Abstract
Two pandemics of respiratory distress diseases associated with zoonotic introductions of the species Severe acute respiratory syndrome-related coronavirus in the human population during 21st century raised unprecedented interest in coronavirus research and assigned it unseen urgency. The two viruses responsible for the outbreaks, SARS-CoV and SARS-CoV-2, respectively, are in the spotlight, and SARS-CoV-2 is the focus of the current fast-paced research. Its foundation was laid down by studies of many corona- and related viruses that collectively form the vast order Nidovirales. Comparative genomics of nidoviruses played a key role in this advancement over more than 30 years. It facilitated the transfer of knowledge from characterized to newly identified viruses, including SARS-CoV and SARS-CoV-2, as well as contributed to the dissection of the nidovirus proteome and identification of patterns of variations between different taxonomic groups, from species to families. This review revisits selected cases of protein conservation and variation that define nidoviruses, illustrates the remarkable plasticity of the proteome during nidovirus adaptation, and asks questions at the interface of the proteome and processes that are vital for nidovirus reproduction and could inform the ongoing research of SARS-CoV-2.
Keywords: Comparative genomics, Coronaviruses, Evolution, Nidoviruses, Proteome
1. From SARS-CoV-2 to nidoviruses and back
The loss of many hundreds of thousands human lives to the COVID-19, the severe respiratory disease and its complications, raised unprecedented interest in its causative agent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) of the species Severe acute respiratory syndrome-related coronavirus [[1], [2], [3], [4]] as well as other coronaviruses. The state-of-the art of coronavirus research is in the spotlight and scrutinized by scientists, stakeholders as well as the public. It reveals a group of remarkable RNA viruses of extraordinary genome sizes and complex biology, which characterization was largely advanced by a relatively small group of dedicated researchers until 2020 (e.g. see reviews in [[5], [6], [7]]). Also apparent, are considerable gaps in our understanding of the fundamentals of coronavirus infection that complicate developing safe and efficient vaccines and antivirals against COVID-19. Mobilization of resources and expertise across many fields of science are addressing both the immediate challenges of the COVID-19 pandemic (e.g. [[8], [9], [10]] and advancement of our knowledge of coronaviruses (e.g. [[11], [12], [13], [14], [15], [16]]).
The observed very low variation of SARS-CoV-2, during the virus transmission in human population since December 2019 [[17], [18], [19]], is one of the silver linings that emerged from the research so far. It lends support to a belief that prospective therapeutics directed against the currently circulating virus variants will remain effective in the future. However, we also know that RNA viruses can evolve fast, could SARS-CoV-2 be an exception even if its replication includes proofreading?
In this regard, it may be informative to look at the virus genomic variation beyond the current pandemic and consider relatives of SARS-CoV-2 that collectively form the order Nidovirales [20]. The natural variation of nidoviruses has realized over a considerable, although poorly defined, timeframe in vastly different vertebrate and invertebrate hosts; its scale far exceeds mutation ranges observed for SARS-CoV-2 and tested in experiments with SARS-CoV-2 and few other selected nidoviruses. Characterization of various patterns of natural protein variation reveal constraints on evolution and may also inform about potential of the respective proteins to evolve under directional selection pressure of antivirals. Comparative genomics of corona- and related viruses have successfully guided experimental research on the nidovirus proteome, which is critical for identifying targets of antivirals in SARS-CoV-2. This article provides a primer on the recognized commonalities of nidoviruses and reviews cases of outstanding genome conservation and the variation that underlies the biology of nidoviruses and informs about the potential to change. It attests for numerous connections between research on diverse nidoviruses and the ongoing characterization of SARS-CoV-2 including efforts to contain the pandemic with antivirals.
1.1. Origin of Nidovirales name and nidovirus identity
The most distinguished characteristic of nidoviruses, as recognized early in the course of research on coronaviruses and arteriviruses, is the production of a set of subgenomic (sg) mRNAs that is 5′- and 3′-nested relative to genomic RNA. This shared characteristic provided a basis for the order’s name: nidus means nest in Latin [21]. This name is retained despite several later described nidoviruses appear to have only 3′-nested set of sg RNAs [22,23]. Nidoviruses were also found to share a genome organization and expression mechanisms. Yet practically, new viruses are assigned to nidoviruses using comparative sequence analysis and considering phylogeny affinity in most conserved protein domains that form a unique synteny [24].
1.2. Nidovirus diversity and taxonomy
Nidoviruses possess positive-sense, non-segmented linear RNA genomes in the unprecedented large size range of 12–41 kb. They replicate in the cytoplasm and have genomes packaged into enveloped virions that may vary in shape, depending on the virus lineage [24,25]. SARS-CoV-2 is a variant of one of about a hundred known nidovirus species [4]. These viruses form the order Nidovirales that was established in 1996 by merging two families of viruses infecting vertebrates, Coronaviridae and Arteriviridae [[26], [27]]; the former was recently split in two, elevating the original subfamily Torovirinae to the new family Tobaniviridae [28]. First invertebrate nidoviruses, comprising family Roniviridae [22], were identified only 20 years ago, although currently the order includes already 8 families of vertebrate and 6 families of invertebrate nidoviruses, with majority including only a single species [[29], [30], [31], [32], [33]][34].
With the exponential growth of the number of available nidovirus genome sequences, the number of known nidovirus species began to grow accordingly, although their formal classification within the taxonomy framework may lag behind. Likewise, the gap between the newly identified and the few experimentally characterized nidoviruses is also rapidly increasing. The latter group includes arteriviruses: equine arteritis virus (EAV) and porcine reproduction respiratory syndrome viruses (PRRSV), and coronaviruses: human coronavirus 229E (HCoV-229E), transmissible gastroenteritis virus (TGEV), mouse hepatitis virus (MHV), Middle East respiratory syndrome coronavirus (MERS-CoV) and avian infectious bronchitis virus (IBV), in addition to SARS-CoV and SARS-CoV-2. Also, the limited characterization of several tobaniviruses, and invertebrate mesoniviruses and roniviruses, often isolated from ‘exotic’ hosts, was important for understanding generalities and host- and lineage-dependent specifics of nidoviruses, and for the validation of many models of comparative genomics. Since viruses of the subfamily Orthocoronavirinae (formerly Coronavirinae) and the family Arteriviridae are most frequently sampled, they were predominantly used to characterize patterns of conservation and evolution at subfamily and family levels, respectively.
1.3. Canonical multi-ORF organization of nidovirus genome: three functional regions and their expression
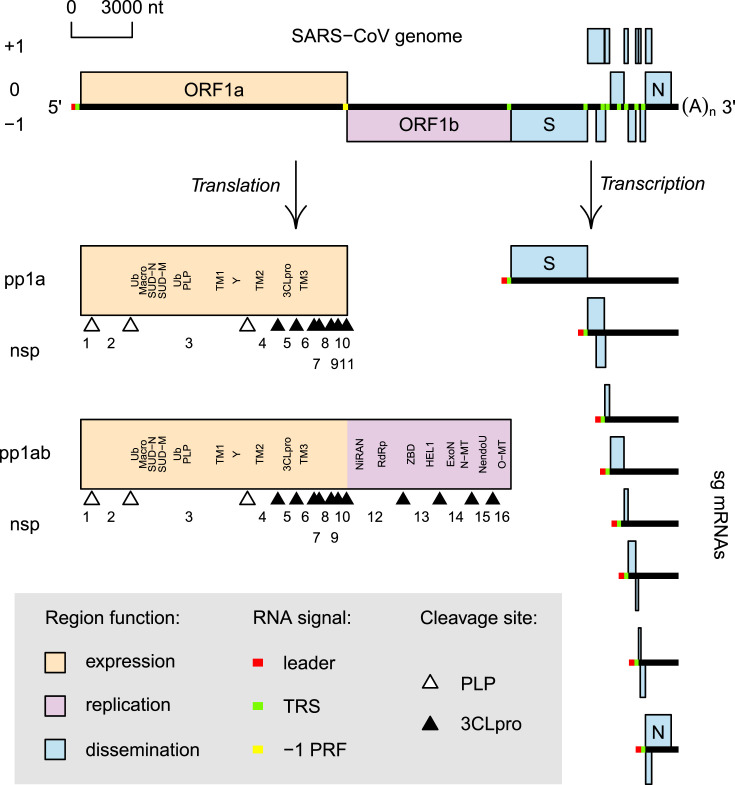
All known coronaviruses, including SARS-CoV-2, as well as other nidoviruses, whose genomes were sequenced from 1987 up to 2016, are characterized by a conserved multicistronic genome organization including multiple open reading frames (ORFs) (Fig. 1 ). The two largest and slightly overlapping ORFs, 1a and 1b, occupy the 5′-terminal two-thirds of the genome and encode non-structural proteins (nsps) that are derived by autoproteolytic processing from polyproteins 1a and 1ab, pp1a and pp1ab, encoded by ORF1a and jointly ORF1a/ORF1b, respectively [35]. The number of nsps vary from 12 to 16 in arteriviruses and coronaviruses, respectively; comparable number may be produced in other lesser characterized nidoviruses. Together these two ORFs are often referred to as the replicase gene, although ORF1a and ORF1b and their products are chiefly responsible for the control of genome expression and replication, respectively. Consequently, considering them as two separate major regions facilitates functional and evolutionary analyses of nidoviruses [36]. The 3′-terminal region of the genome contains multiple smaller ORFs (3′ORFs), the number of which varies considerably among nidoviruses and which encode structural and, in some nidoviruses, accessory proteins. This third region is chiefly responsible for virus dissemination [36] and, in the Coronaviridae, may vary in respect to ORF composition even between representatives of the same species, as we learned from the comparison of SARS-CoV and SARS-CoV-2 [37]. Untranslated regions (UTRs) are present at the 5′- and 3′-ends of the genome, and may also be found between ORFs in the 3′ORFs region. The genomic 5′-end is believed to be capped [23,38,39], and 3′-end of the genome is polyadenylated [40,41].
Fig. 1.
SARS-CoV genome organization and expression. Genome (top), products of genome translation (left) and transcription (right) are shown. ORFs and polyprotein regions are colored according to their predominant function (see inset). Genome ORFs are depicted in their frame, with ORF1a frame set to zero. For each sg mRNA, only ORFs believed to be translated from it are shown, without indicating their frame relative to ORF1a. For genome and sg mRNAs, RNA signals are indicated by color (see inset). For polyproteins, autoproteolytic processing scheme (see inset) and selected protein domains (see text for abbreviations) are specified. The NC_004718.3 record was used to prepare this figure. Note that sg mRNA 3.1 [55] is not shown; the most N-terminal ubiquitin (Ub) and Macro domains are separated by acidic, structurally disordered region of ∼70 aa [56,57]. SUD-N and SUD-M, N-terminal and Middle domains of SARS-CoV Unique Domain, respectively [58,59]; Y, Y domain [57].
Nsps assemble into a membrane-bound replication-transcription complex (RTC) that mediates genome replication and synthesis of sg mRNAs (transcription) [[42], [43], [44]] for expression of the 3′ORFs (Fig. 1). Transcription involves a discontinuous synthesis of minus strand sg-size RNAs that employs short conserved sequences known as the body and leader transcription-regulating sequence (bTRS and lTRS) located upstream of a 3′ORF and ORF1a, respectively [21], although some nidoviruses may not use lTRS [23,45]. Most sg mRNA species are monocistronic and serve to translate only their 5′-most ORF, but some sg mRNA species are polycistronic [21,46,47]. Expression from separate sg mRNAs allows the regulation of the abundance of the respective structural and accessory proteins relative to each other and nsps [[48], [49]].
The assembly of a virus particle is a multistage process that includes the encapsidation of a viral genome by multiple copies of a nucleocapsid protein, and the wrapping of the nucleoprotein complex by a host membrane, carrying viral structural proteins. The wrapping is coupled with budding into the lumen of the endoplasmic reticulum (ER) or Golgi complex, and followed by transportation of the virus particles to the plasma membrane through the secretory pathway, culminating in their release from the cell [50,51].
The multi-ORF genome organization, called hereafter canonical, is coupled with conserved expression mechanisms of transcription and translation, controlling the relative quantities of functionally different proteins in the infected cell. Specifically, pp1a proteins are synthesized in higher quantities than pp1ab unique proteins, due to −1 programmed ribosomal frameshifting (PRF) directing a fraction of the ribosomes from ORF1a to ORF1b translation of the incoming and newly synthesized genomic RNAs [52]. In contrast, the 3′ORFs are expressed from a separate set of sg mRNAs in a differential mRNA-specific manner at a later point in time, although the actual complexity of this regulation is just emerging [[21], [53], [54], [16]].
This apparent coupling of the genome organization with the differential expression seems to be functionally sensible. The least expressed ORF1b-encoded proteins include core enzymes of the RTC, such as RNA-dependent RNA polymerase (RdRp) and superfamily 1 Helicase (HEL1), that are required in relatively minute molar quantities to catalyze synthesis of diverse virus RNAs and other RNA-dependent reactions [24]. They are assisted by other ORF1a-encoded subunits in the RTC which are produced in larger molar quantities compared to the ORF1b-encoded core enzymes for the reasons that are yet to be fully understood but would be compatible with either structural role or less efficient enzymatic activity or plentitude reactions they may be involved in. Likewise, the 3′ORFs may be expressed most actively to provide subunits of virus particles or proteins that may modulate virus-host interaction, with some proteins being multifunctional.
1.4. Non-canonical ORF organizations of recently discovered nidoviruses
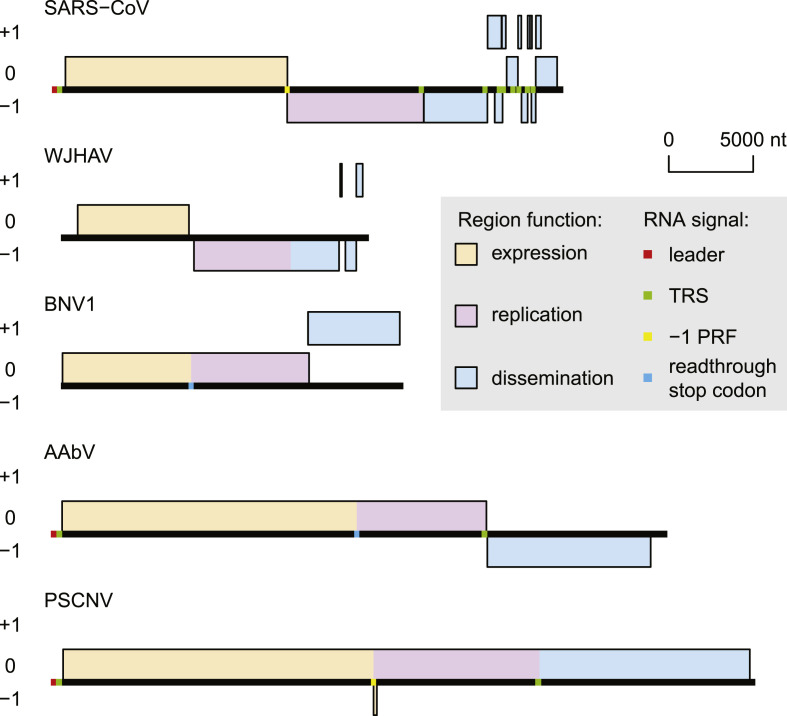
Canonical multi-ORF organization, including two large overlapping ORFs for the replicase, was long considered a defining characteristic of nidoviruses [20], since it was invariantly found in diverse viruses that: 1) infect either vertebrate or invertebrate hosts, 2) have vastly different genome sizes, in the range from 12 to 34 kb, and 3) are separated by evolutionary distances comparable to the most distant in the Tree of Life [36]. Yet, we learned recently that this conclusion was premature and, presumably, due to poor sampling of viruses in the two genome size ranges: between 16 and 20 kb, and larger than 34 kb, respectively. Comparative genomic analysis of recently identified four highly divergent nidoviruses, Wuhan Japanese halfbeak arterivirus (WJHAV; 18.2 kb) [60], Beihai nido-like virus 1 (BNV1; 20.3 kb) [60], Aplysia abyssovirus 1 (AAbV; 35.9 kb) [30,31] and planarian secretory cell nidovirus (PSCNV; 41.1 kb) [32] revealed surprising plasticity of nidoviruses at the two critical junctions separating the three major genomic regions: between ORF1a and ORF1b, and ORF1b and 3′ORFs, respectively (Fig. 2 ). In the WJHAV genome, ORF1b is fused with a gene encoding putative glycoprotein [60], presumably a structural protein. Both BNV1 and AAbV contain two ORFs, a 5′-terminal ORF combining ORF1a- and ORF1b-like regions, and a single 3′-terminal ORF encoding structural protein domains [30,31,61]. The PSCNV genome has a single large ORF, which is an equivalent of ORF1a, ORF1b and 3′ORFs fused together [32]. This PSCNV ORF is exceptionally large: it encodes a 13,556 aa polyprotein that is 58–67% larger than the largest single- or multi-ORF polyproteins of other viruses. These nidoviruses could be considered non-canonical. Collectively they account for a large share of nidoviruses at the family level: four versus ten that have the canonical multi-ORFs organization. In contrast, they are substantially underrepresented at the species level.
Fig. 2.
Nidoviruses with canonical (SARS-CoV) and non-canonical genome ORFs organization. WJHAV, Wuhan Japanese halfbeak arterivirus (MG600020.1), species Halfbeak nidovirus 1, family Nanhypoviridae; BNV1, Beihai nido-like virus 1 (KX883629.1), species Turrinivirus 1, family Medioniviridae; AAbV, Aplysia abyssovirus 1 (GBBW01007738.1), species Aplysia abyssovirus 1, family Abyssoviridae; PSCNV, planarian secretory cell nidovirus (MH933735.1), species Planidovirus 1, family Mononiviridae. ORFs are positioned according to their reading frame, with the most 5′-terminal depicted ORF set as zero. ORF regions are colored according to their main function assignment (see inset). Genome signals, described by the discoverers of each virus, are indicated by color (see inset).
The discovery of non-canonical nidoviruses raises the intriguing question about genome expression mechanisms used by these viruses and specifically about whether they are able to maintain region-specific stoichiometry of viral proteins described for canonical nidoviruses. It seems that despite differences in the ORFs organization, the end result of genome expression may be quite similar in both canonical and non-canonical nidoviruses.
Computational analysis of the genome sequences of BNV1, AAbV and PSCNV shows how these non-canonical nidoviruses may attenuate translation of the ORF1b-like region to achieve a non-equimolar ratio of nonstructural proteins that are encoded in ORF1a and ORF1b of canonical nidoviruses. Both BNV1 and AAbV have ORF1a-like and ORF1b-like regions residing in the same reading frame and separated by a single stop codon (Fig. 2) [30,31,61]. If a readthrough of this stop codon only occurs in a fraction of translation events, proteins encoded in the ORF1a-like region would be expressed in a higher quantity compared to proteins encoded in the ORF1b-like region. This type of regulation was documented for the attenuation of RdRp (nsP4) of alphaviruses [62]. The PSCNV single-ORF genome includes a predicted −1 PRF site immedaitely upstream of the ORF1b-like region with a potential to divert translation of a fraction of ribosomes to a tiny 39 nt ORF in an alternative reading frame (Fig. 2). As a result, the ORF1a-like compared to the ORF1b-like region of the PSCNV genome would be expressed more frequently upon translation of the genomic RNA [32]. The main difference between the −1 PRF-directed mechanisms in canonical nidoviruses and in PSCNV is that −1 PRF directs translation into ORF1b in the former but diverts it from ORF1b-like region in the latter. A similar mechanism was already demonstrated for diverting a fraction of ribosomes during translation of ORF1a in the nsp2 region of some arteriviruses [63].
Likewise, non-canonical nidoviruses may be similar to the canonical nidoviruses in the use of sg mRNAs for the expression of the 3’-terminal genome region. Such evidence was obtained for the single 3′ORF of AAbV [30] and the 3′ORFs-like region of the PSCNV genome [32]; based on similarity with canonical nidoviruses, this hypothesis may be extended to the single 3′ORF of BNV1 and the three small 3′ORFs of WJHAV, although these viruses are yet to be studied in this respect (Fig. 2). In both the PSCNV and AAbV, potential leader and body TRSs were identified by comparative genomics as large repeats in the 5′UTR and upstream of the genome region predicted to encode structural proteins, respectively (Fig. 2). A sharp increase in coverage of the genome by RNA-seq reads was observed at the body TRS of both PSCNV and AAbV, consistent with the downstream region being a subject of transcription. Existence of PSCNV sg mRNA species, expected to be expressed when the identified TRSs are employed, was confirmed in a 5′-RACE experiment. Importantly, if translation of the PSCNV sg mRNA species is initiated at its most 5′-terminal start-codon, it would result in production of a polyprotein identical to the C-terminus of the giant polyprotein expressed from the PSCNV genome [32]. Thus, predicted structural proteins of PSCNV may be expressed from both genome and sg mRNA, unlike their counterparts in canonical nidoviruses.
Interestingly, the PSCNV may not be the only non-canonical nidovirus, structural proteins of which are synthesized from both genome and sg mRNA. The WJHAV potentially encodes a glycoprotein in the unusually long 3′-terminus of its ORF1b that is located downstream of the otherwise terminal 2′-O-methyltransferase (O-MT) locus [60,34]. While the WJHAV was not analyzed in this respect, this genome organization is compatible with both genome and sg mRNA directing synthesis of the glycoprotein (Fig. 2). Production of certain structural proteins from both genome and sg mRNA can also be envisioned for some of the canonical nidoviruses, such that stop-codon of ORF1b and start-codon of the downstream structural ORF are in-frame and separated by few codons in their genomes. If a readthrough of the ORF1b stop-codon would occur, with the stop-codon being decoded by a suppressor tRNA [52], it would lead to a continuation of translation, resulting in the production of pp1ab fused with a structural protein. For example, SARS-CoV ORF1b and ORF2, encoding S protein, belong to the same reading frame and are separated by just two codons downstream of the ORF1b termination codon (Fig. 1). No evidence for such suppression has been reported for this or other viruses of the species Severe acute respiratory syndrome-related coronavirus.
Unlike the expression of structural proteins from individual ORFs, observed in most known nidoviruses, expression of multiple structural proteins from a single ORF, predicted for non-canonical BNV1, AAbV and PSCNV (Fig. 2), would require processing of the structural polyprotein by host and/or viral proteases, unless their structural domains function in the context of a single polyprotein. Accordingly, a chymotrypsin-like serine protease domain was detected in the structural polyprotein sequences of BNV1 and AAbV [30,31], and potential cleavage sites of cellular proteases furin and signal peptidase were identified in the C-terminal region of the PSCNV polyprotein [32].
1.5. All nidoviruses share seven protein domains that are vital to control of genome replication and expression
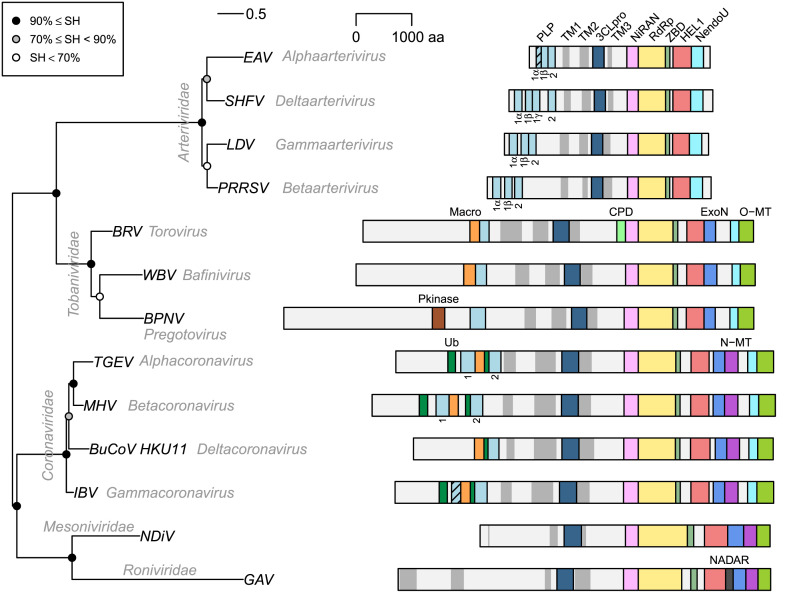
Six domains, transmembrane domain 2 (TM2), 3C-like protease (3CLpro), transmebrane domain 3 (TM3), RdRp, Zn-binding domain (ZBD) and HEL1, were delineated in pp1a/pp1ab soon after the first sequence of the coronavirus genome was released [[64], [65], [66], [67], [68]]. Subsequently, they were found to be universally conserved in all nidoviruses [26,[69], [70], [71]] (Fig. 3 ), although unlike other universally conserved domains, TM2 and TM3 have no residues invariant in all nidoviruses.
Fig. 3.
Phylogeny and pp1ab domain organization of selected nidoviruses. Representatives of 13 lineages of nidoviruses that infect vertebrates (three virus families) and invertebrates (two virus families) are depicted. Names of taxa, families and genera, are indicated in grey italic font, and names of viruses are given as acronyms: EAV, equine arteritis virus; SHFV, simian hemorrhagic fever virus; LDV, lactate dehydrogenase-elevating virus; PRRSV, porcine reproduction respiratory syndrome virus; BRV, Breda virus; WBV, white bream virus; "BPNV, ball python nidovirus;" TGEV, porcine transmissible gastroenteritis virus; MHV, mouse hepatitis virus; BuCoV_HKU11, bulbul coronavirus HKU11; IBV, avian infectious bronchitis virus; NDiV, Nam Dinh virus; GAV, gill-associated virus. Midpoint rooted phylogeny was reconstructed based on Viralis multiple sequence alignment [92] of the conserved core of RdRp, using IQ-Tree 1.5.5 [93] with automatically selected the rtREV + F + I + G4 evolutionary model. To estimate branch support, SH-like approximate likelihood ratio test with 1000 replicates was conducted. Polyproteins pp1ab are shown as light grey bars; they are autoproteolytically processed to nsps that were identified only for few nidoviruses and omitted here (see also Fig. 1). TM domains are shown as dark grey bars; TM helices were predicted by TMHMM2.0c [94] and clustered if separated by less than 300 aa (less than 180 aa for arteri- and tobaniviruses). Other selected domains, whose coordinates were obtained from the Viralis database [92], are shown as colored bars; proteolytically inactive PLP domains are indicated by stripes on bars; indices of PLP domains are specified below the bars. "Pkinase, protein kinase [25]; CPD, cyclic phosphodiestarase known also as 2′,5′-phosphodiesterase, 2′PDE [58,95]; NADAR, domain involved in the utilization of NAD and ADP-ribose derivatives [96]; for other domains, see Fig. 1 and the text."
Many more protein domains were delineated in further studies but none were conserved across the entire order until the discovery of the nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain in the ORF1b, which remained undetected for almost twenty years since the sequencing of the first ORF1b due to most pronounced divergence [72] (Fig. 3). It is the only enzymatic core domain that has no apparent ortholog in other known RNA viruses, although it uses a kinase fold [73] that is likely shared with protein kinases identified in few nidoviruses [25,74]. With the identification of the NiRAN domain, one hallmark of nidoviruses – universally conserved replicative domains encoded in a certain order (synteny) – expanded to include seven domains: (TM2)-3CLpro-(TM3)-NiRAN-RdRp-ZBD-HEL1. This domain constellation remains the most reliable characteristic that readily distinguishes the ever-growing diversity of nidoviruses from other viruses. Its presence allows to demarcate a monophyletic nidovirus cluster in phylogenetic trees including also other viruses.
In agreement with their nidovirus-wide conservation, all domains of the synteny, when tested in experiments, proved to be essential for nidovirus replication [72,[75], [76], [77], [78]].
1.6. Plasticity of the most conserved replicative proteins of nidoviruses
While remaining under a strong purifying selection, most conserved replicative domains have accepted rare or unique substitutions of key residues in nidoviruses that are among most divergent phylogenetically and in respect to other characteristics. Namely, PSCNV, singularly representing the suborder Monidovirineae [33], contains a number of remarkable substitutions in three enzymatic domains of the synteny, 3CLpro, NiRAN and RdRp [32]. A substrate pocket of PSCNV 3CLpro contains Val residue in place of His residue absolutely conserved in other nidoviruses [35]; the substitution was predicted to confer an unusual substrate specificity that remains to be established for the enzyme. 3CLpro is also known for repeated toggling between the nucleophile Cys and Ser residues of its catalytic center [[35], [79]]. It is observed between virus families and its implications for the nidovirus life cycle remains unknown. PSCNV NiRAN has a substitution of one out of the seven residues absolutely conserved in nidoviruses previously characterized. The RdRps of PSCNV, as well as WPDV [80], have a Gly-Asp-Asp (GDD) signature in its catalytic motif C, instead of Ser-Asp-Asp (SDD) signature characteristic for nidoviruses [24]. These observations indicate plasticity of active sites of three key enzymes, possibly related to functional diversification and their coupling. It reveals considerable potential of nidoviruses adapt at the molecular level even when changes concern the vital functions and most constrained enzymes, which are among most immediate targets of prospective antivirals.
1.7. Do nidoviruses use cognate enzymes to catalyze RNA capping?
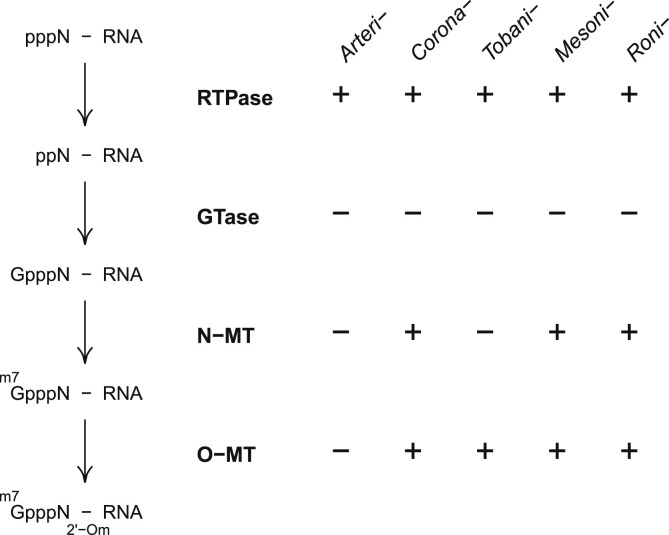
The most conserved domains of nidoviruses either control genome expression by autoproteolytic processing of pp1a/pp1ab (3CLpro) or genome replication (RdRp and HEL1, assisted by NiRAN and ZBD, respectively) or possibly both (TM2 and TM3). Intriguingly, no such ubiquitous link is evident between the protein domain conservation and control of translation of virus RNAs, including the incoming virion RNA, that launches virus-specific biosynthetic processes in the infected cell. Such control might include encoding a full complement of enzymes of an RNA capping pathway, like it was documented for a highly diverse monophyletic group of RNA viruses, historically known as alpha-like virus supergroup and currently comprising the class Alsuviricetes [[81], [82], [83]]. Indeed, coronavirus HEL1, residing in nsp13, possesses RNA 5′-triphosphotase (RTPase) activity that may catalyze the first reaction of the RNA capping pathway [84,85] (Fig. 4 ). In addition, two other ORF1b-encoded enzymes, guanine-N7-methyltransferase (N-MT) and O-MT, may catalyze the third and fourth reactions of the conventional mRNA capping pathway [[86], [87], [88], [89]]. N-MT and O-MT reside in coronaviruses nsp14 and nsp16 (Fig. 1), respectively, and they are colinear in the pp1ab polyproteins of mesoni- and roniviruses, whose nsps are yet to be described fully (Fig. 3) [71,88,58]. However, contrary to their assumed essential involvement in the mRNA capping, these enzymes are not conserved in all nidoviruses (Fig. 4). Specifically, tobaniviruses encode O-MT, but appear to lack N-MT or at least its catalytic domain, while both N-MT and O-MT are missing in arteriviruses [71,90]. It was proposed that the N-MT function in tobaniviruses may be complemented by a putative MT encoded in ORF1a [91]. However, this enzyme was not found in some tobaniviruses and its specificity remains unknown [74]. Additionally, the enzyme catalyzing the second reaction of the capping pathway, guanylyltransferase (GTase), has not been identified in any nidovirus [89], although NiRAN was proposed as a possible candidate [72]. Since nidoviruses are unlikely to subvert the capping machinery of eukaryotic hosts that functions in the nucleus, it remains unresolved how they synthesize the 5′-end cap [23,38,39], which controls translation initiation and protects the RNA molecule from degradation [97]. This uncertainty leaves open also the question about the natural targets of N-MT and O-MT, and methylation of other substrates than the 5′-terminal nucleotides remains a valid option [58].
Fig. 4.
Capping pathway and enzymes in relation to the proteome of nidoviruses. The conventional mRNA capping pathway is shown on the left, with the enzymes catalyzing the respective four reactions listed in bold. Further to the right, presence of these enzymes in viruses of five nidovirus families, each designated by its prefix, is listed (see Fig. 3 for phylogeny and pp1ab domain organization). RTPase, 5′-triphosphotase; GTase, guanylyl transferase; N-MT, guanine-N7-methyltransferase; O-MT, 2′-O-methyltransferase. In m7GpppN2’-Om notation, m7G stands for 7-methylguanosine, p stands for phosphate, N2’-Om stands for the 5′-terminal nucleoside of the RNA molecule, methylated at the ribose-2′-O position. For details, see text.
1.8. Hotspots of nidovirus evolution: duplications in pre-TM2 region
Most large-scale genomic changes in nidoviruses, including variation in respect to the N-MT and O-MT domains discussed above, can be attributed to aberrant homologous and nonhomologous recombination, the mechanisms behind deletions, duplications and gene acquisitions in RNA viruses [98,99]. These evolutionary events are most frequently observed in the two regions of nidovirus genome controlling nidovirus-host interactions: pre-TM2 region of ORF1a and 3′ORFs, as was documented in the past (e.g. [100,101]). Several notable examples of deletions, duplications and gene acquisitions, mapping to these genome regions, were described in recent years and summarized below.
One of the most common mechanisms of genome and protein innovation is the generation of tandem repeats. Possibly due to fast evolution, adjacent and highly similar tandem repeats were rarely observed in the genomes of RNA viruses. They were reported in an nsp3 region between Ubiquitin (Ub) and papain-like protease (PLP1) domains (Fig. 3) in several coronaviruses, including human coronavirus HKU1 (HCoV-HKU1, species Human coronavirus HKU1, genus Betacoronavirus) [102,103] and duck-dominant coronavirus (DdCoV; species Duck coronavirus 2714, genus Gammacoronavirus) [104]. The analyzed HCoV-HKU1 isolates encode from 2 to 17 perfect, and from 1 to 4 imperfect copies of the acidic NDDEDVVTGD repeat. Four analyzed isolates of DdCoV all harbored five almost-identical copies of a 23 aa charged residue-rich repeat.
Interestingly, also arteriviruses contain repeats positioned in close proximity to each other in a similar location of pre-TM2 region: three copies of the PxPxPR motif, separated by ∼10 aa, were identified within the HVR domain of EAV and WPDV [80]. At least one copy of this motif was also found within the Hinge or HVR domain of almost all other arteriviruses. PxPxPR motifs may be recognized by cellular Src homology 3 (SH3) protein domains, implicated in signal transduction [105]. The same function was previously suggested for the canonical SH3-binding motifs PxxP detected in the nsp2 sequence of PRRSV-1 [106]. Given the small size of PxPxPR motifs and their scattered position within the fast-evolving Hinge and HVR domains of arteriviruses, they might have emerged by either point mutation fixed by selection, or duplication followed by diversification.
Remarkably, the pre-TM2 region is also the location of the only described tandem repeats in the newly discovered invertebrate nidovirus PSCNV [32]. Two tandem repeats of 67 and 66 aa are separated by 3 aa and share 41.1% identity. No homologs of these repeats, which could hint to their function, were identified in other viruses or elsewhere.
The described tandem repeats are most likely to have emerged by duplication. The PSCNV genome encodes also the potential leader and body TRSs (see above) that are exceptionally ∼60 nt long, share 86% nucleotide sequence identity, and are separated by 28,327 nt. They might have emerged as a result of duplication, but incremental extension of ancestral elements with insertions under positive selection (convergence) of genome expansion remains a credible scenario as well. A shorter ancestral genome might have already had TRSs at the respective positions in the genome, as is typical for nidoviruses. Expansion of the genome would have necessitated TRSs extension: short motifs identical to TRSs can be encountered in a long genome just by chance, compromising quality genome expression. Consequently, gradual expansion of the genome could have promoted coordinated extension of TRSs through the convergence mechanism. Expansion of virus sampling in the PSCNV clad could help in resolving the evolutionary history of this intriguing similarity between exceptionally long TRS(-like) elements.
1.9. Could the host NF-kB pathway be universally targeted by all nidoviruses?
Another major source of genome and protein innovation is domain acquisition from other species. Many domains found in subsets of nidoviruses (Fig. 3) may have been acquired through this mechanism [107]. The study of PSCNV was particularly insightful in this respect since it expanded the previously known proteome repertoire of nidoviruses or even larger groups of viruses [32].
A domain acquisition by a nidovirus must be reconciled with many constraints acting on genome, RNome and proteome that remain mostly unknown [36]. It seems plausible that, if fixed, a newly acquired domain provides a fitness advantage by improving control over a process critical for either virus reproduction per se or modulation of host environment to facilitate virus reproduction, or both. For instance, it was proposed that the acquisition of the 3’-5’ exoribonuclease (ExoN) domain [108] improved the RTC control over progeny quality through the proofreading of RNA synthesis and allowed expansion of the nidovirus genome over 20 kb during evolution [71]; it provides a plausible explanation for the universal domain presence in all known nidoviruses with genomes of 20 kb or larger (Fig. 3). It is more uncertain whether a similar broad generalization is feasible when a clear-cut correlation between the domain presence and a characteristic may not be immediately evident in many nidoviruses. In this case, one of the options is that nidoviruses might have recruited different domains to regulate a common process, and analysis of a domain acquisition in a selected virus may offer a specific window into the process.
For instance, counteracting innate immunity response may be vital for nidoviruses to proliferate in wide range of hosts. It was reported that MERS-CoV targets the NF-κB pathway of the innate immunity using accessory protein 4b that is apparently conserved only in virus species of subgenus Merbecovirus, genus Betacoronavirus [109]. This pathway is targeted by many RNA viruses, including potentially SARS-CoV-2 [110]. Indeed, the planarian nidovirus PSCNV, which belongs to a different suborder than MERS-CoV and SARS-CoV-2, was proposed to target the NF-κB system by using an ANK-dependent pathway [32] in a manner shared by large dsDNA viruses [[111], [112], [113]].
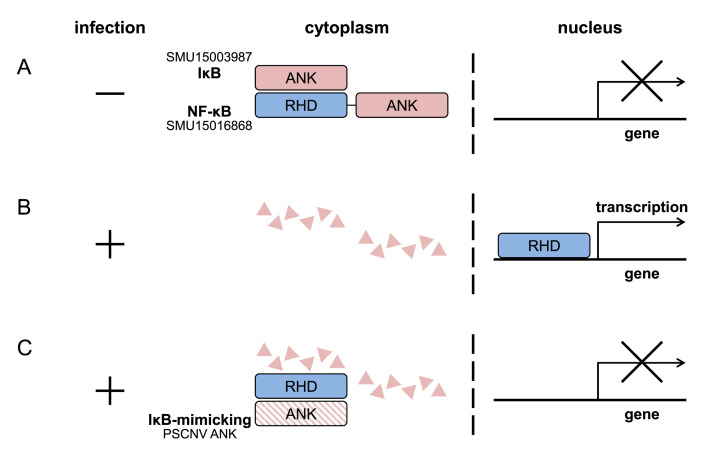
The ANK domain is ubiquitous in proteins of diverse cellular organisms, and dsDNA viruses with large genomes (>200 kb), but PSCNV is the first and the only currently known RNA virus that encodes ANK domain [32,114]. The PSCNV ANK domain clusters phylogenetically with ANK domains of a pair of host proteins, SMU15016868 and SMU15003987, indicating that the PSCNV ancestor might have been acquired from an ancestor of the host, flatworm Schmidtea mediterranea. The two ANK-containing proteins of the PSCNV host have domain architectures suggestive of their interaction: SMU15016868 is characteristic for NF-κB protein, N-RHD-ANK-C (RHD is a Rel homology domain), while SMU15003987 is characteristic for its inhibitor IκB, N-ANK-C [115]. Based on studies of several viruses [111,112], the NF-κB protein is expected to reside in the cytoplasm, bound by inhibitors, its own ANK domain and protein IκB, in the absence of a viral infection (Fig. 5 A). A viral infection would trigger degradation of NF-κB inhibitors, allowing NF-κB transcription factor to enter the nucleus and modulate gene expression to promote an antiviral immune response (Fig. 5B). Different viruses evolved diverse counteracting measures, and PSCNV may recruit cognate ANK as a IκB-mimicking protein, retaining a NF-κB transcription factor in the cytoplasm after the degradation of its inhibitors, and thus downregulating the immune response (Fig. 5C) [32]. This model is testable and also we could expect to learn whether other nidoviruses target the NF-κB pathway as well, which seems plausible. Our understanding of control of innate immunity by nidoviruses will improve if the future studies could also explain why PSCNV appears to resemble large DNA viruses rather than a fellow nidovirus MERS-CoV (and presumably others), which phylogenetically, genetically and size-wise are much more similar.
Fig. 5.
Proposed roles of PSCNV ANK and its host homologs in modulation of antiviral immune response. (A) In the non-infected cells, NF-ĸB protein (SMU15016868) resides in the cytoplasm, bound by inhibitors: its own ANK domain and protein IκB (SMU15003987). (B) In response to viral infection, inhibitors are degraded, allowing the NF-κB transcription factor to enter the nucleus and modulate gene expression to promote antiviral immune response. (C) IκB-mimicking viral protein (PSCNV ANK) may retain the NF-κB transcription factor in the cytoplasm after its inhibitors were degraded, thus downregulating the immune response.
2. Concluding remarks
The identification of SARS-CoV-2 was prompted by the outbreak of the infectious disease, unlike discovery of many nidoviruses in recent years by phenotype-free metagenomics- and transcriptomic-based research. SARS-CoV-2 is a variant of the known species which is another major difference compared to other newly described nidoviruses that prototype new species. This dominance of the disease-free virus species discovery is likely to continue and accelerate even further in the future; it is vital for advancement of the field. Beneficiaries of the massive nidovirus discovery are numerous and range from research on the ecology of nidoviruses to the in-depth characterization of selected nidoviruses of recognized societal importance, like SARS-CoV-2. Each newly described nidovirus decreases bias of our knowledge that is most critical to address for achieving a comprehensive understanding of nidovirus biology. Functional annotation of new nidoviruses by comparative genomics is based on the knowledge already accumulated in the field. Likewise, each novel nidovirus sheds a new light on known nidoviruses, putting state-of-the-art of experimental and computational research to a test of the lab of nature. Comparative genomics expands knowledge about the proteome and RNome of nidoviruses and their possible functions. It reveals what can and not be changed over the life of multitude of virus generations in many hosts and under many recurrent and fluctuating selection forces whose specifics remain to be described. And it also highlights paralells between distantly related nidoviruses even in the genomic regions that are poorly conserved. This insight may become particularly relevant when we try to interrogate SARS-CoV-2 with compounds to limit or even eliminate the virus transmission in the human population. How this coronavirus may adapt to these challenges is a big unknown, but it would be foolish to underestimate its plasticity and resilience, as comparative genomics of known nidoviruses attests in this review.
Acknowledgements
The authors are grateful to all collaborators whose work is cited in this review. Its reference list was compiled to illustrate raised points, and we regret for many omissions, especially concerning the fast growing literature on SARS-CoV-2. We thank Katya Dalgaard for the expert proofreading. Research in the AEG lab was partly supported by the EU Horizon2020 EVAg 653316 project and the LUMC MoBiLe program.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., for China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Li M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T.K., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G., Shi Z.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S.G. Siddell (Ed.), The Coronaviridae, in: The Viruses, H. Fraenkel-Conrat, R.R. Wagner (Eds.), Plenum Press, New York, 1995, pp. XVIII-418.
- 6.Perlman S, Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; 2008. pp. XVI–433. [Google Scholar]
- 7.Ziebuhr J., (Ed.) Coronaviruses. Adv. Virus Res. 2016;96:i–310. [Google Scholar]
- 8.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaender S., Mar K.B., Michailidis E., Kratzel A., Boys I.N., V’kovski P., Fan W.C., Kelly J.N., Hirt D., Ebert N., Stalder H., Kleine-Weber H., Hoffmann M., Hoffmann H.H., Saeed M., Dijkman R., Steinmann E., Wight-Carter M., McDougal M.B., Hanners N.W., Pohlmann S., Gallagher T., Todt D., Zimmer G., Rice C.M., Schoggins J.W., Thiel V. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol. 2020;5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon A., Selisko B., Le N.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B., Peersen O., Canard B. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff G., Limpens R., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grunewald K., Koster A.J., Snijder E.J., Barcena M. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Muhlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 13.Peng Q., Peng R.C., Yuan B., Zhao J.R., Wang M., Wang X.X., Wang Q., Sun Y., Fan Z., Qi J.X., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Wu J.Q., Wang H.F., Gao Y., Liu Q.J., Mu A., Ji W.X., Yan L.M., Zhu Y., Zhu C., Fang X., Yang X.B., Huang Y.C., Gao H.L., Liu F.J., Ge J., Sun Q.Q., Yang X.N., Xu W.Q., Liu Z.J., Yang H.T., Lou Z.Y., Jiang B., Guddat L.W., Gong P., Rao Z.H. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome, Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cagliani R., Forni D., Clerici M., Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dearlove B., Lewitus E., Bai H.J., Li Y.F., Reeves D.B., Joyce M.G., Scott P.T., Amare M.F., Vasan S., Michael N.L., Modjarrad K., Rolland M. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. U.S.A. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Groot R.J., Cowley J.A., Enjuanes L., Faaberg K.S., Perlman S., Rottier P.J.M., Snijder E.J., Ziebuhr J., Gorbalenya A.E. In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Elsevier Academic Press; Amsterdam: 2012. Order Nidovirales; pp. 785–795. [Google Scholar]
- 21.Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense ... ? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 22.Cowley J.A., Walker P.J. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Vliet A.L.W., Smits S.L., Rottier P.J.M., de Groot R.J. Discontinuous and non-discontinuous subgenornic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F., Kincaid A., Gordon M., Porter B.F., Baumgartner W., Stahl S., Kelley K., Towner J.S., DeRisi J.L. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in Python regius. mBio. 2014;5 doi: 10.1128/mBio.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 28.Ziebuhr J., Baric R.S., Baker S.C., de Groot R.J., Drosten C., Gulyaeva A., Haagmans B.L., Neuman B.W., Perlman S., Poon L.L.M., Sola I., Gorbalenya A.E. Proposal 2017.013S.A.v1. Reorganization of the family Coronaviridae into two families, Coronaviridae (including the current subfamily Coronavirinae and the new subfamily Letovirinae) and the new family Tobaniviridae (accommodating the current subfamily Torovirinae and three other subfamilies), revision of the genus rank structure and introduction of a new subgenus rank. ICTVonline. 2017 https://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/animal-ssrna-viruses/7996 [Google Scholar]
- 29.Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P.T., Morita K., Snijder E.J., Gorbalenya A.E. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bukhari K., Mulley G., Gulyaeva A.A., Zhao L., Shu G., Jiang J., Neuman B.W. Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology. 2018;524:160–171. doi: 10.1016/j.virol.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debat H.J. bioRxiv; 2018. Expanding the Size Limit of RNA Viruses: Evidence of a Novel Divergent Nidovirus in California Sea Hare, with a ∼35.9 Kb Virus Genome. [DOI] [Google Scholar]
- 32.Saberi A., Gulyaeva A.A., Brubacher J.L., Newmark P.A., Gorbalenya A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddell S.G., Walker P.J., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M., Rubino L., Sabanadzovic S., Sanfaçon H., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Additional changes to taxonomy ratified in a special vote by the international committee on taxonomy of viruses (october 2018) Arch. Virol. 2019;164:943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- 34.Gorbalenya A.E., Brinton M.A., de Groot R.J., Gulyaeva A.A., Lauber C., Neuman B.W., Ziebuhr J. ICTVonline; 2019. Proposal 2019.023S Create Five New Families and a New Suborder of Vertebrate Viruses in the Order Nidovirales.https://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/animal-ssrna-viruses/9495 [Google Scholar]
- 35.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 36.Lauber C., Goeman J.J., Parquet M.D., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai M.M., Patton C.D., Stohlman S.A. Further characterization of mRNA’s of mouse hepatitis virus: presence of common 5’-end nucleotides. J. Virol. 1982;41:557–565. doi: 10.1128/jvi.41.2.557-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagripanti J.L., Zandomeni R.O., Weinmann R. The cap structure of simian hemorrhagic fever virion RNA. Virology. 1986;151:146–150. doi: 10.1016/0042-6822(86)90113-3. [DOI] [PubMed] [Google Scholar]
- 40.Lai M.M., Brayton P.R., Armen R.C., Patton C.D., Pugh C., Stohlman S.A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J. Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanberlo M.F., Horzinek M.C., Vanderzeijst B.A.M. Equine arteritis virus-infected cells contain 6 polyadenylated virus-specific rnas. Virology. 1982;118:345–352. doi: 10.1016/0042-6822(82)90354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hemert M.J., van den Worm S.H.E., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denison M.R. Seeking membranes: positive-strand RNA virus replication complexes. PLoS Biol. 2008;6:2098–2100. doi: 10.1371/journal.pbio.0060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Barcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3’- coterminal subgenomic mRNAs that do not possess 5’-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- 46.Jendrach M., Thiel V., Siddell S. Characterization of an internal ribosome entry site within mRNA 5 of murine hepatitis virus. Arch. Virol. 1999;144:921–933. doi: 10.1007/s007050050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baric R.S., Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nedialkova D.D., Gorbalenya A.E., Snijder E.J. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogue B.G., Machamer C.E. In: Coronavirus Structural Proteins and Virus Assembly. Perlman S., Gallagher T., Snijder E.J., editors. ASM Press; Washington, DC: 2008. pp. 179–200. (Nidoviruses). [Google Scholar]
- 51.Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 52.Firth A.E., Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irigoyen N., Firth A.E., Jones J.D., Chung B.Y.W., Siddell S.G., Brierley I. High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling, PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di H., Madden J.C., Morantz E.K., Tang H.Y., Graham R.L., Baric R.S., Brinton M.A. Expanded subgenomic mRNA transcriptome and coding capacity of a nidovirus. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E8895–E8904. doi: 10.1073/pnas.1706696114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain S., Pan J., Chen Y., Yang Y.L., Xu J., Peng Y., Wu Y., Li Z.Y., Zhu Y., Tien P., Guo D.Y. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:5288–5295. doi: 10.1128/Jvi.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan J.Z., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-Unique Domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. Plos Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K., Wang W., Eden J.S., Shen J.J., Liu L., Holmes E.C., Zhang Y.Z. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 61.Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X., Qin X.C., Li J., Cao J.P., Eden J.S., Buchmann J., Wang W., Xu J.G., Holmes E.C., Zhang Y.Z. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–545. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 62.Strauss J.H., Strauss E.G. The Alphaviruses: gene expression, replication and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang Y., Treffers E.E., Li Y.H., Tas A., Sun Z., van der Meer Y., De Ru A.H., van Veelen P.A., Atkins J.F., Snijder E.J., Firth A.E. Efficient-2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boursnell M.E.G., Brown T.D.K., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 65.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodgman T.C. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 67.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorbalenya A.E., Koonin E.V. Birnavirus RNA polymerase is related to polymerases of positive strand RNA viruses. Nucleic Acids Res. 1988;16:7735. doi: 10.1093/nar/16.15.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- 70.Zirkel F., Kurth A., Quan P.L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C., Junglen S. mBio. Vol. 2. ASM; 2011. An Insect Nidovirus Emerging from a Primary Tropical Rainforest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nga P.T., Parquet M.D., Lauber C., Parida M., Nabeshima T., Yu F.X., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gulyaeva A.A., Sigorskih A.I., Ocheredko E.S., Samborskiy D.V., Gorbalenya A.E. LAMPA, LArge Multidomain Protein Annotator, and its application to RNA virus polyproteins. Bioinformatics. 2020;36:2731–2739. doi: 10.1093/bioinformatics/btaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Dinten L.C., Rensen S., Gorbalenya A.E., Snijder E.J. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and Is essential for virus replication. J. Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanitz M., Blanck S., Heine A., Gulyaeva A.A., Gorbalenya A.E., Ziebuhr J., Diederich W.E. Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus. Virology. 2019;533:21–33. doi: 10.1016/j.virol.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gulyaeva A., Dunowska M., Hoogendoorn E., Giles J., Samborskiy D., Gorbalenya A.E. Domain organization and evolution of the highly divergent 5’ coding region of genomes of arteriviruses, including the novel possum nidovirus. J. Virol. 2017;91 doi: 10.1128/JVI.02096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rozanov M.N., Koonin E.V., Gorbalenya A.E. Conservation of the putative methyltransferase domain: a hallmark of the ’Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 82.Koonin E.V., Dolja V.V., Krupovic M., Varsani A., Wolf Y.I., Yutin N., Zerbini F.M., Kuhn J.H. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020;84 doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahola T., Kaariainen L. Reaction in Alphavirus Messenger-Rna Capping - Formation of A Covalent Complex of Nonstructural Protein Nsp1 with 7-Methyl-Gmp. Proc. Natl. Acad. Sci. U.S.A. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with Severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5 ’-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Decroly E., Imbert I., Coutard B., Bouvet M.L., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D.Y. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. Vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lehmann K.C., Hooghiemstra L., Gulyaeva A., Samborskiy D.V., Zevenhoven-Dobbe J.C., Snijder E.J., Gorbalenya A.E., Posthuma C.C. Arterivirus nsp12 versus the coronavirus nsp16 2 ’-O-methyltransferase: comparison of the C-terminal cleavage products of two nidovirus pp1ab polyproteins. J. Gen. Virol. 2015;96:2643–2655. doi: 10.1099/vir.0.000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferron F., Debat H.J., Shannon A., Decroly E., Canard B. A N7-guanine RNA cap methyltransferase signature-sequence as a genetic marker of large genome, non-mammalian Tobaniviridae. NAR Genom. Bioinform. 2020;2 doi: 10.1093/nargab/lqz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorbalenya A.E., Lieutaud P., Harris M.R., Coutard B., Canard B., Kleywegt G.J., Kravchenko A.A., Samborskiy D.V., Sidorov I.A., Leontovich A.M., Jones T.A. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antivir. Res. 2010;87:95–110. doi: 10.1016/j.antiviral.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 95.Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A.E., Silverman R.H., Weiss S.R. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1073/pnas.1706696114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Souza R.F., Aravind L. Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions. Mol Biosyst. 2012;8:1661–1677. doi: 10.1039/c2mb05487f. [DOI] [PubMed] [Google Scholar]
- 97.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lai M.M.C. RNA recombination in animal and plant-viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gorbalenya A.E., Koonin E.V., Lai M.M.C. Putative papain-related thiol proteases of positive-strand RNA viruses - identification of rubivirus and aphthovirus proteases and delineation of A novel conserved domain associated with proteases of rubivirus, alpha- and coronaviruses. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Godeny E.K., de Vries A.A., Wang X.C., Smith S.L., de Groot R.J. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 1998;72:862–867. doi: 10.1128/jvi.72.1.862-867.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Wong H.L., Poon R.W.S., Cai J.J., Luk W.K., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Woo P.C.Y., Lau S.K.P., Yip C.C.Y., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhuang Q.Y., Wang K.C., Liu S., Hou G.Y., Jiang W.M., Wang S.C., Li J.P., Yu J.M., Chen J.M. Genomic analysis and surveillance of the coronavirus dominant in ducks in China. PloS One. 2015;10 doi: 10.1371/journal.pone.0129256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kowanetz K., Szymkiewicz I., Haglund K., Kowanetz M., Husnjak K., Taylor J.D., Soubeyran P., Engstrom U., Ladbury J.E., Dikic I. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J. Biol. Chem. 2003;278:39735–39746. doi: 10.1074/jbc.M304541200. [DOI] [PubMed] [Google Scholar]
- 106.Ropp S.L., Wees C.E.M., Fang Y., Nelson E.A., Rossow K.D., Bien M., Arndt B., Preszler S., Steen P., Christopher-Hennings J., Collins J.E., Benfield D.A., Faaberg K.S. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 2004;78:3684–3703. doi: 10.1128/JVI.78.7.3684-3703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gorbalenya A.E. In: Nidoviruses. Perlman S., Gallagher T., Snijder E.J., editors. ASM Press; Washington: 2008. Genomics and Evolution of the Nidovirales; pp. 15–28. [Google Scholar]
- 108.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3 ’-> 5 ’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Canton J., Fehr A.R., Fernandez-Delgado R., Gutierrez-Alvarez F.J., Sanchez-Aparicio M.T., Garcia-Sastre A., Perlman S., Enjuanes L., Sola I. MERS-CoV 4b protein interferes with the NF-kappa B-dependent innate immune response during infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ribero M.S., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Falabella P., Varricchio P., Provost B., Espagne E., Ferrarese R., Grimaldi A., de E.M., Fimiani G., Ursini M.V., Malva C., Drezen J.M., Pennacchio F. Characterization of the IkappaB-like gene family in polydnaviruses associated with wasps belonging to different Braconid subfamilies. J. Gen. Virol. 2007;88:92–104. doi: 10.1099/vir.0.82306-0. [DOI] [PubMed] [Google Scholar]
- 112.Tait S.W., Reid E.B., Greaves D.R., Wileman T.E., Powell P.P. Mechanism of inactivation of NF-kappa B by a viral homologue of I kappa b alpha. Signal-induced release of i kappa b alpha results in binding of the viral homologue to NF-kappa B. J. Biol. Chem. 2000;275:34656–34664. doi: 10.1074/jbc.M000320200. [DOI] [PubMed] [Google Scholar]
- 113.Camus-Bouclainville C., Fiette L., Bouchiha S., Pignolet B., Counor D., Filipe C., Gelfi J., Messud-Petit F. A virulence factor of myxoma virus colocalizes with NF-kappaB in the nucleus and interferes with inflammation. J. Virol. 2004;78:2510–2516. doi: 10.1128/JVI.78.5.2510-2516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Al-Khodor S., Price C.T., Kalia A., Abu K.Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gilmore T.D., Wolenski F.S. NF-kappaB: where did it come from and why? Immunol. Rev. 2012;246:14–35. doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]