Abstract
The current study aimed to analyze the genotype-phenotype relationship in patients with variants of zinc finger E box-binding homeobox 2 (ZEB2), which is a gene encoding a homeobox transcription factor known to be mutated in Mowat Wilson syndrome (MWS). Whole genome sequencing (WGS) was performed in 530 children, of whom 333 had epilepsy with or without developmental delay and 197 developmental delay alone. Pathogenic variants were identified and verified using Sanger sequencing, and the disease phenotypes of the corresponding patients were analyzed for features of MWS. WGS was performed in 333 children with epilepsy, with or without developmental delays or intellectual disability and 197 children with developmental delay alone. A total of 4 unrelated patients were indicated to be heterozygous for truncating mutations in ZEB2. A total of three of these were nonsense mutations (novel Gln1072X and recurrent Trp97X and Arg921X), and one was a frameshift mutation (novel Val357Aspfs*15). The mutations have occurred de novo as confirmed by Sanger sequence comparisons in patients and their parents. All 4 patients exhibited signs of MWS, whereby the severity increased the closer a mutation was located to the amino terminus of the protein. The results suggest that the clinical outcome in MWS depends on the relative position of the truncation in the ZEB2 gene. A number of interpretations of this genotype/phenotype association are discussed in the present study.
Keywords: Mowat-Wilson Syndrome, zinc finger E box-binding homeobox 2, whole genome sequencing, nonsense mutations, frameshift mutation
Introduction
Mowat-Wilson syndrome [MWS; Phenotype Mendelian Inheritance in Man (MIM) no. 235730] was first described in 1998 as a rare condition characterized by dysmorphic facial features, delayed motor development, intellectual disability and multisystem involvement (~1 among 50,000-70,000 live births) (1). The typical facial features of MWS include hypertelorism, horizontal wedge-shaped eyebrows with medial flaring, deep-set eyes, a broad nasal bridge, a prominent nasal tip, uplifted earlobes with a central depression, a prominent, pointed chin and prognathism (2). Affected individuals display moderate to severe intellectual disability, and >70% of individuals with MWS develop epilepsy (3-5). Multisystem structural anomalies include Hirschsprung disease (HSCR), genitourinary anomalies (hypospadias and renal tract anomalies), congenital heart defects and eye and brain abnormalities, including agenesis of the corpus callosum, ventriculomegaly and hippocampal and white matter abnormalities (6-8). In addition, individuals with MWS may develop microcephaly and a short stature as well as musculoskeletal anomalies (5,9,10). Due to this broad phenotypic spectrum and the varying degrees of disease severity, clinical diagnosis of MWS is often challenging but is facilitated by an analysis of the underlying genetic cause.
MWS has been demonstrated to be associated with de novo heterozygous loss-of-function alleles of zinc finger E box-binding homeobox2 (ZEB2), which is a gene that was previously named ZFHX1B or SMAD interacting protein 1 (SIP1; Gene/Locus MIM no. 605802) and is located on chromosome 2q22.3 (11,12). ZEB2 spans ~70 kb, comprises 10 exons and 9 introns, and encodes ZEB2, also known as SIP1 or SMAD interacting protein 1, which has been indicated to be associated with regulating the TGF-β/BMP/SMAD signaling cascade (12,13). Exon 1 is non-coding, exon 2 contains the initiation codon and exon 10 encodes the homeodomain and the stop codon (3). ZEB2 is expressed in the majority of human tissues and is critical for the development and migration of neural-crest cells (14), heart septation and midline development during early embryogenesis (15).
The ZEB2 protein contains a number of functional domains, including a nucleosome remodeling and deacetylase-interaction motif, one zinc-finger (ZF) cluster in the amino terminal region (N-ZF), a SMAD binding domain, a homeodomain, a C-terminal binding protein interacting domain, and one ZF cluster in the carboxyl terminal region (C-ZF) (16).
ZEB2 acts as a transcriptional repressor, and one of its characterized targets is the E-cadherin promoter, with the ZEB2 C-ZF being necessary for proper DNA-binding and transactivation activities (17). Given the complex domain structure of the protein, it is likely that besides N- and C-ZF, other domains may also serve important roles in target gene regulation and consequently embryonic development.
Up to date, >220 pathogenic variants of ZEB2 have been associated with MWS, including point mutations, deletions, duplications and large chromosomal rearrangements according to the Human Gene Mutation Database (HGMD) (18). Previous studies have demonstrated that patients with ZEB2 deletions and truncations exhibit similar phenotypic severities, but those with larger deletions have more severe phenotypes compared with those with smaller deletions (19,20). It is still unclear, however, to what degree the variable MWS phenotypes are influenced directly by the nature of the corresponding ZEB2 mutation. The analysis of genotype/phenotype associations in MWS is therefore required. For the majority of children with MWS, the facial features are not obvious during the early postnatal period but start to develop with increasing age (4). Therefore, early on, the identification of a ZEB2 mutation may be the only way to confirm a suspicion of MWS and to distinguish MWS from other genetic disorders such as Goldberg-Shprintzen megacolon syndrome (Phenotype MIM no. 609460), with which MWS shares some clinical features but which is caused by variants of a different gene.
The current study described the identification of two previously described and two novel de novo mutations in 4 unrelated children and associate their specific mutations with their distinct phenotypes.
Materials and methods
Patients
The current study assessed two cohorts of patients from the Shenzhen Children's Hospital (Shenzhen, China) between October 2016 and December 2017. The inclusion criteria were: i) Diagnosis of epilepsy based on International League Against Epilepsy criteria (ILAE2017) (21); ii) patients presented with developmental delays or intellectual disabilities; and iii) age <18 years. The exclusion criterion was that individuals with acquired brain injuries, including head trauma, brain tumors, central nervous system infections, immune encephalitis and cerebrovascular pathological changes. Cohort A included 333 patients (sex, 197 males and 136 females) who presented with epilepsy and whose ages ranged from 1 month to 17 years (median age, 4.4 years). Of these, 209 also exhibited developmental delays or intellectual disabilities. Cohort B included 197 patients (sex, 112 males and 85 females) who presented with developmental delays or intellectual disabilities but not epilepsy (age range, 3 months to 16 years; median age, 5.6 years). DNA samples from the patients and their parents were collected. The four patients in whom ZEB2 mutations were identified underwent a general medical examination, which included physical examination, doppler ultrasound of the heart and urinary system, abdominal X-ray, electrocardiogram, gas chromatography-mass spectrometry (GC/MS) (22), which has been widely used in metabolomics analyses of biofluid samples, tandem mass spectrometry (MS/MS) (23), which is the fundamental platform technology for proteomics, and as a major technology for peptide sequencing. Both GC/MS and MS/MS were used in the screening of metabolic diseases in the present study. In addition, a detailed clinical assessment of their epilepsy that included electroencephalogram (24) and MRI brain studies (25). Their developmental and general medical history and medical records were reviewed. Written informed consent was obtained from all parents or legal guardians of the patients. The current study was approved by the Ethics Committee of the Shenzhen Children's Hospital (reference no. 2017014).
Whole genome sequencing, variant identification and validation
Genomic DNA was isolated from probands' peripheral leukocyte with the QIAamp DNA Blood Mini Kit (Qiagen GmbH), according to the manufacturer's protocol. Leukocyte DNA was also obtained from the parents of the four probands. Sample collection from the adult participants was performed in Shenzhen Children's Hospital (Shenzhen, China) at the same time with the probands between December 12th, 2016 and May 5th, 2017. Totally, eight parents (sex, four males and four females) participated in the study (age range, 30-40 years; median age, 34 years). Whole genome sequencing was conducted using the BGISEQ-500 platform (cat. no. 1000005478; BGI Group) performing paired-end, 100 bp (PE100) sequencing as described previously (26). Deep sequencing data were aligned to the reference GRCh37 build (hg19), and variants were called using the Edico Dragen analysis pipeline (27). The Edico Genome's Dragen Bio-IT Platform is based on the Dragen Bio-IT Processor, a bioinformatics application-specific integrated reference-based mapping, aligning, sorting, de-duplication and variant calling system (28). Variants were annotated using bcfanno (v1.4; https://github.com/shiquan/bcfanno) in Frequency data [The Exome Aggregation Consortium (29), the Genome Aggregation Database (GnomAD; v2.1.1) (30), and 1000Genomes (G1000) (31)] and gene-disease data [ClinVar (32), Clinical genomic database (33), Online MIM (OMIM) (34) and HGMD (35)]. The predictive programs SIFT (v 5.2.2) (36), Polyphen2 (v2.2.2) (37), MutationTaster (NCBI 37/Ensembl 69) (38) and PROVEAN (v1.1.5) (39) were used to access the pathogenic potential of the variants. Human Phenotype Ontology (HPO) items were used to prioritize candidate genes using Phenolyzer (40), which incorporates a list of gene-disease databases, compiled from several data sources, including OMIM, Orphanet (41), ClinVar and Genome Wide Association Studies Catalog (42). Rare variants [<1% minor allele frequency in G1000, ExAC-East Asian population (EAS), and GnomAD-exome-EAS] in HPO candidate genes were classified into five categories (pathogenic, likely pathogenic, uncertain significance, likely benign, and benign) according to American College of Medical Genetics and Genomics (ACMG) Guidelines (43). Sanger sequencing was performed to validate the variants and for parental segregation testing.
Primers for the amplification of the four ZEB2 variants using PCR were designed using Primer Premier 6.0 (http://www.premierbiosoft.com; Table I) and synthesized by BGI Tech Solutions Co., Ltd., who also performed the Sanger sequencing. The source of DNA was from peripheral blood leukocyte. The DNA polymerase was supplied by NEBNext® High-Fidelity 2X PCR Master Mix (cat. no. M0541; New England Biolabs, Inc.). Thermocycling conditions were: Initial denaturation at 98˚C for 30 sec, followed by 35 cycles of 98˚C for 10 sec; the annealing temperature was 66˚C for primer pair of P1, P3 and P4 and 63˚C for primer pair of P2 for 30 sec. Extension temperature was 72˚C for 30 sec per kb. Final extension temperature was 72˚C for 5 min.
Table I.
Primers for the amplification of four ZEB2 variants by PCR.
| Patient | Variant | Primer (5'-3') |
|---|---|---|
| P1 | c.290G>A (p.Trp97X) | Forward: GATGTAACTGCCGCAATGTGA |
| Reverse: GGGGTGGCTGATGTTTCTCA | ||
| P2 | c.1067_1068insAGACG (p.Val357Aspfs*15) | Forward: AGTGCCACTAAACCCGTGTG |
| Reverse: TGTCCTCCCAGGGCAGATAA | ||
| P3 | c.2761C>T(p.Arg921X) | Forward: AGCCCACTGATGGTTTTA |
| Reverse: GAGCCTCTGAACTTGACTTT | ||
| P4 | c.3214C>T(p.Gln1072X) | Forward: GCCTTCTTTCTCGTGCTCCT |
| Reverse: CCATCGATTAGACCGGGGTG |
ZEB2, zinc finger E box-binding homeobox2; P, patient.
Multiple sequence alignments were generated for homologous ZEB2 sequences to evaluate conservation using T-Coffee (44). Alignments for ZEB2 were generated using the following sequences: Homo sapiens, NP_055610.1; Mus musculus, NP_056568.2; Rattus norvegicus, NP_001028873.1; Gallus gallus, NP_001305395.1; Pan troglodytes, XP_001158120.1 isoform X1; Canis lupus familiaris, XP_005632021.2; Bos taurus, NP_001069660.2; Danio rerio, NP_001232895.1; Equus caballus, XP_005601530.2; and Sus scrofa and XP_020932163.1.
Results
Demographics and genotypic features
WGS was performed in 530 patients with epilepsy, developmental delays and intellectual disabilities alone or in combination, as aforementioned. Among these, 4 (~0.75 %) unrelated patients, ranging in age from 2 years and 4 months to 6 years and 5 months, who harbored heterozygous de novo pathogenic variants of ZEB2 were identified.
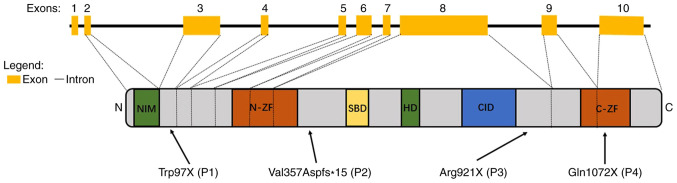
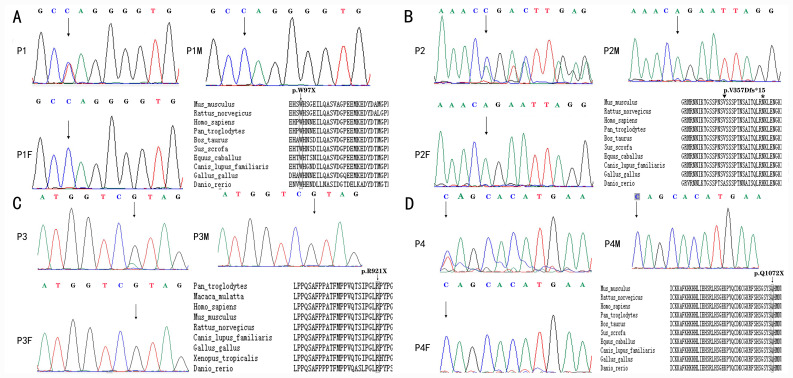
Each of the four distinct mutations introduced premature stop codons, either directly as a consequence of a codon change to a stop codon, or indirectly as a consequence of a frame shift. In patients 1, 2 and 3, the stop codons were located outside the known protein domains (NM_014795.3:c.290G>A/p.Trp97X; NM_014795.3: c.1067_1068insAGACG/p.Val357Aspfs*15; and NM_014795.3:c.2761C>T/p.Arg 921X, respectively) while in patient 4, the stop codon was located in the C-ZF domain (NM_014795.3:c.3214C>T/p.Gln1072X; Fig. 1). A comparison of the patients' mutations with the sequences of their corresponding parents by Sanger sequencing indicated that the mutations occurred de novo (data not shown). Of the four mutations, the Arg921X mutation of patient 3 had previously been reported in a number of patients (2,4,45). Furthermore, the Trp97X mutation of patient 1 had been reported in one Chinese population (46), while the other mutations were, to the best of our knowledge, novel. The electropherograms of the four mutations in ZEB2 along with the amino acid conservation in the regions of the mutations in several vertebrates are presented in Fig. 2. None of these mutations were found in G1000 and ExAC-EAS databases. Each of the mutations produced a stop codon at a location that was 100% conserved at the amino acid level across a number of mammalian and avian species. Bioinformatic tools (Polyphen2, SIFT, MutationTaster and PROVEAN) were used to predict that if the truncated proteins were indeed produced, they would be altered in both structure and function compared with the corresponding portions of the wild type protein. According to the rules of ACMG, all of the four mutations aforementioned were considered pathogenic.
Figure 1.
Schematic representation of the ZEB2 exons and protein structure. The figure represents the functional domains of ZEB2 and the corresponding exon distributions: NIM, N-ZF, SBD, HD, CID and C-ZF. Arrows indicate the localization of the mutations in patients, with the numbers P1-4 representing patients 1, 2, 3 and 4, respectively. ZEB2, zinc finger E box-binding homeobox2; NIM, nucleosome remodeling and deacetylase-interaction motif; N-ZF, zinc-finger cluster in the amino terminal region; SBD, SMAD binding domain; HD, homeodomain; CID, C-terminal binding protein interacting domain; C-ZF, zinc-finger cluster in the carboxyl terminal region.
Figure 2.
Electropherograms of patients with Mowat-Wilson syndrome with ZEB2 pathogenic variants. Heterozygous truncating mutations (nonsense or frameshift mutations) resulting in premature termination of the ZEB2-encoded protein are shown. (A) Patient 1, exon 3, c.290G>A (p.Trp97X). (B) Patient 2, exon 8, c.1067_1068insAGACG (p. Val357Aspfs*15). Inverted triangle on top of the V, where the insertion is located; * at the N, where the stop is. (C) Patient 3, exon 8, c.2761C>T (p. Arg921X). (D) Patient 4, exon 10, c.3214C>T (p. Gln1072X). Each figure (A-D) also indicates the conservation among vertebrates of the sequence regions where the respective premature stop codons are located. Rectangles indicated the location of the nonsense mutation in the location. Triangle and * were used to indicate the location of frameshift mutation, insertion and the stop codon. The ZEB2 accession numbers are as follows: Homo sapiens, NP_055610.1; Mus musculus, NP_056568.2; Rattus norvegicus, NP_001028873.1; Gallus gallus, NP_001305395.1; Pan troglodytes, XP_001158120.1 isoform X1; Canis lupus familiaris, XP_005632021.2; Bos taurus, NP_001069660.2; Danio rerio, NP_001232895.1; Equus caballus, XP_005601530.2; and Sus scrofa, XP_020932163.1.
Spectrum of phenotypic features
The four patients demonstrated a wide spectrum of phenotypic features (Table II). Patient 1 exhibited classical features of MWS, including typical facial features, delayed motor development, intellectual disability, epilepsy, hypotonia, microcephaly, frontal cortical atrophy, HSCR and hypospadias. Patient 2 presented with distinctive facial characteristics, delayed motor development, intellectual disability, hypotonia, microcephaly, ventriculomegaly and constipation. Patient 3 had typical facial features, delayed motor development, intellectual disability, epilepsy, hypotonia, microcephaly, myelin dysplasia and constipation. Patient 4 presented with relatively milder symptoms that, although included distinctive facial features, delayed motor development, intellectual disability and epilepsy, lacked the more severe symptoms aforementioned for the other three patients.
Table II.
Clinical features and ZEB2 mutations in patients with MWS.
| Patient characteristic | P1 | P2 | P3 | P4 |
|---|---|---|---|---|
| Age | 3 y | 2 y 4 m | 3 y 2 m | 6 y 5 m |
| Sex | M | F | M | M |
| Exon affected by the mutation | 3 | 8 | 8 | 10 |
| Mutation | c.290G>A | c.1067_1068insAGACG | c.2761C>T | c.3214C>T |
| Novel | No | Yes | No | Yes |
| AA | p. Trp97X | p. Val357Aspfs*15 | p. Arg921X | p. Gln1072X |
| Inheritance | De novo | De novo | De novo | De novo |
| CFA | HT, BNB, PNT, UECD, PC | HT, BNB, PNT, UECD, PC | HT, DSE, BNB, PNT, PC | HT, BNB, PNT, UECD, PC |
| WA | -a | -a | 3 y | 2 y |
| ID | ++ | ++ | ++ | + |
| Speech | - | - | - | FW |
| Epilepsy (medication received) | + | - | + (VPA) | + (VPA, LEV) |
| Disposition | Happy | Happy | Happy | Happy |
| Hypotonia | + | + | + | - |
| MC | + | + | + | - |
| BA | FCA | ETV | MD | - |
| HSCR | + | - | - | - |
| CO | - | + | + | - |
| Other organ malformations | Hypospadias | - | - | - |
aNot able to walk at the time of assessment. y, year; m, month; M, male; F, female; AA, amino acid; CFA, characteristic facial appearance; HT, Hypertelorism; BNB, broad nasal bridge; PNT, prominent nasal tip; UECD, uplifted earlobes with a central depression; PC, pointed chin; DSE, deep-set eyes; WA, walking at age; ID, intellectual disability (++ severe, + moderate); FW, few words; VPA, valproate; LEV, levetiracetam; MC, microcephaly; BA, brain anomaly; FCA, frontal cortical atrophy; ETV, enlargement of the third ventricle; MD, myelin dysplasia; HSCR, Hirschsprung disease; CO, constipation; ZEB2, zinc finger E box-binding homeobox2; MWS, Mowat-Wilson syndrome; P, patient.
At the time of diagnosis, all 4 patients exhibited some signs of distinctive facial features of MWS, including a broad nasal bridge, a prominent and pointed chin, a prominent nasal tip, and uplifted earlobes with a central depression. As an example, the characteristic facial appearance of patient 2 is presented in Fig. S1. Patients 1 and 2 are still unable to walk, patient 3 started to walk at ~3 years, but the less severely affected patient 4 was able to walk at ~2 years. The date at which these observations are still valid for patient 1 and 2 was July 30th, 2020; for patient 3 and 4 this was June 25th, 2020. Similar to the differential delay in the ability to walk, the intellectual disabilities were severe in patients 1-3 but moderate in patient 4. Among the four patients, only patient 4 could speak at age 3 years and this patient was able to point at objects kept out of reach. At 6 years, patient 4 could speak short sentences with two to five words, and language comprehension was only moderately impaired. Patient 1 presented with epilepsy at the age of 2, but the seizures stopped after half a year even without the use of antiepileptic drugs. The epilepsy of patient 3 and 4 was easily controlled with one or two types of antiepileptic drugs, where patient 3 was treated with valproate (20 mg/kg/d) and patient 4 was treated with valproate (25 mg/kg/d) and levetiracetam (30 mg/kg/d). Patients 1, 2 and 3 displayed hypotonia and microcephaly. MRI indicated frontal cortical atrophy in patient 1, ventriculomegaly in patient 2 and mild myelin dysplasia in patient 3. In patient 4, however, the MRI was normal. None of the patients had congenital heart disease, but patient 1 exhibited signs of HSCR and displayed hypospadias while the others exhibited no signs of multisystem involvement. The disease severity was based on a comprehensive evaluation of the phenotypes, including multisystem involvements, differential delay in the ability to walk and speak, and MRI findings. In general, patient 1 presented with the most severe symptoms and patient 4 with the mildest. Patient 3 had no signs of HSCR, not affected by hypospadias and demonstrated less severe MRI findings than either patient 1 or patient 2. Patient 2 demonstrated no signs of multisystem involvements and epilepsy and hence was less severely affected than patient 1.
Discussion
The wide phenotypic spectrum and varying degrees of disease severity render a clinical diagnosis of MWS difficult, especially when the typical facial features are not clearly present at an early age. Therefore, none of the 4 patients in the current study were identified to carry truncating ZEB2 mutations and had not been clearly diagnosed as exhibiting MWS despite displaying signs of the disorder. In fact, it was only after identification of mutations in ZEB2 that a diagnosis was made.
The detailed clinical assessment showed considerable patient-to-patient differences in disease manifestations, which ranged from mild in patient 4 to severe in patient 1. While patient 4 presented with mild delayed motor development, intellectual disability and epilepsy, but no multisystem involvements, patient 1 exhibited severely delayed motor development, severe intellectual disability, epilepsy, hypotonia, microcephaly, frontal cortical atrophy, HSCR and hypospadias. The disease severity in patients 2 and 3 was milder compared with that in patient 1 but more severe compared with that in patient 4. The molecular genetic analysis revealed that patient 4 had a truncating mutation in the ultimate exon 10, which encodes a highly conserved cluster of zinc fingers. This mutation resembles one described by Ivanovski et al (47) (c.3031delA, p. S1011Afs*64), which was associated with mild to moderate intellectual disability, no seizures and absence of HSCR (47).
As premature stop codons in the ultimate exon do not lead to nonsense-mediated decay of mRNA and may not alter mRNA stability or translatability (48), it can be assumed that the truncated protein accumulated in this patient. This protein retains a number of protein- and DNA-interacting domains but lacks part of the domain that is associated with target gene interaction. Notwithstanding the fact that MWS-associated ZEB2 alleles seemingly represent loss-of-function alleles or haploinsufficiency, it is still theoretically possible that a truncated protein lacking a DNA interaction domain may act in a dominant-negative fashion. Such dominant-negative action, however, may be partial and not devastating for various reasons, for instance due to the fact the truncated protein is less stable or the retained wild type mRNA and protein are upregulated (49,50). Further elucidation of this would require a deeper molecular analysis of expression and activity of the mutant ZEB2.
In contrast to patient 4, patients 1, 2 and 3 all exhibited coding region truncations originating in internal exons (exons 3 and 8) where premature stop codons usually lead to nonsense-mediated decay of the corresponding mRNAs and hence a severe reduction or total absence of polypeptides that otherwise may be derived from them (47). This would mean that the corresponding alleles are all equally nonfunctional null-alleles, leaving their carriers with ZEB2 protein derived from just one wild type copy of the gene. The corresponding symptoms would be consistent with an earlier report on gene deletions that had established haploid insufficiency of ZEB2 in MWS (51). Previous studies have revealed no obvious genotype-phenotype associations in patients with MWS except in cases where large deletions are associated with more severe phenotypes (20,52). A recent study indicated that milder clinical presentations can be identified with variant ZEB2 proteins that are predicted to preserve some functionality (47). Likewise, the present study revealed that the severity of the corresponding disease phenotypes would seem to depend on where exactly the truncation is localized, being most severe in patient 1 carrying the allele with the truncation localized closest to the amino terminus and being mildest in patient 4 with the truncation closer to the carboxyl terminus. It is conceivable that the expression level of the wild type allele may be influenced by the nature of the mutant allele, thereby potentially enabling partial compensation of the loss of protein from the mutant allele (53). It is equally conceivable, however, that alternative splicing events of the mutant mRNA may lead to altered mRNAs that can, or cannot, be properly translated depending on the allele. In addition, disease severities may also be influenced by varying genetic backgrounds of the different patients. Each of these mechanisms, alone or in combination, may then influence the phenotypes associated with a given allele (54).
Alternatively, it is possible that the mutant mRNAs of patients 1, 2, and 3, as suggested for the mRNA of patient 4, are not subject to nonsense-mediated decay and are translated, leading to accumulation of the truncated proteins. If so, it would appear that the smaller the resulting polypeptide is, the more severe the phenotype. This may be consistent with the earlier notion that while ZEB2 deletions lead to severe disease, intragenic mutations that are predicted to preserve some ZEB2 protein functionality lead to milder clinical manifestations (47), supporting the notion that not all alleles are necessarily complete loss-of-function alleles. Larger proteins, such as those only truncated in exon 8 (as in patients 2 and 3), would likely preserve more functionality and may lead to less severe phenotypes than those truncated in exon 3 (as in patient 1). None of the domains, however, in which the proteins of patients 1, 2 and 3 are truncated, have been recognized to carry specific functions despite their general sequence conservation. To explain the allele-specific disease manifestations, other molecular or cell biological properties of the corresponding polypeptides should be investigated, such as differences in specific half-lives or nuclear/cytoplasmic distributions.
In conclusion, the current study described 4 patients with de novo ZEB2 mutations, two novel and two recurrent, that were associated with features of MWS. Indeed, it was only through a molecular genetic analysis that a clear diagnosis became possible in these patients due to the rarity and phenotypic variability of MWS. The analysis revealed the possibility of an association between the nature of the truncating mutation and the phenotype. The results suggested that not all ZEB2 alleles found in MWS are complete loss-of-function alleles. In the current study, the number of patients assessed was small and additional experiments, including experiments in animal models, may be necessary to identify the mechanisms by which each allele is responsible for its distinct associated set of symptoms.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The current study was supported by the Science and Technology Innovation Commission of Shenzhen (grant no. KJYY20151116165726645) and Sanming Project of Medicine in Shenzhen (grant no. SZSM201812005).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL and JG designed the study. FW designed the study and administered the project. DZ completed the recruitment of the patients, collection of the clinical data, analysis of the data, and draft of the manuscript. LW, JD and HX performed data analysis and interpretation. TZ, ZY and QD performed data analysis and provided technical assistance. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The current study was approved by the Ethics Committee of the Shenzhen Children's Hospital (Shenzhen, China; reference no. 2017014). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments. Written informed consent was obtained from all parents or legal guardians of the patients.
Patient consent for publication
Written patient consent for publication was obtained from patients' legal guardians.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Evans E, Einfeld S, Mowat D, Taffe J, Tonge B, Wilson M. The behavioral phenotype of Mowat-Wilson syndrome. Am J Med Genet A. 2012;158A:358–366. doi: 10.1002/ajmg.a.34405. [DOI] [PubMed] [Google Scholar]
- 2.Zweier C, Thiel CT, Dufke A, Crow YJ, Meinecke P, Suri M, Ala-Mello S, Beemer F, Bernasconi S, Bianchi P, et al. Clinical and mutational spectrum of Mowat-Wilson syndrome. Eur J Med Genet. 2005;48:97–111. doi: 10.1016/j.ejmg.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Garavelli L, Mainardi PC. Mowat-Wilson syndrome. Orphanet J Rare Dis. 2007;2(42) doi: 10.1186/1750-1172-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garavelli L, Zollino M, Mainardi PC, Gurrieri F, Rivieri F, Soli F, Verri R, Albertini E, Favaron E, Zignani M, et al. Mowat-Wilson syndrome: Facial phenotype changing with age: Study of 19 Italian patients and review of the literature. Am J Med Genet A. 2009;149A:417–426. doi: 10.1002/ajmg.a.32693. [DOI] [PubMed] [Google Scholar]
- 5.Cordelli DM, Garavelli L, Savasta S, Guerra A, Pellicciari A, Giordano L, Bonetti S, Cecconi I, Wischmeijer A, Seri M, et al. Epilepsy in Mowat-Wilson syndrome: Delineation of the electroclinical phenotype. Am J Med Genet A. 2013;161A:273–284. doi: 10.1002/ajmg.a.35717. [DOI] [PubMed] [Google Scholar]
- 6.Coyle D, Puri P. Hirschsprung's disease in children with Mowat-Wilson syndrome. Pediatr Surg Int. 2015;31:711–717. doi: 10.1007/s00383-015-3732-x. [DOI] [PubMed] [Google Scholar]
- 7.Bourchany A, Giurgea I, Thevenon J, Goldenberg A, Morin G, Bremond-Gignac D, Paillot C, Lafontaine PO, Thouvenin D, Massy J, et al. Clinical spectrum of eye malformations in four patients with Mowat-Wilson syndrome. Am J Med Genet A. 2015;167:1587–1592. doi: 10.1002/ajmg.a.36898. [DOI] [PubMed] [Google Scholar]
- 8.Garavelli L, Ivanovski I, Caraffi SG, Santodirocco D, Pollazzon M, Cordelli DM, Abdalla E, Accorsi P, Adam MP, Baldo C, et al. Neuroimaging findings in Mowat-Wilson syndrome: A study of 54 patients. Genet Med. 2017;19:691–700. doi: 10.1038/gim.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mowat DR, Wilson MJ, Goossens M. Mowat-Wilson syndrome. J Med Genet. 2003;40:305–310. doi: 10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adam MP, Schelley S, Gallagher R, Brady AN, Barr K, Blumberg B, Shieh JT, Graham J, Slavotinek A, Martin M, et al. Clinical features and management issues in Mowat-Wilson syndrome. Am J Med Genet A. 2006;140:2730–2741. doi: 10.1002/ajmg.a.31530. [DOI] [PubMed] [Google Scholar]
- 11.Cacheux V, Dastot-Le Moal F, Kaariainen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D, Goossens M. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet. 2001;10:1503–1510. doi: 10.1093/hmg/10.14.1503. [DOI] [PubMed] [Google Scholar]
- 12.Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, et al. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet. 2001;27:369–370. doi: 10.1038/86860. [DOI] [PubMed] [Google Scholar]
- 13.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: DeltaEF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghoumid J, Drevillon L, Alavi-Naini SM, Bondurand N, Rio M, Briand-Suleau A, Nasser M, Goodwin L, Raymond P, Yanicostas C, et al. ZEB2 zinc-finger missense mutations lead to hypomorphic alleles and a mild Mowat-Wilson syndrome. Hum Mol Genet. 2013;22:2652–2661. doi: 10.1093/hmg/ddt114. [DOI] [PubMed] [Google Scholar]
- 18. HGMD® Professional 2020.2 Trial version. http://hgmdtrial.biobase-international.com/hgmd/pro/gene.php?gene=ZEB2. Accessed September 2, 2020. [Google Scholar]
- 19.Cerruti Mainardi P, Pastore G, Zweier C, Rauch A. Mowat-Wilson syndrome and mutation in the zinc finger homeo box 1B gene: A well defined clinical entity. J Med Genet. 2004;41(e16) doi: 10.1136/jmg.2003.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihara N, Yamada K, Yamada Y, Miura K, Kato J, Kuwabara N, Hara Y, Kobayashi Y, Hoshino K, Nomura Y, et al. Clinical and molecular analysis of Mowat-Wilson syndrome associated with ZFHX1B mutations and deletions at 2q22-q24.1. J Med Genet. 2004;41:387–393. doi: 10.1136/jmg.2003.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, Hirsch E, Jain S, Mathern GW, Moshé SL, et al. ILAE classification of the epilepsies: Position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emwas AH, Al-Talla ZA, Yang Y, Kharbatia NM. Gas chromatography-mass spectrometry of biofluids and extracts. Methods Mol Biol. 2015;1277:91–112. doi: 10.1007/978-1-4939-2377-9_8. [DOI] [PubMed] [Google Scholar]
- 23.Yan Y, Kusalik AJ, Wu FX. Recent developments in computational methods for de novo peptide sequencing from tandem mass spectrometry (MS/MS) Protein Pept Lett. 2015;22:983–991. doi: 10.2174/0929866522666150821113127. [DOI] [PubMed] [Google Scholar]
- 24.Krumholz A. Which EEG protocol is best in young adults with possible epilepsy? Nat Clin Pract Neurol. 2007;3:128–129. doi: 10.1038/ncpneuro0400. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Rios C, McAndrews MP, Logan W, Krings T, Lee D, Widjaja E. MRI in the evaluation of localization-related epilepsy. J Magn Reson Imaging. 2016;44:12–22. doi: 10.1002/jmri.25269. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, et al. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017;6:1–9. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7(100) doi: 10.1186/s13073-015-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edico Genomes DRAGEN Platform. http://edicogenome.com/dragen-bioit-platform/. Accessed February 5, 2018. [Google Scholar]
- 29.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karczewski KJ, Franciol LC, Tiao G, Cummings BB, Alföld J, Wang Q, Collins RL, Laricchia KM, Gann A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. doi: 10.1038/nature15393. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA and Abecasis GR: A global reference for human genetic variation. Nature 526: 68-74, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44 (D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon BD, Nguyen AD, Bear KA, Wolfsberg TG. Clinical genomic database. Proc Natl Acad Sci USA. 2013;110:9851–9855. doi: 10.1073/pnas.1302575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33 (Database Issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The human gene mutation database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 39.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(e46688) doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Robinson PN, Wang K. Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: The Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 42.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42 (Database Issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 45.Yamada Y, Nomura N, Yamada K, Matsuo M, Suzuki Y, Sameshima K, Kimura R, Yamamoto Y, Fukushi D, Fukuhara Y, et al. The spectrum of ZEB2 mutations causing the Mowat-Wilson syndrome in Japanese populations. Am J Med Genet A. 2014;164A:1899–1908. doi: 10.1002/ajmg.a.36551. [DOI] [PubMed] [Google Scholar]
- 46.Ho S, Luk HM, Chung BH, Fung JL, Mak HH, Lo IFM. Mowat-Wilson syndrome in a Chinese population: A case series. Am J Med Genet A. 2020;182:1336–1341. doi: 10.1002/ajmg.a.61557. [DOI] [PubMed] [Google Scholar]
- 47.Ivanovski I, Djuric O, Caraffi SG, Santodirocco D, Pollazzon M, Rosato S, Cordelli DM, Abdalla E, Accorsi P, Adam MP, et al. Phenotype and genotype of 87 patients with Mowat-Wilson syndrome and recommendations for care. Genet Med. 2018;20:965–975. doi: 10.1038/gim.2017.221. [DOI] [PubMed] [Google Scholar]
- 48.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 49.Sheppard D. Dominant negative mutants: Tools for the study of protein function in vitro and in vivo. Am J Respir Cell Mol Biol. 1994;11:1–6. doi: 10.1165/ajrcmb.11.1.8018332. [DOI] [PubMed] [Google Scholar]
- 50.Veitia RA, Caburet S, Birchler JA. Mechanisms of mendelian dominance. Clin Genet. 2018;93:419–428. doi: 10.1111/cge.13107. [DOI] [PubMed] [Google Scholar]
- 51.Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- 52.Zweier C, Temple IK, Beemer F, Zackai E, Lerman-Sagie T, Weschke B, Anderson CE, Rauch A. Characterisation of deletions of the ZFHX1B region and genotype-phenotype analysis in Mowat-Wilson syndrome. J Med Genet. 2003;40:601–605. doi: 10.1136/jmg.40.8.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 54.Peng J. Gene redundancy and gene compensation: An updated view. J Genet Genomics. 2019;46:329–333. doi: 10.1016/j.jgg.2019.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.