Supplemental Digital Content is available in the text.
Background.
Nonalcoholic fatty liver disease (NAFLD) and its progressive form nonalcoholic steatohepatitis (NASH) are a growing problem globally and recur even after liver transplant (LT). We aim to characterize the gut dysbiosis in patients who developed recurrent NAFLD compared with patients without recurrence following LT.
Methods.
Twenty-one patients who received LT for NASH and had a protocol liver biopsy performed beyond 1-y post-LT were included prospectively (January 2018–December 2018). Genomic DNA extraction, next-generation sequencing, and quantitative PCR analysis were performed on stool samples collected within 1.1 ± 1.6 y from time of liver biopsy.
Results.
Recurrent NAFLD was noted in 15 of the 21 included patients. Stool microbiome analysis at the genus level showed significant loss of Akkermansia and increasing Fusobacterium associated with NAFLD recurrence. Quantitative PCR analysis revealed significantly decreased relative abundance of Firmicutes in patients with NAFLD activity scores (NASs) ≥5 as compared with patients with lower NAS scores, whereas Bacteroidetes were significantly increased with higher NAS (P < 0.05). Firmicutes (P = 0.007) and Bifidobacterium group (P = 0.037) were inversely correlated, whereas Bacteroidetes (P = 0.001) showed a positive correlation with higher hepatic steatosis content. The Firmicutes/Bacteroidetes ratios were higher in patients without NAFLD or NASH as compared with patients diagnosed with NAFLD or NASH at the time of sample collection.
Conclusions.
Akkermansia, Firmicutes, and Bifidobacterium may play protective roles in the development of recurrent NAFLD in LT recipients, whereas Fusobacteria and Bacteroidetes may play pathogenic roles. These findings highlight the potential role of the “gut-liver” axis in the pathogenesis of NAFLD recurrence after LT.
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease worldwide. It is a clinical hepatic manifestation of insulin resistance and is strongly associated with metabolic syndrome and its associated risk factors, such as obesity, type 2 diabetes, dyslipidemia, and hypertension.1 NAFLD in its progressive form manifests as nonalcoholic steatohepatitis (NASH), characterized by hepatic steatosis, lobular inflammation, and hepatocellular damage with or without fibrosis. NASH is expected to increase by 63% between 2015 and 2030.2 With a worldwide increase in the prevalence of obesity, NASH is an increasingly common liver problem affecting global public health and healthcare costs. Up to one-third of the United States population is affected by NAFLD, with the majority (70%–90%) presenting with simple steatosis, of which 20%–30% of patients go on to develop NASH,3 the second most common indication for liver transplantation (LT) in the United States and projected to become the leading indication by 2025.4
LT is the only treatment for advanced NASH cirrhosis. However, NAFLD can frequently recur post-LT in patients with the pretransplant diagnosis of NASH. NAFLD has been reported in rates of 30%–100% within 3–5 y post-LT, with rates of recurrent NASH varying from 8% to 18%.5 Currently, there is a paucity of information available on the risk factors for recurrent disease and its natural history, and there is virtually no treatment available for this progressive illness following LT.5 Together, this justifies the need for a “bench to bedside” research study for NAFLD in the transplant setting.
Recent studies have noted that LT improves gut microbiota diversity and dysbiosis compared with pre-LT baseline but residual dysbiosis remains.6 Additionally, progression of NAFLD may be influenced by changes in the intestinal microbiome.7,8
Our aim is to identify specific gut microbial diversity of patients who develop recurrent NAFLD/NASH following LT.
MATERIALS AND METHODS:
Study Location and Ethical Clearance
This is a prospective observational study conducted at the James D. Eason Transplant Institute of Methodist University Hospital, an affiliate of the University of Tennessee Health Sciences Center, Memphis, TN. The study included adult LT recipients (age > 18) with NASH as an indication for liver transplant who had a liver biopsy performed at 1-y after their LT. Participants were enrolled between January 2018 and December 2018. Patients with prior bariatric surgery or recent acute cellular rejection or patients on chronic antibiotics were excluded by prescreening. All participants signed an informed consent before being enrolled in the study. Their demographic, clinical, and laboratory data were collected prospectively by review of their electronic medical record.
The protocol for the study was approved a priori by the University of Tennessee Institutional Review Board (Study Protocol # 15-03891-XP UM).
Stool Sample Collection
Study subjects were provided the commode stool collection containers (Fisher Catalog No.02-544-208, Fisher Scientific, Pittsburgh, PA) and simplified microbiome sampling instructions (derived from Human Microbiome Project HMP Protocol # 07-001, version 12.0, 2010). Written instructions, a prepaid FedEx package, and packaging materials for refrigerated shipping were provided to the subjects for sending samples back to laboratory at their convenience. Twenty-four patients consented for the study of which 2 patients were excluded as they did not return the stool samples. All stool samples were collected from LT recipients who had a liver biopsy done at least 1 y after their LT. If the interval between stool collection and the liver biopsy was >1 y, persistent hepatic steatosis was confirmed with an imaging modality such as abdominal computed tomography (CT) scan (Liver/Spleen HU density < 1), MRI (MRI-PDFF), or ultrasound-based quantification of hepatic steatosis the liver (controlled attenuation parameter score of Fibroscan). Stool samples were collected a mean of 1.1 ± 1.6 y from time of liver biopsy. Seventy-one percent of the stool samples were collected at or within 1 y of the liver biopsy, 15% within 2 y, and in the final 3 patients at 3, 4, and 6 y. Absence or persistence of hepatic steatosis was confirmed with an imaging modality such as abdominal CT scan, MRI, or ultrasound of the liver in all the patients if the interval between the liver biopsy and the time of stool collection is >1 y. Of the 6 patients who had no steatosis on liver biopsy, 2 had longer than 6 mo interval between stool collection, and liver biopsy. Both these patients had no steatosis on imaging studies, 1 based on CT and the other based on MR Elastography. Additionally, there is essentially no significant difference in the demographic and clinical data at the time of liver biopsy and stool collection (Table S1, SDC, http://links.lww.com/TXD/A289).
The liver biopsy specimens were reviewed by a single experienced hepato-pathologist (D.K.) who was blinded of the clinical information of the patient at the time of the liver biopsy and graded/staged per nonalcoholic steatohepatitis clinical research network (CRN) scoring system,9 and Ishak scoring system.10 The pathologist defined the biopsy as suboptimal if the biopsy sample is either small (≤10 mm) or had histological artifacts that made scoring difficult. All but 1 sample was considered suboptimal by the pathologist. The NAFLD activity score (NAS) was calculated as per NASH-CRN criteria with score ranging from 0 to 8 according to the sum of the scores for steatosis (0–3), lobular inflammation (0–3), and ballooning (0–2). Cases were grouped into no (NAS 0), mild (NAS 1–2), moderate (3–4), and severe activity (5–8). Diagnostic categorization into NAFLD without features of NASH, borderline steatohepatitis, and definite steatohepatitis was made based on standard histological criteria.11 Steatosis was defined as presence of at least 5% macrovesicular steatotic hepatocytes. Mild steatosis was defined as 5%–33% steatosis, moderate steatosis 33%–66%, and severe steatosis was defined as >66% macrovesicular steatosis. Isolated steatosis was defined as the presence of steatosis alone without features suggesting steatohepatitis. Multiple additional features were also reviewed in the liver biopsy including the presence or absence of mega-mitochondria, acidophil bodies, and distribution of fat (zonal, azonal, or panacinar) and the presence or absence of microvesicular steatosis, cholestasis, bile duct injury, and acute or chronic cellular rejection. Portal fibrosis was also staged according to the Ishak method.10 Liver biopsy adequacy was assessed, and suboptimal biopsies were defined as those that were either small (≤10 mm) or had histological artifacts that impeded scoring. Data on the donor liver biopsy were acquired from the Organ Procurement and Transplantation Network database and were reviewed for macrosteatosis%.
DNA Extraction
Genomic DNA was extracted from the stool samples using the PowerFecal DNA extraction kit (MO BIO Laboratories, Carlsbad, CA), following the manufacturer’s protocol. The extracted genomic DNA samples were quantified spectrophotometrically using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE).
Next-generation Sequencing
To evaluate the microbial ecology of samples, 16S universal Eubacterial primers 530F-926R targeting V4-V5 variable region of the 16S rRNA gene were utilized on the Illumina MiSeq for amplicon sequencing. A single-step 30 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA) was used under the following conditions: 94oC for 3 min, followed by 35 cycles of 94oC for 30 s; 53oC for 40 s, and 72oC for 1 min; after which a final elongation step at 72oC for 5 min was performed. After the completion of PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA). Then, the pooled and purified PCR product was used to prepare Illumina DNA library. Sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX) on a MiSeq following the manufacturer’s guidelines.12-14
Sequencing data were processed and analyzed using Quantitative Insights into Microbial Ecology 1.9.1. Sequences were first demultiplexed, then denoised and clustered into sequence variants before rarefaction to a depth of 3000 sequences. Representative bacterial sequences were aligned via PyNAST, taxonomy assigned using the RDP Classifier. Processed data were then imported into Calypso 8.84 for further analysis and data visualization.15 The Shannon Index was used to quantify alpha diversity (intersample).16 The Bray Curtis analysis was used to quantify beta diversity (intrasample), and the differences were compared using Anosim. To quantify relative abundance of taxa between groups, we utilized ANOVA adjusted using the Bonferroni correction and false discovery rate for multiple comparisons. We used linear discriminant analysis of effect size to test for significance and perform high-dimensional biomarker identification.17 Network analysis was generated from Spearman’s correlations. Positive correlations with a false discovery rate-adjusted, P < 0.05 were presented as an edge.
Quantitative PCR
To evaluate the changes in relative abundance of following human gut microbial genera quantitative PCR (qPCR) analyses were performed following published parameters for Firmicutes,18 Bacteroidetes,18 certain Clostridium clusters,19,20 lactic acid bacteria,19 and bifidobacterial.19 The relative abundance of the specific genera was evaluated by using the 2−ΔΔCt method by normalizing the 16S threshold to the host DNA amplification signal.21
Posttransplant Immunosuppression
Our institute utilized a steroid-free immunosuppression protocol, which consisted of rabbit antithymocyte globulin used for induction immunosuppression. This was administered as 2 doses of 1.5 mg/kg. The first induction dose was administered during the anhepatic phase, which was defined as the time period from physical removal of the liver to recirculation of the graft. This was followed by a second dose on post-LT day 2. Starting on post-LT day 1, mycophenolate mofetil at 1000 mg (or mycophenolic acid at 720 mg BID) was administered 2 times a day for 3 mo. Mycophenolate mofetil dose adjustments were made for gastrointestinal side effects and/or development of cytopenia. The initiation of tacrolimus was delayed for 3–7 d and started when the serum creatinine was <2.0 mg/dL. An mammalian target of rapamycin inhibitor was used instead of tacrolimus if the recipient’s serum creatinine remained >2.0 mg/dL beyond posttransplant day 7. Goal trough levels for tacrolimus and sirolimus during the first 3 mo postoperatively were 5–8 and 8–10 ng/dL, respectively. The trough level was individualized beyond 3 mo but generally kept within 4–6 ng/dL and was further lowered to 2–4 ng/mL beyond 1 y.
At the time of stool collection, 80.9% (N = 17) of patients were on tacrolimus therapy, with 13 of these patients in the recurrence of NAFLD subgroup. 9.5% (N = 2) of patients were on everolimus and another 9.5% (N = 2) were on rapamycin. One patient on everolimus and 1 patient on rapamycin presented with recurrent NAFLD. 19.1% (N = 4) of patients were on mycophenolate mofetil at the time of collection, with 2 of these patients in the recurrence of NAFLD subgroup.
RESULTS
Demographic and Clinical Characteristics
In this cohort, 23 patients who received LT for NASH were enrolled. Of the 23 patients, 2 were excluded because stool samples were not being collected. In the final cohort of 21 patients, mean age at time of transplant was 59 ± 7 y, mean MELD score was 19.2 ± 6.4. All recipients were Caucasian, with 52% (n = 11) males. All patients received LT for primary indication of NASH. Demographic and clinical data at the time of transplant are reported in Table 1.
TABLE 1.
Pretransplant demographic and clinical data
| Variables | Pretransplant |
|---|---|
| All patients | |
| N = 21 | |
| Age | 59 ± 7 |
| Male gender | 52.4% male (N = 11) |
| Race (White) | 100% white (N = 21) |
| Diabetes | 47.6% (N = 10) |
| BMI (kg/m2) | 32.1 ± 6.6 |
| AST (IU/mL) | 98.5 ± 127.2 |
| ALT (IU/mL) | 55.5 ± 47.0 |
| T. Bili (mg/dL) | 5.0 ± 5.7 |
| ALP (IU/mL) | 156.6 ± 93.9 |
| Albumin (g/dL) | 3.0 ± 0.6 |
| Cr (mg/dL) | 1.4 ± 0.7 |
| Hgb (g/dL) | 11.3 ± 2.1 |
| WBC (×103/uL) | 5.8 ± 3.9 |
| INR | 1.8 ± 0.6 |
| Triglycerides (mg/dL) | 123.8 ± 52.7 |
| Cholesterol (mg/dL) | 149.3 ± 42.2 |
| HDL (mg/dL) | 37.7 ± 13.2 |
| LDL (mg/dL) | 90.1 ± 35.5 |
| HbA1c (%) | 5.3 ± 1.3 |
| MELD | 19.2 ± 6.4 |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; Cr, creatinine; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; Hgb, hemoglobin; INR, international normalized ratio; LDL, low-density lipoprotein; T. Bili, total bilirubin; WBC, white blood cell count.
Twenty-one patients had liver biopsies performed at 1-y post-LT. Six patients had no evidence of steatosis on biopsy; these patients were categorized into the nonrecurrence subgroup. Fifteen patients had some degree of steatosis and were categorized into the recurrence of NAFLD subgroup. Demographic and clinical data at the time of stool collection are summarized in Table 2. When comparing patients with recurrence of NAFLD to patients with no evidence of biopsy proven NAFLD at the time of stool collection, significant differences were noted in aspartate aminotransferase (P = 0.01), alanine aminotransferase (P = 0.04), and triglycerides (P = 0.02). Thirteen patients had diabetes at the time of stool collection, compared with 10 patients at the time of LT. Compared with the pretransplant period, patients had significantly higher hemoglobin A1c (6.4% versus 5.3%, P = 0.02) and triglycerides (213 versus 123.8, P = 0.008) at the time of stool collection (Table S2, SDC, http://links.lww.com/TXD/A289).
TABLE 2.
Demographic and clinical data at the time of stool collection
| Variables | At the time stool collection | P | ||
|---|---|---|---|---|
| All patients | No Recurrent | Recurrent | ||
| NAFLD | NAFLD | |||
| N = 21 | N = 6 | N = 15 | ||
| Age | 63 ± 7 | 62 ± 8 | 63 ± 8 | 0.82 |
| Male gender | 11 (53) | 4 (67) | 7 (47) | 0.64 |
| Race (White) | 21 (100) | 6 (100) | 15 (100) | 1 |
| Diabetes | 13 (62) | 3 (50) | 10 (67) | 0.63 |
| BMI (kg/m2) | 33 ± 8 | 38 ± 11 | 31 ± 6 | 0.15 |
| AST (IU/mL) | 38 ± 21 | 22 ± 7 | 44 ± 21 | 0.01 |
| ALT (IU/mL) | 51 ± 32 | 32 ± 10 | 58 ± 35 | 0.04 |
| T. Bili (mg/dL) | 0.7 ± 0.4 | 1 ± 0.6 | 0.6 ± 0.4 | 0.15 |
| ALP (IU/mL) | 95 ± 42 | 74 ± 42 | 103 ± 40 | 0.29 |
| Albumin (g/dL) | 4 ± 0.4 | 4 ± 0.32 | 4.14 ± 0.45 | 0.56 |
| Cr (mg/dL) | 1.2 ± 0.4 | 1 ± 0.2 | 1.17 ± 0.47 | 0.64 |
| Hgb (g/dL) | 13.4 ± 2.1 | 14.5 ± 0.9 | 13 ± 2.3 | 0.18 |
| WBC (x103/uL) | 5.8 ± 2.6 | 6.7 ± 1.9 | 5.5 ± 2.8 | 0.16 |
| INR | 1.04 ± 0.1 | 0.97 ± 0.1 | 1.05 ± 0.1 | 0.15 |
| Triglycerides (mg/dL) | 213 ± 109 | 120 ± 43 | 246 ± 107 | 0.02 |
| Cholesterol (mg/dL) | 164 ± 38 | 155 ± 19 | 167 ± 43 | 0.62 |
| HDL (mg/dL) | 38 ± 8 | 41 ± 8 | 37 ± 8 | 0.26 |
| LDL (mg/dL) | 91 ± 39 | 93 ± 19 | 90 ± 44 | 0.89 |
| HbA1c (%) | 6.4 ± 1.1 | 6.0 ± 1.3 | 6.5 ± 1.0 | 0.38 |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; Cr, creatinine; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; Hgb, hemoglobin; INR, international normalized ratio; LDL, low-density lipoprotein; NAFLD, nonalcoholic fatty liver disease; T. Bili, total bilirubin; WBC, white blood cell count.
Histological Summary of Liver Biopsies
Histological analyses of liver biopsies performed at or beyond 1-y posttransplant are summarized in Table 3. No patients in either subgroup had bridging fibrosis or cirrhosis on biopsy. Only 4 patients had definite NASH, all of which were in the recurrence of NAFLD subgroup. Overall, NAS score was significantly higher in the recurrence of NAFLD group compared to the nonrecurrence subgroup (P = 0.003). In the recurrence of the NAFLD subgroup, 33.3% of patients (N = 5) had mild NAFLD activity, 40% of patients (N = 6) had moderate NAFLD activity, and 26.7% of patients (N = 4) had severe NAFLD activity. Data on donor liver biopsy were available in 15 of the 23 included patients. Four of the 15 had mild macrosteatosis, 1 with 10%–20% macrosteatosis, 3 with 5%–10% steatosis. Of these 4 with macrosteatosis in the donor liver, 3 had significant hepatic steatosis on their post-LT liver biopsy.
TABLE 3.
Distribution of histological characteristics in the liver transplant recipients at 1-y follow-up with and without recurrent NAFLD based on NASH CRN staging criteria
| Parameters | One-year protocol liver biopsy | |||
|---|---|---|---|---|
| All patients | NAFLD-No | NAFLD-Yes | P | |
| N = 21 | N = 6 | N = 15 | ||
| Steatosis (% of cohort) | N(%) | N(%) | N(%) | |
| None [<5%] | 6 (28.6) | 6 (100) | 0 | .0001 |
| Mild [5%–33%] | 7 (33.3) | 0 | 7 (46.7) | |
| Moderate [33%–66%] | 4 (19.0) | 0 | 4 (26.7) | |
| Severe [>66%] | 4 (19.0) | 0 | 4 (26.7) | |
| Lobular inflammation | ||||
| Absent | 7 (33.3) | 3 (50.0) | 4 (26.7) | .46 |
| <2 foci/field | 12 (57.1) | 3 (50.0) | 9 (60.0) | |
| 2–4 foci/field | 2 (9.5) | 0 | 2 (13.3) | |
| >4 foci/field | 0 | 0 | 0 | |
| Cytological ballooning | ||||
| None | 13 (61.9) | 6 (100) | 7 (46.7) | .08 |
| Few | 7 (33.3) | 0 | 7 (46.7) | |
| More than few | 1 (4.8) | 0 | 1 (6.7) | |
| Portal inflammation | ||||
| None | 13 (61.9) | 3 (50.0) | 10 (66.7) | .29 |
| Mild | 5 (23.8) | 1 (16.7) | 4 (26.7) | |
| More than mild | 3 (14.3) | 2 (33.3) | 1 (6.7) | |
| Mallory bodies | ||||
| None | 19 (9.0) | 6 (100) | 13 (86.7) | 1.0 |
| Present | 2 (9.5) | 0 | 2 (13.3) | |
| NAFLD activity score | ||||
| None [NAS 0]a | 4 (19.0) | 4 (66.7) | 0 | 0.003 |
| Mild [NAS 1–2] | 7 (33.3) | 2 (33.3) | 5 (33.3) | |
| Moderate [NAS 3–4] | 6 (28.6) | 0 | 6 (40.0) | |
| Severe [NAS 5–8] | 4 (19.0) | 0 | 4 (26.7) | |
| NASH diagnosis category | ||||
| Definite NASH | 4 (19.0) | 0 | 4 (26.7) | 0.28 |
| Fibrosis and NASH CRN | ||||
| Stage 0 [absent] | 12 (57.1) | 4 (66.7) | 8 (53.3) | 0.50 |
| Stage 1 | 3 (14.3) | 0 | 3 (20.0) | |
| Stage 2 [zone 3 perisinusoidal fibrosis with portal/periportal fibrosis] | 6 (28.6) | 2 (33.3) | 4 (26.7) | |
| Stage 3 [bridging fibrosis] | 0 | 0 | 0 | |
| Stage 4 [cirrhosis] | 0 | 0 | 0 | |
CRN, clinical research network; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; NASH, nonalcoholic steatohepatitis.
Gut Microbiome by Hepatic Steatosis Score
Patient stool microbiota were assembled and analyzed based on NASH CRN scoring criteria and metadata. A heatmap of the 50 most abundant genera in all samples with associated biopsy scoring is shown in Figure 1A. Stool samples were first analyzed by Steatosis Grade, in which the percentage of fat was binned into 4 groups (<5%, 5%–33%, 34%–66%, and ≥67%). At the genus level, no significant differences were observed across groups in the alpha diversity, assessed with the Shannon Index, Richness, and Chao1 (Figure 1B). Microbial beta diversity analysis with Bray Curtis PCoA clustering demonstrated the degree of fat along PCoA1 (17%), in which higher fat percentages clustered left and lower levels clustered right, although Anosim did not reach significance based on the current sample size (Figure 1C). However, the taxonomic composition of stool microbial communities varied across fat CRN groups (Figure 1D), wherein a stepwise decrease in the orders Verrucomicrobiales (light brown) and increased levels of Fusobacteriales (red) and Enterobacteriales (pink) were associated with higher fat. At the genus level, the loss of Akkermansia (order Verrucomicrobiales) and an emergence of Fusobacterium, Proteus, Trabulsiella, and Microvirgula were observed with the greatest degree of biopsy steatosis (Figure 1E).
FIGURE 1.
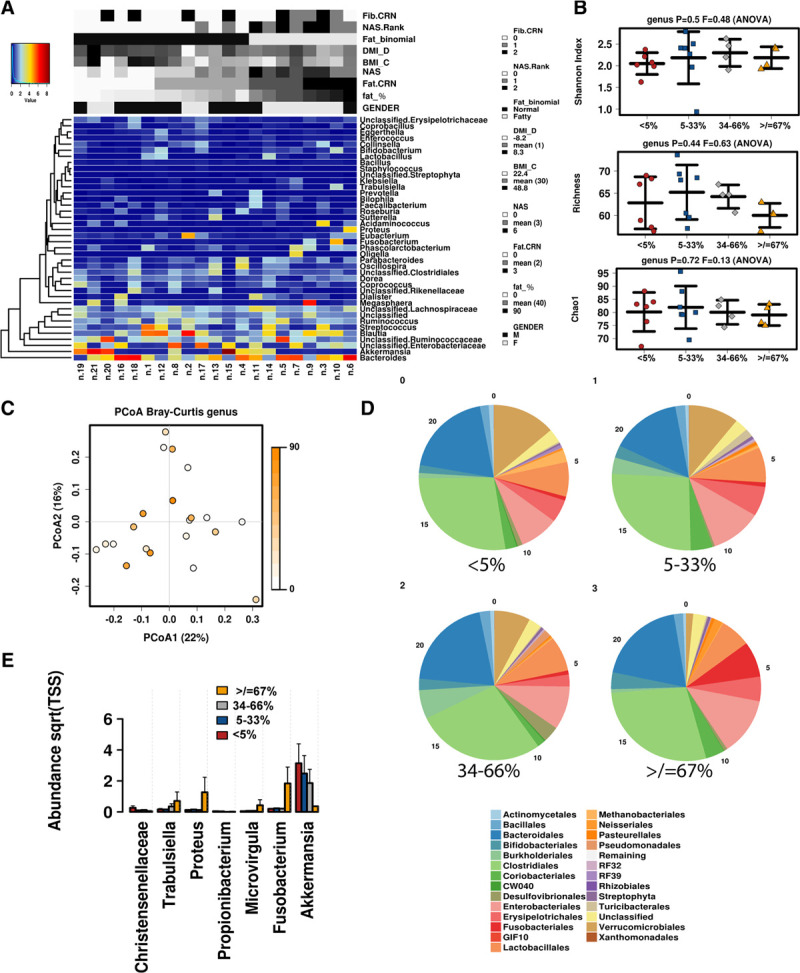
Microbiome associated with steatosis scores. A, Heatmap of the most abundant taxa detected by metagenomics is displayed with associated biopsy metadata. B, Alpha diversity determined by Shannon Index, Richness, and Chao1. C, Principal component analysis based on the associated biopsy fat percentage. D, Relative abundance of taxa at the order level by degree of steatosis. E, Abundance of select genera displayed by steatosis groups.
Gut Microbiome by Liver NAS Rank
We next examined microbiome composition based on biopsy NAS score categories, in which the relative composition of the microbiome at the family level is shown in Figure 2A. Similar to the steatosis grade, no significant differences were observed in alpha diversity across groups, including Shannon Index, Richness, and Chao1 (Figure 2B); however, significant differences were found in community membership across groups. At the genus level, a loss of Clostridiales, Propionibacterium, and Rikenella was observed with increasing NAS scoring, whereas Veillonella, Faecalibacterium, and Bilophila were elevated (Figure 2C). At the OTU level, a significant increase in Lachnospiraceae, Faecalibacterium, and Dialister occurred with increasing NAS (Figure 2D), whereas Dorea, Rikenellaceae, and Propionibacterium were decreased.
FIGURE 2.
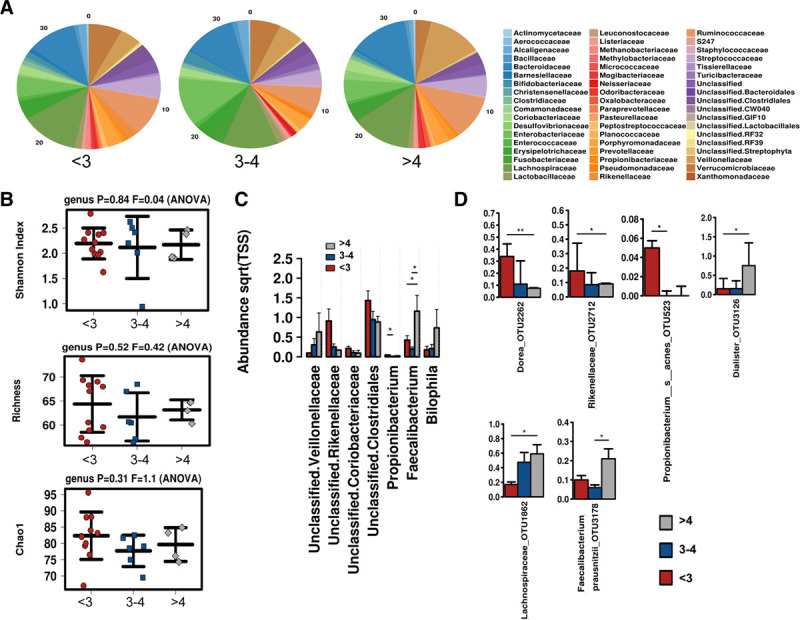
Microbiome associated with NAS scores. A, Relative abundance of microbial communities at the family level. B, Alpha diversity determined by Shannon Index, Richness, and Chao1. C, Differentially abundant taxa at the family and genus and (D) significantly altered OTUs across NAS groups. NAS, NAFLD activity scores; OTU.
Gut Microbiome by Hepatic Steatosis Binomial
Next, microbiome samples were clustered into 2 binomial groups based on biopsy fatty versus normal clustering. Relative abundances of the microbiota community at the family level are displayed between groups in Figure 3A. Similar to the NAS score and steatosis grade, no differences were observed in alpha diversity, including Shannon Index, Richness, and Chao1, based on Fat as a binomial (Figure 3B). Greater levels of Phascolarctobacterium, Fusobacterium, Trabulsiella, Bilophila, and Leuconostocaceae were observed in fatty samples, whereas Rikenellaceae and Coriobacteriaceae were observed in normal samples (Figure 3C). Use of linear discriminant analysis of effect size was utilized to detect a specific enrichment of taxa between groups, in which normal samples displayed greater Aerococcaceae and Propionibacterium, whereas fatty associated samples displayed greater Bilophila and Veillonellaceae (Figure 3C).
FIGURE 3.
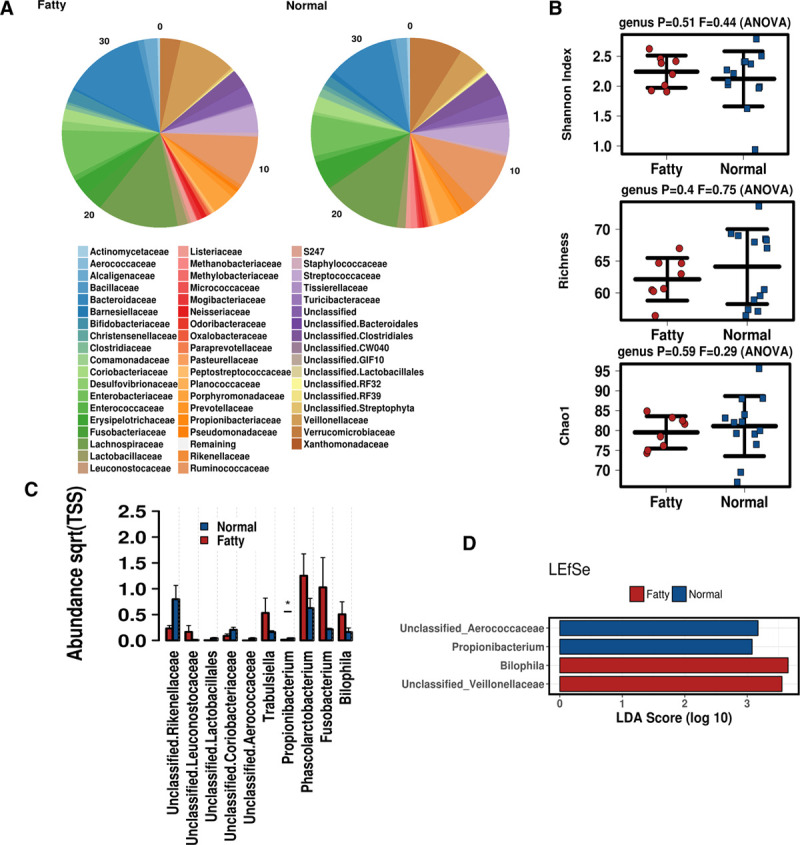
Microbiome associated with hepatic steatosis as a binomial. A, Relative abundance of the microbial communities at the family level. B, Alpha diversity determined by Shannon Index, Richness, and Chao1. C, Differentially abundant taxa at the family and genus level. D, Linear discriminant analysis of effect size was employed to determine differences in the microbiome associated with fatty vs normal biopsies.
Gut Microbiome by Liver Fib CRN Score
Finally, stool microbiome was analyzed based on biopsy fibrosis stage, for Stages 0, 1, and 2. Relative abundance of microbial communities at the family level is shown in Figure 4A. Similar to other biopsy scoring criteria, no statistically significant differences were observed for alpha diversity (Figure 4B); however, Richness and Chao1 indicated the presence of fibrosis may be associated with decreasing levels of diversity. Consistent with this concept, stage 2 fibrosis was associated with the significant loss of several taxa, but none that were significantly elevated. Specifically, there was a significant loss of Lachnospiraceae, Coprococcus, Ruminococcus, and Bacillus associated with stage 2 fibrosis (Figure 4C and D). No patients in the current cohort had stage 3 or 4 fibrosis.
FIGURE 4.
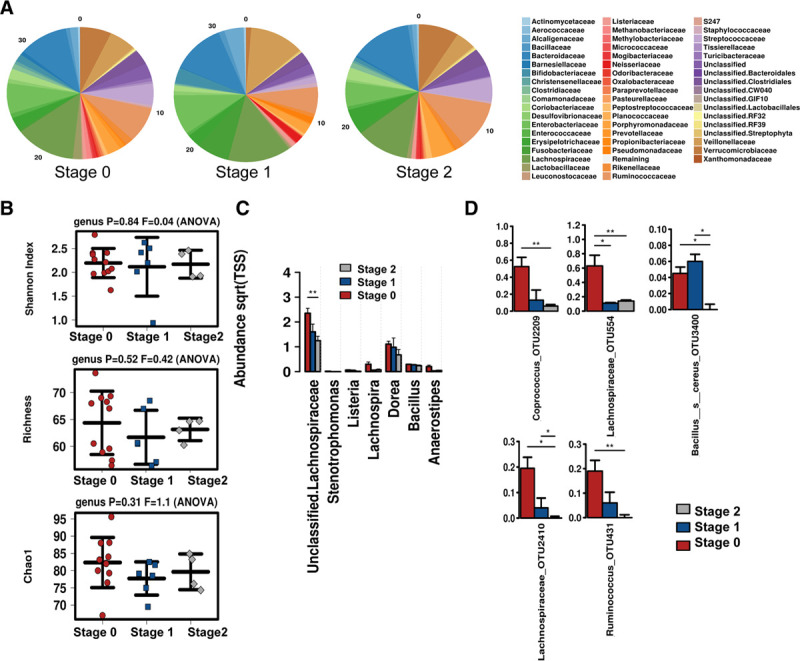
Microbiome associated with degree of biopsy Fibrosis stage. A, Relative abundance of the microbial communities at the family level. B, Alpha diversity shown by Shannon Index, Richness, and Chao1. C, Differentially abundant taxa at the family and genus and (D) significantly altered OTUs across fibrosis stages.
Status of Major Human Fecal Bacterial Groups Determined by qPCR
The 16S normalized qPCR analysis revealed that the relative abundance of Firmicutes was significantly (P < 0.05) decreased in patients with NAS scores of ≥5 (Figure 5A) as compared with patients with lower NAS scores (ie, 4 or below). On the contrary, Bacteroidetes were found to be significantly increased with higher NAS scores (P < 0.05). The relative abundance of Firmicutes and Bacteroidetes showed a strong correlation with liver fat percentage. Firmicutes were reversely correlated (r = −0.57, P = 0.007), whereas Bacteroidetes showed a positive correlation (r = 0.66, P = 0.001) with liver fat content (Figures S1–S10, SDC, http://links.lww.com/TXD/A289). The Bifidobacterium group also showed a reverse correlation (P = 0.037) with fat content (Figure 5B). Among the different Clostridium clusters, Clostridium cluster XIVab, Clostridium cluster XIVa, and Clostridium cluster IV increased in relative abundance in patients with higher NAS scores and higher liver fat content (Figure 5A and B). The relative abundance of Firmicutes was lower, whereas Bacteroidetes were higher in patients with NASH and NAFLD when compared to no-NASH or no-NAFLD patients, respectively (Figure 5C and D). The Firmicutes/Bacteroidetes ratios were found to be higher in patients without NAFLD or NASH as compared to patients diagnosed with NAFLD and NASH at the time of sample collection (Figure 6). The Random Forest (RF) model ranked different bacterial groups in terms of their association with NAS scores, steatosis grade, NAFLD diagnosis, and NASH diagnosis (Figure S11, SDC, http://links.lww.com/TXD/A289). The RF model revealed that Bacteroidetes and Clostridium cluster IV were the top 2 groups that were associated with the prediction of NAS status of patients (Figure S11A, SDC, http://links.lww.com/TXD/A289). Likewise, Bacteroidetes and Firmicutes were the top predictors of steatosis-grade–based grouping of the patients (Figure S11B, SDC, http://links.lww.com/TXD/A289). Moreover, the results from the RF model of qPCR data demonstrated Clostridium cluster IV and Bacteroidetes were associated with the prediction of NASH (Figure S11C, SDC, http://links.lww.com/TXD/A289), whereas Bacteroidetes and Clostridium cluster XIVab were found to be strongly associated with the prediction of NAFLD (Figure S11D, SDC, http://links.lww.com/TXD/A289).
FIGURE 5.
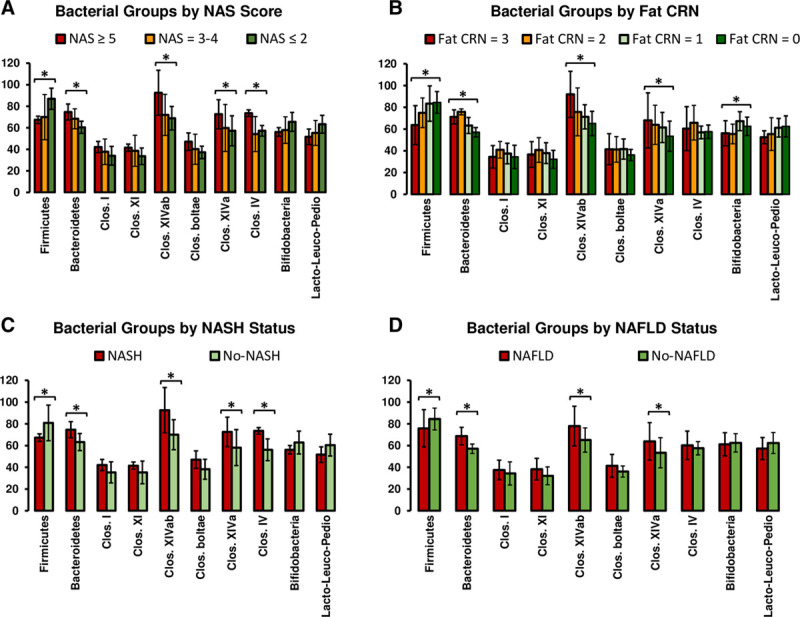
Relative abundance of fecal bacteria of liver transplant recipients. The stool samples were analyzed by qPCR. The results depict relative abundance of target bacteria normalized by universal 16S rRNA gene abundance representing all bacteria in a sample. Values are presented as mean ± SD of 2 experiments done in duplicates. Columns; mean; bars, SD. Columns with (*) indicate significantly different (P = 0.05) relative abundance values. CRN, clinical research network; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; NASH, nonalcoholic steatohepatitis; qPCR, quantitative PCR.
FIGURE 6.
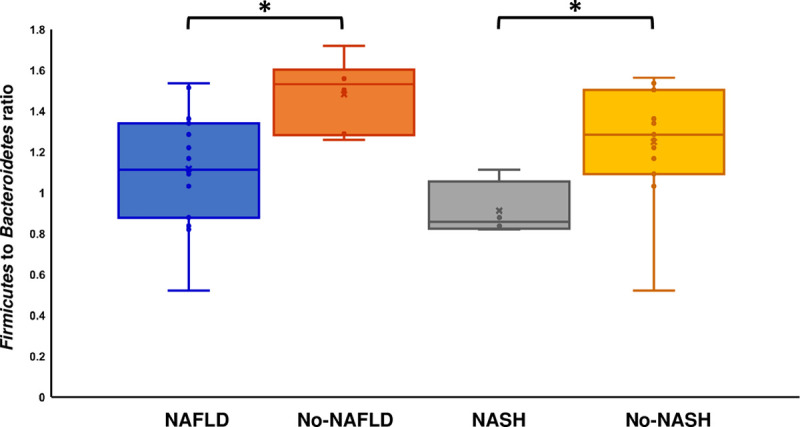
Firmicutes-to-Bacteroidetes ratio in the NASH and NAFLD patients. The Box and Whisker plot represents the ratios of the 16S rRNA gene normalized relative abundances of Firmicutes to Bacteroidetes. Values are presented as mean ± SD of 2 experiments done in duplicates. Columns; mean; bars, SD. Columns with (*) indicate significantly different (P = 0.05) values. NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis.
DISCUSSION
Our study provides qualitative microbiome data in patients with biopsy-proven recurrent NAFLD in patients with NASH requiring LT. In this cross-sectional study, we have noted a potential association of gut microbial diversity in developing recurrent NAFLD (based on liver biopsy beyond 1 y after their liver transplant). We noted a specific incremental trend in Akkermansia muciniphila with decreasing hepatic fat severity. On the other hand, an incremental trend for Fusobacteria was noted with high fat content based on liver histology. A significant loss of Lachnospiraceae and Bifidobacterium was noted in the gut microbiome as stage of fibrosis and percentage of fat content increased, respectively. When analyzing changes in relative abundance of the major phyla in the gut microbiome as determined by qualitative PCR, a significantly lower relative abundance of Firmicutes and higher relative abundance of Bacteroidetes ratio was noted in patients with greater fat percentage at 1-y liver biopsy. Overall, the Firmicutes-to-Bacteroides ratio was lower in patients with greater fat percentage on liver biopsy.
A. muciniphila is a mucin-degrading bacterium present in the mucous layer of the large intestine that has been reported to reverse fat mass gain, metabolic endotoxemia, adipose tissue inflammation, and insulin resistance associated with high-fat induced metabolic disorders.22,23 It has been suggested that it may play a protective role in gut-barrier function by improving the integrity of intestinal tight junctions through production of several bioactive lipids.24 Although A. muciniphila has been shown to be inversely correlated to obesity and metabolic syndrome,25,26 a study from Turkey recently showed that NASH patients had significantly decreased A. muciniphila in their gut microbiome when compared to healthy controls, even after adjusting for the body mass index and age.27 Thus, A. muciniphila may have a body mass index-independent effect on NAFLD progression. A. muciniphila also increases intestinal levels of endocannabinoids that control inflammation, the gut barrier, and gut peptide secretion.25 Notably, patients with ulcerative colitis have been shown to have lower levels of A. muciniphila,28,29 which further supports the bacterium’s anti-inflammatory properties. Loss of the protective function of Akkermansia due to decreasing abundance across the progression of disease may contribute to further dysbiosis and inflammation that augments the development and progression of steatosis. As the direct relationship between gut microbiota composition and the development of steatosis remains to be elucidated, the gold standard diagnostic tool for NASH remains liver biopsy.9 However, future studies with larger cohorts may show that qualitatively analyzing the gut microbiome could lead to earlier prediction of recurrent NAFLD development in post-LT patients.
Fusobacterium is capable of producing short-chain fatty acids (SCFAs)30,31 and are associated with increased expression and activation of inflammatory markers.32,33 Several studies have suggested Fusobacterium are upregulated in colorectal cancer, and increased abundance may serve as a marker for tumorigenesis and intestinal inflammation, with increased fecal SCFAs serving as a marker for increased bacterial metabolites resulting from dysbiosis.34-37 Fusobacterium has been shown to predominate in NASH patients when compared to NAFLD patients.38 The rising abundance of Fusobacterium seen in patients with increasing levels of steatosis may point to increased inflammation contributing to progression of disease. SCFAs that are products of fermentation by gut microbiome such as acetate, propionate, and even butyrate have been shown to influence T-cell differentiation, thus leading to an inflammatory cascade of events, 1 that could lead to disease progression of NAFLD.39 Therefore, SCFAs have been shown to play both an anti-inflammatory role and a proinflammatory role. Various studies have suggested that this broad spectrum of inflammatory activity may be concentration-dependent; that is, a low-concentration of SCFAs induces an anti-inflammatory response, whereas higher concentrations induce a proinflammatory response.40,41 Last, NAFLD patients have been shown to exhibit a higher level of fecal SCFA levels when compared to healthy controls and a positive correlation between SCFA levels and T-cell differentiation was also shown.42 There is also evidence in support of specific SCFAs such as gut microbiota metabolite sodium butyrate to attenuate progression of steatohepatitis in the animal model.43
Bacteroidetes is capable of producing SCFAs and is thought to predominate in overweight and obese subjects44 well as in NASH patients,45 although there are conflicting studies.46-48 Regardless, increased abundance of Bacteroidetes in patients with recurrence of NAFLD further suggests that the progression of obesity and post-LT recurrent NAFLD may share similar microbiome signatures. Firmicutes, on the other hand, has been known to have decreased relative abundance in the microbiome of diabetic49 and obese patients.44 Similarly, a 2013 study concluded that improvement of steatosis in NASH patients leads to decreased relative abundance of Firmicutes.50 In the current study, the decreased Firmicutes in recurrent NAFLD patients could be attributed to decreased Lachnospiraceae, a family within Firmicutes, that has been shown to have decreased abundance in NASH patients and obese patients.7 Decreased firmicutes has been noted in patients receiving LT 5–9 mo after transplant, although the study showed no association with fat content on liver biopsy.6
A decreased Firmicutes-to-Bacteroidetes ratio has been implicated in the pathogenesis of NAFLD51 and NASH,52 with a higher relative abundance of Bacteroidetes also being seen in NASH patients. This study showed increased Bacteroidetes are associated with development of NASH, independent of diabetes and metformin use.52 Studies have shown an increase in serum alcohol could contribute to the pathogenesis of NASH, which may be impacted by an increase in alcohol-producing bacteria. In the current study, we have shown that the relative abundance of Firmicutes was significantly (P < 0.05) decreased and the Bacteroidetes were significantly increased with higher NAS scores (NAS > 5). Loomba et al have reported as the disease progresses from mild/moderate NAFLD to advanced fibrosis, the Proteobacteria phylum has a statistically significant increase in abundance while the Firmicutes phylum decrease. The study by Loomba et al53 also have noted significantly lower Ruminococcus obeum CAG: 39, in addition to R. obeum, and Eubacterium rectale in advanced fibrosis than mild/moderate NAFLD. Current study is unable to assess the relative abundance of Firmicutes and the Bacteroidetes in relation to fibrosis as we have no patients with advanced fibrosis (stage 3 or 4). We did however find significant loss of Lachnospiraceae, Coprococcus, Ruminococcus, and Bacillus associated with stage 2 fibrosis. Additionally, the relative abundance of Firmicutes and Bacteroidetes showed a strong correlation with liver fat percentage, a finding similar to published studies in the nontransplant setting.52
In our study, higher levels of Bifidobacterium noted in patients with less fat content on liver biopsy could explain the protective roles it may have on the progression of NAFLD and obesity, as suggested by a recent 2018 study.54 However, the sample group analyzed by Nobili et al54 were pediatric patients with NAFLD in a nontransplant setting. Bifidobacterium belongs to the phylum Actinobacteria has been shown to be decreased in NASH and obese patients.7
The strength of the study is in its prospective design, and reliance on protocol-defined liver biopsy in the diagnosis of recurrent NAFLD. The limitations of the current study include small sample size and its single-center design, which may present inherent bias with regards to patient selection. This is a cross-sectional study and considering variability of microbiome and complexity associated with transplant model, a single sample per patient is possibly not ideal. However, having paired biopsies is a major strength. Additionally, our center uses a steroid-free immunosuppression protocol that may have an affect early on the gut bacterial diversity. The impact of immunosuppression on gut diversity is not analyzable in this small pilot study. The study also does not include a pretransplant baseline stool collection; hence, changes in the bacterial communes across transplant were not assessed.
The study of dysbiosis following LT is an emerging field of interest, especially in patients with NAFLD. To the best of our knowledge, this is the first study highlighting the potential role of gut microbiome in the pathogenesis of recurrent NAFLD and NASH in LT recipients. Despite limitations due to small sample size, the study clearly mirrors several important findings that merit further studies.
The highlights of the study include a potential protective role for Akkermansia muciniphila, Firmicutes, and Bifidobacterium in the development of recurrent NAFLD, whereas Fusobacteria and Bacteroidetes showed an association with recurrent NAFLD. The low ratio of Firmicutes to Bacteroidetes in our study reaffirms its potential role in the pathogenesis of NAFLD in posttransplant patients. These findings will be of great interest to the scientific community, and if confirmed in future studies, may help identify patients at increased risk for NAFLD following liver transplantation, potentially leading to a therapeutic strategy to protect patients from recurrent NASH.
Supplementary Material
Footnotes
Published online 10 November, 2020.
This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, and by the Methodist Health Care Foundation, Methodist University Hospital, Memphis.
The authors declare no conflicts of interest.
S.K.S. conceptualized the manuscript. P.B. and S.K.S. collected the samples. P.B., D.H., S.D., and V.K.M. collected the data. D.E.K. performed the histological analysis. P.B. and D.H. performed the stool sample analysis. S.K.S., P.B., and J.F.P. did the data and statistical analysis. S.K.S., S.D.K., and R.H. drafted the initial draft of the manuscript. All authors participated in intellectual input, data interpretation, critical revision, and approval of the final manuscript.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Bonora E, Targher G. Increased risk of cardiovascular disease and chronic kidney disease in NAFLD. Nat Rev Gastroenterol Hepatol. 2012; 9:372–381 [DOI] [PubMed] [Google Scholar]
- 2.Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018; 67:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D, Kim WR, Kim HJ, et al. Association between noninvasive fibrosis markers and mortality among adults with nonalcoholic fatty liver disease in the United States. Hepatology. 2013; 57:1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlton MR, Burns JM, Pedersen RA, et al. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011; 141:1249–1253 [DOI] [PubMed] [Google Scholar]
- 5.Samji NS, Verma R, Keri KC, et al. Liver transplantation for nonalcoholic steatohepatitis: pathophysiology of recurrence and clinical challenges. Dig Dis Sci. 2019; 64:3413–3430 [DOI] [PubMed] [Google Scholar]
- 6.Bajaj JS, Fagan A, Sikaroodi M, et al. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 2017; 23:907–914 [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013; 57:601–609 [DOI] [PubMed] [Google Scholar]
- 8.Schwimmer JB, Johnson JS, Angeles JE, et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology. 2019; 157:1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleiner DE, Brunt EM, Van Natta M, et al. ; Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005; 41:1313–1321 [DOI] [PubMed] [Google Scholar]
- 10.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995; 22:696–699 [DOI] [PubMed] [Google Scholar]
- 11.Kleiner DE, Brunt EM. Nonalcoholic fatty liver disease: pathologic patterns and biopsy evaluation in clinical research. Semin Liver Dis. 2012; 32:3–13 [DOI] [PubMed] [Google Scholar]
- 12.Higgins D, Pal C, Sulaiman IM, et al. Application of high-throughput pyrosequencing in the analysis of microbiota of food commodities procured from small and large retail outlets in a U.S. metropolitan area—A pilot study. Food Res Int. 2018; 105:29–40 [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee N, Bartelli D, Patra C, et al. Microbial diversity of source and point-of-use water in Rural Haiti—A pyrosequencing-based metagenomic survey. PLoS One. 2016; 11:e0167353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee N, Dowd SE, Wise A, et al. Diversity of bacterial communities of fitness center surfaces in a U.S. metropolitan area. Int J Environ Res Public Health. 2014; 11:12544–12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zakrzewski M, Proietti C, Ellis JJ, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017; 33:782–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacchetti De Gregoris T, Aldred N, Clare AS, et al. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011; 86:351–356 [DOI] [PubMed] [Google Scholar]
- 19.Furet JP, Firmesse O, Gourmelon M, et al. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009; 68:351–362 [DOI] [PubMed] [Google Scholar]
- 20.Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004; 70:6459–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasberger H, Gao J, Nagao-Kitamoto H, et al. Increased expression of DUOX2 is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology. 2015; 149:1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019; 25:1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneeberger M, Everard A, Gómez-Valadés AG, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015; 5:16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, Plovier H, Van Hul M, et al. Endocannabinoids–at the crossroads between the gut microbiota and host metabolism. Nat Rev Endocrinol. 2016; 12:133–143 [DOI] [PubMed] [Google Scholar]
- 25.Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013; 110:9066–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017; 23:107–113 [DOI] [PubMed] [Google Scholar]
- 27.Özkul C, Yalinay M, Karakan T, et al. Determination of certain bacterial groups in gut microbiota and endotoxin levels in patients with nonalcoholic steatohepatitis. Turk J Gastroenterol. 2017; 28:361–369 [DOI] [PubMed] [Google Scholar]
- 28.Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010; 105:2420–2428 [DOI] [PubMed] [Google Scholar]
- 29.Rajilić-Stojanović M, Shanahan F, Guarner F, et al. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013; 19:481–488 [DOI] [PubMed] [Google Scholar]
- 30.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014; 5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basson A, Trotter A, Rodriguez-Palacios A, et al. Mucosal interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front Immunol. 2016; 7:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Yang J, Feng Q, et al. Compositional and functional analysis of the microbiome in tissue and saliva of oral squamous cell carcinoma. Front Microbiol. 2019; 10:1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019; 18:1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saffarian A, Mulet C, Regnault B, et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. MBio. 2019; 10:e01315–e01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012; 22:292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Cai S, Ma Y. Association between Fusobacterium nucleatum and colorectal cancer: progress and future directions. J Cancer. 2018; 9:1652–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn KJ, Baxter NT, Schloss PD. Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere. 2016; 1:e00102–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep. 2018; 8:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Herck MA, Weyler J, Kwanten WJ, et al. The differential roles of t cells in non-alcoholic fatty liver disease and obesity. Front Immunol. 2019; 10:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kespohl M, Vachharajani N, Luu M, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T Cells. Front Immunol. 2017; 8:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim CH, Park J, Kim M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw. 2014; 14:277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rau M, Rehman A, Dittrich M, et al. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J. 2018; 6:1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D, Pan Q, Xin FZ, et al. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol. 2017; 23:60–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwiertz A, Taras D, Schäfer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010; 18:190–195 [DOI] [PubMed] [Google Scholar]
- 45.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016; 63:764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 48.Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013; 58:120–127 [DOI] [PubMed] [Google Scholar]
- 49.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010; 5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong VW, Tse CH, Lam TT, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis–a longitudinal study. PLoS One. 2013; 8:e62885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gkolfakis P, Dimitriadis G, Triantafyllou K. Gut microbiota and non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2015; 14:572–581 [DOI] [PubMed] [Google Scholar]
- 52.Sobhonslidsuk A, Chanprasertyothin S, Pongrujikorn T, et al. The association of gut microbiota with nonalcoholic steatohepatitis in Thais. Biomed Res Int. 2018; 2018:9340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017; 25:1054–1062.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nobili V, Putignani L, Mosca A, et al. Bifidobacteria and lactobacilli in the gut microbiome of children with non-alcoholic fatty liver disease: which strains act as health players?. Arch Med Sci. 2018; 14:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


