Summary
Amino acid-induced lysosomal mTORC1 localization through the Rag GTPases is a critical step for its activation by Rheb GTPase. However, how the mTORC1 interacts with Rheb on the lysosome remains elusive. We report that amino acids enhance the polyubiquitination of Rheb (Ub-Rheb), which shows a strong binding preference for mTORC1 and supports its activation, while the Ub-Rheb is subjected to subsequent degradation. Mechanistically, we identified ATXN3 as a Ub-Rheb deubiquitinase whose lysosomal localization is blocked by the active Rags in response to amino acid stimulation. Consistently, cells lacking functional Rags on the lysosome accumulate Ub-Rheb, and blockade of its degradation instigates robust lysosomal mTORC1 localization and its activation without the Ragulator-Rag system. Thus, polyubiquitination of Rheb is an important post-translational modification, which facilitates the binding of mTORC1 to Rheb on the lysosome and is another crosstalk between the amino acid and growth factor signaling for mTORC1 activation.
eTOC Blurb
Yao et al. demonstrate that amino acids enhance the polyubiquitination of lysosomal Rheb (Ub-Rheb) which has a strong binding preference for mTORC1 and supports amino acid-induced mTORC1 activation. Mechanistically, we identified that the Ub-Rheb is deubiquitinated by ATXN3, of which lysosomal localization is enhanced by the inactive Rag GTPases.
Graphical Abstract
Introduction
Mechanistic target of rapamycin complex 1 (TORC1) is primarily activated by amino acids and growth factors through two distinct small GTPases, Rags and Rheb, respectively, on the lysosomal membrane (Saxton and Sabatini, 2017; Wullschleger et al., 2006). Upon amino acid stimulation, the Rag heterodimer is activated and recruits mTORC1 to the lysosomal membrane (Kim et al., 2002; Sancak et al., 2010; Sancak et al., 2008). The Ragulator, a lysosome-anchored pentameric protein complex, plays key roles in not only tethering the Rag heterodimer to the lysosomal membrane but also activating Rag heterodimer by acting as components of guanine nucleotide exchange factor (GEF) complex with the arginine-primed amino acid transporter (SLC38A9) (Sancak et al., 2010; Shen and Sabatini, 2018). The ablation of p18/LAMTOR1, a membrane-binding component of Ragulator, disrupts the lysosomal Ragulator and dissociates the Rag heterodimer from lysosomal membranes (Sancak et al., 2010; Yao et al., 2016). Recent studies demonstrated that amino acids in the lumen of the lysosome, such as arginine, trigger the activation of the Ragulator/SLC38A9 GEF complex likely through the vacuolar ATPase (vATPase) activity (Rebsamen et al., 2015; Wang et al., 2015; Wyant et al., 2017; Zoncu et al., 2011). Furthermore, cytosolic amino acids inhibit GATOR1, a specific GTPase activating protein (GAP) complex for RagA/B, through cytosolic amino acid-sensing proteins such as Sestrins and CASTOR1, thereby supporting the activation of the Rag heterodimer (Bar-Peled et al., 2013; Chantranupong et al., 2016; Wolfson et al., 2016). On the lysosomal membrane, the growth factor-induced PI3K-Akt pathway enhances Rheb small GTPase activity, which, in turn, directly activates mTORC1 (Long et al., 2005; Sancak et al., 2007). Upon growth factor stimulation, active Akt phosphorylates TSC2, the GAP for Rheb, and induces its dissociation from the lysosome, thereby providing a permissive condition for Rheb activation (Castro et al., 2003; Garami et al., 2003; Inoki et al., 2003; Inoki et al., 2002; Manning et al., 2002; Menon et al., 2014; Potter et al., 2002; Stocker et al., 2003; Tee et al., 2002; Tee et al., 2003; Zhang et al., 2003). Notably, TSC2 also interacts with inactive RagA (i.e., the GDP-bound RagA) on the lysosome under amino acid-starved conditions. Inactive RagA-dependent lysosomal localization of the TSC complex is required for complete mTORC1 inhibition under amino acid-starved conditions (Demetriades et al., 2014). Thus, both the growth factor and amino acid signaling cooperatively stimulate or inhibit Rheb activity by regulating lysosomal TSC2 localization. However, the molecular mechanisms by which lysosomal mTORC1 recruited by the active Rag heterodimer finds and interacts with Rheb on the lysosomal membrane remain elusive.
ATXN3 is a deubiquitinase belonging to the Machado Joseph Disease Protease deubiquitinase class (Takiyama et al., 1993). Mutations in ATXN3 gene with the aberrant expansion of CAG repeat causes an autosomal dominantly inherited neurodegenerative disease, Spinocerebellar Ataxia Type 3, which is characterized by the progressive impairment in motor movement due to polyQ-mediated neurotoxicity (Berke et al., 2004; Jung et al., 2009). In addition, wild-type ATXN3 plays a key role in autophagy induction by deubiquitinating and stabilizing Beclin1, an essential autophagy initiator (Ashkenazi et al., 2017).
In this study, we found that lysosomal Rheb is polyubiquitinated in response to amino acid stimulation. Importantly, the polyubiquitinated Rheb (Ub-Rheb) shows a strong binding preference for mTORC1 and facilitates amino acid-induced mTORC1 activation, although the Ub-Rheb is subjected to the lysosome- and proteasome-mediated degradation. We identified that ATXN3 acts as a deubiquitinase for Rheb. Mechanistically, the inactive Rags recruit ATXN3 to the lysosome to maintain lysosomal Rheb in a hypo-ubiquitinated state under amino acid-starved conditions, whereas active Rags release ATXN3 from the lysosome in response to amino acids. Thus, poly-ubiquitination of Rheb acts as a positive signal that facilitates Rheb interaction with mTORC1 on the lysosome and contributes to amino acid-induced mTORC1 activation.
Results
Amino acid replenishment induces poly-ubiquitination of lysosomal Rheb, which displays a strong binding preference for mTOR and supports mTORC1 activation.
The GTP-bound active Rheb directly interacts with and activates mTORC1 on the lysosome. In addition to its GTP-GDP cycling, the activity of Rheb is also regulated by its phosphorylation and ubiquitination (Harraz et al., 2016; Zheng et al., 2011). Although the ubiquitination of Rheb inhibits its function by simply destabilizing the protein or facilitating its interaction with TSC2 (Deng et al., 2019), the GAP for Rheb, we found that in cells replenished with amino acids for 15 min, levels of immunoprecipitated (IPed) endogenous Rheb migrated in the high molecular weight (HMW) ranges (above 250 kDa) were increased (Figure 1A and S1A). However, levels of the expected molecular weight of Rheb protein (16 kDa) were not changed by amino acid replenishment. The ablation of endogenous Rheb eliminated the HMW bands detected by the Rheb antibody in both lysate and IPed samples (Figure S1B), suggesting that these HMW proteins were likely polyubiquitinated Rheb (Ub-Rheb). In fact, levels of the HMW Rheb species were significantly accumulated by MG-132 treatment, and the HA-tagged ubiquitin was effectively incorporated into these HMW Rheb species (Figure 1B). Furthermore, levels of HMW Rheb were significantly reduced by expressing the ubiquitin K0 mutant, in which all the lysine residues of ubiquitin protein were replaced with arginines, indicating that the HMW Rheb is polyubiquitinated (Figure S1C). Interestingly, the blockade of lysosomal function with chloroquine (CQ) also accumulated the Ub-Rheb (Figure S1D). These observations indicate that upon amino acid stimulation, the accumulated Ub-Rheb is subjected to both proteasome- and lysosome-mediated degradation. The accumulation of Ub-Rheb is specific to amino acids as growth factors such as insulin stimulation did not cause Ub-Rheb accumulation (Figure 1A and S1E).
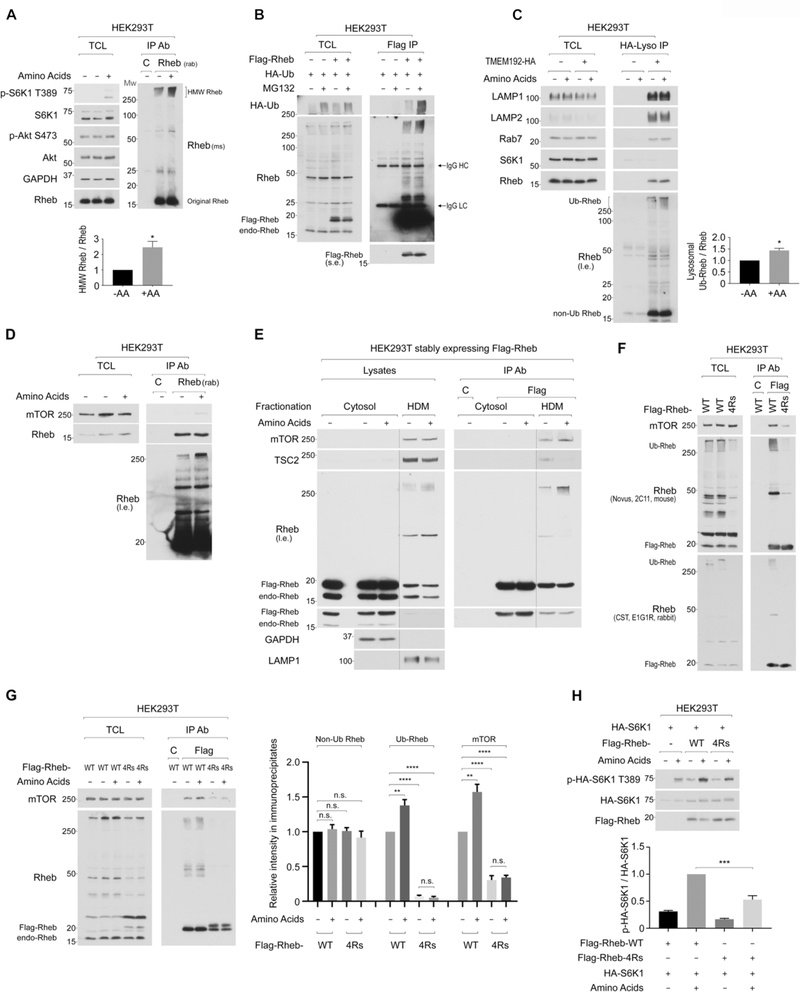
Figure 1. Amino acid replenishment increases the level of polyubiquitinated Rheb on the lysosome, which supports amino acid-induced mTORC1 activation.
1A. Amino acid replenishment increases levels of high-molecular-weight (HMW) Rheb species. Endogenous Rheb was IPed with the rabbit monoclonal Rheb antibody (E1G1R) and detected by the mouse monoclonal Rheb antibody (2C11). The intensity of HMW-Rheb species (>250 kD) was quantified and expressed as the ratio of HMW-Rheb/original Rheb. *p<0.05, mean±SEM, n=4
1B. HMW Rheb species are subjected to proteasome-mediated degradation. Cells transfected with Flag-Rheb and HA-ubiquitin were treated with MG-132 (10 μM) for 2 hr in the presence of amino acids in the medium.
1C. Amino acid replenishment increases levels of lysosomal Ub-Rheb. Lysosomes were immunopurified, and levels of lysosomal Ub-Rheb were quantified as Fig. 1A. *p<0.05, mean±SEM, n=3
1D. Levels of mTOR co-IPed with endogenous Rheb positively correlate with levels of Ub-Rheb, but not ubiquitin-free Rheb. Endogenous Rheb was IPed, and levels of Ub-Rheb, ubiquitin-free Rheb, and co-IPed mTOR were monitored.
1E. Levels of mTOR co-IPed with membrane-bound Rheb positively correlate with levels of Ub-Rheb, but not ubiquitin-free Rheb. Cells were fractionated into a cytosolic and heavy-density fraction (HDM). Flag-Rheb in each fraction was IPed and eluted with Flag peptides.
1F. Ub-Rheb displays a strong binding preference for mTORC1. Flag-Rheb or Flag-Rheb-4Rs was transiently expressed, and levels of Rheb ubiquitination and co-IPed mTOR with Flag-Rheb were monitored.
1G. Ub-Rheb accounts for the enhancement of Rheb-mTOR interaction in response to amino acid stimulation. Cells were transiently expressed with the indicated Rheb. Levels of IPed non-Ub Rheb, Ub-Rheb, and co-IPed mTOR were monitored. **p<0.01, ****p<0.0001, mean±SEM, n=4. n.s. denotes not significant.
1H. Ub-Rheb facilitates amino acid-induced mTORC1 activation. The indicated Flag-Rheb was co-expressed with HA-S6K1 in HEK293T cells. Levels of phosphorylated S6K1 were monitored and quantified. ***p<0.001, mean±SEM, n=4
The lysosomal Rheb is subjected to polyubiquitination as levels of Ub-Rheb but not ubiquitin-free Rheb in the purified lysosomes were increased by amino acid replenishment (Figure 1C). As expected, amino acid replenishment enhanced the interaction between endogenous Rheb and mTOR (Figure 1D). Interestingly, levels of Ub-Rheb, but not ubiquitin-free Rheb, positively correlated with the amount of mTOR co-IPed with Rheb. Amino acid stimulation also increased the level of Ub-Rheb and enhanced Rheb-mTOR interaction in the high-density cell fraction (HDM), which contains lysosome (Figure 1E). These results raise the possibility that the ubiquitination of Rheb may have an important role in supporting the interaction between mTORC1 and Rheb on the lysosome. To examine this hypothesis, we attempted to map the major ubiquitination sites of Rheb. By searching publicly accessible PTM Database and examining various combinations of lysine to arginine mutations in Rheb, we identified that the substitution of four lysine residues with arginine (4Rs: K109, 135, 151, 178R) significantly reduced levels of Ub-Rheb (Figure 1F, and S1F). In support of our hypothesis, the Rheb-4Rs mutant largely lost its binding preference for mTOR, as levels of Ub-Rheb, but not ubiquitin-free Rheb, correlated with the amount of mTOR co-IPed with Rheb (Figure 1F). It is noteworthy that the bacterially-produced wild-type Rheb and Rheb-4Rs mutant, which have no ubiquitin-related modifications, displayed a similar binding preference for mTOR in vitro (Figure S1G). Moreover, the wild-type Rheb and the Rheb-4Rs mutant were loaded with a similar level of GTP in HEK293T cells (Figure S1H), indicating that these K to R mutations of Rheb are not likely to affect its tertiary structure or activity.
Previous studies demonstrated that the interaction between Rheb and mTOR was independent of Rheb’s GTP/GDP-loading status in vivo (Long et al., 2005; Sun et al., 2008), and the GDP-bound Rheb D60I mutant displayed a higher binding affinity to mTOR compared to wild-type or GTP-bound Rheb mutants in vivo. Consistent with these observations, we also observed that the Rheb D60I mutant showed a stronger binding preference for mTOR than that of wild-type or GTP-bound active Rheb mutants (S16H and Q64L) (Figure S1I). Importantly, among those Rhebs, Rheb G60I displayed the highest level of its ubiquitination (Figure S1I), suggesting that the level of ubiquitination of Rheb rather than its activity accounts for its association with mTOR in vivo.
The effect of amino acids in enhancing the ubiquitination of Rheb and its binding to mTOR was diminished in the Rheb-4Rs mutant (Figure 1G), suggesting that amino acids promote the Rheb-mTOR interaction by enhancing the polyubiquitination of at least the four lysine residues. Furthermore, cells expressing the Rheb-4Rs mutant were compromised in effectively inducing mTORC1 activity by amino acids compared to those expressing wild-type Rheb (Figure 1H, S1J, and S1K). In contrast, levels of amino acid-induced lysosomal mTOR localization were comparable in cells expressing wild-type Rheb or Rheb-4Rs mutant (Figure S1L). Together these observations suggest that the polyubiquitination of Rheb facilitates its interaction with mTORC1 on the lysosome and mTORC1 activation.
ATXN3, a deubiquitinase, interacts with Rheb and decreases levels of Ub-Rheb.
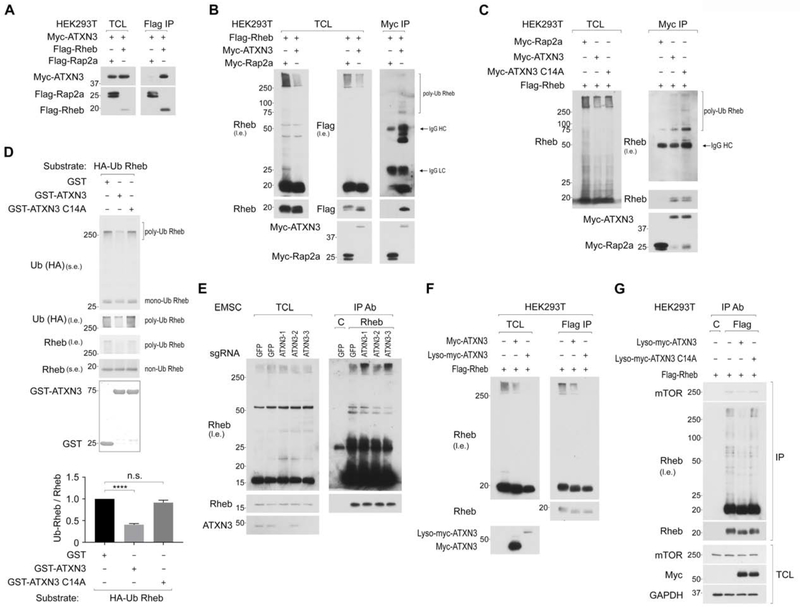
To explore the molecular mechanisms by which the ubiquitination of lysosomal Rheb is regulated, we searched ubiquitin regulators such as deubiquitinases and ubiquitin ligases of which expression has been observed in the proteome of purified lysosomes (Wyant et al., 2018). Importantly, we identified that Rheb interacted with ATXN3, a deubiquitinase, which preferentially catalyzes the long and mixed ubiquitin chains (Chai et al., 2004; Winborn et al., 2008). Flag-Rheb but not Flag-Rap2a, which is also expressed on the lysosome (Wyant et al., 2018), specifically co-IPed Myc-ATXN3 (Figure 2A). Reciprocally, Myc-ATXN3 co-IPed both ubiquitin-free Flag-Rheb and Ub-Flag-Rheb (Figure 2B). Notably, the expression of Myc-ATXN3 but not its enzyme-inactive mutant (C14A) reduced levels of Ub-Rheb in the lysate (Figure 2B and 2C). Furthermore, the ATXN3 C14A mutant co-IPed more Ub-Rheb than wild-type ATXN3 (Figure 2C), suggesting that the binding of inactive ATXN3 to the Ub-Rheb may prevent its deubiquitination in the cells or during the purification of the ATXN3-Rheb complex. In fact, higher levels of Ub-Rheb were co-IPed with ATXN3 when N-Ethylmaleimide, a deubiquitinase inhibitor, was included in the lysis and purification buffers (Figure S2A), supporting the idea that the Ub-Rheb that binds to ATXN3 was subjected to deubiquitination during the isolation processes. In support of this notion, prolonged incubation of the lysate facilitated the deubiquitination of the Ub-Rheb co-IPed with wild-type ATXN3. However, such incubation did not facilitate the deubiquitination of the Ub-Rheb co-IPed with the ATXN3 C14A mutant (Figure S2B and S2C). As expected, the four lysine residues of Rheb, which are responsible for the polyubiquitination of Rheb, are important for the binding of Ub-Rheb with ATXN3 (Figure S2C). Furthermore, the wild-type ATXN3 but not the ATXN3 C14A mutant purified from bacteria deubiquitinated Ub-Rheb in vitro (Figure 2D). In contrast to the effect of ATXN3 overexpression on the decrease in Rheb ubiquitination, the ablation of endogenous ATXN3 using three distinct sgRNAs, of which two (1 and 3) of them effectively silenced ATXN3 expression, increased levels of Ub-Rheb (Figure 2E), while amino acids failed to increase levels of Ub-Rheb in cells lacking endogenous ATXN3 (Figure S2D). Notably, the Lyso-ATXN3, which constitutively localizes on the lysosomal membrane (Nada et al., 2009; Sancak et al., 2010), decreased levels of Ub-Rheb more effectively compared to the wild-type ATXN3 (Figure 2F), suggesting that ATXN3 likely deubiquitinates Ub-Rheb on the lysosomal membrane to decrease the affinity of lysosomal Rheb to mTORC1 (Figure 2G).
Figure 2. ATXN3 interacts with Rheb and reduces levels of Ub-Rheb.
2A. Rheb interacts with ATXN3. Cells co-expressed with Myc-ATXN3 and Flag-Rheb or Flag-Rap2a as control were subjected to Flag IP, followed by Flag peptide elution. Levels of co-IPed Myc-ATXN3 were monitored.
2B. ATXN3 interacts with both ubiquitin-free Rheb and Ub-Rheb and decreases levels of Ub-Rheb expression. Cells co-expressed with Flag-Rheb and Myc-ATXN3 or Myc-Rap2a as control were subjected to Myc IP. The arrow indicates IgG.
2C. ATXN3 C14A mutant precipitates more Ub-Rheb than wild-type ATXN3. Myc-ATXN3, Myc-ATXN3 C14A mutant, or Myc-Rap2a as control, was transiently co-expressed with Flag-Rheb in HEK293T cells. Experiments were performed as Fig. 2B.
2D. ATXN3 deubiquitinates Ub-Rheb in vitro. Ub-Rheb purified from HEK293T cells were used for the reaction with the indicated GST-ATXN3 purified from bacteria. ****p<0.0001, mean±SEM, n=8
2E. The ablation of endogenous ATXN3 accumulates Ub-Rheb. The effect of ATXN3 ablation on the level of Ub-Rheb was shown.
2F. Lyso-ATXN3 deubiquitinates Ub-Rheb more effectively compared to wild-type ATXN3. Cells co-expressed with Flag-Rheb and Myc-ATXN3 or Lyso-Myc-ATXN3 were subjected to Flag IP.
2G. DUB activity of ATXN3 determines levels of Ub-Rheb and Rheb-mTOR interaction. Cells co-expressed with Flag-Rheb and Lyso-Myc-ATXN3 or Lyso-Myc-ATXN3 C14A were subjected to Flag IP.
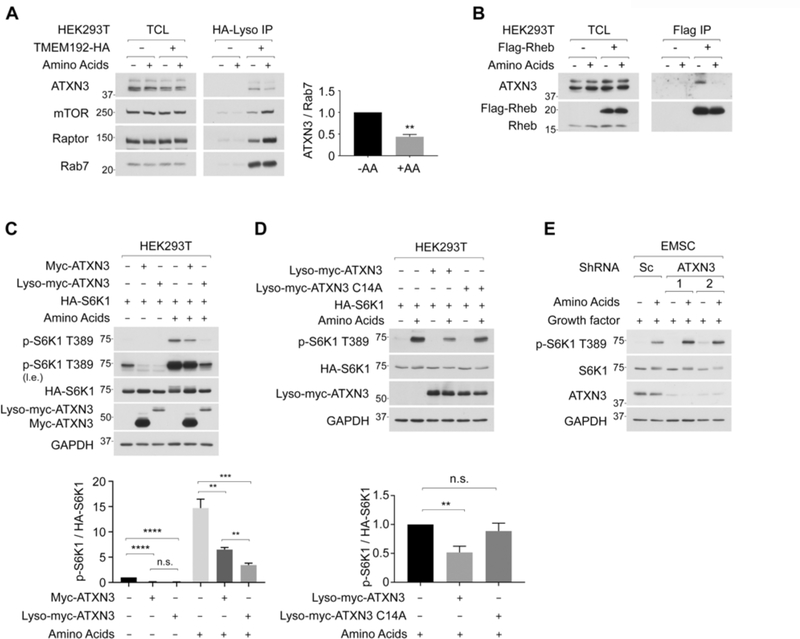
Amino acid starvation renders ATXN3 to localize on the lysosome, where it deubiquitinates Rheb and inhibits mTORC1 activity.
Interestingly, we found that levels of endogenous ATXN3 in the purified lysosome were significantly decreased, while levels of mTORC1 were increased in response to amino acid stimulation, suggesting that amino acid replenishment inhibits lysosomal localization of ATXN3 (Figure 3A). As a consequence, the interaction between Rheb and ATXN3 was reduced by amino acid stimulation (Figure 3B and S3A). The interaction of Rheb with the Lyso-ATXN3 was significantly strengthened compared to that with wild-type ATXN3 under amino acid-replete conditions as Flag-Rheb co-IPed similar levels of wild-type ATXN3 and Lyso-ATXN3 even though the expression of Lyso-ATXN3 was much lower than wild-type ATXN3 (Figure S3B).
Figure 3. Amino acid stimulation suppresses lysosomal localization of ATXN3, thereby inhibiting its interaction with Rheb and activating mTORC1.
3A. Amino acid replenishment inhibits lysosomal ATXN3 localization. Levels of endogenous ATXN3, mTORC1, and Rab7, a lysosomal resident protein, were monitored in the purified lysosome. Levels of lysosomal ATXN3 were quantified as the ratio of ATXN3/Rab7. **p<0.01, mean±SEM, n=3
3B. Amino acid replenishment decreases the interaction between Rheb and endogenous ATXN3. Cells stably expressing Flag-Rheb were subjected to Flag IP, and co-IPed endogenous ATXN3 was shown.
3C. Lyso-ATXN3 inhibits mTORC1 activity in a manner independent of amino acid availability. Cells were co-expressed with HA-S6K1 and Myc-ATXN3 or Lyso-Myc-ATXN3. **p<0.01, ***p<0.001, ****p<0.0001, mean±SEM, n=5
3D. Lyso-ATXN3 inhibits mTORC1 through its deubiquitinase activity. Cells were co-expressed with HA-S6K1 and Lyso-Myc-ATXN3 or Lyco-Myc-ATXN3 C14A. Data were expressed as Fig. 3D. **p<0.01, mean±SEM, n=4
3E. The ablation of endogenous ATXN3 enhances amino acid-induced mTORC1 activation. Two distinct shRNAs were used to knockdown ATXN3. Levels of endogenous S6K1 phosphorylation were monitored.
Next, we monitored the effect of wild-type ATXN3 or Lyso-ATXN3 on mTORC1 activity under amino acid-starved or -replete conditions. Importantly, while the wild-type ATXN3 only moderately inhibited mTORC1 activity (Figure 3C and S3C) without affecting Akt activity (Figure S3D), the Lyso-ATXN3 strongly suppressed mTORC1 activity under amino acid-replete conditions (Figure 3C, right three lanes). It is noteworthy that, under amino acid-starved conditions, both wild-type and the Lyso-ATXN3 effectively inhibited the remaining mTORC1 activity to a similar extent (Figure 3C, left three lanes). We also confirmed that the deubiquitinase activity of ATXN3 is required for its inhibitory effect on mTORC1 activity (Figure 3D). In contrast, the ablation of endogenous ATXN3 increased mTORC1 activity in response to amino acid stimulation (Figure 3E). Furthermore, the ablation of ATXN3 conferred mTORC1 activity and lysosomal mTOR localization partially resistant to amino acid starvation (Figure S3E and S3F). These observations indicate that amino acids induce the dissociation of ATXN3 from the lysosome, which relieves the lysosomal Ub-Rheb from ATXN3-dependent deubiquitination, thereby enhancing the accumulation of Ub-Rheb and mTORC1 activity.
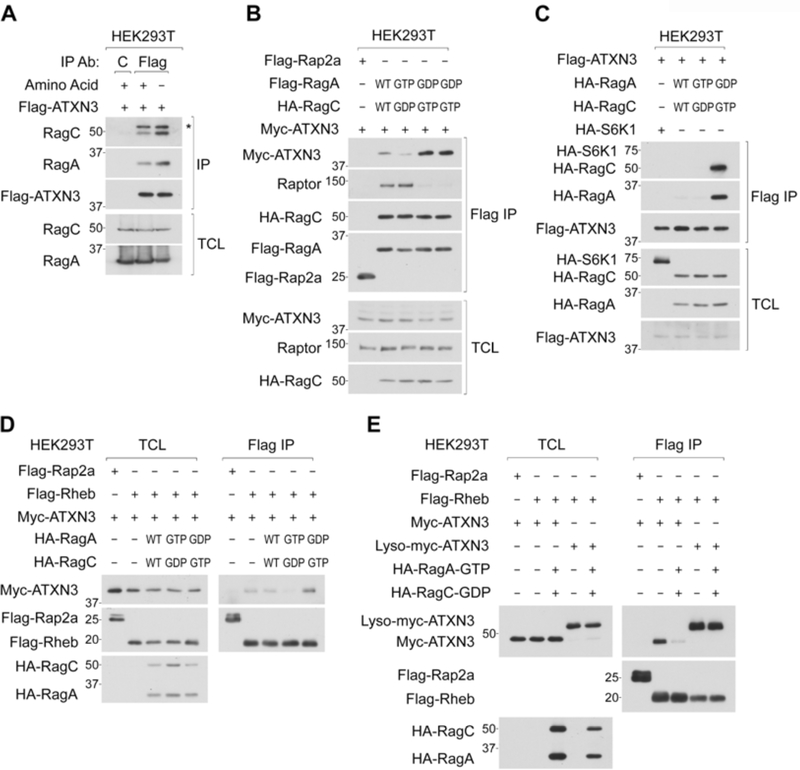
The inactive Rag heterodimer recruits ATXN3 to the lysosome.
Amino acids-regulated lysosomal ATXN3 localization prompted us to investigate the possible role of Rag small GTPases, which control the lysosomal localization of mTORC1, TFEB, TSC2, and folliculin, in the regulation of ATXN3 trafficking. Both active and inactive Rags play critical roles in tethering cytosolic proteins to the lysosome: while the active Rag heterodimer (e.g., GTP-RagA/GDP-RagC) displays a strong binding preference for Raptor (mTORC1) or TFEB, the inactive Rag heterodimer (e.g., GDP-RagA/GTP-RagC) preferentially interacts with TSC2 or folliculin (Demetriades et al., 2014; Petit et al., 2013; Sancak et al., 2008; Settembre et al., 2012; Tsun et al., 2013).
Interestingly, as amino acid stimulation decreased levels of ATXN3 expression on the lysosome (Figure 3A), it also decreased the interaction between ATXN3 and Rag heterodimer (Figure 4A and S4A). Furthermore, ATXN3 showed a much stronger binding preference for the inactive Rag heterodimer compared to the wild-type or active Rag heterodimer, which preferentially interacts with Raptor (Figure 4B, 4C, and S4B). These observations raise the possibility that under amino acid-starved conditions, the inactive Rag heterodimer renders ATXN3 to localize on the lysosome and supports ATXN3-dependent deubiquitination of lysosomal Rheb. In fact, while the interaction between Rheb and ATXN3 was increased in cells expressing the inactive Rag heterodimer, it was diminished by the active Rag heterodimer (Figure 4D). Importantly, the Lyso-ATXN3 constitutively interacted with Rheb even in the cells expressing the active Rag heterodimer (Figure 4E). Moreover, ATXN3-induced deubiquitination of Ub-Rheb was further facilitated by inactive Rag heterodimer under amino acid-replete conditions (Figure S4C). These observations support the idea that ATXN3 translocates to the lysosome and interacts with Ub-Rheb for its deubiquitination in a manner dependent on the inactive Rag heterodimer in response to amino acid starvation.
Figure 4. Active Rag small GTPases suppress lysosomal localization of ATXN3 and its interaction with Rheb.
4A. Amino acid starvation enhances the interaction between ATXN3 and RagA/C. Cells stably expressing Flag-ATXN3 were starved for amino acids for 2 hr and subjected to Flag IP. * denotes IgG heavy chain.
4B and 4C. Inactive Rag heterodimer preferentially interacts with ATXN3. Cells transiently co-expressed with Myc-ATXN3 and the indicated Flag-Rag heterodimers were subjected to Flag IP (B). Reciprocal IP experiments were performed (C).
4D. Inactive Rag heterodimer enhances the interaction between ATXN3 and Rheb. Cells transiently co-expressed with the indicated proteins were subjected to Flag IP.
4E. Lyso-ATXN3 constitutively interacts with Rheb in a manner independent of Rag activity. Cells transiently expressed with the indicated proteins were subjected to Flag IP.
Ub-Rheb is accumulated, and the blockade of Ub-Rheb degradation stimulates mTORC1 activity in Ragulator or RagA/B deficient cells.
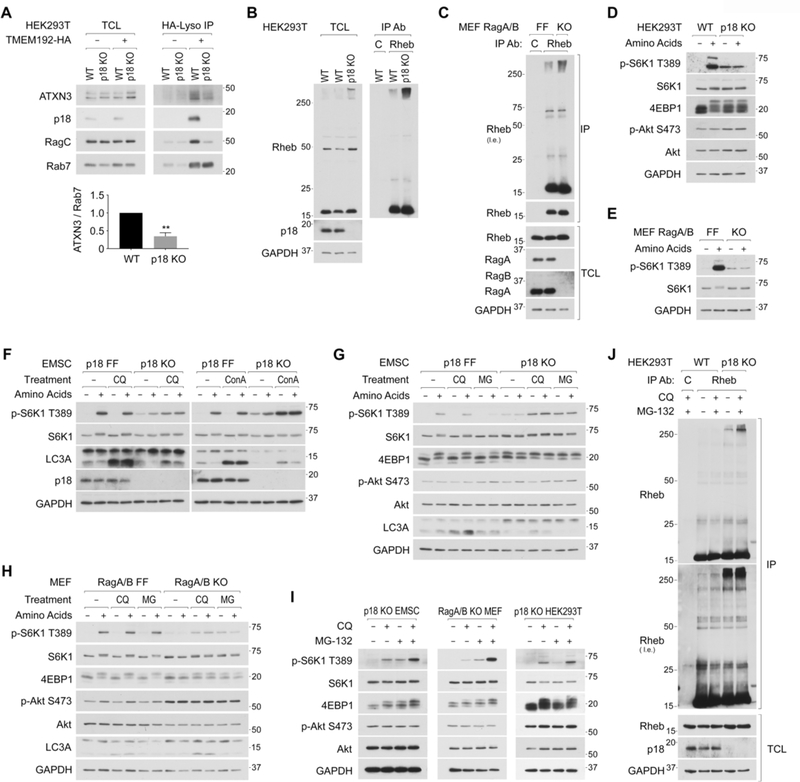
The lysosomal Ragulator complex, comprised of p18/LAMTOR1 and other four subunits, provides the lysosomal docking site for Rag heterodimer (Sancak et al., 2010). In Ragulator deficient cells such as p18 KO cells, Rags are located away from lysosomes. In support of the finding that the inactive Rags tethers ATXN3 to the lysosome, levels of lysosomal ATXN3 was significantly reduced in the purified lysosomes from p18 KO cells (Figure 5A), further demonstrating an essential role of the Ragulator-Rag complex for the lysosomal localization of ATXN3. Consistent with the observations that ATXN3 induced deubiquitination of Rheb (Figure 2), levels of Ub-Rheb were significantly increased in p18 KO (Figure 5B) or RagA/B double KO (Figure 5C) cells compared to those in their control cells. Although amino acid-induced acute and robust mTORC1 activation was absent due to a lack of essential lysosomal recruitment machinery for mTORC1 in p18 KO (Figure 5D) and Rag A/B double KO (Figure 5E) cells, these cells displayed amino acid-insensitive higher basal mTORC1 activity. We speculated that these amino acid-independent mTORC1 activity might due to the higher level of lysosomal Ub-Rheb, which has a strong binding affinity to mTOR.
Figure 5. The accumulation of Ub-Rheb renders lysosomal mTORC1 localization and its activation in Ragulator deficient cells.
5A. Levels of lysosomal ATXN3 are reduced in p18 deficient cells. Levels of endogenous ATXN3 and indicated proteins were monitored in the purified lysosomes. Levels of lysosomal ATXN3 were quantified as the ratio of ATXN3/Rab7. **p<0.01, mean±SEM, n=3
5B and 5C. Ub-Rheb is accumulated in p18 KO HEK293T and RagA/B KO MEF cells. Endogenous Rheb was IPed from p18 KO HEK293T cells (5B) and RagA/B KO MEF cells (5C).
5D and 5E. Ragulator or RagA/B deficient cells display constitutive and amino acid-independent mTORC1 activity. mTORC1 activity in p18 KO HEK293T cells (5D) and RagA/B KO MEF cells (5E) were shown.
5F. Blocking lysosome activity enhances mTORC1 activity in Ragulator deficient cells independent of amino acid availability. The indicated cells were starved for amino acids in the presence or absence of a lysosomal inhibitor, Chloroquine (CQ, 50 μM) or Concanamycin A (ConA, 5 μM) for 50 min, then replenished with amino acids for another 10 min.
5G. Blocking proteasome activity also enhances mTORC1 activity in Ragulator deficient cells independent of amino acid availability. Similar experiments were performed as Fig. 5F using a proteasome inhibitor (MG-132; 20 μM).
5H. Blocking lysosome or proteasome activity enhances mTORC1 activity in RagA/B deficient cells independent of amino acid availability. Wild-type (RagA/B FF) or RagA/B KO MEF cells were treated as Fig. 5G.
5I Blocking both lysosome and proteasome function additively enhances mTORC1 activity in Ragulator or RagA/B deficient cells. The indicated cells were starved for amino acids for 15 min, then treated with Chloroquine (CQ; 50 μM), MG-132 (MG; 20 μM), or Chloroquine and MG-132 (CQ+MG) for another 45 min.
5J. Blocking both proteasome and lysosome function further accumulates Ub-Rheb in Ragulator deficient cells. p18 KO HEK293T cells were treated as Fig. 5I.
In addition to proteasome-mediated protein degradation, the lysosome-dependent proteolysis also plays an important role in supplying newly synthesized amino acids such as arginine, which stimulates the SLC38A9-Ragulator-Rag system, consequent lysosomal mTORC1 localization, and its activation in wild-type cells (Wyant et al., 2017). Furthermore, it has also been reported that vATPase activity itself plays an essential role in stimulating the Ragulator-Rag activity, as concanamycin A (ConA), a vATPase inhibitor, effectively inhibits amino acid-induced mTORC1 activation in wild type HEK293T cells (Zoncu et al., 2011). Intriguingly, while CQ or ConA treatment effectively blocked lysosomal function as judged by the blockade of LC3A-II degradation, they showed little effect on mTORC1 activity in wild-type EMSC cells (p18 FF) (Figure 5F).
Thus, the effect of inhibition of vATPase activity on the suppression of mTORC1 activity is cell-type dependent. Interestingly, treatments with ConA or CQ, which do not inhibit vATPase activity but neutralizes lysosomal luminal pH, strongly enhanced mTORC1 activity in p18 KO EMSC (Figure 5F, 5G, and S5A), p18 KO MEF (Figure S5B), and Rag A/B KO MEF cells (Figure 5H) in a manner independent of Akt phosphorylation or amino acid availability. Bafilomycin A1, another vATPase inhibitor, also activated mTORC1 activity in p18 KO EMSC cells (Figure S5A). These observations suggest that the blockade of lysosome-mediated proteolysis underlies mTORC1 activation in cells lacking the Ragulator-Rag system. Furthermore, analogous to the lysosomal inhibitors, proteasome inhibition with MG-132 or other proteasome inhibitors (ALLN and PS-341) led to the enhancement of mTORC1 activity in p18 KO EMSC (Figure 5G and S5C) or RagA/B KO MEF cells (Figure 5H) in a manner independent of amino acid availability. As both the proteasome and lysosome are capable of digesting the Ub-Rheb (Figure 1B and S1D) and other ubiquitinated proteins (Geisler et al., 2010; Hicke and Riezman, 1996; Li et al., 2015), the effects of simultaneous lysosome and proteasome inhibition (CQ+MG treatment) on the enhancement of mTORC1 activity were additive in p18 KO EMSCs, RagA/B KO MEF, and p18 KO HEK293T cells under amino acid-starved conditions (Figure 5I). We confirmed that levels of Ub-Rheb were further increased by CQ+MG treatment in p18 KO HEK293T (Figure 5J). It is noteworthy that CQ+MG-induced mTORC1 activation did not require amino acid inputs; however, it still requires inputs from growth factors to stimulate mTORC1 activity in cells lacking the Ragulator-Rag system (Figure S5D, S5E, and S5F). These observations raise the possibility that the accumulation of lysosomal Ub-Rheb is sufficient to render lysosomal mTORC1 localization and its activation due to its strong binding preference for mTORC1 in Ragulator or RagA/B deficient cells.
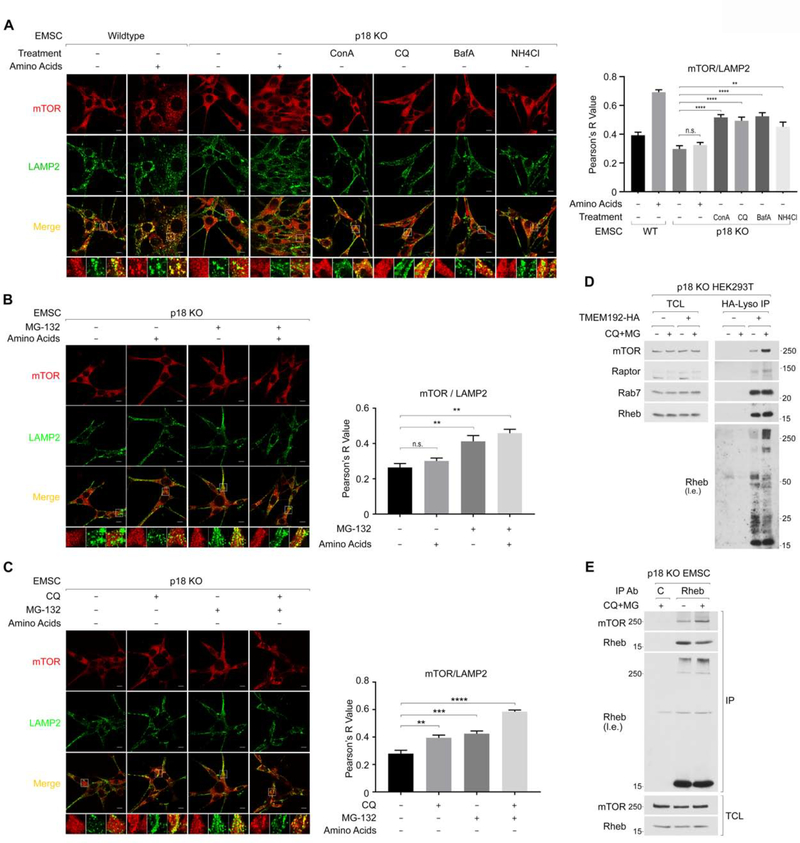
Inhibition of lysosome or proteasome function induces lysosomal mTOR localization in cells lacking the Ragulator-Rag system.
In support of our hypothesis, inhibition of either lysosome or proteasome function increased lysosomal mTOR localization in a manner independent of amino acids in p18 KO EMSC (Figure 6A and 6B) or p18 KO MEF cells (Figure S6A). Consistent with the observations that lysosome and proteasome inhibition (CQ+MG treatment) additively enhance mTORC1 activity (Figure 5I), simultaneous inhibition of both lysosome and proteasome function additively enhanced lysosomal mTOR localization in 18 KO EMSC (Figure 6C) or RagA/B KO MEF cells (Figure S6B). Furthermore, CQ+MG treatment largely enhanced levels of mTORC1 and Ub-Rheb but not ubiquitin-free Rheb in the immunopurified lysosomes from p18 KO HEK293T cells (Figure 6D). As expected, CQ+MG treatment enhanced the interaction between endogenous Rheb and mTOR in 18 KO EMSC cells. Again, levels of Ub-Rheb, but not ubiquitin-free Rheb, correlated the amount of mTOR co-IPed with Rheb (Figure 6E).
Figure 6. The inhibition of lysosome- or proteasome-mediated cellular degradation machinery induces lysosomal mTORC1 localization in Rag-Ragulator deficient cells independent of amino acid availability.
6A. Blocking lysosome function induces lysosomal mTOR localization in p18 KO EMSC cells. The indicated cells were starved for amino acids for 15 min, then treated with a vehicle (0.1% DMSO) or the indicated lysosomal inhibitors (Concanamycin A (ConA, 5 μM), Chloroquine (CQ, 50 μM), Bafilomycin A1 (BafA, 1 μM), or NH4Cl (20 mM)) for another 45 min. Before fixing the cells, the cells were treated with a 1x amino acid mixture for 10 min as indicated (+AA). Pearson’s correlation coefficient, R values were determined for quantifying the colocalization of mTOR and LAMP2. The scale bar indicates 10 μm. **p< 0.01, ****p<0.0001 compared to p18 KO EMSC cells with vehicle treatment in the absence of amino acids. mean ± SEM, n (number of p18 KO cells analyzed) : (vehicle-AA, vehicle+AA, ConA-AA, CQ-AA, BafA-AA, NH4Cl-AA)=(16, 10, 9, 9,10,6)
6B. Blocking proteasome function also induces lysosomal mTORC1 localization. Similar experiments were performed by using MG-132 as Fig. 6A. **p< 0.01 compared to vehicle treatment in the absence of amino acids. mean ± SEM, n: (vehicle −AA, vehicle +AA, MG-AA, MG+AA)=(9, 8, 12, 14)
6C. Blocking both lysosome and proteasome function additively induces lysosomal mTOR localization. p18 KO EMSC cells were treated as Fig. 6A. **p< 0.01, ***p<0.001, ****p<0.0001 compared to vehicle treatment. mean ± SEM, n: (Veh, CQ, MG, CQ+MG)=(13,30,31,29)
6D. Blocking both lysosome and proteasome function accumulates Ub-Rheb and mTORC1 on the lysosome in Ragulator deficient cells. Lysosomes were immunopurified from p18 KO HEK293T cells in the presence or absence of CQ+MG treatment under amino acid starvation conditions.
6E. Blocking lysosome and proteasome function increases levels of Ub-Rheb and the interaction between Rheb and mTOR in p18 KO EMSC cells. p18 KO EMSC cells were starved for amino acids for 50 min then treated with or without CQ+MG for another 75 min.
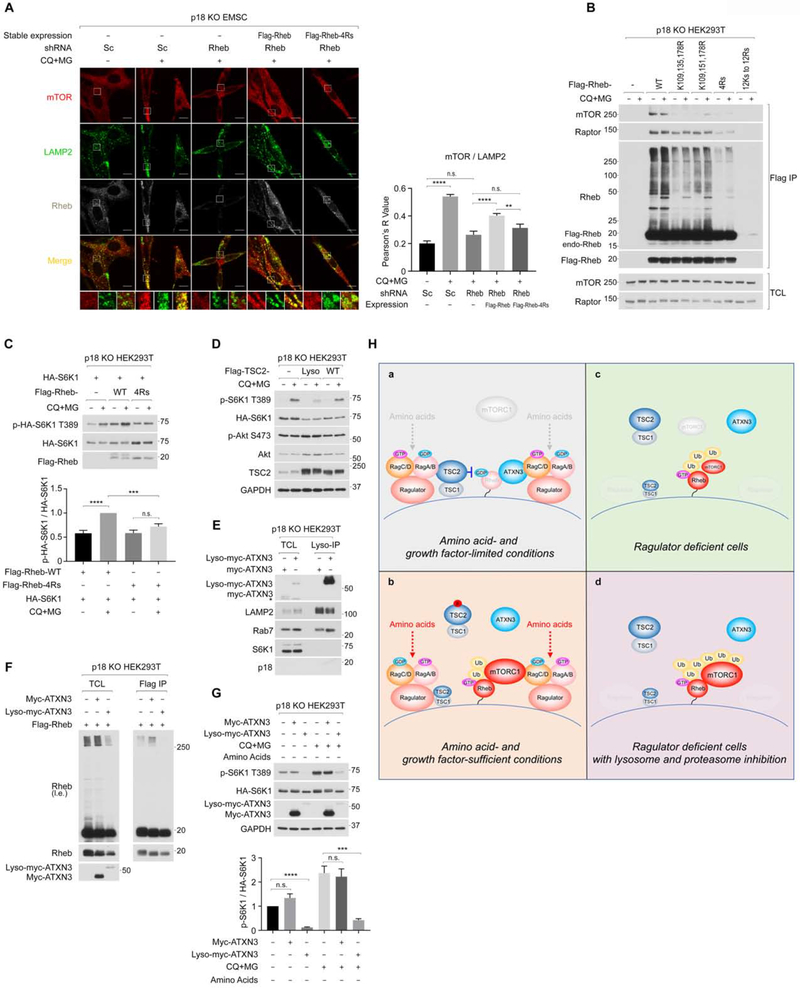
Ub-Rheb is essential for mTORC1 activation in p18 knockout cells.
Although ablation of Rheb did not affect amino acid-induced lysosomal localization of mTOR in wild-type EMSC cells (Figure S7A), either FTI-227 treatment, a farnesyltransferase inhibitor, which blocks the membrane localization of Rheb, or the ablation of Rheb effectively inhibited CQ+MG-induced lysosomal mTORC1 localization (Figure 7A, S7B, and S7D) and its activation (Figure S7C and S7E) in p18 KO cells. Re-expressing exogenous Flag-Rheb, which is resistant to the shRNA for endogenous Rheb, significantly restored lysosomal mTOR localization (Figure 7A) and its activation (Figure S7F) induced by CQ+MG treatment in p18 KO EMSC cells. In contrast, re-expressing exogenous Rheb-4Rs, which is defective in poly-ubiquitination (Figure 7B and S7G), failed to restore lysosomal mTOR localization in p18 KO cells (Figure 7A). Moreover, the Rheb-4Rs mutant largely lost its binding preference for both mTOR and Raptor, as levels of Ub-Rheb, but not ubiquitin-free Rheb, correlated with the amount of mTORC1 co-IPed with Rheb (Figure 7B). Note that the effect of CQ+MG treatment on the interaction between mTORC1 and overexpressed Rheb, which was significantly ubiquitinated under basal conditions, were mitigated. Importantly, the ectopic expression of wild-type Rheb, but not Rheb-4Rs mutant, enhanced CQ+MG treatment-induced mTORC1 activation in p18 KO HEK293T cells (Figure 7C). In addition, HEK293T cells exclusively expressing the Rheb-4Rs mutant were significantly compromised in the activation of mTORC1 by the CQ+MG treatment (Figure S7H). While the CQ+MG treatment-induced mTORC1 activation was effectively suppressed by overexpressing Lyso-TSC2, a functional TSC2 constitutively expressed on the lysosome (Menon et al., 2014), wild-type TSC2, whose lysosomal expression depends on the Ragulator-Rag system, failed to effectively inhibit mTORC1 activity in p18 KO cells (Figure 7D) (Demetriades et al., 2014). These observations indicate that the accumulation of lysosomal Ub-Rheb triggers lysosomal mTORC1 localization and its activation without the Rag-Ragulator system, highlighting the molecular mechanism that the Ub-Rheb captures mTORC1 on the lysosome.
Figure 7. Poly-ubiquitination of Rheb contributes to lysosomal mTOR localization and its activation in Ragulator deficient cells.
7A. Ub-Rheb is required for lysosomal mTOR localization induced by the CQ+MG treatment. The indicated cells were treated as Fig. 6E, and levels of lysosomal mTOR localization quantified as Fig. 6A. **p<0.01, ****p<0.0001, mean ± SEM, n (control/Veh, control/CQ+MG, Rheb KD/CQ+MG, Rheb KD + Flag-Rheb/CQ+MG, Rheb KD + Flag-Rheb-4Rs/CQ+MG) = (21, 28, 16, 28, 15)
7B. Ub-Rheb displays a strong binding preference for mTORC1. p18 KO HEK293T cells were expressed with the indicated Rheb and treated as Fig. 6E and subjected to Flag IP. Note that the Rheb 12Rs mutant did not express in the cells.
7C. Rheb-4Rs mutant fails to support CQ+MG-induced mTORC1 activation. Cells co-expressed HA-S6K1 and Flag-Rheb or Flag-Rheb-4Rs were starved for amino acids for 60 min and then treated with CQ+MG for 75 min. ***p<0.001, ****p<0.0001, mean±SEM, n=4
7D. Blocking lysosomal Rheb activity diminishes CQ+MG treatment-induced mTORC1 activation. The lysosome-tethering TSC2 (Lyso-TSC2) or wild-type TSC2 was transiently expressed with HA-S6K1 in p18 KO HEK293T cells.
7E. Lyso-ATXN3 but not wild-type ATXN3 expresses on the lysosome in p18 KO cells. Lysosomes were immunopurified from the cells expressing Myc-ATXN3 or Lyso-Myc-ATXN3. * denotes non-specific bands.
7F. Lyso-ATXN3 but not wild-type ATXN3 effectively decreases levels of Ub-Rheb in p18 KO cells. Cells co-expressed with Flag-Rheb and Myc-ATXN3 or Lyso-Myc-ATXN3 were subjected to Flag IP.
7G. Lyso-ATXN3 but not wild-type ATXN3 effectively inhibits CQ+MG-induced mTORC1 activation in p18 KO cells. Cells were co-expressed with HA-S6K1 and the indicated ATXN3. ***p< 0.001, ****p< 0.0001, mean±SEM, n=4
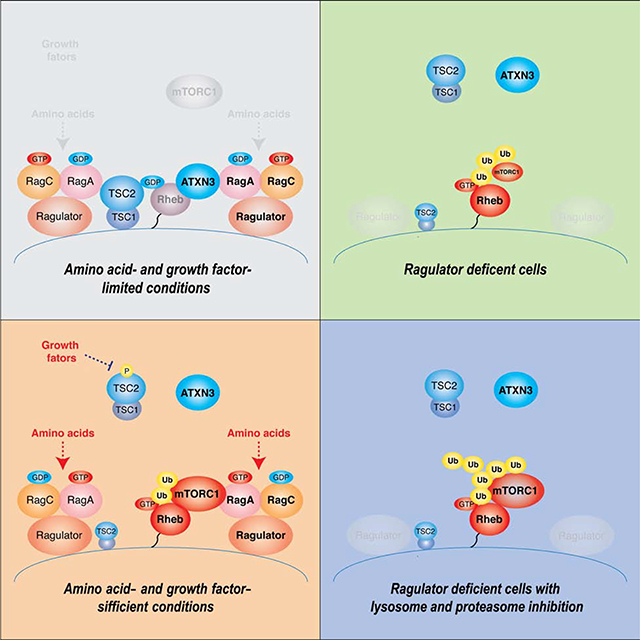
7H. Hypothetical model for the regulation and the role of Ub-Rheb in the activation of mTORC1 signaling on the lysosome. Ub-Rheb-dependent mTORC1 regulation in wild-type cells under amino acid- and growth factor-starved (7a) or -replete conditions (7b), and in Ragulator deficient cells in the absence (7c) or presence (7d) of lysosomal/proteasome blockade was depicted.
Finally, we examined if the Lyso-ATXN3 is able to inhibit CQ+MG-induced Ub-Rheb accumulation and mTORC1 activation in Ragulator-deficient cells, in which lysosomal localization of wild-type ATXN3 is compromised due to a lack of lysosomal Rags (Figure 7E). Accordingly, while the expression of wild-type ATXN3 had little effect on CQ+MG-induced Ub-Rheb accumulation, the expression of Lyso-ATXN3 abolished Ub-Rheb in p18 KO HEK293T cells (Figure 7F). Furthermore, the Lyso-ATXN3 but not wild-type ATXN3 effectively inhibited CQ+MG-induced mTORC1 activation in p18 KO HEK293T cells (Figure 7G). Taken together, these data indicate that in response to amino acid stimulation, the Ragulator-Rag system plays key roles in not only recruiting mTORC1 to the lysosome but also repelling ATXN3 from the lysosome, which leads to the accumulation of lysosomal Ub-Rheb, a process important in supporting Rheb interaction with mTORC1 on the lysosome and facilitating mTORC1 activation.
Discussion
The study described here demonstrated a key role of Rheb polyubiquitination for its interaction with mTORC1 on the lysosome and provided the molecular mechanism by which Rheb ubiquitination is regulated by amino acid availability and contributes to amino acid-induced mTORC1 activation. Our studies identified that ATXN3 is a specific substrate of the inactive Rag heterodimer and localizes on the lysosome and deubiquitinates Ub-Rheb under amino acid-insufficient conditions (Figure 7H–a and 7H–b). Thus, in addition to the TSC complex (Demetriades et al., 2014), which directly inhibits the activity of Rheb, the inactive Rag heterodimer also anchors ATXN3 on the lysosome to suppress Rheb ubiquitination, keeping its affinity to mTORC1 at lower levels. This mechanism is evident in cells lacking the Ragulator-Rag system in which the lysosomal expression of ATXN3 and TSC2 are mitigated. In these cells, levels of Ub-Rheb were increased regardless of amino acid availability, and the Ub-Rheb renders mTORC1 activity constitutive, although the level of mTORC1 activity is moderate due to a lack of the lysosomal Rag heterodimer, a potent mTORC1 recruiter to the lysosome (Figure 7H–c). However, further accumulation of the Ub-Rheb by inhibition of cellular degradation systems triggers strong lysosomal mTORC1 localization and its activation in Ragulator or RagA/B deficient cells (Figure 7H–d). The above mechanisms may explain why levels of mTORC1 activity were also higher in the primary RagA KO MEF or Rag A/B KO MEF cells compared to their wild-type cells under amino acid-starved conditions (Efeyan et al., 2014; Kim et al., 2014).
Key questions that arise from our study include whether the Ub-Rheb constitutes the active form of Rheb, and, if so, how the Ub-Rheb fits in the binding pocket of mTOR kinase to allosterically activate its kinase activity as previously shown in the cryo-EM reconstitution studies (Yang et al., 2017). Despite considerable efforts, we failed to isolate Ub-Rheb from ubiquitin-free Rheb specifically (data not shown). Intriguingly, overexpressed and ubiquitinated wild-type Flag-Rheb, but not polyubiquitin-defective Flag-Rheb-4Rs, co-IPed with endogenous ubiquitin-free Rheb (Figure 7B and S1F). These data suggest that the Ub-Rheb may form functional heteromultimers with ubiquitin-free Rheb. Interestingly, a previous report demonstrated that viral infection triggers ubiquitin-mediated functional aggregation of mitochondrial antiviral-signaling protein (MAVS), which propagates anti-viral signaling cascades (Hou et al., 2011). Although further studies are required to reveal the oligomerization state of Ub-Rheb, we posit that such a higher-order assembly of Rheb may increase its density on the lysosomal membrane and help sequester mTORC1 on the lysosome surface in response to amino acid stimulation in wild-type cells.
Deng. et al. recently reported that the mono-ubiquitinated Rheb by RNF152, which also ubiquitinates RagA, induced a negative impact on growth factor-induced mTORC1 activation (Deng et al., 2019). The study proposed that the mono-ubiquitination of lysine 8 of Rheb promotes its interaction with TSC2 and inhibits the activity of Rheb, thereby inhibiting cellular mTORC1 activity. However, the study did not monitor the high-molecular-weight polyubiquitinated Rheb. In our studies, levels of polyubiquitinated endogenous Rheb were not significantly altered in response to growth factors such as insulin stimulation (Figure S1E). Furthermore, the Rheb mutant bearing K8R mutation did not decrease the levels of polyubiquitinated Ub-Rheb (Figure S7G), indicating that the lysine 8 is dispensable for amino acid-induced Ub-Rheb accumulation. In this regard, further experiments will be required to investigate the functional difference in distinct types of Rheb ubiquitination.
A recent genetic study demonstrated that the loss of functional ATXN3 in C. elegans aggravated the mortality of worms in response to starvation (Herzog et al., 2020). Interestingly, the study revealed that ATXN3 facilitates autophagy in a manner independent of Beclin1, a known substrate of ATXN3 (Ashkenazi et al., 2017). These observations suggest that ATXN3 supports cellular autophagy through multiple mechanisms (Durcan et al., 2011).
Finally, we anticipate that Ub-Rheb-mediated lysosomal mTORC1 retention and its activation may contribute to the disease-associated aberrant mTORC1 activity, which may include TSC and lysosome-storage diseases. These disorders associate with impaired lysosome-mediated proteolysis. In fact, loss of functional TSC complex renders lysosomal mTORC1 localization and mTORC1 activity partially resistant to amino acid starvation in a Rheb dependent manner (Demetriades et al., 2014). Furthermore, a recent study reported that amino acid-independent lysosomal mTORC1 localization and its aberrant activation in several lysosome storage diseases (LSDs) (Bartolomeo et al., 2017). We posit that amino acid-insensitive lysosomal mTORC1 localization and its inappropriate activation caused by the accumulation of the Ub-Rheb may also be implicated in the pathogenesis of cellular dysfunctions in LSDs.
Limitations
The data presented in this study indicated that polyubiquitination of Rheb positively contributes to the Rheb-dependent mTORC1 activation by facilitating the interaction of Rheb with mTORC1 on the lysosome. Although we mapped four responsible lysine residues for Rheb polyubiquitination, it remains largely elusive which ubiquitination sites exert key roles in the interaction between Ub-Rheb and mTORC1. Moreover, it remains an open question as to the chain linkages involved, whether homotypic, mixed or branched, and the length of chains required for facilitating the Ub-Rheb-mTORC1 interaction. Future studies are warranted to clarify the molecular nature and actions of the Ub-Rheb for the activation processes of mTORC1 on the lysosome.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ken Inoki (inokik@umich.edu).
Material Availability
All unique and stable reagents generated in this study are available from the Lead Contact.
Data and Code Availability
Original raw data is available at: https://data.mendeley.com/datasets/t7vxvsxfmb/draft?a=abd5b86d-f969-4801-94f7-331500bead1d
EXPRIMENTAL MODEL AND SUBJECT DETAILS
Cultured cells
HEK293T and MEF cells were cultured as previously described (Yao et al., 2016). The mouse mesenchymal stem cells (EMSC) were cultured in DMEM/F12 (#11330–032, Invitrogen) containing 15 % FBS (Corning, #35–015-CV), 100 μl Primocin (#ant-pm-1, Invivogen,) per 50 ml media, and 10 ng/mL recombinant murine FGF-basic (#450–33, Peprotech). The EMSC cells were obtained as previously reported (Rim et al., 2005). Briefly, the ear tissues collected from p18 floxed mice were quickly washed with 75% ethanol and incubated in a sterile HBSS with antibiotics and digested in the collagenase solution at 37°C. The digested tissue suspension was filtered through a 100 μm cell strainer (#352350, BD Biosciences) and centrifuged at 1600 rpm for 10 min. The cell pellet was re-suspended in one ml of the red blood cell lysis buffer (#R7757, Sigma) for 1 min, and the lysis was stopped by adding 15 ml of the cell culture medium. After centrifuging at 1600 rpm for 10 min, the cell pellet was re-suspended with the culture medium and seeded to one well of a 6-well-plate. p18 KO EMSC cells were generated by the infection of p18 Flox/Flox (FF) EMSC cells with the Adeno-CRE virus.
METHOD DETAILS
Cell treatment
For amino acid starvation, cells were rinsed twice with pre-warmed Hank’s Balanced Salt Solution (Invitrogen), and incubated with amino acid-free DMEM/F12 (US Biological) containing 2% dialyzed FBS for 50 min. For amino acid re-stimulation, 50X amino acid stock solution (Sigma) was added to the medium to the final concentration of 1X for 15 min unless stated otherwise. For the treatment of RagA/B KO MEF or p18 KO EMSC cells, cells were amino acid-starved for 15 min, then the final concentration of chloroquine (CQ, 50 μM) and/or MG-132 (MG, 20 μM) were added to the amino acid-free medium for 45 min. For the treatment of p18 KO HEK293T cells, cells were amino acid-starved for 60 min, and then chloroquine and MG-132 were added to the amino acid-free medium for another 75 min.
Lentivirus generation and infection
The protocol of virus packaging and infection was described as previously (Yao et al., 2016). Briefly, for the lentivirus packaging, the pLKO, or pLenti-CRISPR, or pLJM1 vector (10 μg) was transfected into HEK293T cells with the lentiviral packaging plasmid psPAX2 (15 μg) and envelope plasmid pMD2.G (8 μg). The condition media containing the lentivirus were collected every 24 hr until 72 hr post-transfection. The media containing lentivirus were concentrated by ultracentrifugation at 23,000 rpm for 90 min. Viral pellets were re-suspended in 1/300 of the original volume. For the lentivirus infection, cells were seeded 24 hr before infection. The second-day post-infection, viral-infected cells were selected by puromycin.
Cell fractionation
Cell fractionation was performed as previously reported (Menon et al., 2014). Briefly, sub-confluent cells from two 15 cm culture dishes were washed with cold PBS, scraped into cold PBS, pelleted by centrifugation at 800 g for 2 min at 4 °C, and re-suspended in 300 μl cold hypotonic lysis buffer (10 mM HEPES, pH 7.2, 10 mM KCl, 1.5 mM MgCl2, 20 mM NaF, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 250 mM sucrose, 100 mM N-ethylmaleimide with freshly added protease inhibitors). Cells were mechanically lysed by drawing 4 times through a 23 G needle and then centrifuged at 500 g for 10 min at 4 °C, yielding a post-nuclear supernatant (PNS). The PNS was centrifuged at 20,000 g for 2 hr to separate the soluble supernatant (light density membrane/cytosolic fraction) from the insoluble pellet (heavy density membrane fraction). The light density membrane/cytosolic fraction was centrifuged at 100,000g for one hr to further separate the light density membrane and cytosolic fraction. The pellets were re-suspended in RIPA buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 0.1 % SDS, 1 % Na deoxycholate, 5 % glycerol, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 50 mM NaF, 100 mM N-Ethylmaleimide and 1X protease inhibitors) and used as the light density membrane fraction. Protein concentrations of each fraction were equalized prior to the analyses for immunoblotting or immunoprecipitation.
CRISPR-Cas9 mediated gene knockout
The 20-nucleotide guide sequence targeting genes of interest were designed using the CRISPR design tool (http://crispr.mit.edu/). The guide RNAs (sgRNA) encoding target nucleotides were cloned into a bicistronic expression vector (Lenti-CRISPR v2, a gift from Feng Zhang [Addgene plasmid no. 52961]) (Sanjana et al., 2014) containing human codon-optimized Cas9 and the RNA components. As a control, a sgRNA sequence targeting GFP was developed. The Lenti-CRISPR vectors with sgRNAs were transfected into HEK293T cells with the lentiviral packaging plasmid psPAX2 and envelope plasmid pMD2.G. The virus was collected and concentrated for infection. 24 hr post-infection, cells were selected with puromycin for another 48 hr. 7 days post-infection, the cells were used for the experiments.
Lysosome immune-purification
The purification of lysosomes was performed as previously reported (Abu-Remaileh et al., 2017). Briefly, cells stably expressing TMEM192–3×HA (#102930, addgene) were established by a standard lentivirus infection protocol in either wild-type or p18 knockout HEK293T cells. Approximately 35 million cells were rinsed with PBS and scraped into one ml of KPBS buffer (136 mM KCl, 10 mM KH2PO4, pH 7.25) and centrifuged at 1000 g for 2 min at 4 °C. The pelleted cells were re-suspended in the KPBS buffer with a protease inhibitor and deubiquitinase inhibitor NEM [100 mM], and homogenized to disrupt the cells. After 2 min centrifugation at 1000 g, the supernatant was incubated with 100 μl HA antibody-conjugated magnetic beads (#88836, Pierce) at 4 °C. Beads were washed 4 times with KPBS buffer, and IPed lysosomes were dissolved in RIPA buffer containing protease and deubiquitinase inhibitors. The beads extract containing the dissolved lysosome proteins were examined by western blot.
Immunoblotting
Cells were lysed in NP-40 cell lysis buffer (1 % NP-40, 40 mM HEPES pH 7.5, 120 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 10mM sodium pyrophosphate, 1 mM EDTA, 1× Protease Inhibitor Cocktail (Roche. Cat.NO. 11873580001)). Lysates were then boiled in SDS sample buffer (20 mM Tris pH 6.8, 2% SDS, 0.01% bromophenol blue, 10% glycerol, 5% 2-mercaptoethanol) and subjected to SDS-PAGE and immunoblotting. The SDS-PAGE gel was running in Tris-Glycine running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS) at the voltage of 100V, and then the proteins were transferred to PVDF membrane in the transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) at constant 350 mA for 3 hours at 4 °C. After incubation with the primary antibody overnight at 4 °C and the secondary antibody for 2 hours at room temperature, the membrane was washed four times with 1 × TTBS buffer (20 mM Tris HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20 (w/v)) before applying to enhanced chemiluminescence (Cat#RPN2232, GE).
Immunostaining
Cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature. Fixed cells were washed twice with PBS. For Rheb/p-S6 staining, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and blocked with 0.5 x Odyssey blocking buffer (LI-COR Bioscience) in PBS for 1 hr. The primary antibody was diluted in 0.5 x Odyssey blocking buffer and 0.2 % Triton X-100-containing PBS with the following dilution: Rheb (Cat. NO. H00006009-M01, 1:200, Novus Biologicals); p-S6 S235/6 (#4857, 1:500, Cell Signaling). Cells were incubated with the primary antibody at 4 °C for overnight. After washed 3 times with PBS, the cells were incubated with the secondary antibody diluted (1:500) in the 0.5 X Odyssey blocking buffer and 0.2 % Triton X-100-containing PBS. For mTOR/LAMP2 staining, cells were permeabilized and blocked with 2 % BSA in 0.1 % Saponin-containing PBS for 1 hr, and then incubated with the primary antibody diluted in the blocking buffer at the following ratios: mTOR (#2983, 1:300, Cell Signaling); LAMP2 (H4B4 for human cells, 1:500, Developmental Studies Hybridoma Bank); LAMP2 (#ab13524 for mouse cells, 1:300, Abcam) at 4 °C for overnight. After washed 3 times with PBS, the cells were incubated with the secondary antibody diluted (1:500) in 2 % BSA and 0.1 % Saponin-containing PBS. After rinsed 3 times with PBST, the cells on the cover glass were mounted on microscope slides with ProLong Gold anti-fade reagent (Cat. NO. P36930, Thermo Fisher Scientific). Images were taken using a Leica TCS SP5 confocal microscope with a 63x oil immersion objective and processed using Photoshop software (Adobe). All the scale bars in the images indicate 10 μm. Image J software was used to calculate the Overlap value between different channels.
Immunoprecipitation
For the detection of endogenous ubiquitinated Rheb, cells were lysed in RIPA lysis buffer (1 % NP-40, 0.1 % SDS, 0.5 % sodium deoxycholate, 50 mM Tris pH 7.5, 150 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1 mM EDTA, 100 mM N-Ethylmaleimide, 1× Protease Inhibitor Cocktail) and rotated at 4 °C for 10 min, and then the lysates were centrifuged at 14,000 rpm for 15 min. The supernatants of the lysates were subjected to immunoprecipitation with the Rheb antibody at 4 °C for 3 hr, and the antibody-protein conjugates were precipitated with 40 μl of protein G beads. For the immunoprecipitation with the Flag antibody-conjugated agarose c Flag-Rheb was eluted with 200 μg/ml Flag peptide-containing lysis buffer. To examine the interaction between Rheb and ATXN3 or Rags and ATXN3, cells were lysed in 1× TX-100 lysis buffer (20 mM HEPES-KOH pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 % Triton X-100, with protease inhibitor and 20 mM NEM) and incubated with the Flag antibody-conjugated agarose beads at 4 °C for 1 hr, then eluted with 200 μg/ml Flag-peptide (Cat. NO. LT12022, LifeTein) containing lysis buffer.
GST Fusion Protein Purification
pGEX-6P1 plasmids encoding GST, GST-ATXN3, or GST-ATXN3 C14A mutant were transformed into BL21 E. coli cells. The expression of GST proteins was induced with 0.3 mM Isopropyl-β-D-thiogalactopyranoside for 4 hrs at 30 °C. Cells were centrifuged and re-suspended in ice-cold phosphate-buffered saline containing 0.5% Triton X-100, and lysed by sonication, and high-speed centrifuged for 15 min to remove debris. The lysates were incubated with Glutathione agarose for 2 hr at 4°C. After washing, the recombinant proteins were eluted with 10 mM reduced L-Glutathione.
In-vitro deubiquitination assay
Flag-Rheb containing polyubiquitinated Rheb was purified from HEK293T cells cotransfected with HA-Ub for 48 hr. The cells were treated with a proteasome inhibitor, MG-132 (10 μM) for 4 hr before harvest. Flag-Rheb was IPed with anti-Flag affinity beads (Sigma, A2220) and eluted with 200 μg/ml Flag peptide. Because Ub-Rheb becomes insoluble in detergent-free buffers (data not shown), 1% Triton X-100 was added to the conventional detergent-free deubiquitination reaction buffer (50 mM HEPES pH 7.5, 0.5 mM EDTA, 100 μg/ml ovalbumin, 1 mM DTT) (Burnett et al., 2003), and used this buffer for throughout of Ub-Rheb preparation including cell lysis, IP, elution, and reaction. In vitro deubiquitination assay was initiated by mixing the purified Flag-Rheb with 2 μg bacteria-derived recombinant GST-ATXN3, GST-ATXN3 C14A, or GST and incubated at 37°C for 16 hr.
In vitro DUB run-on assay
Flag-Rheb (or Flag-Rheb-4Rs) and Myc-ATXN3 (or Myc-ATXN3 C14A) were transiently expressed in HEK293T cells. 48 hr post-transfection, cells were harvested in the lysis buffer (20 mM Tris-HCl pH 6.8, 137 mM NaCl, 1 mM EGTA, 1 % Triton X-100, 1 mM DTT, 10 % glycerol and protease inhibitors cocktail) supplemented with 100mM N-Ethylmaleimide (NEM) when indicated. To examine levels of the Ub-Rheb co-IPed with Myc-ATXN3, 2 μg of goat Myc antibody (A190–104A) was added to 1 mg of lysates and incubated for 120 min at 4 °C. Then, 30 μl of protein G beads slurry was added and incubated another 60 min. The beads were washed, and the bound proteins were eluted in 1 × Laemmli buffer. For the in vitro DUB run-on assay, cells were lysed in the lysis buffer without NEM and, the lysate was incubated with the pre-bound Myc antibody-G beads for the indicated time period at 4 °C.
QUANTIFICATION AND STASTICAL ANALYSIS
Data are expressed as a mean ± SEM. The significance of differences in colocalization experiments was determined using an unpaired two-tailed Student’s t-test, assuming equal variance. p values of less than 0.05 were considered statistically significant: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-p-S6K1 T389 | Cell Signaling Technology | Cat#9205; RRID:AB_330944 |
| Rabbit monoclonal anti-S6K1 (clone 49D7) | Cell Signaling Technology | Cat#2708; RRID:AB_390722 |
| Rabbit polyclonal anti-p-S6 S240/4 | Cell Signaling Technology | Cat#2215; RRID:AB_331682 |
| Rabbit monoclonal anti-p-S6 S235/6 (clone 91B2) | Cell Signaling Technology | Cat#4857; RRID:AB_2181035 |
| Rabbit monoclonal anti-S6 (clone 5G10) | Cell Signaling Technology | Cat#2217; RRID:AB_331355 |
| Rabbit monoclonal anti-4EBP1 (clone 53H11) | Cell Signaling Technology | Cat#9644; RRID:AB_2097841 |
| Rabbit monoclonal anti-p-Akt S473 (clone 193H12) | Cell Signaling Technology | Cat#4058; RRID:AB_331168 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat#9272; RRID:AB_329827 |
| Rabbit monoclonal anti-p18/LAMTOR1 (clone D11H6) | Cell Signaling Technology | Cat#8975; RRID:AB_10860252 |
| Rabbit monoclonal anti-RagA (clone D8B5) | Cell Signaling Technology | Cat#4357; RRID:AB_10545136 |
| Rabbit polyclonal anti-RagC | Cell Signaling Technology | Cat#3360; RRID:AB_2180068 |
| Rabbit monoclonal anti-mTOR (clone 7C10) | Cell Signaling Technology | Cat#2983; RRID:AB_2105622 |
| Rabbit monoclonal anti-Raptor (clone 24C12) | Cell Signaling Technology | Cat#2280; RRID:AB_561245 |
| Rabbit monoclonal anti-Rheb (clone E1G1R) (for IP) | Cell Signaling Technology | Cat#13879; RRID:AB_2721022 |
| Rabbit monoclonal anti-LC3A (clone D50G8) | Cell Signaling Technology | Cat#4599; RRID:AB_10548192 |
| Rabbit monoclonal anti-Rab7 (clone D95F2) | Cell Signaling Technology | Cat#9367; RRID:AB_1904103 |
| Mouse monoclonal anti-Rheb (clone 2C11) (for WB) | Novus Biologicals | Cat#H00006009-M01; RRID:AB_1112097 |
| Rabbit polyclonal anti-LAMP1 | Abcam | Cat#ab24170; RRID:AB_775978 |
| Rat monoclonal anti-LAMP2 (clone GL2A7) | Abcam | Cat#ab13524; RRID:AB_2134736 |
| Mouse monoclonal anti-Flag (clone M2) | Sigma-Aldrich | Cat#1804; RRID:AB_262044 |
| Mouse monoclonal anti-FLAG®M2 Affinity Gel | Sigma-Aldrich | Cat#A2220; RRID:AB_10063035 |
| Mouse monoclonal anti-Myc (clone 9E10) | Biolegend | Cat#626802; RRID:AB_2148451 |
| Mouse monoclonal anti-HA (clone 16B12) | Biolegend | Cat#901503; RRID:AB_2565005 |
| Goat polyclonal anti-Myc | Bethyl laboratory | Cat#A190-104A; RRID:AB_66864 |
| Mouse monoclonal anti-ATXN3 (clone 1H9) | MilliporeSigma | Cat#MAB5360; RRID:AB_2129339 |
| Mouse monoclonal anti-GAPDH (clone 6C5) | MilliporeSigma | Cat#MAB374; RRID:AB_2107445 |
| Rabbit polyclonal anti-Raptor | MilliporeSigma | Cat#09-217; RRID:AB_612103 |
| Rabbit polyclonal anti-ATXN3 | MilliporeSigma | Cat#GTX115032; RRID:AB_10621230 |
| Alexa Fluor 594-goat polyclonal anti-rabbit IgG | Thermo Fisher Scientific | Cat#A11012; RRID:AB_141359 |
| Alexa Fluor 488-goat polyclonal anti-rat IgG | Thermo Fisher Scientific | Cat#A11006; RRID:AB_141373 |
| Alexa Fluor 488-goat polyclonal anti-mouse IgG | Thermo Fisher Scientific | Cat#A11029; RRID:AB_138404 |
| Alexa Fluor 594-goat polyclonal anti-mouse IgG | Thermo Fisher Scientific | Cat#R37121; RRID:AB_2556549 |
| Anti-goat IgG (H+L), HRP | Thermo Fisher Scientific | Cat#81-1620; RRID: AB_2534006 |
| Anti-rabbit IgG HRP Linked Whole Ab | GE Healthcare | Cat#NA934; RRID:AB_772206 |
| Anti-mouse IgG HRP Linked Whole Ab | GE Healthcare | Cat#NA931; RRID:AB_772210 |
| Bacterial and Virus Strains | ||
| One Shot™ Stbl3™ Chemically Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
| Ad5 CMV-cre | University of Michigan Biomedical Research Core | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bortezomib | Cell Signaling Technology | Cat#2204 |
| ALLN | Sigma-Aldrich | Cat#A6185 |
| Amino acid mixtures | Sigma-Aldrich | Cat#R7131 |
| Chloroquine | Sigma-Aldrich | Cat#C6628 |
| FTI-277 trifluoroacetate salt | Sigma-Aldrich | Cat#F9803 |
| L-Glutathione reduced | Sigma-Aldrich | Cat#G4251 |
| MG-132 | Sigma-Aldrich | Cat#C2211 |
| N-Ethylmaleimide | Sigma-Aldrich | Cat#E3876 |
| Saponin | Sigma-Aldrich | Cat#S4521 |
| Bafilomycin A | A.G. Scientific | Cat#B-1183 |
| Concanamycin A | A.G. Scientific | Cat#C-1057 |
| Amino acid-free DMEM/F12 medium | US biological Life Sciences | Cat#D9811-01 |
| Dialyzed Fetal Bovine Serum | Thermo Fisher Scientific | Cat#26400044 |
| Pierce™ Anti-HA Magnetic Beads | Thermo Fisher Scientific | Cat#88836 |
| Pierce™ Glutathione Agarose | Thermo Fisher Scientific | Cat#16100 |
| Prolong Gold antifade reagent with DAPI | Thermo Fisher Scientific | Cat#P36931 |
| Protein G Sepharose 4 Fast Flow | GE Healthcare | Cat#17061801 |
| Flag peptide | LifeTein | Cat#12022 |
| Recombinant murine FGF-basic | Peprotech | Cat#450-33 |
| Primocin | Invivogen | Cat#ant-pm-1 |
| Deposited Data | ||
| Original raw data | This paper | https://data.mendeley.com/datasets/t7vxvsxfmb/draft?a=abd5b86d-f969-4801-94f7-331500bead1d |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | CRL-11268; RRID:CVCL_1926 |
| Mouse: mesenchymal stem cell (EMSC) p18 flox/flox | This paper | N/A |
| Mouse: MEF RagA/B double flox/flox | Dr. Kun-Liang Guan | (Kim et al., 2014) |
| Mouse: MEF p18 Rev & p18 KO | Dr. Masato Okada | (Nada et al., 2009) |
| Oligonucleotides | ||
| sgRNA targeting GFP 5’-CTTGTCACTACTTTCGGTTA | This paper | N/A |
| sgRNA target human Rheb 5’-GTTTTCTATGGTTGGATCGT | This paper | N/A |
| sgRNA targeting human p18/LAMTOR1 5’- CTGCTACAGCAGCGAGAACG | This paper | N/A |
| sgRNA targeting human ATXN3 (1) 5’-GCAATTGAGGATAATTCCAC | This paper | N/A |
| sgRNA targeting human ATXN3 (2) 5’-CTGTCATCCATATTTCCAGA | This paper | N/A |
| sgRNA targeting mouse ATXN3 (1) 5’- GCAATTGAGGATAGCTCCAC | This paper | N/A |
| sgRNA targeting mouse ATXN3 (2) 5’- CTCCTTGCAATAGGTTATTC | This paper | N/A |
| sgRNA targeting mouse ATXN3 (3) 5’- AGGAGAGTATTTTAGCCCTG | This paper | N/A |
| shRNA targeting human Rheb mRNA 5’- TCTGTTAACCTGAAAGATA | This paper | N/A |
| shRNA targeting mouse Rheb mRNA 5’- GGAAAGTCCTCATTGACAATT | This paper | N/A |
| shRNA targeting mouse ATXN3 mRNA (1) 5’- GCAAGCAGTGGTTTAACTTGA | This paper | N/A |
| shRNA targeting mouse ATXN3 mRNA (2) 5’- GCAGATGATCAAGGTCCAACA | This paper | N/A |
| Recombinant DNA | ||
| pLKO1-Sc (Scramble shRNA) | (Sarbassov et al., 2005) | Addgene plasmid #1864 |
| Lenti-CRISPR v2 | (Sanjana et al., 2014) | Addgene plasmid #52961 |
| psPAX2 | Dr. Didier Trono | Addgene plasmid #12260 |
| pMD2.G | Dr. Didier Trono | Addgene plasmid #12259 |
| pLJM1-Flag-Raptor | (Sancak et al., 2010) | Addgene plasmid #26633 |
| pcDNA3-HA-Ubiquitin | (Kamitani et al., 1997) | Addgene plasmid #18712 |
| pGEX-6P1-Ataxin3Q22 | (Winborn et al., 2008) | Addgene plasmid #22117 |
| pGEX-6P1-Ataxin3Q22 C14A | (Winborn et al., 2008) | Addgene plasmid #22113 |
| pcDNA3-Flag-RagA | Dr. Kun-Liang Guan | (Kim et al., 2008) |
| pcDNA3-HA-RagA | Dr. Kun-Liang Guan | (Kim et al., 2008) |
| pcDNA3-HA-RagC | Dr. Kun-Liang Guan | (Kim et al., 2008) |
| pLJM1-Flag-Rheb | This paper | N/A |
| pLJM1-Flag-Rheb (nucleotide C102T) | This paper | N/A |
| pLJM1-Flag-Rheb-4Rs | This paper | N/A |
| pLJM1-Flag-Rheb-4Rs (nucleotide C102T) | This paper | N/A |
| pRK7-Flag-ATXN3 | This paper | N/A |
| pRK7-Myc-ATXN3 | This paper | N/A |
| pRK7-Lyso-Flag-ATXN3 | This paper | N/A |
| pRK7-Lyso-Myc-ATXN3 | This paper | N/A |
| pRK7-Lyso-Myc-ATXN3 C14A | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Adobe Photoshop CS6 | Adobe | https://www.adobe.com/products/photoshop.html |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Amino acids stimulate mTORC1 activity in part by enhancing Rheb ubiquitination.
Polyubiquitinated Rheb displays a strong binding preference for mTORC1.
ATXN3 interacts with and deubiquitinates Rheb on the lysosome.
The inactive Rags recruits ATXN3 to the lysosome upon amino acid starvation.
Acknowledgment
The RagA/B knockout MEFs were kindly provided by Dr. Kun-liang Guan (UCSD) (Jewell et al., 2015). The lyso- and WT-TSC2 plasmids were a gift from Dr. Brendan Manning (Harvard University). We thank Dr. John Kim and Ayaka Inoki (Johns Hopkins University) for their critical reading and discussions. We also thank Dr. Bing Ye and Dr. Yukiko Yamashita (U of Michigan) for technical assistance. The studies were supported by grants from the NIH (DK083491, GM110019, DK62876), and the DOD (TS140055), Core Services from the Nutrition and Obesity Research Center (DK089503), and Rogel Cancer Center Research Grant (U of Michigan).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, and Sabatini DM (2017). Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, Squitieri F, Hardenberg MC, Imarisio S, Menzies FM, et al. (2017). Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomeo R, Cinque L, De Leonibus C, Forrester A, Salzano AC, Monfregola J, De Gennaro E, Nusco E, Azario I, Lanzara C, et al. (2017). mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J Clin Invest 127, 3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke SJ, Schmied FA, Brunt ER, Ellerby LM, and Paulson HL (2004). Caspase-mediated proteolysis of the polyglutamine disease protein ataxin-3. J Neurochem 89, 908–918. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, and Pittman RN (2003). The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet 12, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GJ, and Quilliam LA (2003). Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem 278, 32493–32496. [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, and Paulson HL (2004). Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem 279, 3605–3611. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, and Teleman AA (2014). Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Chen L, Zhao L, Xu Y, Peng X, Wang X, Ding L, Jin J, Teng H, Wang Y, et al. (2019). Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res 29, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM, Kontogiannea M, Thorarinsdottir T, Fallon L, Williams AJ, Djarmati A, Fantaneanu T, Paulson HL, and Fon EA (2011). The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum Mol Genet 20, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Schweitzer LD, Bilate AM, Chang S, Kirak O, Lamming DW, and Sabatini DM (2014). RagA, but not RagB, is essential for embryonic development and adult mice. Dev Cell 29, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, and Thomas G (2003). Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11, 1457–1466. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, and Springer W (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12, 119–131. [DOI] [PubMed] [Google Scholar]
- Harraz MM, Tyagi R, Cortes P, and Snyder SH (2016). Antidepressant action of ketamine via mTOR is mediated by inhibition of nitrergic Rheb degradation. Mol Psychiatry 21, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog LK, Kevei E, Marchante R, Bottcher C, Bindesboll C, Lystad AH, Pfeiffer A, Gierisch ME, Salomons FA, Simonsen A, et al. (2020). The Machado-Joseph disease deubiquitylase ataxin-3 interacts with LC3C/GABARAP and promotes autophagy. Aging Cell 19, e13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, and Riezman H (1996). Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277–287. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, and Chen ZJ (2011). MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, and Guan KL (2003). Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, and Guan KL (2002). TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4, 648–657. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, and Guan KL (2015). Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Xu K, Lessing D, and Bonini NM (2009). Preventing Ataxin-3 protein cleavage mitigates degeneration in a Drosophila model of SCA3. Hum Mol Genet 18, 4843–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, and Yeh ET (1997). Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272, 28557–28562. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, and Sabatini DM (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, and Guan KL (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Park HW, Sciarretta S, Mo JS, Jewell JL, Russell RC, Wu X, Sadoshima J, and Guan KL (2014). Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun 5, 4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Rong Y, Chuang YS, Peng D, and Emr SD (2015). Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell 57, 467–478. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, and Avruch J (2005). Rheb binds and regulates the mTOR kinase. Curr Biol 15, 702–713. [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, and Cantley LC (2002). Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10, 151–162. [DOI] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, and Manning BD (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, and Okada M (2009). The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J 28, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, and Ferguson SM (2013). Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202, 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, and Xu T (2002). Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 4, 658–665. [DOI] [PubMed] [Google Scholar]
- Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. (2015). SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rim JS, Mynatt RL, and Gawronska-Kozak B (2005). Mesenchymal stem cells from the outer ear: a novel adult stem cell model system for the study of adipogenesis. FASEB J 19, 1205–1207. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, and Sabatini DM (2007). PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25, 903–915. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Sabatini DM (2018). Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci U S A 115, 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, and Hafen E (2003). Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5, 559–565. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, and Chen J (2008). Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A 105, 8286–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiyama Y, Nishizawa M, Tanaka H, Kawashima S, Sakamoto H, Karube Y, Shimazaki H, Soutome M, Endo K, Ohta S, et al. (1993). The gene for Machado-Joseph disease maps to human chromosome 14q. Nat Genet 4, 300–304. [DOI] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, and Blenis J (2002). Tuberous sclerosis complex-1 and −2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A 99, 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, and Blenis J (2003). Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13, 1259–1268. [DOI] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, and Sabatini DM (2013). The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, and Paulson HL (2008). The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem 283, 26436–26443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, and Sabatini DM (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, and Hall MN (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, and Sabatini DM (2017). mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 171, 642–654 e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, and Pavletich NP (2017). Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Wang J, Yoshida S, Nada S, Okada M, and Inoki K (2016). Role of Ragulator in the Regulation of Mechanistic Target of Rapamycin Signaling in Podocytes and Glomerular Function. J Am Soc Nephrol 27, 3653–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, and Pan D (2003). Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5, 578–581. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang YH, Wu XN, Wu SQ, Lu BJ, Dong MQ, Zhang H, Sun P, Lin SC, Guan KL, et al. (2011). Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol 13, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, and Sabatini DM (2011). mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original raw data is available at: https://data.mendeley.com/datasets/t7vxvsxfmb/draft?a=abd5b86d-f969-4801-94f7-331500bead1d