Summary:
The Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) is a chloride channel central to the development of secretory diarrhea and cystic fibrosis. The oldest CFTR ortholog identified is from dogfish shark which retains similar structural and functional characteristics to the mammalian protein, thereby highlighting CFTR’s critical role in regulating epithelial ion transport in vertebrates. However, the identification of an early CFTR ortholog with altered structure/function would provide critical insight into the evolution of epithelial anion transport. Here, we describe the earliest known CFTR, expressed in sea lamprey (Petromyzon marinus), with unique structural features, activation/inhibition kinetics, and altered single channel conductance compared to human CFTR. Our data provide the earliest evolutionary evidence of CFTR, offering insight regarding changes in gene and protein structure that underpin evolution from transporter to anion channel. Importantly, these data provide a unique platform to enhance our understanding of vertebrate phylogeny over a critical period of evolutionary expansion.
Keywords: ABC transporters, molecular evolution, lamprey, vertebrates, CFTR, phosphorylation, channel
Graphical Abstract

eTOC blurb
CFTR is the only member of the ABC transporter superfamily with ion channel function. Here, Cui et al. provide the earliest evidence of CFTR in lamprey with altered genetic, structural, and functional features compared to recent orthologs, providing key insights into the evolution of function from transporter to anion channel.
Introduction:
The ATP binding cassette (ABC) superfamily is an ancient class of transporters which is ubiquitously expressed in cells and whose members transport an array of substrate across membranes(Wilkens, 2015). This superfamily is found in both prokaryotes and eukaryotes, playing central roles in fundamental cellular functions(Linton, 2007). One of the most recent evolutionary members of the ABC superfamily is the cystic fibrosis transmembrane conductance regulator (CFTR) (Dean and Annilo, 2005). Rather than bearing known function as a transporter, per se, CFTR (ABCC7) is unique in functioning as an ATP-gated chloride channel in the epithelia of a variety of organs (Li et al., 2007) and is the only member with an expanded regulatory (R) domain bearing consensus sites for protein kinase A- (PKA-) dependent phosphorylation (Sheppard and Welsh, 1999). Excessive CFTR activation via toxins in the gastrointestinal tract is critical to the development of a devastating secretory diarrhea observed in cholera, a major cause of worldwide morbidity and mortality(Clemens et al., 2017). In contrast, loss of function mutations in CFTR are associated with cystic fibrosis (CF), which impacts epithelial function in a variety of organs(Rowe et al., 2005). In humans, loss of CFTR function leads to defects in lung mucociliary transport that result in bacterial colonization, airway inflammation, bronchiectasis, and eventual respiratory failure(Mall and Hartl, 2014).
The physiology of clinical secretory diarrhea and CF underscore the importance of CFTR in maintenance of epithelial homeostasis. Recent models of mutant CFTR in other mammalian systems validate the critical role of CFTR in epithelial function(Fisher et al., 2011). CFTR orthologs have been found in a variety of pre-mammalian organisms including amphibians and bony fish(Sebastian et al., 2013). To date, the oldest validated CFTR ortholog is that of the dogfish shark (arising ~150 million years ago (MYA)) where it is heavily expressed and active in the distal bowel(Marshall et al., 1991). This CFTR ortholog arose after an important gene duplication event in the vertebrate lineage(Vandepoele et al., 2004), likely from an ancient ABCC4 transporter(Jordan et al., 2008), suggesting the possibility that functional CFTR may only be found in gnathostomes. However, the recent discovery of putative ABCC sequences in sea lamprey indicates the potential for CFTR to pre-date this critical window of vertebrate evolution and the gnathostome branch(Ren et al., 2015). In this study, we examine an ancient CFTR ortholog found in sea lamprey, which harbors key structural and functional characteristics and highlight both differences and similarities from jawed vertebrate CFTR.
Results:
Lamprey CFTR diverges in sequence from other CFTRs, lacking both PKA-dependent phosphorylation sites and the canonical phenylalanine 508 residue
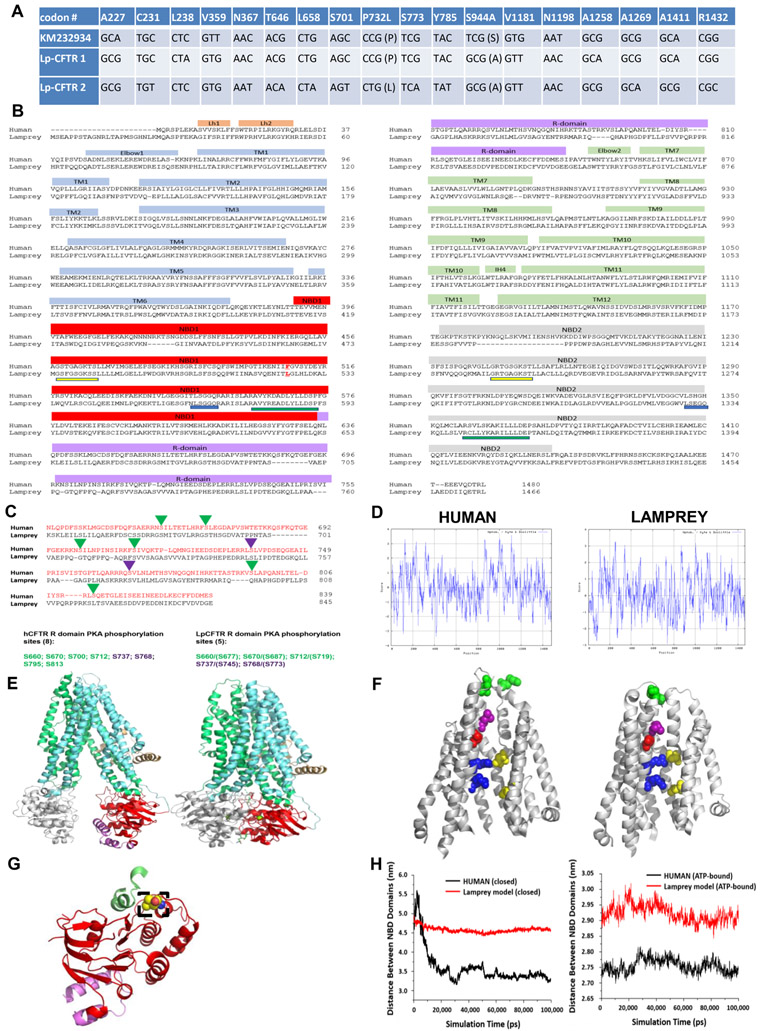
We generated two independent lamprey CFTR (Lp-CFTR) cDNA clones (expressed from each allele) using total pooled RNA from lamprey intestine tissue. Each cDNA clone bears a number of distinct SNPs relative to a published putative sequence (Ren et al., 2015). Two amino acid differences were observed between the three lamprey sequences examined here (Figure 1A), with >99% DNA identity overall. Sequence analysis indicates 72% identity and 85% homology in amino acid sequence between dogfish shark and human orthologs (Marshall et al., 1991) and 46% identity and 65% homology between lamprey and human orthologs (Figure 1B). Of note, Lp-CFTR displayed an extended 23 amino acid N-terminal sequence and did not exhibit an otherwise invariant phenylalanine at the position equivalent to F508 in human CFTR (hCFTR), which is deleted in a majority of CF patients(Rowe et al., 2005). There are also a reduced number of consensus sites for PKA-mediated phosphorylation in the R domain of Lp-CFTR relative to hCFTR, with eight sites identified by NMR in hCFTR (Bozoky et al., 2017) and five predicted sites in Lp-CFTR (Figure 1C). The two inhibitory sites are preserved in the lamprey but two of the sites proposed to play significant roles in PKA-mediated stimulation, S795 and S813 (Wilkinson et al., 1997), are not present in the lamprey; the stimulatory site at S700 in hCFTR also is missing in the lamprey ortholog.
Figure 1: Lamprey CFTR (Lp-CFTR) demonstrates key similarities and differences in sequence and structure compared to human CFTR (hCFTR).
A: SNPs identified in Lp-CFTR. The Lp-CFTR gene was cloned from lamprey cDNA, sequenced, and compared to a published putative sequence (KM232934). Eighteen SNPs were identified with 2 amino acid changes between the three Lp-CFTR sequences.
B: Amino acid sequence of Lp-CFTR aligned with that of hCFTR. Key major domains are delineated above alignment. Lp-CFTR has an extended N-terminus, a leucine at the position corresponding to F508 in hCFTR (delineated by red letter and underline), intact Walker A domains (yellow underline) and Walker B domains (green underline) in both NBD1 and NBD2, but conserved signature sequence (blue lines) in NBD1 only.
C: Lp-CFTR has a reduced number of consensus PKA phosphorylation sites. The R-domains of Lp-CFTR and hCFTR were aligned and predicted phosphorylation sites were identified for activation (green arrowheads) and inhibition (purple arrowheads), reflecting a reduced number of consensus PKA phosphorylation sites in Lp-CFTR.
D: Similar hydropathy predicted from lamprey and human CFTR sequences. CFTR sequences were compared using the Kyte-Doolittle scale for hydropathy via ExPasy, with increased hydrophilicity as negative values and increased hydrophobicity as positive values.
E: Structural homology models of Lp-CFTR. Lp-CFTR was modeled using the cryo-EM structures of CFTR in the closed state, left, and in the ATP-bound nearly open state, right. Domains colored as in Figure 1B. The segment of the R-domain visible in the closed state (purple) is not shown in the nearly open state.
F: Structure of the pore domain in the homology models of Lp-CFTR. Lp-CFTR was modeled using CFTR cryo-EM structures in closed (left) and ATP-bound nearly open (right) conformations, demonstrating development of a pore; residues 1101-1153 deleted in order to reveal the inner pore. Of note, F337 (L360 in Lp-CFTR, purple) occludes the pore in the closed state and S341 (S364, red) is at the narrowest spot in the pore interior although not yet rotated into the pore axis. R347 interacts with D924 (R370 and D927, upper blue-yellow pair) in both states, while R352 interacts with D993 (R375 and D996, bottom blue-yellow pair) only in the nearly open state. Of note, a key interaction between D110 in ECL1 and K892 in ECL4 observed in hCFTR in the closed state is not enabled in Lp-CFTR (N134 with R896, green pair, partially occluded in the view of the ATP-bound near-open state).
G: NBD1 of Lp-CFTR bears a key substitution. Lp-CFTR modeled using the CFTR cryo-EM structures demonstrated folding of NBD1 similar to that of human CFTR. NBD1 is noted in red ribbon and a portion of the R-domain is in purple ribbon. The lamprey substitution for phenylalanine 508, leucine 525, is noted in dashed box region and is positioned to interact with intracellular loop 4 (green ribbon).
H: NBD domains are more distant from each other in Lp-CFTR compared to the human ortholog: Comparison of distances between the centers of mass of NBD1 and NBD2 of human (black) and lamprey (red) CFTR conformations obtained from 100 ns MD simulations. (A) Closed hCFTR (5uak.pdb) and the corresponding Lp-CFTR homology model. (B) ATP-bound nearly-open hCFTR (6msm.pdb) and the corresponding Lp-CFTR homology model.
See also Data Figure S1 and Figure S1
Lamprey CFTR has overall hydropathy similar to that of human CFTR but has notable changes in predicted pore structure
The human and lamprey CFTR orthologs display similarities in hydropathy (Figure 1D), indicating potential for the development of a pore in the latter. Utilizing recently published Cryo-EM structural models of zebrafish and human CFTR proteins in a closed state and human CFTR in a nearly open state (Liu et al., 2017; Zhang and Chen, 2016; Zhang et al., 2018), we generated homology models of the lamprey protein in each state (Figure 1E and Data Figure S1). Lp-CFTR is predicted to fold in a conformation consistent with the location of the hCFTR pore, including key residues linked to chloride transport lining the pore (McCarty and Zhang, 2001) and development of a pore structure (Cui et al., 2012; Liu and Dawson, 2011; Rahman et al., 2013), although neither state is fully open to permeation (Figure 1F). The structure of Lp-CFTR’s NBD1 appears intact despite the substitution of leucine for the critical phenylalanine at position 508; this residue is thought to interact with intracellular loop 4 in hCFTR (Serohijos et al., 2008) (Figure 1G) but replacement with leucine at this site in hCFTR altered single channel gating, as recently demonstrated (Sorum et al., 2017).
Both homology models were subjected to molecular dynamics simulations over 100 ns, as described in Methods. Both hCFTR and Lp-CFTR dynamic models were found to be robust; hCFTR in the closed state relaxed much more than did the Lp-CFTR homology model (ΔRMSD >7.4 Ǻ for hCFTR compared to <5 Ǻ for Lp-CFTR, relative to their starting positions) (Figure S1). The distance between centers of mass in the NBDs at the end of the simulation was ~3.25 A for hCFTR but >4.5 Ǻ for Lp-CFTR in the closed state. Similarly, NBD-NBD distance at the end of simulation for the nearly open state was ~2.85 Ǻ for Lp-CFTR but <2.8 for hCFTR (Figure 1H). These results are consistent with the interpretation that Lp-CFTR prefers to adopt a more inward facing conformation than hCFTR, with NBDs further apart than in hCFTR.
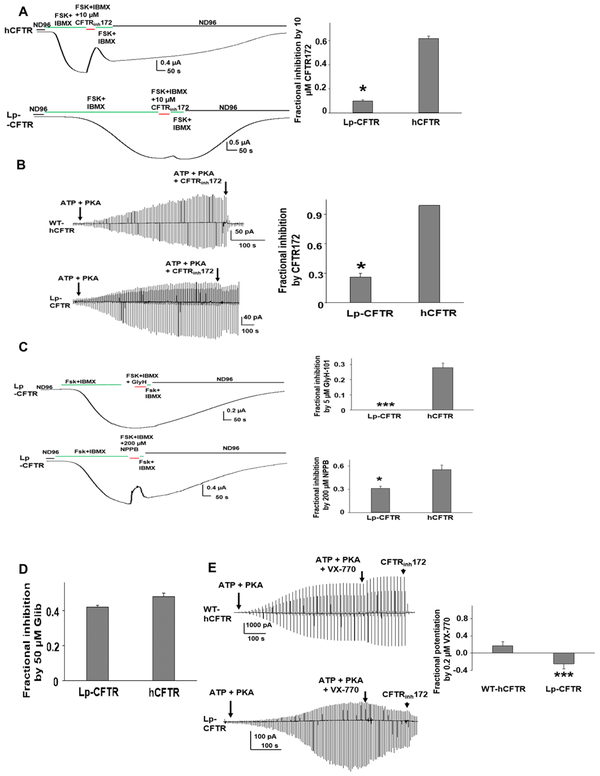
Lamprey CFTR has reduced rate of activation compared to human CFTR, is poorly blocked by known CFTR inhibitors, and is not stimulated by the CFTR potentiator VX-770
We expressed Lp-CFTR (variant 1) and wild type hCFTR in an oocyte system that roughly approximates the ambient body temperature of larval sea lamprey (17°C). Using two-electrode voltage clamp we found that macroscopic currents in Lp-CFTR were activated by PKA, much like hCFTR, although the rate of activation in whole cell experiments appeared lower in Lp-CFTR compared to hCFTR (Figure 2A, left panel). Further, by utilizing an ATP step-down protocol, we observed a notable decrease in steady-state ATP-dependent activation by Lp-CFTR (Figure S2). Lp-CFTR also demonstrated a reduced sensitivity to the channel gating inhibitor CFTRINH172 (Figure 2A, right panel), which was verified utilizing inside-out oocyte macropatches following activation with ATP and PKA (Figure 2B). Lp-CFTR also was insensitive to the hCFTR pore blocker GlyH-101, and was only weakly blocked by the broad-spectrum pore inhibitor NPPB (Figure 2C). In contrast to the results with CFTRINH172 and GlyH-101, Lp-CFTR was equally sensitive to blockade by the less specific CFTR inhibitor, gliblenclamide (Figure 2D). We also examined if a clinically available CFTR modulator (VX-770), found to potentiate hCFTR, is capable of augmenting Lp-CFTR function. Figure 2E demonstrates that although hCFTR is potentiated, Lp-CFTR shows no enhancement of function and in fact was inhibited to a significant degree; this makes Lp-CFTR the only CFTR ortholog found to be completely insensitive to potentiation by VX-770 (Cui et al., 2016). A recent structural study identified a potential hotspot for binding of VX-770 within transmembrane helix #8 of hCFTR. Mutagenesis studies combined with a binding assay showed that affinity for VX-770 was affected most by mutation at residues S308, F312, and F931 (F. Liu et al, 2019). Of these, only F312 is conserved in Lp-CFTR, suggesting that the failure of VX-770 to potentiate Lp-CFTR may be due to lost interactions between the compound and the lamprey protein at residues equivalent to S308 and F931.
Figure 2: Lp-CFTR has limited chloride conduction and is poorly responsive to both CFTR potentiators and inhibitors.
A: Whole oocyte characterization of Lp-CFTR function. Like hCFTR, Lp-CFTR can be activated by 10 μM Forskolin (FSK) with 1 mM isobutylmethylxanthine (IBMX) but exhibits distinctly slower activation. CFTRinh172 significantly blocked hCFTR (upper panel), with markedly diminished effect on Lp-CFTR (lower panel). Summary data are shown (right panel), data shown as mean±SEM; *= p< 0.05 via unpaired t-test, n=10 for hCFTR; n=4 for Lp-CFTR.
B: Macropatch characterization of Lp-CFTR. Representative inside-out macropatch currents of hCFTR (upper panel) and Lp-CFTR (lower panel). Channels were activated by 127.6 U/mL PKA and 1 mM ATP until currents reached plateau. A voltage-ramp protocol was applied every 5 s. CFTR currents were blocked with 10 μM CFTRinh172. Summary data are shown in right panel, data shown as mean±SEM. *= p<0.05 compared to hCFTR via unpaired t-test; n=4 for each group.
C: Lp-CFTR is not inhibited by GlyH-101 and is poorly sensitive to NPPB. Lp-CFTR demonstrates reduced sensitivity to inhibition by the CFTR pore blockers GlyH-101 (GlyH, 5uM, upper panel) and NPPB (200 μM) in whole oocyte recordings. Summary data are shown in right panels, data shown as mean±SEM. *** P<0.001; * P<0.05 compared to hCFTR via unpaired t-test. n=4 for Lp-CFTR, n=7 for hCFTR.
D: Inhibition of Lp-CFTR by glibenclamide. Glibenclamide (50 μM) blocked hCFTR and Lp-CFTR to a similar extent in inside-out macropatches; no statistical difference between groups via unpaired t-test. Data shown as mean±SEM; n=6 for each group.
E: Lp-CFTR is insensitive to potentiation by VX-770. Representative currents of WT-hCFTR (top panel) and Lp-CFTR (bottom panel) were recorded in inside-out macropatches in presence of 1 mM ATP and 127.6 U/mL PKA with or without 0.2 μM VX-770. A voltage-ramp protocol was applied every 5 s. CFTR currents were blocked with 10 μM CFTRinh172. Summary data for fractional potentiation are shown in right panel (Fractional potentiation by VX-770 = (I(ATP + PKA + VX-770)/ I(ATP + PKA) −1), data shown as mean±SEM; n = 4 –7 each. ***, p < 0.001 compared to hCFTR via unpaired t-test.
See also Figure S2
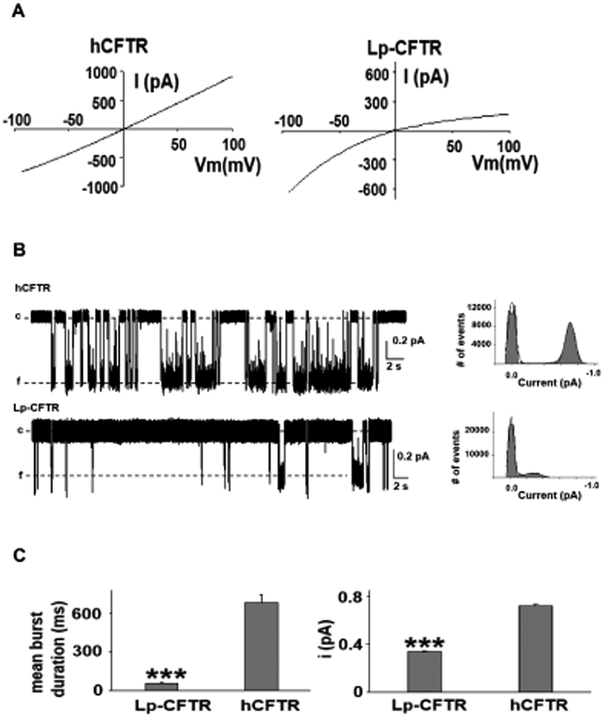
Lamprey CFTR demonstrated a notable inward rectification of current, reduced single channel stability, and altered single channel conductance
Lp-CFTR’s structural and functional features highlighted its potential for a unique current-voltage relationship. We utilized inside-out macropatch recordings of Lp-CFTR in symmetrical chloride conditions and demonstrated a notable inward rectification of macroscopic current relative to hCFTR (Figure 3A), a finding not observed in other CFTR orthologs. Further, we conducted single channel recording of Lp-CFTR to better assess the dynamics of this unique channel. We observed that Lp-CFTR had reduced open single channel stability (Figure 3B), leading to both a significantly reduced mean burst duration and single-channel amplitude in lamprey versus the human ortholog (Figure 3C).
Figure 3: Lamprey CFTR inwardly rectifies and has altered single channel kinetics.
A: Notable inward rectification of lamprey CFTR. Representative current-voltage (IV) curves of hCFTR (left panel) and Lp-CFTR (right panel) recorded in inside-out macropatches in symmetrical chloride conditions demonstrated inward rectification of Lp-CFTR current; similar results seen in n=5 for hCFTR and n=7 for Lp-CFTR.
B: Single channel recordings exhibit reduced channel stability. (Left) Traces of Lp-CFTR and hCFTR (at VM = −100 mV, activated with 1 mM MgATP and 127.6 U/mL PKA) demonstrated significant differences in opening frequency (c=closed, f=open). (Right) Representative all points histograms over the recording periods show reduced open conductance in Lp-CFTR. Solid lines in the histograms represent fits to a Gaussian function.
C: Lamprey CFTR demonstrates altered single channel behavior. Lp-CFTR exhibit shorter open burst duration (left) and lower single channel amplitude (right) compared to hCFTR in inside-out patches. Data shown as mean±SEM. ***=p< 0.001 vs. hCFTR, n= 6 for Lp-CFTR. hCFTR data (n=10) were cited from a previous publication (Cui et al., 2014). VM = −100 mV.
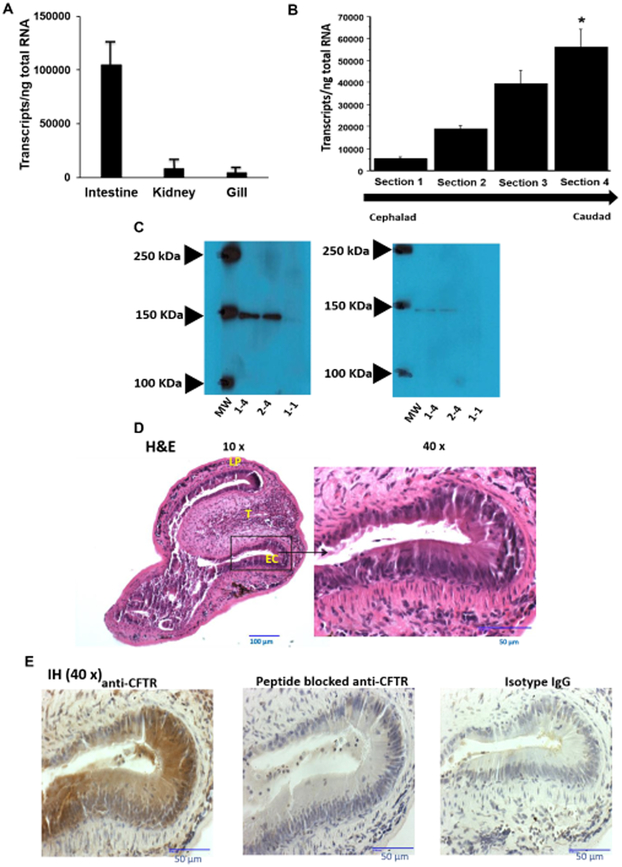
CFTR is expressed at highest levels in distal gastrointestinal tissue of the sea lamprey
We next inquired if Lp-CFTR was expressed in organs which have been previously shown to have increased expression of other CFTR orthologs (Rowe et al., 2005). We generated real time quantitative PCR (RT-qPCR) primers for Lp-CFTR and isolated RNA from lamprey kidney, gill, and intestine (Figure 4A), demonstrating expression in all tissues but increased Lp-CFTR in the intestine. We next isolated intestinal tissue from larval lamprey and divided the tract into 4 equal sections from cephalad (near mouth, termed section 1) to caudad (near cloacae, termed section 4) from each segment. RT-qPCR demonstrated increased CFTR RNA expression in section 4 relative to other intestinal regions (Figure 4B). A polyclonal antibody directed to Lp-CFTR used in Western blot detected a single band in tissues from section 4 (~150 kDa band, Figure 4C, left panel), consistent with RT-qPCR results and paralleling CFTR expression observed in the dogfish shark (Marshall et al., 1991) and human (Strong et al., 1994). We depleted the antibody signal with peptide competition, demonstrating specificity (Figure 4C, right panel). We also generated transverse sections of the lamprey GI tract, identifying key structural features in the caudal portion of the tract (Aghaallaei et al., 2016) (Figure 4D). Incubation of the unstained sections with the Lp-CFTR antibody demonstrated significant staining in GI tract epithelia which was lost following peptide competition (Figure 4E).
Figure 4: Lamprey CFTR is highly expressed in the distal gastrointestinal (GI) tract of the lamprey.
A: CFTR transcripts are found in various organs in the lamprey. RNA was isolated from intestine, kidney, and gill from seven different larval lampreys and quantified for Lp-CFTR via SYBR-green RT-qPCR (each lamprey organ run in triplicate), data shown as mean+SD; p<0.001 via ANOVA for all groups, p<0.001 for intestine vs. kidney and p<0.001 for intestine vs. gill via Tukey’s post-test. Of note, no difference was observed in 40S rRNA between tissues.
B: CFTR transcripts are increased in the distal GI tract. RNA was isolated from different sections of the GI tract (cephalad (Section 1) to caudad (Section 4)) from 7 different larval lamprey and quantified for Lp-CFTR via SYBR-green RT-qPCR (each lamprey section run in triplicate), data shown as mean+SD. p<0.01 via ANOVA for all groups, * p<0.01 Section 4 vs. Section 1 via Tukey’s post-test. Of note, no difference was observed in 40S rRNA between sections.
C: CFTR expression in lamprey GI tract. Representative Western blot (from six separate lampreys) with rabbit anti-lamprey CFTR antibody. Blot of Section 4 GI tract tissue from two separate lampreys (numbers 1-4 and 2-4), and a representative GI tract tissue of the Section 1 of a lamprey (number 1-1), demonstrating predicted ~150 kDa band at higher density in Section 4 vs. Section 1 (left panel). Antibody depletion with competing peptide demonstrating loss of 150 kDa band in all samples (right panel).
D Transverse section of lamprey GI tract. Image of the caudal section (Section 4) representative of four separate lampreys. Lampreys were dissected transversally and stained with H&E. At 10x magnification, the lamina propria (LP) and typhosole (T) were identified with lining epithelial cells (EC). At 40x, the ECs exhibit features of simple columnar epithelium with darkly stained nuclei and brush border.
E: CFTR is expressed in lamprey GI tract. Image of the caudal region of the intestine (Section 4) representative of images from four separate lampreys. Lampreys were dissected transversely and stained with either anti-CFTR antibody (1:3000 dilution), isotype control IgG (1:3000 dilution), or peptide blocked anti-CFTR antibody (1:3000 dilution), and imaged at 40x over a region of epithelial cells. Of note, there is significant Lp-CFTR staining of intestinal epithelia that is greatly reduced with peptide block and not observed with isotype IgG control.
Discussion
Numerous fundamental questions remain unanswered concerning structure/activity relationships for CFTR and other ABC transporters. One important question is how CFTR, which is unique in the ABC superfamily, has evolved to serve as a regulated epithelial ion channel. Identification of Lp-CFTR as a CFTR ortholog is due to its similarity with known CFTRs including exhibiting key structural domains (including a putative R-domain), homology to known CFTRs, and PKA- and ATP-dependent chloride conductance. Despite these features, Lp-CFTR also exhibits both structural and functional dissimilarity compared to hCFTR, delineating key evolutionary features that provide insight into development towards more effective chloride conductance. For example, both hCFTR and Lp-CFTR have intact ABC signature sequence, Walker A, and Walker B regions in NBD1 and a catalytic glutamate in NBD2, indicating potential for effective ATP hydrolysis at the first ATP binding site. However, the NBD2 signature sequence in Lp-CFTR breaks with the conserved sequence by one residue, and may represent evolutionary transition toward the jawed vertebrate orthologs, which have two non-conserved residues in this region (Muallem and Vergani, 2009). Moreover, substitution of leucine in the 508 equivalent position in Lp-CFTR presents an interesting observation since many ancient ABC transporters (including all ABCC4 orthologs) have a phenylalanine in this position, as does every known CFTR ortholog from dogfish shark through human (Figure S3) (Ostedgaard et al., 2007). This result suggests that the leucine substitution is a lamprey-specific adaptation which may provide a specific advantage for survival in freshwater.
Lp-CFTR represents a significant departure from the relatively high amino acid identity and homology observed among CFTR orthologs from jawed vertebrates, suggesting the possibility of a function independent of anion transport—and more akin to ATP-dependent solute transport. The homology model of Lp-CFTR in the closed state was compared to the cryo-EM structure of hCFTR (Liu et al., 2017). Although many key stabilizing interactions of the pore appear intact in Lp-CFTR (i.e., R347 with D924, R352 with D924, R352 with D993), others are not present in the pore of Lp-CFTR (e.g., D110 interacting with K892 to maintain a closed configuration) (Cotten and Welsh, 1999; Cui et al., 2013; Cui et al., 2014; Infield et al., 2016). These results provide evidence for altered pore dynamics in Lp-CFTR and serve as background for evolution at key amino acids within the pore observed in later species, which confer improved pore stability and function. Our single channel results provide clear evidence that although the Lp-CFTR channel does not open frequently, channel activation leads to a fully-open state, albeit with greatly reduced single-channel conductance and open duration.
A recent observation in the human airway has identified ionocytes with markedly increased expression of CFTR (Montoro et al., 2018; Plasschaert et al., 2018) and suggests the potential of specific epithelial cell populations which are enriched for Lp-CFTR. Epithelial cells with high expression in tissues from other species (including fish) raise the possibility of similar phenomena for CFTR in the lamprey intestine. Future studies would be necessary to determine whether gene expression signatures in individual lamprey epithelial cells may delineate Lp-CFTR-rich ionocyte-type populations. Increased Lp-CFTR expression within the intestinal tract relative to kidney and gill also suggest the possibility that early CFTR orthologs may have a critical role in both bicarbonate and chloride transport. As case in point, a recent study (LaRusch et al., 2014) suggested specific residues in hCFTR for which mutations selectively cause a defect in bicarbonate conductance and are associated with chronic pancreatitis, without an apparent impact on pulmonary function. Those residues in hCFTR, termed CFTRBD sites, include: (a) R74, R75, R170, L967, and R1162, in the N-terminus and intracellular loops (ICLs); (b) L997 and D115 in the pore region; and (c) S1235 and D1270 on the surface of NBD2. While findings such as these continue to undergo further assessment, it was of interest to determine whether the same residues are conserved across evolution from lamprey to human. When we inspected a multiple sequence alignment for CFTR orthologs from human, mouse, chicken, frog, dogfish shark, and lamprey, six of nine putative bicarbonate transport-related positions exhibited at least conservation of biochemical character (e.g., basic charge) if not full identity across all species. Conservation of numerous CFTRBD residues in Lp-CFTR suggests that the lamprey orthologs may play a role in bicarbonate transport—in addition to chloride secretion—thereby mediating pH regulation within the intestinal tract.
Lp-CFTR displayed slowed whole-cell activation relative to hCFTR, likely due in part to a reduced number of sites for PKA-mediated activation. Interestingly, all eight of the hCFTR R-domain PKA-dependent phosphorylation sites are preserved in the dogfish shark (Marshall et al., 1991), highlighting a significant increase in the molecular complexity in CFTR from jawless to jawed vertebrates. The observation that pan-CFTR pore blockers such as GlyH101 and NPPB are not effective inhibitors of Lp-CFTR underscores its unique pore architecture, while the inhibition of Lp-CFTR by the broad ABCC/ABCB inhibitor glibenclamide (Bessadok et al., 2011) suggests that a common structural feature of ABC proteins has remained intact. Additionally, the strong inward rectification of the Lp-CFTR makes it unique among known CFTR orthologs and suggests alterations in the charge of amino acids lining the pore relative to hCFTR. The failure of VX-770 to potentiate Lp-CFTR at a concentration that has clear effect on hCFTR is interesting, as Lp-CFTR is the only CFTR ortholog shown to be insensitive to VX-770-mediated potentiation. While it is not clear how this finding may relate to CFTR evolution, Lp-CFTR may represent a useful tool for the identification of potentiator binding sites in the human ortholog.
It is known that CFTR—like most other eukaryotic gene products—can accommodate point mutations and maintain physiologic function. Our present studies with lamprey CFTR substantiate this perspective, but suggest a paradox (Hill et al., 2014; Plyler et al., 2015). In particular, our finding that a majority (89%) of the SNPs in the three lamprey CFTR coding sequences (Figure 1a) are synonymous is difficult to reconcile with the traditional viewpoint of random SNP formation over an evolutionary time frame. Non-synonymous SNPs are expected to be formed far more frequently on a stochastic basis in the eukaryotic genome, and the present studies suggest that non-synonymous mutations in CFTR have had such a profound effect on survival or reproductive fitness that they have been ‘weeded out’ of the lamprey CFTR gene pool. Such an assertion is in marked contrast to the well-established plasticity of human CFTR in vitro and in vivo. These findings therefore point to very robust mechanisms driving SNP formation that are distinctly (and predominantly) non-random (Hill et al., 2014; Plyler et al., 2015).
Our findings delineate the earliest known CFTR ortholog, arising ~450 MYA (Smith et al., 2013), establishing that CFTR pre-dates the divergence of gnathostomes and likely evolved from ancient ABCC4 sequences (Figure S4). The lamprey ortholog provides a snapshot of the molecular evolution of CFTR; key domains are intact, but the channel lacks several features that are fixed in all jawed vertebrate representatives. Overall phylogenic analyses, as well as the presence of canonical ABC transporter features, suggest that many differences between lamprey and jawed vertebrate CFTR are vestiges of evolutionary transition from organic anion transporter to specialized chloride and bicarbonate channel, while other characteristics (such as replacement at F508) are more likely to reflect later adaptation to the specific environment of the organism. These results provide evidence of a similarly activated but poorly efficient ancient CFTR with altered kinetics and channel properties. As a result, the identification of Lp-CFTR provides a critical link between ancient ABC transporters and the hCFTR ortholog, which bears a more highly evolved function as a regulated chloride channel (Sebastian et al., 2013).
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Amit Gaggar, MD PhD (agaggar@uabmc.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary lamprey tissue
Larval Petromyzon marinus lampreys (M2 development phase, gender undetermined at this developmental stage), collected by the staff of the U.S. Geological Survey Hammond Bay Biological Station or by the survey crew of U.S. Fish and Wildlife Service Ludington Biological Station (Ludington, MI, USA), were kept in plastic tanks (98 × 54 × 48 cm, length × width × height) with flow-through water. These animals were transferred to Michigan State University (MSU) where they underwent euthanasia and dissection. All animal handling procedures were approved by the Institutional Animal Care and Use Committee at Michigan State University. Primary lamprey kidney, gill, and alimentary tissue was excised from metamorphic stage 2 (M2, premetamorphic) larval lampreys (n=17 total animals). Intestines were also further cut into four equal-length sections, with Section 1 being the most cephalad and the Section 4 the most caudal. Some of this tissue was fixed and placed in paraffin block for future staining (n=4 lampreys). Remainder of tissue was either made into lysate in RIPA buffer for Western Blot with lamprey-directed CFTR antibody (n=6 lampreys) or RNA was extracted and stored in RNA-Later and then utilized for RT-qPCR (n=7 lampreys).
Lamprey and human CFTR oocyte experiments
Stage V oocytes (from female Xenopus laevis) were from Xenopus1 (Dexter, MI). These oocytes were injected with cRNA encoding human CFTR (hCFTR) or lamprey CFTR (Lp-CFTR). For single channel and macropatch recordings 0.01 ng-10 ng cRNA was injected. Experiments were performed 2-4 days following injection. Extracellular solution for inside-out single channel and macropatch experiments contained (in mM): 150 NMDG-Cl, 5 MgCl2, and 10 TES, pH 7.4. Intracellular solution contained 150 NMDG-Cl, 1.1 MgCl2, 2 Tris-EGTA, and 10 TES, pH 7.4. hCFTR and Lp-CFTR were activated with 1 mM Mg ATP and PKA (protein kinase A, 127.6 U/mL; Promega). Standard two-electrode voltage clamp technique (TEVC) was used for studying whole oocyte currents. Electrodes were filled with 3M KCl and pipette resistances measured 0.4-1.5 MΩ in standard ND96 bath solution that contained (in mM): 96 NaCl, 2 KCl, 1 MgCl2, and 5 HEPES (pH 7.5).
METHOD DETAILS
Cloning lamprey CFTR
Lamprey cDNA was prepared from a total RNA preparation from intestinal tissue, using Superscript IV reverse transcriptase (Thermo Fisher) following manufacturer’s suggested protocol. Full length lamprey CFTR cDNA was PCR amplified using KOD hot start DNA polymerase (EMD Millipore) per manufacturer’s suggested protocol. The primer sequences used for PCR were: forward: 5’ ATGCAAGCTTGGATCAGCCGCATGAGTTCAT 3’; reverse: 5’ TTCCACTAGTGATATCTCTAGAGGCTCGCCAACAAAACTG 3’. PCR products were cloned into the pCR4-blunt TOPO vector (Thermo Fisher). cDNA clones were fully sequenced and compared to the published putative lamprey ABCC7 (CFTR) sequence (KM232934).
Predicted CFTR hydropathy
The amino acid sequences for both human and lamprey CFTR were assessed by ExPasy and used to generate hydropathy plots based on the Kyte-Dolittle scale.
Development of lamprey CFTR structural model
The model of Lp-CFTR in the inward facing conformation was built based on the available cryo-EM_structures of human and zebrafish CFTR (PDB codes: 5UAK and 5UAR, respectively(Zhang and Chen, 2016)’(Liu et al., 2017)). The sequence of lamprey CFTR was aligned to the templates using the align2d python script in MODELLER v.9.17(Sali and Blundell, 1993). After visual inspection of the alignment, models were generated using the default parameters in MODELLER, except for the degree of MD refinement, which was set to slow MD annealing (“refine.slow”). The resulting models were ranked according the following parameters: (i) Discrete optimized protein energy (DOPE) score that was calculated using the evaluate model python script in MODELLER v.9.17; (ii) the stereochemical quality of the models that was checked using PROCHECK(Laskowski et al., 1993); and (iii) the backbone RMSD of the model to the templates. Similar steps were then taken to generate the homology model of Lp-CFTR in the outward facing conformation, beginning with the available cryo-EM structure of human CFTR in the phosphorylated, ATP-bound state (PDB code: 6MSM; Zhang et al., 2018)
Generation of lamprey-directed CFTR antibody
A peptide sequence of the C-terminal region of the lamprey (TLRHSIKHISLQE, amino acids 1441-1453, a sequence unique to Lp-CFTR) was generated and conjugated to KLH. This was injected into rabbits and 6 weeks later, rabbit sera were collected and antibody (clone 3813) was affinity purified (New England Peptide, Boston MA).
Western blot
Tissue lysates from Sections 1 and 4 of lamprey intestine (n=6 lampreys) were loaded onto nitrocellulose membranes, blocked in TBS (pH 7.4) containing 5% BSA for 1 h and probed with 2.1 μg/ml anti-lamprey CFTR antibody followed by anti-rabbit IgG. Immunoblots were then developed using ECL chemiluminescence kits (Pierce). For antibody depletion experiments, the primary antibody was incubated with recombinant TLRHSIKHISLQE peptide for 12 hours at 4°C.
Real-time quantitative PCR (RT-qPCR)
Oligonucelotide primers were designed with consideration of GC repeat density and potential micro-RNA binding sites. RT-qPCR was performed using the SYBR Green system (ThermoFisher). Each RT-qPCR reaction contained 2 μl cDNA (40 ng) and 8 μl SYBR Green Universal PCR master mix with forward and reverse primers (900 nM each). Amplification plots were analyzed using an ABI 7900 HT RT-PCR system (ThermoFisher). 40S ribosomal RNA was used as internal standard and confirmed not to change in expression level among treatment groups. Sequence for the standard oligo and primers (underlined) were as follows (Note: the 3’ primer is reverse complementarity). 5’ ACAGAGAAATCTCGCA GTCAGAAAA CCGCCCCCACCTGCTCACCGCGATTCGGCGGTGCTTCCT3’.
Immunohistochemistry of gastrointestinal tissues
Serial sections (5 μm) were cut from the formalin fixed, paraffin embedded tissue blocks and floated onto charged glass slides (Super-Frost Plus, Fisher Scientific, Pittsburgh, PA) and followed by drying overnight at 60°C. Hemotoxylin and eosin (H&E) stained sections were obtained from tissue blocks. All sections for immunohistochemistry were deparaffinized and hydrated using graded concentrations of ethanol to deionized water. The tissue sections were subjected to antigen retrieval by Proteinase K (Sigma cat# P6556, 1:50 dilution in TE-CaCl2 buffer pH 8) for 15 min in a 37°C oven. Following a ntigen retrieval, all sections were washed gently in deionized water, then transferred to 0.05 M Tris-based solution in 0.15 M NaCl with 0.1% v/v Triton-X-100, pH 7.6 (TBST). Endogenous peroxidase was blocked with 3% hydrogen peroxide for 20 min. To further reduce nonspecific background staining, slides were incubated with 3% normal goat serum (Sigma) for 30 min at RT. All slides then were incubated at 4°C overnight with anti-CFTR antibody clone 3813 in three conditions: Anti-CFTR, 1:3000; Anti-CFTR + peptide (specific peptide targeting CFTR antibody), and IgG control antibody, 1:3000. After washing with TBST, sections were then incubated with the Goat Anti-Rabbit IgG H&L secondary antibody conjugated with HRP (Abcam #ab6721, 1:800.) Diaminobenzidine (DAB; Scy Tek Laboratories, Logan, UT) was used as the chromagen and hematoxylin (Richard-Allen Scientific, Kalamazoo, MI) as counterstain.
Maximum parsimony analysis
The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
Statistical analyses were performed using Prism 6.0 (Graphpad, La Jolla CA). Normality was assumed for testing. For groups of two, two-tailed paired or unpaired t-tests were utilized, dependent upon data type. For multiple comparisons, we performed one-way ANOVA with Tukey’s post-test for defined groups. Data are displayed as mean +/− standard error of the mean for Figure 2 and 3, and mean +/− standard deviation for Figure 4. Values of p<0.05 were considered statistically significant.
DATA AND CODE AVAILABILITY
cDNA sequences
Two clones were fully sequenced and compared to the published putative lamprey ABCC7 sequence (KM232934). These sequences are deposited in GenBank (accession numbers: MN216026 and MN216027)
Structural homology models
The published article includes the .pdb files for the homology models of both the closed and nearly-opened, ATP-bound lamprey CFTR (Data Figure S1).
Supplementary Material
Data Figure S1. (separate files): Structures of Lamprey CFTR from homology models (.pdb files), related to Figure 1
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal antibody against Lamprey CFTR | This paper | New England Peptide (Boston MA):Item # 3813 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Adenosine Triphosphate | G. Cui et al, 2013. | SigmaAldrich(St. Louis, MO): CAS# 56-65-5 |
| Protein Kinase A | G. Cui et al, 2016. | Promega (Madison, WI): CAS# 142008-29-5 |
| Forskolin | D. Infield et al, 2016 | SigmaAldrich (St. Louis, MO): CAS# 66575-29-9 |
| GlyH-101 | G. Cui et al, 2014 | EMD Millipore (Burlington, MA): CAS# 328541-79-3 |
| Gliblenclamide | G. Cui et al, 2012 | SigmaAldrich (St. Louis, MO): CAS#:10238-21-8 |
| CFTRINH-172 | D. Infield et al, 2016 | Cayman Chem (Ann Arbor, MI): CAS# 307510-92-5 |
| VX-770 | G. Cui et al, 2016. | Selleckchem (Houston, TX): CAS# 873054-44-5 |
| Lp-CFTR peptide | This paper | New England Peptide (Boston, MA): TLRHSIKHISLQE, amino acids 1441-1453 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Genbank (Lp-CFTR sequences) | This paper | MN216026 and MN216027 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oocytes | Xenopus1 (Dexter, MI) | Class V Xenopus laevis oocytes |
| Petromyzon marinus | Hammond Bay or Luddington Bio. Station | Larval M2 lamprey |
| Oligonucleotides | ||
| Primers for cloning lamprey CFTR 5’ATGCAAGCTTGGATCAGCCGCATGAGTTCAT 3’ |
This paper | Integrated DNA Technologies |
| Primers for RT-qPCR for lamprey CFTR 5’ACAGAGAAATCTCGCAGTCAGAAAACCGCCCCCA CCTGCTCACCGCGATTCGGCGGTGCTTCCT3’ |
This paper | Integrated DNA Technologies |
| Recombinant DNA | ||
| hCFTRcDNA | Provided by D. Gadsby | G. Cui et al, 2016. |
| Lp-CFTR cDNA | This paper | |
| Software and Algorithms | ||
| MODELLER | Sali, A., Blundell, T.L., 1993 | https://salilab.org/modeller/ |
| PROCHECK | Laskowski, R.A. et al., 1993 | https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/ |
| MEGA7 | Kumar, S et al., 2016. | https://megasoftware.net/ |
| Other | ||
| Homology model of lamprey CFTR (closed and nearly-open state) | This paper | Supplemental Figure 1 |
Highlights.
Lamprey bear the oldest CFTR ortholog (Lp-CFTR), highly expressed in intestine
Lp-CFTR has canonical CFTR structural domains and exhibits channel activity
Lp-CFTR anion channel function differs from recent CFTR orthologs in many ways
Lp-CFTR provides insights into evolution of function in this critical ABC protein
Acknowledgements:
We would like to thank Dr. Dezhi Wang at UAB Pathology Core Research Laboratory for help in histology, Drs. K. Kirk and J. Koff for their careful examination of this manuscript, Drs. Z. Plyer and I. Thornell for helpful discussions. This research is supported by the Great Lakes Fishery Commission (WL), NIH (HL102371 (AG), 5R01-DK056481-07 (NM)), CF Foundation (MCCART17G0 (NM) and SENDER13XX0 (HS)), American Heart Association (16SDG27040000 (XX)), BSF (Grant number 2013391 (HS)), Veterans Administration Merit Review (1 I01 BX001756 (AG)), and the Ismail Moustafa Scholar Fund (AG).
Footnotes
Data and materials availability: All data is available in the main text or the supplementary materials Correspondence and request for material should be addressed to AG (agaggar@uabmc.edu).
Competing Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes:
- Aghaallaei N, Gruhl F, Schaefer CQ, Wernet T, Weinhardt V, Centanin L, Loosli F, Baumbach T, and Wittbrodt J (2016). Identification, visualization and clonal analysis of intestinal stem cells in fish. Development 143, 3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessadok A, Garcia E, Jacquet H, Martin S, Garrigues A, Loiseau N, Andre F, Orlowski S, and Vivaudou M (2011). Recognition of sulfonylurea receptor (ABCC8/9) ligands by the multidrug resistance transporter P-glycoprotein (ABCB1): functional similarities based on common structural features between two multispecific ABC proteins. J Biol Chem 286, 3552–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozoky Z, Ahmadi S, Milman T, Kim TH, Du K, Di Paola M, Pasyk S, Pekhletski R, Keller JP, Bear CE, et al. (2017). Synergy of cAMP and calcium signaling pathways in CFTR regulation. Proc Natl Acad Sci U S A 114, E2086–E2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens JD, Nair GB, Ahmed T, Qadri F, and Holmgren J (2017). Cholera. Lancet 390, 1539–1549. [DOI] [PubMed] [Google Scholar]
- Cotten JF, and Welsh MJ (1999). Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR. Evidence for disruption of a salt bridge. J Biol Chem 274, 5429–5435. [DOI] [PubMed] [Google Scholar]
- Cui G, Freeman CS, Knotts T, Prince CZ, Kuang C, and McCarty NA (2013). Two salt bridges differentially contribute to the maintenance of cystic fibrosis transmembrane conductance regulator (CFTR) channel function. J Biol Chem 288, 20758–20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Khazanov N, Stauffer BB, Infield DT, Imhoff BR, Senderowitz H, and McCarty NA (2016). Potentiators exert distinct effects on human, murine, and Xenopus CFTR. Am J Physiol Lung Cell Mol Physiol 311, L192–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Rahman KS, Infield DT, Kuang C, Prince CZ, and McCarty NA (2014). Three charged amino acids in extracellular loop 1 are involved in maintaining the outer pore architecture of CFTR. J Gen Physiol 144, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Song B, Turki HW, and McCarty NA (2012). Differential contribution of TM6 and TM12 to the pore of CFTR identified by three sulfonylurea-based blockers. Pflugers Arch 463, 405–418. [DOI] [PubMed] [Google Scholar]
- Dean M, and Annilo T (2005). Evolution of ATP-Binding Cassette (ABC) transporter superfamily in vertebrates. Annual Review of Genomics and Human Genetics 6, 123–142. [DOI] [PubMed] [Google Scholar]
- Fisher JT, Zhang Y, and Engelhardt JF (2011). Comparative biology of cystic fibrosis animal models. Methods Mol Biol 742, 311–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AE, Plyler ZE, Tiwari H, Patki A, Tully JP, McAtee CW, Moseley LA, and Sorscher EJ (2014). Longevity and plasticity of CFTR provide an argument for noncanonical SNP organization in hominid DNA. pLoS One 9, e109186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infield DT, Cui G, Kuang C, and McCarty NA (2016). Positioning of extracellular loop 1 affects pore gating of the cystic fibrosis transmembrane conductance regulator. Am J Physiol Lung Cell Mol Physiol 310, L403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Kota KC, Cui G, Thompson CH, and McCarty NA (2008). Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporters. Proc Natl Acad Sci U S A 105, 18865–18870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRusch J, Jung J, General IJ, Lewis MD, Park HW, Brand RE, Gelrud A, Anderson MA, Banks PA, Conwell D, et al. (2014). Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS Genet 10, e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, and Thornton JM (1993). PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26, 283–291. [Google Scholar]
- Li C, Krishnamurthy PC, Penmatsa H, Marrs KL, Wang XQ, Zaccolo M, Jalink K, Li M, Nelson DJ, Schuetz JD, et al. (2007). Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell 131, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton KJ (2007). Structure and function of ABC transporters. Physiology (Bethesda) 22, 122–130. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhang Z, Csanady L, Gadsby DC, and Chen J (2017). Molecular Structure of the Human CFTR Ion Channel. Cell 169, 85–95 e88. [DOI] [PubMed] [Google Scholar]
- Liu F et al. (2019). Structural identification of a hotspot on CFTR for potentiation. Science 364(6446): 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, and Dawson DC (2011). Cystic fibrosis transmembrane conductance regulator: temperature-dependent cysteine reactivity suggests different stable conformers of the conduction pathway. Biochemistry 50, 10311–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall MA, and Hartl D (2014). CFTR: cystic fibrosis and beyond. Eur Respir J 44, 1042–1054. [DOI] [PubMed] [Google Scholar]
- Marshall J, Martin KA, Picciotto M, Hockfield S, Nairn AC, and Kaczmarek LK (1991). Identification and localization of a dogfish homolog of human cystic fibrosis transmembrane conductance regulator. J Biol Chem 266, 22749–22754. [PubMed] [Google Scholar]
- McCarty NA, and Zhang ZR (2001). Identification of a region of strong discrimination in the pore of CFTR. Am J Physiol Lung Cell Mol Physiol 281, L852–867. [DOI] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muallem D, and Vergani P (2009). Review. ATP hydrolysis-driven gating in cystic fibrosis transmembrane conductance regulator. Philos Trans R Soc Lond B Biol Sci 364, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Rogers CS, Dong Q, Randak CO, Vermeer DW, Rokhlina T, Karp PH, and Welsh MJ (2007). Processing and function of CFTR-DeltaF508 are species-dependent. Proc Natl Acad Sci U S A 104, 15370–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyler ZE, Hill AE, McAtee CW, Cui X, Moseley LA, and Sorscher EJ (2015). SNP Formation Bias in the Murine Genome Provides Evidence for Parallel Evolution. Genome Biol Evol 7, 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman KS, Cui G, Harvey SC, and McCarty NA (2013). Modeling the conformational changes underlying channel opening in CFTR. PLoS One 8, e74574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Chung-Davidson YW, Yeh CY, Scott C, Brown T, and Li W (2015). Genomewide analysis of the ATP-binding cassette (ABC) transporter gene family in sea lamprey and Japanese lamprey. BMC Genomics 16, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe SM, Miller S, and Sorscher EJ (2005). Cystic fibrosis. N Engl J Med 352, 1992–2001. [DOI] [PubMed] [Google Scholar]
- Sali A, and Blundell TL (1993). Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Sebastian A, Rishishwar L, Wang J, Bernard KF, Conley AB, McCarty NA, and Jordan K (2013). Origin and evolution of the cystic fibrosis transmembrane regulator protein R domain. Gene 523, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, and Riordan JR (2008). Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A 105, 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DN, and Welsh MJ (1999). Structure and function of the CFTR chloride channel. Physiol Rev 79, S23–45. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. (2013). Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45, 415–421, 421e411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorum B, Torocsik B, and Csanady L (2017). Asymmetry of movements in CFTR's two ATP sites during pore opening serves their distinct functions. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong TV, Boehm K, and Collins FS (1994). Localization of cystic fibrosis transmembrane conductance regulator mRNA in the human gastrointestinal tract by in situ hybridization. J Clin Invest 93, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, and Van de Peer Y (2004). Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A 101, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S (2015). Structure and mechanism of ABC transporters. F1000Prime Rep 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DJ, Strong TV, Mansoura MK, Wood DL, Smith SS, Collins FS, and Dawson DC (1997). CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol 273, L127–133. [DOI] [PubMed] [Google Scholar]
- Zhang Z, and Chen J (2016). Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator. Cell 167, 1586–1597 e1589. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu F, and Chen J (2018). Molecular structure of the ATP-bound, phosphorylated human CFTR. Proc Natl Acad Sci U S A 115, 12757–12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Figure S1. (separate files): Structures of Lamprey CFTR from homology models (.pdb files), related to Figure 1
Data Availability Statement
cDNA sequences
Two clones were fully sequenced and compared to the published putative lamprey ABCC7 sequence (KM232934). These sequences are deposited in GenBank (accession numbers: MN216026 and MN216027)
Structural homology models
The published article includes the .pdb files for the homology models of both the closed and nearly-opened, ATP-bound lamprey CFTR (Data Figure S1).