1) was used to make lentiviruses or S-MEN coronavirus like particles with SARS-CoV-2 spikes incorporated in the virus surface.
Lentivirus particles carrying SARS-CoV-2 spikes were produced by pseudotying an HIV-1 core with full-length spike proteins. To generate lentivirus particles that carry only one double tagged S on average among all the S trimers on the virus surface, plasmids encoding wildtype pCMV-S, pCMV-S Q3-1 A4-1, and pCMV delta R8.2 were transfected at a ratio of 20:1:21, respectively. Under these conditions, the vast majority of virus particles carry wildtype spikes. Among the small portion of virus particles containing tagged S, more than 95% will carry one dually tagged protomer while the other two protomers remain wildtype. The same strategy has been deployed in our previous studies (Alsahafi et al., 2019; Lu et al., 2019a; Ma et al., 2018; Munro et al., 2014). 293T cells were transfected using FuGENE 6 with above indicated plasmids to express spikes on the HIV-1. When virus particles were generated that contained the disulfide bridge between S383C and D985C S, both plasmids, the plasmid encoding the spike protein, as well as plasmids encoding the dually tagged spike protein contained the S383C and D985C modification. To generate S-MEN particles, plasmids encoding wildtype pCMV-S, dual-tagged pCMV-S Q3-1 A4-1, pLVX-M (SARS-CoV-2 membrane protein expressing plasmid), an pLVX-E (SARS-CoV-2 envelope expressing plasmid, and pLVX-N (SARS-CoV-2 nucleocapsid expressing plasmid) were transfected at a ratio of 20:1:21:21:21. pLVX plasmids are gifts from Nevan Krogan, UCSF.
Lentivirus or S-MEN particles carrying dually tagged S proteins were harvested 40 h post-transfection, filtered through syringe filter with 0.45 μm pore size, and sedimented through a 15% sucrose cushion at 25,000 rpm for 2 h. The virus pellets were then re-suspended in 50 mM pH 7.5 HEPES buffer supplied with 10 mM MgCl2 and 10 mM CaCl2.
Cryo-electron Tomography
6 nm gold tracer was added to the concentrated S-decorated HIV-1 lentivirus and S-MEN particles viruses at 1:3 ratio, and 5 μl of the mixture was placed onto freshly glow discharged holey carbon grids for 1 min. Grids were blotted with filter paper, and rapidly frozen in liquid ethane using a homemade gravity-driven plunger apparatus. Cryo-grids were imaged on a cryo-transmission electron microscope (Titan Krios, Thermo Fisher Scientific) that was operated at 300 kV, using a Gatan K3 direct electron detector in counting mode with a 20 eV energy slit. Tomographic tilt series between −51° and +51° were collected by using SerialEM (Mastronarde, 2005) in a dose-symmetric scheme (Hagen et al., 2017) with increments of 3°. The nominal magnification was 64,000 X, giving a pixel size of 1.346 Å on the specimen. The raw images were collected from single-axis tilt series with accumulative dose of ∼50 e− per Å2. The defocus was −3 μm and 8 frames were saved for each tilt angle. Frames were motion-corrected using Motioncorr2 (Zheng et al., 2017) to generate drift-corrected stack files, which were aligned using gold fiducial makers by IMOD/etomo (Mastronarde and Held, 2017). Tomograms were reconstructed by weighted back projection and tomographic slices were visualized with IMOD.
Fluorescently Labeling Spikes Embedded on Lentivirus Particles
Virus particles were labeled through site-specifically enzymatic labeling, as previously described (Lu et al., 2019a; Munro et al., 2014). Transglutaminase transferred Cy3B(3S) from the cadaverine conjugate to the central glutamine residue of the Q3 (GQQQLG) tag in S1. The AcpS enzyme mediated the addition of the Cy5 derivative (LD650-CoA) to the serine residue of the A4 tag (DSLDMLEM). For these reactions to occur, Cy3B(3S)-cadaverine (0.5 μM, Lumidyne Technologies), LD650-CoA (0.5 μM, Lumidyne Technologies), transglutaminase (0.65 μM, Sigma Aldrich), and AcpS (5 μM, home-made) were added to the above suspensions of viruses. The labeling reaction mix was incubated at room temperature overnight. PEG2000-biotin (0.02 mg/mL, Avanti Polar Lipids) was added to the reaction mix and incubated for 30 min at room temperature with rotation. Free dyes and lipids were then purified away from labeled virus particles by ultracentrifugation for one h at 4°C at 40,000 rpm over a 6%–18% Optiprep (Sigma Aldrich) gradient. Purified virus particles were stored at −80°C for future use in smFRET imaging.
smFRET Imaging Data Acquisition
All smFRET imaging data acquisition was performed on a home-built prism-based total internal reflection fluorescence (TIRF) microscope. Lentiviruses or S-MEN coronavirus-like particles carrying fluorescently tagged spike protein, as well as lipid-biotin in the viral membranes, were immobilized on polyethylene glycol (PEG)-passivated, streptavidin-coated quartz slides for imaging. The evanescent field was generated at the interface between the quartz slide and the virus sample solution by prism-based total internal reflection, with laser excitation from a single-frequency 532-nm laser. Donor fluorophores labeled on viruses were directly excited by the generated evanescent field. Fluorescence signals from both donor and acceptor fluorophores were collected through a 1.27-NA 60 x water-immersion objective (Nikon). Collected signals were optically separated by passing through a 650 DCXR dichroic filter (Chroma) mounted on MultiCam LS image splitter (Cairn Research). Separated donor (ET590/50, Chroma) and acceptor fluorescent (ET690/50, Chroma) signals were simultaneously recorded on two synchronized ORCA-Flash4.0 V3 sCMOS cameras (Hamamatsu) at 25 frames per second for 80 s. Unless otherwise noted, all virus samples were imaged in pH 7.4, 50 mM Tris buffer with 50 mM NaCl, a cocktail of triplet-state quenchers, and an oxygen-scavenger system (Aitken et al., 2008). Where indicated, the conformational effects of different ligands/antibodies/plasma on SARS-CoV-2 spike were tested by pre-incubating fluorescently labeled viruses for 90 min at room temperature before imaging in the continued presence of the ligands. In experiments with two different ligands presented, the second ligand (hACE2) was added to first-ligand-bound viruses and incubated similarly. The specific concentration of ligands incubated with viruses in smFRET imaging are as follows: 200 μg/mL hACE2 (human hACE2); 200 μg/mL dimeric hACE2, 50 μg/mL trypsin; 200 μg/mL CR3022; 200 μg/mL VHH72; 200 μg/mL H4; 200 μg/mL 2-4; 200 μg/mL 2-43; 100 fold dilution of a SARS-CoV-2 positive patient plasma.
Virus Capture Assay
The assay was modified from a previous published method (Ding et al., 2019) and recently described in detail elsewhere (Ding et al., 2020). Briefly, pseudoviral particles were produced by transfecting 2 × 106 HEK293T cells with pNL4.3 Luc R-E- (3.5 μg), plasmids (Beaudoin-Bussières et al., 2020; Prévost et al., 2020) encoding for SARS-CoV-2 Spike or SARS-CoV-1 Spike (3.5 μg) protein and VSV-G (pSVCMV-IN-VSV-G, 1 μg) using the standard calcium phosphate protocol. Forty-eight h later, supernatant-containing virion was collected and cell debris was removed through centrifugation (1,500 rpm for 10 min). To immobilize antibodies on ELISA plates, white MaxiSorp ELISA plates (Thermo Fisher Scientific) were incubated with 5 μg/mL of antibodies or 1:500 diluted plasma in 100 μL phosphate-buffered saline (PBS) overnight at 4°C. Unbound antibodies or plasma were removed by washing the plates twice with PBS. Plates were subsequently blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature. After two washes with PBS, 200 μL of virus-containing supernatant was added to the wells. For sACE2 competition, 10 μM sACE2 was incubated with the supernatant at 37°C for 1 h before adding to the coated wells. After 4 to 6 h incubation, supernatant was removed and the wells were washed with PBS 3 times. Viral capture by any given antibody was visualized by adding 10 × 104 SARS-CoV-2-resistant Cf2Th cells (not shown) in full DMEM medium per well. Forty-eight h post-infection, cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) and three freeze-thaw cycles. An LB941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15 mM MgSO4, 15 mM KH2PO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM D-luciferin potassium salt (Prolume).
Flow Cytometry Analysis of Cell-Surface Staining
Using the standard calcium phosphate method, 10 μg of Spike expressor (SARS-CoV-1, SARS-CoV-2, NL63, 229E, OC43) (Prévost et al., 2020) and 2 μg of a green fluorescent protein (GFP) expressor (pIRES-GFP) was transfected into 2 × 106 293T cells. At 48 h post transfection, 293T cells were stained with plasma from SARS-CoV-2-infected or uninfected individuals (1:250 dilution). The percentage of transfected cells (GFP+ cells) was determined by gating the living cell population based on the basis of viability dye staining (Aqua Vivid, Invitrogen). Samples were acquired on a LSRII cytometer (BD Biosciences, Mississauga, ON, Canada) and data analysis was performed using FlowJo vX.0.7 (Tree Star, Ashland, OR, USA). Alternatively, convalescent plasma was incubated with 20 μg/mL of SARS-CoV-2 RBD prior cell-surface staining in order to compete for RBD-specific antibodies.
ELISA (Enzyme-Linked Immunosorbent Assay)
The SARS-CoV-2 RBD ELISA assay used was recently described (Beaudoin-Bussières et al., 2020; Prévost et al., 2020). Briefly, recombinant SARS-CoV-2 S RBD proteins (2.5 μg/mL), or bovine serum albumin (BSA) (2.5 μg/mL) as a negative control, were prepared in PBS and were adsorbed to plates (MaxiSorp; Nunc) overnight at 4°C. Coated wells were subsequently blocked with blocking buffer (Tris-buffered saline [TBS] containing 0.1% Tween20 and 2% BSA) for 1 h at room temperature. Wells were then washed four times with washing buffer (Tris-buffered saline [TBS] containing 0.1% Tween20). CR3022 mAb (50 ng/mL) or serial dilutions of plasma from SARS-CoV-2-infected or uninfected donors (1/100; 1/250; 1/500; 1/1000; 1/2000; 1/4000) were prepared in a diluted solution of blocking buffer (0.1% BSA) and incubated with the RBD-coated wells for 90 min at room temperature. Plates were washed four times with washing buffer followed by incubation with anti-IgG secondary Abs (diluted in a diluted solution of blocking buffer [0.4% BSA]) for 1 h at room temperature, followed by four washes. HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning oxidizing and luminol reagents (Perkin Elmer Life Sciences). Light emission was measured with a LB941 TriStar luminometer (Berthold Technologies). Signal obtained with BSA was subtracted for each plasma and was then normalized to the signal obtained with CR3022 mAb present in each plate.
Quantification and Statistical Analyses
Statistical Analyses
Statistical significance (in Figures 3F, 4B, and S4C) was analyzed using GraphPad Prism version 8.0.2 by applying unpaired Student’s t test to obtain p values. p values lower than 0.05 were considered statistically significant. P values were indicated as ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
smFRET Data Analysis
smFRET data analysis was performed using MATLAB (MathWorks)-based customized SPARTAN software package (Juette et al., 2016). For each labeled virus recorded over 80 s, the background signal at the single-molecule level was first identified based on the fluorophore bleaching point and subtracted. Donor and acceptor fluorescence intensity time-trajectory or traces were then extracted for each recorded virus and corrected for donor to acceptor crosstalk. The energy transfer efficiency (termed as FRET in graphs) from the donor to the neighboring acceptor was evaluated as FRET = IA/(γID+IA), where ID and IA are the fluorescence intensities of donor and acceptor, respectively, and γ is the correlation coefficient compensating for the difference in quantum yields and detection efficiencies of donor and acceptor.
FRET efficiencies of each FRET pair report on the relative distance between the donor and the acceptor over time and ultimately reveal the conformational dynamics of host molecules (spikes on the virus in our case) in real-time. Viruses lacking donor or acceptor fluorophores, containing multiple labeled protomers, or containing more than one labeled spike on a single virus were automatically excluded from further analysis. FRET traces (FRET values as a function of real-time) were then manually selected if they displayed sufficient signal-to-noise (S/N) ratio and anti-correlated fluctuations in donor and acceptor fluorescence intensity between clearly defined FRET states, which are indicative of fully active molecules. N (or N m), number of traces/molecules as indicated in each FRET histogram, was compiled into the associated FRET histogram. Based on visual inspection of traces that revealed direct observations of state-to-state transitions, FRET histograms were fitted into the sum of four Gaussian distributions using the least-squares fitting algorithm in MATLAB. FRET histograms represent mean ± SEM, determined from three randomly assigned populations of FRET traces, as indicated in corresponding figure legends. Each Gaussian represents one FRET-indicated conformational state, and the area under each Gaussian curve estimates each state’s occupancy. The occupancy of each state was used to evaluate the difference in free energies between states i and j, according to ΔG° ij = -k B Tln(P i /P j ), where P i and P j are the ith and jth state’s occupancy, respectively, k B is the Boltzmann constant, and T is the temperature in kelvin. Relative state occupancies are presented as mean ± SEM, derived from histograms, and the determining parameters are listed in. Table S1.
The idealization of each FRET trace using a 4-state Hidden Markov Model was performed in the SPARTAN software package using a segmental K-means algorithm (McKinney et al., 2006). The state-to-state transition tracing, indicative of the locations and the frequencies of transitions, was displayed in a transition density plot (termed as TDP). Dwell times (the time during which a molecule occupies one specific conformation before transitioning to any other conformations) distributions were compiled into survival probability plots and were fitted to the sum of two exponential distributions (y = A 1 exp –k1t + A 2 exp –k2t). The transition rates were weighted from averaging the two rate constants by their amplitudes.
Acknowledgments
We thank Dr. Craig Wilen for sharing Huh7.5 cells; Dr. Nevan Krogan for sharing M, E, and N expression plasmids; Dr. Marc Johnson for sharing pCMV-S Δ19 and HIV-1 GagPol-InGluc plasmids; and MLV In-GLuc and Dr. David Derse for sharing HIV In-GLuc plasmids. We also thank M. Gordon Joyce (U.S. MHRP) for the monoclonal antibody CR3022. This work was supported by NIAID, NIH grant R01 AI150560 to W.M. and S.C.B.; by NIGMS, NIH R01 GM098859 to S.C.B.; by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH to P.D.K.; and by Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation, the Fondation du CHUM, as well as by the Canadian Institutes of Health Research, via the Immunity Task Force and a foundation grant to A.F. A.F. is the recipient of a Canada Research Chair on Retroviral Entry RCHS0235 950-232424. R.G. is supported by a MITACS Accélération postdoctoral fellowship. J.P., G.B.B., and S.P.A. are supported by CIHR graduate fellowships.
Author Contributions
M.L. and W.M. designed the studies. M.L. performed mutagenesis, virus infectivity assays, generated fluorescently labeled viruses, performed smFRET imaging, and analyzed the data. P.D.U. designed and performed virus-cell fusion, cell-cell fusion, and sACE2 inhibition assays. W.L. generated tomograms of virus particles. W.L., J.R.G., and P.D.U. assisted with virus-like particles. D.Z. built the prism-TIRF microscope with assistance from M.L. D.S.T. and S.C.B. supported and advised on the development and implementation of single-molecule instrumentation, data acquisition, and analysis in the Mothes lab. B.Z., T.Z., J.G., L.L., D.D.H., and P.D.K. provided various antibodies and scientific inputs into experiments and manuscript editing. W.S. and J.R.M. provided monomeric and dimeric hACE2. A.L. and A.F. processed all the blood samples and prepared patient plasma. S.D. performed virus capture assay. R.G., S.D., and A.L. performed neutralization experiments. G.G.G. performed ELISA experiments. J.P. and S.P.A. performed flow cytometry experiments. M.L. and W.M. wrote the manuscript with input from all authors.
Declaration of Interests
S.C.B. has an equity interest in Lumidyne Technologies.
Published: November 13, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.11.001.
Supplemental Information
References
- Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N., Ding S., Richard J., Markle T., Brassard N., Walker B., Lewis G.K., Kaufmann D.E., Brockman M.A., Finzi A. Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J. Virol. 2015;90:2993–3002. doi: 10.1128/JVI.02973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N., Bakouche N., Kazemi M., Richard J., Ding S., Bhattacharyya S., Das D., Anand S.P., Mothes W., Lifson J. An Asymmetric Opening of HIV-1 Envelope Mediates Antibody-Dependent Cellular Cytotoxicity. Cell Host Microbe. 2019;25:578–587. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. MBio. 2020;11:194639. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D.K., Govindan R., Nikić-Spiegel I., Krammer F., Lemke E.A., Munro J.B. Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell. 2018;174:926–937.e12. doi: 10.1016/j.cell.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D.K., Bulow U., Diehl W.E., Durham N.D., Senjobe F., Chandran K., Luban J., Munro J.B. Conformational changes in the Ebola virus membrane fusion machine induced by pH, Ca2+, and receptor binding. PLoS Biol. 2020;18:e3000626. doi: 10.1371/journal.pbio.3000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Gasser R., Gendron-Lepage G., Medjahed H., Tolbert W.D., Sodroski J., Pazgier M., Finzi A. CD4 Incorporation into HIV-1 Viral Particles Exposes Envelope Epitopes Recognized by CD4-Induced Antibodies. J. Virol. 2019;93:e01403–e01419. doi: 10.1128/JVI.01403-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Laumaea A., Benlarbi M., Beaudoin-Bussières G., Gasser R., Medjahed H., Pancera M., Stamatatos L., McGuire A.T., Bazin R., Finzi A. Antibody binding to SARS-CoV-2 S glycoprotein correlates with but does not predict neutralization. Viruses. 2020;12:1214. doi: 10.3390/v12111214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen W.J.H., Wan W., Briggs J.A.G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Müller M.A., Drexler J.F., Glende J., Erdt M., Gützkow T., Losemann C., Binger T., Deng H., Schwegmann-Weßels C. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS ONE. 2013;8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Sherer N.M., Heidecker G., Derse D., Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7:e1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized pseudotyping conditions for the SARS-COV-2 spike glycoprotein. J. Virol. 2020;94 doi: 10.1128/JVI.01062-20. e01062-e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Juette M.F., Terry D.S., Wasserman M.R., Altman R.B., Zhou Z., Zhao H., Blanchard S.C. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods. 2016;13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020 doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. bioRxiv. 2020 doi: 10.1101/2020.06.23.167064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R., Duan H., Sheng Z., Xu K., Acharya P., Chen X., Cheng C., Dingens A.S., Gorman J., Sastry M., NISC Comparative Sequencing Program Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell. 2019;178:567–584.e19. doi: 10.1016/j.cell.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li Z., Li W., Lu M., Bess J., Jr., Chao C.W., Gorman J., Terry D.S., Zhang B., Zhou T., Blanchard S.C. Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat. Struct. Mol. Biol. 2020;27:726–734. doi: 10.1038/s41594-020-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Ting A.Y. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lodge R., Lalonde J.P., Lemay G., Cohen É.A.J.T.E.J. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Ma X., Castillo-Menendez L.R., Gorman J., Alsahafi N., Ermel U., Terry D.S., Chambers M., Peng D., Zhang B. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature. 2019;568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Ma X., Mothes W. Illuminating the virus life cycle with single-molecule FRET imaging. Adv. Virus Res. 2019;105:239–273. doi: 10.1016/bs.aivir.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui I., Zhou X.X., Lim S.A., Elledge S.K., Solomon P., Rettko N.J., Zha B.S., Kirkemo L.L., Gramespacher J.A., Liu J. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. bioRxiv. 2020 doi: 10.1101/2020.05.21.109157. [DOI] [Google Scholar]
- Ma X., Lu M., Gorman J., Terry D.S., Hong X., Zhou Z., Zhao H., Altman R.B., Arthos J., Blanchard S.C. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife. 2018;7:e34271. doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N., Held S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Bowen J.E., Corti D., Veesler D. Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation. Nat. Struct. Mol. Biol. 2020;27:942–949. doi: 10.1038/s41594-020-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney S.A., Joo C., Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J.B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D.R., Koff W.C., Courter J.R., Smith A.B., 3rd, Kwong P.D. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S. Cross-sectional evaluation of humoral responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura J.D., Beloor J., Allen E., Zhang T., Haugh K.A., Uchil P.D., Ochsenbauer C., Kieffer C., Kumar P., Hope T.J., Mothes W. Longitudinal bioluminescent imaging of HIV-1 infection during antiretroviral therapy and treatment interruption in humanized mice. PLoS Pathog. 2019;15:e1008161. doi: 10.1371/journal.ppat.1008161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., VIB-CMB COVID-19 Response Team Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1436–1441. doi: 10.1016/j.cell.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Du Q., Song J., Wang H., Watanabe A., Tanaka Y., Kawaguchi Y., Inoue J.I., Matsuda Z. Cell-cell and virus-cell fusion assay-based analyses of alanine insertion mutants in the distal α9 portion of the JRFL gp41 subunit from HIV-1. J. Biol. Chem. 2019;294:5677–5687. doi: 10.1074/jbc.RA118.004579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Lin A.J., Golan D.E., Walsh C.T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong P., Agosto L.M., Ilinskaya A., Dorjbal B., Truong R., Derse D., Uchil P.D., Heidecker G., Mothes W. Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLoS ONE. 2013;8:e53138. doi: 10.1371/journal.pone.0053138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Teng I.T., Olia A.S., Cerutti G., Gorman J., Nazzari A., Shi W., Tsybovsky Y., Wang L., Wang S. Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes. Cell Rep. 2020;33:108322. doi: 10.1016/j.celrep.2020.108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Olia A.S., Gorman J., Rapp M., Cerutti G., Chuang G.-Y., Katsamba P.S., Nazzari A., Sampson J.M. Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host & Microbe. 2020;28:867–879. doi: 10.1016/j.chom.2020.11.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. The full source code of SPARTAN, which was used for analysis of smFRET data, is publicly available (http://www.scottcblanchardlab.com/software). Custom MATLAB code are available upon request from the Lead Contact.