Abstract
Genetic factors play an important role in the pathogenesis of ischemic stroke. Of these, epigenetic modifications provide a new direction for the study of ischemic stroke pathogenesis. This study aimed to determine the correlation between DNA methylation of the gene encoding S-adenosylhomocysteine hydrolase (AHCY) and the risk of ischemic stroke in 64 ischemic stroke patients and 138 patients with traumatic brain injury (control group). The methylation level of AHCY was analyzed using quantitative methylation-specific polymerase chain reaction. Statistically significant differences in AHCY methylation levels were observed between the case group [medians (interquartile range): 0.13% (0.09%, 0.27%)] and the control group [0.06% (0.00%, 0.17%), p < 0.0001], and these associations remained significant in both male (p = 0.003) and female (p = 0.0005) subjects. A subgroup analysis by age revealed a considerably higher percentage of methylated AHCY in the case group than the control group in all age groups (age < 60 years, p = 0.007; age ≥ 60 years, p < 0.0001). A receiver operating characteristic (ROC) curve analysis revealed a trend toward a role for AHCY methylation as an indicator of risk in all ischemic patients [area under the curve (AUC) = 0.70, p = 0.0001], male patients (AUC = 0.67, p = 0.004), and female patients (AUC = 0.75, p = 0.0002). Our study confirmed a significant association between the AHCY DNA methylation level and the risk of ischemic stroke, suggesting that this gene methylation pattern may be a potential diagnostic marker of ischemic stroke.
Keywords: Ischemic stroke, DNA methylation, S-adenosylhomocysteine hydrolase, sex, age
INTRODUCTION
Stroke is among the three leading causes of disease-related mortality in humans and a common cause of chronic disability in adults [1], and accordingly poses a serious risk to human health and places a heavy burden on society [2]. Stroke can be divided etiologically as ischemic or hemorrhagic stroke [3]. Regarding the former type, common risk factors such as diabetes, hypertension, and hyperlipidemia can explain only a small fraction of the actual risk [4]. Increasingly, studies have shown that genetic factors play key roles in the pathogenesis of ischemic stroke [5,6]. For example, genome-wide association studies have identified several genetic variations that contribute to the risk factors associated with ischemic stroke [7,8]. Other recent studies have indicated that epigenetic changes play important roles in stroke development [9,10], and even that the gap between environmental factors and ischemic stroke may be bridged by an epigenetic mechanism [11].
DNA methylation is an epigenetic mechanism by which the spatial structure of a DNA molecule is transformed to affect the interactions between DNA and proteins. Potentially, DNA hypermethylation could induce the silencing of genes and the loss of gene function [12]. Aberrant DNA methylation has been observed in various vascular developmental diseases, including brain arteriovenous malformations [13], intracranial aneurysms [14], and cerebral infarctions [15]. Furthermore, the DNA methylation level was reported as a key regulator of vascular smooth muscle cell dedifferentiation and vascular remodeling [16].
S-adenosylhomocysteine hydrolase (AHCY) is the only enzyme known to catalyze the hydrolysis of S-adenosyl-L-homocysteine (SAH) to yield homocysteine and adenosine. Hence, this enzyme can relieve the inhibitory activities of SAH on S-adenosyl methionine (SAM)-dependent transmethylation reactions [17,18]. However, dysfunctional AHCY activity can result in serious pathological consequences, such as childhood death [19], Alzheimer’s disease [20], Parkinson’s disease [21], age-related diseases [22], neuroblastoma [23], and large-artery atherosclerotic stroke [24]. Despite these negative outcomes, few studies have investigated the relationship between AHCY and the risk of ischemic stroke. As the homocysteine level is considered an independent predictor of severe neurological impairment in ischemic stroke patients, we hypothesized that DNA methylation of AHCY may play a key role in the pathological development of ischemic stroke. In this case-control study, we explored the role of DNA methylation of AHCY as a risk factor for ischemic stroke in a Chinese population.
MATERIALS AND METHODS
Samples and clinical data
Approval of the study procedure was provided by the Ethics Committee of the Ningbo First Hospital. The investigation included 64 ischemic stroke patients and 138 controls (patients with a traumatic brain injury without any vascular lesions) that visited the Stroke Center, Ningbo First Hospital, between September 2013 and December 2015. In all patients, the fulfillment of the diagnostic criteria based on international standardized definitions was confirmed using magnetic resonance imaging and cranial computed tomography scans. Any subject presenting with a serious liver or kidney disease or cardiovascular disease was excluded from the study.
Peripheral venous blood was collected from all patients in the morning after fasting and stored in tubes containing EDTA (anticoagulant). General information, including sex, age, and the levels of triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), apolipoprotein A (ApoA), apolipoprotein B (ApoB), and apolipoprotein E (ApoE), was collected for all patients. Bioindicators were measured using an automatic biochemical analyzer (AU2700; Olympus Corp., Tokyo, Japan).
SYBR Green-based quantitative methylation-specific polymerase chain reaction (qMSP)
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The DNA bisulfite conversion was assessed using the EZ DNA Methylation-Gold™ Kit (Zymo Research, Orange, CA, USA). The qMSP assay was performed using a LightCycler480 device (Roche Diagnostics, Mannheim, Germany). Human ACTB was used as the internal reference gene to standardize the amount of target DNA. The percentage of methylated reference (PMR) of the genes in each sample was calculated using the 2−ΔΔCt quantification approach [25]. The sequences of specific primers were as follows: AHCY forward, 5´-GGTCGTAATCGGTTGAT-3´ and reverse, 5´-CAATTCCTATTCCCAAACTAAA-3´; ACTB forward, 5´-TGGTGATGGAGGAGGTTTAGTAAGT-3´ and reverse, 5´-AACCAATAAAACCTACTCCTCCCTTAA-3´. The entire protocol for qMSP was previously described [26].
Statistical analysis
Statistical data were analyzed and plotted using SPSS Version 20.0 (IBM Corp, Armonk, NY, USA) and GraphPad Prism version 8.0 (GraphPad Software, La Jolla, CA, USA). Measured data were assessed using the one-sample Kolmogorov–Smirnov test for normality, and normally distributed data were assessed using the independent samples t-test. The data are presented as means ± standard deviations. Nonparametric tests were used to assess data that did not conform to a normal distribution, and these data are expressed as medians (interquartile range). A receiver operating characteristic (ROC) curve analysis was performed to evaluate the sensitivity of AHCY DNA methylation as an indicator of ischemic stroke diagnosis. All statistical tests were bilateral. Statistical significance was defined as a p value < 0.05.
RESULTS
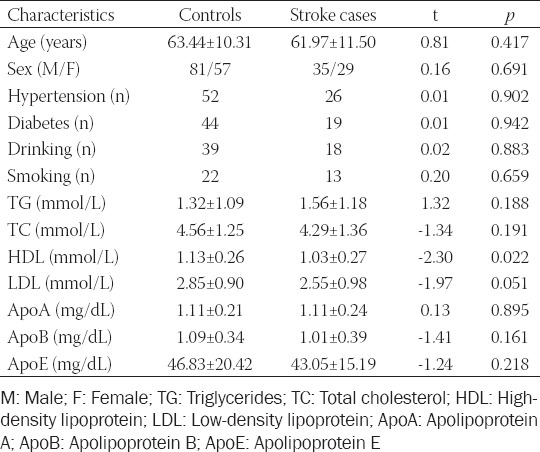
The clinical characteristics of patients in the ischemic stroke and control groups are shown in Table 1. The HDL level was considerably lower in stroke patients (1.03 ± 0.27) than in controls (1.13 ± 0.26, p = 0.022). No significant differences were observed with respect to the other demographic and clinical characteristics (age, sex, hypertension, diabetes, alcohol consumption, smoking, TG, TC, LDL, ApoA, ApoB, and ApoE; p > 0.05) between the two groups.
TABLE 1.
Clinical characteristics of all participants
Target sequences (Chr20: 34303211–34303334) located in the CGI region of AHCY were selected to determine the DNA methylation level (Figure 1). As shown in Figure 2, the PMR level of AHCY was significantly higher in ischemic stroke patients [0.13% (0.09%, 0.27%)] than in controls [0.06% (0.00%, 0.17%), p < 0.0001]. A subgroup analysis stratified by sex similarly demonstrated significantly higher PMR levels of AHCY in both male [0.12% (0.09%, 0.24%)] and female ischemic stroke patients [0.13% (0.10%, 0.33%)] relative to controls [0.06% (0.00%, 0.16%), p = 0.003 and 0.05% (0.00%, 0.17%), p = 0.0005, respectively]. A subgroup analysis stratified by age similarly revealed a significantly higher PMR level of AHCY in the ischemic stroke patients relative to the controls, regardless of age group [<60 years, 0.13% (0.10%, 0.29) vs. 0.07% (0.00%, 0.17%), p = 0.007; age ≥60 years, 0.13% (0.07%, 0.27%) vs. 0.04% (0.00%, 0.17%), p < 0.0001; Figure 3).
FIGURE 1.
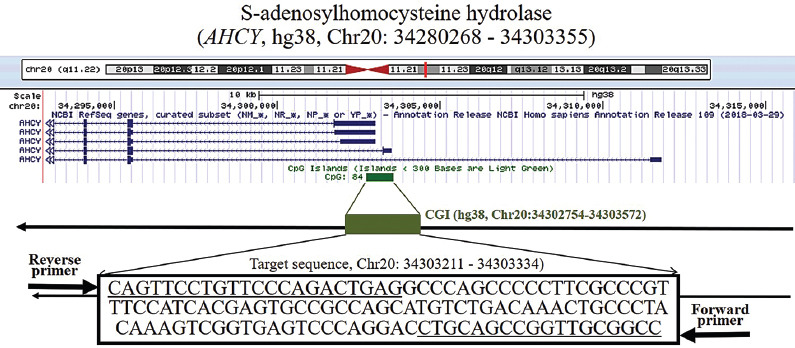
Characteristics of target sequences in S-adenosylhomocysteine hydrolase (AHCY)
FIGURE 2.
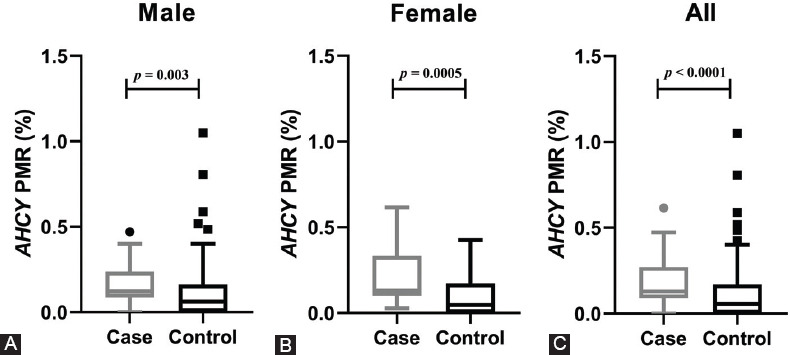
Comparison of S-adenosylhomocysteine hydrolase (AHCY) methylation levels between cases and controls in a sex-stratified subgroup analysis. (A) The percentage of methylated reference (PMR) levels of AHCY in male ischemic cases [0.12% (0.09%, 0.24%)] were higher than those in male controls [0.06% (0.00%, 0.16%), p = 0.003]. (B) The PMR levels of AHCY in female ischemic cases [0.13% (0.10%, 0.33%)] were higher than those in female controls [0.05% (0.00%, 0.17%), p = 0.0005]. (C) The PMR level of AHCY in all ischemic stroke cases [0.13% (0.09%, 0.27%)] was significantly higher than in the controls [0.06% (0.00%, 0.17%), p < 0.0001].
FIGURE 3.
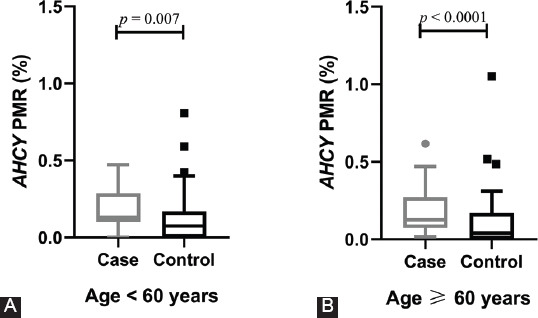
Comparison of S-adenosylhomocysteine hydrolase (AHCY) methylation levels in a subgroup analysis stratified by age. (A) In the <60 years age group, the percentage of methylated reference (PMR) levels of AHCY were higher in the ischemic cases than in the controls [0.13% (0.10%, 0.29) vs. 0.07% (0.00%, 0.17%), p = 0.007]. (B) In the ≥60 years age group, the PMR levels of AHCY were higher in the ischemic cases than that in the controls [0.13% (0.07%, 0.27%) vs. 0.04% (0.00%, 0.17%), p < 0.0001].
As shown in Figure 4, a ROC analysis demonstrated a trend toward AHCY methylation as an indicator of the ischemic stroke risk in all ischemic patients [area under the curve (AUC) = 0.70, p = 0.0001), as well as in male (AUC = 0.67, p = 0.004) and female patients (AUC = 0.75, p = 0.0002).
FIGURE 4.
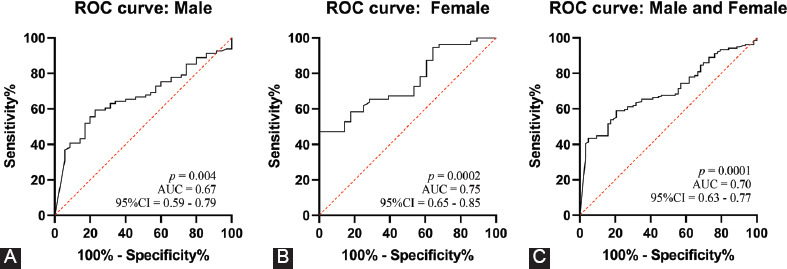
Receiver operating characteristic (ROC) curve analysis of the association between S-adenosylhomocysteine hydrolase (AHCY) DNA methylation and ischemic stroke. (A) In male patients, AHCY methylation tended to indicate a diagnosis of ischemic stroke [area under curve (AUC) = 0.67, 95% confidence interval (CI) = 0.59–0.79, p = 0.004]. (B) In female patients, AHCY methylation also tended to indicate a diagnosis of ischemic stroke (AUC = 0.75, 95% CI = 0.65–0.85, p = 0.0002). (C) In all patients, AHCY methylation tended to indicate a diagnosis of ischemic stroke (AUC = 0.70, 95% CI = 0.63–0.77, p = 0.0001).
DISCUSSION
The development of the field of epigenetics has elicited significant interest in the correlations between human diseases and epigenetic factors. Particularly, epigenetic modifications provide a new direction for studies of the pathogenesis of ischemic stroke. Although epigenetic changes can result in heritable changes in gene expression, the underlying DNA sequence is not altered. Rather, these modifications enable a more selective expression or suppression of genes. DNA methylation is a highly studied epigenetic mechanism that plays an important role in regulating gene expression. As noted previously, abnormal DNA methylation can induce related diseases such as stroke [27], cardiovascular disease [28], and malignancies [29]. However, the role of DNA methylation in the pathogenesis of ischemic stroke is yet to be fully elucidated.
In this study, we evaluated the potential association of AHCY methylation with ischemic stroke and determined a significantly higher frequency of this epigenetic modification among ischemic stroke patients relative to controls, irrespective of sex and age. AHCY, which is located on chromosome 20, contains ten exons and encodes AHCY. This broadly expressed enzyme hydrolyzes SAH, which is produced by the demethylation of SAM, a molecule that participates in the methylation of DNA and histones [30]. AHCY catalysis produces homocysteine from the metabolic precursor SAH and, therefore, the inhibition of AHCY can result in an accumulation of SAH in cells. Additionally, SAH is a potent feedback inhibitor of most SAM-dependent transmethylation reactions, including those related to the methylation of cellular components such as DNA, RNA, lipids, proteins, and neurotransmitters [17,31]. Therefore, AHCY methylation may affect AHCY expression and activity, leading to disordered SAH metabolism, with negative impacts on the regulation of biological processes. A previous study demonstrated that mutations in AHCY were associated with prognosis in neuroblastoma patients [32], suggesting that this gene could be considered as a potential prognostic factor in this context [23]. Giusti et al. identified a potentially significant difference in the allelic frequency of the AHCY polymorphism rs819146 between ischemic stroke patients and control subjects according to the recessive model [33]. Furthermore, the AHCY haplotype was reported as a susceptibility factor for abdominal aortic aneurysm, and this association is independent of the homocysteine modulatory role of this enzyme [34]. Therefore, AHCY methylation may regulate the enzymatic activity of AHCY, resulting in increased homocysteine levels in patients with cerebrovascular disease.
Many biomarkers of brain diseases involve DNA methylation. Jakubowski and Labrie [35] reported that DNA methylation may be a novel biomarker with potential etiological relevance in Parkinson’s disease. Thon et al. suggested that the O6-methylguanine-DNA methyltransferase promoter methylation status is crucial in glioblastoma treatment [36]. Moreover, TRIM59 and KLF14 hypermethylation in blood cells may be a valuable predictor of the molecular processes underlying familial Alzheimer’s disease [37]. Recent studies suggested that age-related DNA methylation changes were independent predictors of ischemic stroke outcomes [38,39]. Li et al. [40] demonstrated a significant difference in the AHCY methylation levels between ischemic stroke patients and controls older than 60 years of age. We similarly demonstrated a significant difference in the AHCY methylation levels between ischemic cases and controls, although this result was consistent in all age groups. Our ROC analysis further confirmed that AHCY methylation is a significant predictor of the risk of ischemic stroke.
Our study had some limitations of note. First, our analysis of the association between AHCY DNA methylation and the risk of ischemic stroke focused on only one portion of the CGI. Therefore, our results may not be applicable to all AHCY sequences. Second, we did not analyze the transcription or translation of AHCY. Therefore, a mechanistic study of the association between AHCY methylation and ischemic stroke is needed. Third, the sample size of this study was small. Further evaluations with larger samples are needed to elucidate the relationship between AHCY methylation and ischemic stroke, particularly in various ethnic populations.
CONCLUSION
Our findings confirm a significant correlation between the AHCY DNA methylation level and the risk of ischemic stroke and suggest that AHCY methylation may be a useful diagnostic biomarker of ischemic stroke.
ACKNOWLEDGMENTS
This study was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (LQ17H090002), the Medicine and Health Science and Technology Projects of Zhejiang Province (2018KY665, 2018KY674, 2017KY586, 2017KY588, 2017KY610), and the Ningbo Health Branding Subject Fund (PPXK2018-04).
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Chen Z, Jiang B, Ru X, Sun H, Sun D, Liu X, et al. Mortality of stroke and its subtypes in China:Results from a nationwide population-based survey. Neuroepidemiology. 2017;48(3-4):95–102. doi: 10.1159/000477494. https://doi.org/10.1159/000477494. [DOI] [PubMed] [Google Scholar]
- 2.Shen Y, Chao BH, Cao L, Tu WJ, Wang LD. Stroke center care and outcome:Results from the CSPPC stroke program. Transl Stroke Res. 2019 doi: 10.1007/s12975-019-00727-6. https://doi.org/10.1007/s12975-019-00727-6. [DOI] [PubMed] [Google Scholar]
- 3.Grysiewicz RA, Thomas K, Pandey DK. Epidemiology of ischemic and hemorrhagic stroke:Incidence, prevalence, mortality, and risk factors. Neurol Clin. 2008;26(4):871–95. doi: 10.1016/j.ncl.2008.07.003. vii. https://doi.org/10.1016/j.ncl.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration):A meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11(11):951–62. doi: 10.1016/S1474-4422(12)70234-X. https://doi.org/10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markus HS. Stroke genetics. Hum Mol Genet. 2011;20(R2):R124–31. doi: 10.1093/hmg/ddr345. https://doi.org/10.1093/hmg/ddr345. [DOI] [PubMed] [Google Scholar]
- 6.Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6(2):149–61. doi: 10.1016/S1474-4422(07)70028-5. https://doi.org/10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Wang Z, Liu X, Li Y, Shi C, Wu J, et al. Association of common variants in the IL-33/ST2 axis with ischemic stroke. Curr Neurovasc Res. 2019;16(5):494–501. doi: 10.2174/1567202616666191029112334. https://doi.org/10.2174/1567202616666191029112334. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, Hu SC, Zheng PW, Jin MJ, Tang ML, Chen K, et al. Association between CPR-related genetic variants and risk of ischemic stroke:A nested case-control study. Neurol Res. 2019;41(12):1090–6. doi: 10.1080/01616412.2019.1673286. https://doi.org/10.1080/01616412.2019.1673286. [DOI] [PubMed] [Google Scholar]
- 9.Schweizer S, Meisel A, Märschenz S. Epigenetic mechanisms in cerebral ischemia. J Cereb Blood Flow Metab. 2013;33(9):1335–46. doi: 10.1038/jcbfm.2013.93. https://doi.org/10.1038/jcbfm.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demyanenko S, Uzdensky A. Epigenetic alterations induced by photothrombotic stroke in the rat cerebral cortex:Deacetylation of histone H3, upregulation of histone deacetylases and histone acetyltransferases. Int J Mol Sci. 2019;20(12):e2882. doi: 10.3390/ijms20122882. https://doi.org/10.3390/ijms20122882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassis H, Shehadah A, Chopp M, Zhang ZG. Epigenetics in stroke recovery. Genes (Basel) 2017;8(3):e89. doi: 10.3390/genes8030089. https://doi.org/10.3390/genes8030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. https://doi.org/10.1038/npp2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Zhao J, Sun J, Nie S, Li K, Gao F, et al. Sex-dichotomous effects of NOS1AP promoter DNA methylation on intracranial aneurysm and brain arteriovenous malformation. Neurosci Lett. 2016;621:47–53. doi: 10.1016/j.neulet.2016.04.016. https://doi.org/10.1016/j.neulet.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S, Gao X, Sun J, Lin Z, Huang Y. DNA methylation of the PDGFD gene promoter increases the risk for intracranial aneurysms and brain arteriovenous malformations. DNA Cell Biol. 2017;36(6):436–42. doi: 10.1089/dna.2016.3499. https://doi.org/10.1089/dna.2016.3499. [DOI] [PubMed] [Google Scholar]
- 15.Xiao J, Li X, Yuan Q, Zhang S, Qu K, Wu B, et al. PON1 hypermethylation and PON3 hypomethylation are associated with risk of cerebral infarction. Curr Neurovasc Res. 2019;16(2):115–22. doi: 10.2174/1567202616666190412154407. https://doi.org/10.2174/1567202616666190412154407. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang J, Luan P, Li H, Wang K, Zhang P, Xu Y, et al. The Yin-Yang dynamics of DNA methylation is the key regulator for smooth muscle cell phenotype switch and vascular remodeling. Arterioscler Thromb Vasc Biol. 2017;37(1):84–97. doi: 10.1161/ATVBAHA.116.307923. https://doi.org/10.1161/atvbaha.116.307923. [DOI] [PubMed] [Google Scholar]
- 17.Tehlivets O, Malanovic N, Visram M, Pavkov-Keller T, Keller W. S-adenosyl-L-homocysteine hydrolase and methylation disorders:Yeast as a model system. Biochim Biophys Acta. 2013;1832(1):204–15. doi: 10.1016/j.bbadis.2012.09.007. https://doi.org/10.1016/j.bbadis.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singhal A, Arora G, Sajid A, Maji A, Bhat A, Virmani R, et al. Regulation of homocysteine metabolism by Mycobacterium tuberculosis S-adenosylhomocysteine hydrolase. Sci Rep. 2013;3:2264. doi: 10.1038/srep02264. https://doi.org/10.1038/srep02264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barić I. Inherited disorders in the conversion of methionine to homocysteine. J Inherit Metab Dis. 2009;32(4):459–71. doi: 10.1007/s10545-009-1146-4. https://doi.org/10.1007/s10545-009-1146-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YD, Ke XY, Shen W, Liu Y. Relationship of homocysteine and gene polymorphisms of its related metabolic enzymes with Alzheimer's disease. Chin Med Sci J. 2005;20(4):247–51. [PubMed] [Google Scholar]
- 21.Haghdoost-Yazdi H, Sarookhani M, Faraj A, Fraidouni N, Dargahi T, Yaghoubidoust MH, et al. Evaluation of the association between blood homocysteine concentration and the degree of behavioral symptoms in the 6-hydroxydopamine-induced Parkinsonism in rat. Pharmacol Biochem Behav. 2014;124:297–304. doi: 10.1016/j.pbb.2014.06.020. https://doi.org/10.1016/j.pbb.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Cardoso AL, Fernandes A, Aguilar-Pimentel JA, de Angelis MH, Guedes JR, Brito MA, et al. Towards frailty biomarkers:Candidates from genes and pathways regulated in aging and age-related diseases. Ageing Res Rev. 2018;47:214–77. doi: 10.1016/j.arr.2018.07.004. https://doi.org/10.1016/j.arr.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhong X, Liu Y, Liu H, Zhang Y, Wang L, Zhang H. Identification of potential prognostic genes for neuroblastoma. Front Genet. 2018;9:589. doi: 10.3389/fgene.2018.00589. https://doi.org/10.3389/fgene.2018.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Z, Zhang Z, Zhang H, Hao Y, Zhang J, Liu W, et al. Prognostic value of C-reactive protein and homocysteine in large- artery atherosclerotic stroke:A prospective observational study. J Stroke Cerebrovasc Dis. 2017;26(3):618–26. doi: 10.1016/j.jstrokecerebrovasdis.2016.11.016. https://d oi.org/ 10.101 6/j.jstrok e cerebro vasdis 201611. 0 16. [DOI] [PubMed] [Google Scholar]
- 25.Hu H, Wang T, Pan R, Yang Y, Li B, Zhou C, et al. Hypermethylated promoters of secreted frizzled- related protein genes are associated with colorectal cancer. Pathol Oncol Res. 2019;25(2):567–75. doi: 10.1007/s12253-018-0505-6. https://doi.org/10.1007/s12253-018-0505-6. [DOI] [PubMed] [Google Scholar]
- 26.Ji H, Zhou C, Pan R, Han L, Chen W, Xu X, et al. APOE hypermethylation is significantly associated with coronary heart disease in males. Gene. 2019;689:84–9. doi: 10.1016/j.gene.2018.11.088. https://doi.org/10.1016/j.gene.2018.11.088. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Paré G, Rundek T. DNA methylation predicts stroke outcome better:The epigenetic clock is ticking. Neurology. 2017;89(8):758–9. doi: 10.1212/WNL.0000000000004278. https://doi.org/10.1212/wnl.0000000000004278. [DOI] [PubMed] [Google Scholar]
- 28.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5(7):401–8. doi: 10.1038/nrendo.2009.102. https://doi.org/10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 29.Shen Z, Chen X, Li Q, Ye H, Li J, Zhou C, et al. TGFB2 and BCL2 L11 methylation in male laryngeal cancer patients. Oncol Lett. 2016;12(4):2999–3003. doi: 10.3892/ol.2016.5018. https://doi.org/10.4236/jhepgc.2018.44036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu YF, Wang JX, Zhao Y, Yang Y, Tang W, Ni J, et al. S-adenosyl-L-homocysteine hydrolase inactivation curtails ovalbumin-induced immune responses. J Pharmacol Exp Ther. 2006;316(3):1229–37. doi: 10.1124/jpet.105.093369. https://doi.org/10.1124/jpet.105.093369. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. https://doi.org/10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novak EM, Halley NS, Gimenez TM, Rangel-Santos A, Azambuja AM, Brumatti M, et al. BLM germline and somatic PKMYT1 and AHCY mutations:Genetic variations beyond MYCN and prognosis in neuroblastoma. Med Hypotheses. 2016;97:22–5. doi: 10.1016/j.mehy.2016.10.008. https://doi.org/10.1016/j.mehy.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Giusti B, Saracini C, Bolli P, Magi A, Martinelli I, Peyvandi F, et al. Early-onset ischaemic stroke:Analysis of 58 polymorphisms in 17 genes involved in methionine metabolism. Thromb Haemost. 2010;104(2):231–42. doi: 10.1160/TH09-11-0748. https://doi.org/10.1160/th09-11-0748. [DOI] [PubMed] [Google Scholar]
- 34.Giusti B, Saracini C, Bolli P, Magi A, Sestini I, Sticchi E, et al. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J Med Genet. 2008;45(11):721–30. doi: 10.1136/jmg.2008.057851. https://doi.org/10.1136/jmg.2008.057851. [DOI] [PubMed] [Google Scholar]
- 35.Jakubowski JL, Labrie V. Epigenetic biomarkers for Parkinson's disease:From diagnostics to therapeutics. J Parkinsons Dis. 2017;7(1):1–12. doi: 10.3233/JPD-160914. https://doi.org/10.1007/s00415-016-8355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thon N, Thorsteinsdottir J, Eigenbrod S, Schüller U, Lutz J, Kreth S, et al. Outcome in unresectable glioblastoma:MGMT promoter methylation makes the difference. J Neurol. 2017;264(2):350–8. doi: 10.1007/s00415-016-8355-1. https://doi.org/10.1007/s00415-016-8355-1. [DOI] [PubMed] [Google Scholar]
- 37.Wezyk M, Spólnicka M, Pośpiech E, Pepłońska B, Zbieć-Piekarska R, Ilkowski J, et al. Hypermethylation of TRIM59 and KLF14 influences cell death signaling in familial Alzheimer's disease. Oxid Med Cell Longev. 2018;2018:6918797. doi: 10.1155/2018/6918797. https://doi.org/10.4236/jhepgc.2018.44036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano-Tárraga C, Giralt-Steinhauer E, Mola-Caminal M, Ois A, Rodríguez-Campello A, Cuadrado-Godia E, et al. Biological age is a predictor of mortality in ischemic stroke. Sci Rep. 2018;8:4148. doi: 10.1038/s41598-018-22579-0. https://doi.org/10.1038/s41598-018-22579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriano-Tárraga C, Mola-Caminal M, Giralt-Steinhauer E, Ois A, Rodríguez-Campello A, Cuadrado-Godia E, et al. Biological age is better than chronological as predictor of 3-month outcome in ischemic stroke. Neurology. 2017;89(8):830–6. doi: 10.1212/WNL.0000000000004261. https://doi.org/10.1212/wnl.0000000000004261. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Duan S, Qu K, Bai H, Xiao J, Yuan Q, et al. The relationship between Hcy metabolism related genes methylation and cerebral infarction. Chin J Pract Nerv Dis. 2018;21:1424–30. [Google Scholar]


