Abstract
Objectives
Serologic detection of prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is needed for definition of convalescent plasma donors, for confounding SARS-CoV-2 presentation, and for seroprevalence studies. Reliable serologic assays with independent validation are required.
Methods
Six SARS-CoV-2 antibody assays from Beckman Coulter, Euroimmun (IgG, IgA), Roche, and Siemens (Centaur, Vista) were assessed for specificity (n = 184), sensitivity (n = 154), and seroconversion in a defined cohort with clinical correlates and molecular SARS-CoV-2 results.
Results
Assay specificity was 99% or greater for all assays except the Euroimmun IgA (95%). Sensitivity at more than 21 days from symptom onset was 84%, 95%, 72%, 98%, 67%, and 96% for Beckman Coulter, Centaur, Vista, Roche, Euroimmun IgA, and Euroimmun IgG, respectively. Average day of seroconversion was similar between assays (8-10 d), with 2 patients not producing nucleocapsid antibodies during hospitalization.
Conclusions
SARS-CoV-2 nucleocapsid antibodies may be less reliably produced early in disease than spike protein antibodies. Assessment of convalescent plasma donors at more than 30 days from symptom onset and seroprevalence studies should use assays with defined sensitivity at time points of interest because not all assays detected antibodies reliably at more than 30 days.
Keywords: SARS-CoV-2, COVID-19, Antibody, Serology, Coronavirus
Key Points.
Independent validation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody automated assays has been lacking, and correlation of semiquantitative results among assays is needed.
High-throughput SARS-CoV-2 antibody assays demonstrate expected specificity and sensitivity for clinical use.
Assessment of SARS-CoV-2 antibody status at more than 30 days varies among assays that may significantly affect seroprevalence and convalescent plasma studies.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a human-pathogenic betacoronavirus that is the causative agent of coronavirus disease 2019 (COVID-19). This disease began in Wuhan, China, but rapidly spread worldwide and was ultimately declared a pandemic by the World Health Organization on March 11, 2020. As of July 28, 2020, there have been more than 16.3 million cases of COVID-19 with more than 650,000 deaths worldwide1 and more than 4.2 million cases with more than 147,000 deaths in the United States.2
We are still learning about the immune response to SARS-CoV-2 infection. Studies published to date suggest that IgM and IgA seroconversion occurs at approximately the same time, at days 5 to 14 after symptom onset, whereas IgG seroconversion occurs slightly later, at 10 to 14 days after symptom onset.3-10 In addition, neutralizing antibodies to both viral spike and nucleocapsid proteins have been found in patients.5,10,11 The longevity of this antibody response is also still under investigation. Preliminary work found that the half-lives of antibodies recognizing the nucleocapsid, spike protein, and receptor binding domain of the spike protein are 52, 81, and 83 days, respectively, with an estimated time to negativity of 50% of the seropositive population of 195, 532, and 260 days, respectively.12 More work is needed to further characterize the humoral response to SARS-CoV-2 infection.
There are several clinical reasons to measure SARS-CoV-2–specific antibodies. First, COVID-19 convalescent plasma is currently among the few available treatment options for COVID-19,13,14 and it is necessary to characterize the SARS-CoV-2 antibody response of these individuals before accepting them for plasma donation. They may be used in selected cases to assist in the diagnosis of patients who present later in their disease course or when the standard RNA detection methods are negative but clinical suspicion is high and other diseases have been ruled out. SARS-CoV-2 serologic assays may also be used for epidemiologic studies to determine the anti–SARS CoV-2 seroprevalence of a population and, relatedly, to determine the frequency of asymptomatic infections. Finally, with a concerted effort now focused on developing a SARS-CoV-2 vaccine, measuring antibody responses will be an essential component of determining vaccine efficacy.
To meet these testing needs, it is critical to have robust, standardized tests to detect and characterize SARS-CoV-2 antibody response. Many commercially available tests have been granted emergency use authorization (EUA) by the US Food and Drug Administration. Few published studies have independently evaluated and compared the performance characteristics of these high-throughput assays.15-17 More studies such as these will be essential to compare and potentially aggregate forthcoming results, especially from seroprevalence and vaccination studies.
As a large health care provider, our medical center has multiple instrumentation platforms and thus needed to validate multiple SARS-CoV-2 EUA serologic tests. We assessed the Beckman Coulter SARS-CoV-2 IgG, Euroimmun Anti–SARS-CoV-2 IgA, Euroimmun Anti–SARS-CoV-2 IgG, Roche Elecsys Anti–SARS-CoV-2 Total Antibody, Siemens Centaur SARS-CoV-2 Total Antibody, and Siemens Vista SARS-CoV-2 Total Antibody tests. All except the Roche assay target antibodies against epitopes on the spike protein. In contrast, Roche targets antibodies against nucleocapsid protein. In this study, we examined each assay’s specificity (including cross-reactivity with common coronaviruses) and sensitivity in a cohort of 341 patient samples. Chart review was performed to allow for assay consensus and clinical correlation. To our knowledge, this study is the first to independently assess 6 high-throughput commercial SARS-CoV-2 serologic assays head to head.
Materials and Methods
Specimens
Remnant sera from specimens and data received in the University of Pittsburgh Medical Center (UPMC) clinical laboratory for routine testing between January 1, 2020, and May 31, 2020, were used for the study under the auspices of UPMC Quality Assurance for Clinical Laboratories and the University of Pittsburgh institutional review board study 20040072, in compliance with the World Medical Association Declaration of Helsinki. These convenience sera were used to perform retrospective validation of the assays being considered for patient testing within the UPMC hospital network. Lithium heparin and serum samples with separator gel were centrifuged and stored at 4°C for up to 2 weeks before storage at −20°C. Samples were banked at −20°C for up to 3 months before analysis. Specimens came from critically ill in-patients, patients undergoing presurgical screening, or convalescent plasma donation or as per standard of care. Specimen inclusion for specificity was based on availability of frozen specimens from before SARS-CoV-2 was geographically present in our catchment area. Specimen inclusion for seroconversion required a specimen that was initially negative for SARS-CoV-2 IgG or IgA and for which we had at least 2 subsequent specimens and a known positive SARS-CoV-2 polymerase chain reaction (PCR) result. These specimens were assessed by the Anti–SARS-CoV-2 IgG and Anti–SARS-CoV-2IgA ELISAs (Euroimmun IgG and Euroimmun IgA; PerkinElmer Germany Diagnostics), which we had validated for use as prototype assays. Specimen inclusion for sensitivity was based on SARS-CoV-2 known PCR or clinical status and levels of SARS-CoV-2 IgG and IgA as assessed by the Euroimmun assay. Seroconversion samples were also included in the sensitivity assessment.
SARS-CoV-2 Assays
SARS-CoV-2 antibody assays were performed in UPMC clinical laboratories, which are Clinical Laboratory Improvement Amendments certified, high-complexity laboratories. The sample cohort was deidentified and aliquoted for assessment with the following assays: Beckman Coulter SARS-CoV-2 IgG (AU5800 analyzer; Beckman Coulter), Roche Elecsys Anti–SARS-CoV-2 Total (ecobas 400 analyzer; Roche), Siemens SARS-CoV-2 Total (Centaur XP analyzer; Siemens-C), and Siemens SARS-CoV-2 Total Antibody (Vista 1500 analyzer; Siemens-V). Euroimmun Anti–SARS-CoV-2 IgA and Euroimmun Anti–SARS-CoV-2 IgG were performed manually and read on the Bio-Rad Laboratories iMark Microplate Absorbance Reader. All assays were run according to the manufacturer’s instructions. All assays use units that are generated by comparison to an internal calibrator or standard; when referring to assay results collectively, we refer to these units as index values for simplicity. Beckman Coulter, Siemens-C, and Roche use an index of greater than 1.0 for positivity; Euroimmun IgA and IgG positivity uses an index greater than 1.1; and Siemens-V positivity uses an index greater than 1,000.
Other Laboratory Testing
PCR detection of SARS-CoV-2 RNA was reported in patient charts as standard of care clinical testing, as detected using the Cepheid GeneXpert or a laboratory-developed test based on the Centers for Disease Control and Prevention (CDC) protocol. Measurement of positivity for antibodies against cytomegalovirus, Epstein-Barr virus, HIV, herpes simplex virus types 1 and 2, syphilis, and toxoplasmosis was performed on the BioPlex 2200 (Bio-Rad). Determination of antibodies against hepatitis B and hepatitis C viruses was performed on the Centaur XP (Siemens). Antinuclear antibodies were determined by indirect immunofluorescence on the Helios (Aesku Group), and varicella zoster IgM was assessed using the Wompole ELISA (Abbott). Respiratory viruses including the endemic coronaviruses (NL63, 229E, OC43, HKU1) were tested on the ePlex respiratory pathogen panel (RVPE; GenMark Dx).
Statistical Analysis
Figures and tables were created in Prism Graph Pad (version 8.0) and Excel (Microsoft). Diagnostic sensitivity calculations were calculated on the Anaconda 3 platform using the pandas 1.0.3 library within Python 3.7.7.
Results
Specificity Testing
To assess the specificity of the SARS-CoV-2 antibody assays, we first tested specimens with prior coronavirus (NL63, 229E, OC43, HKU1) molecular positivity for possible antibody cross-reactivity (CoV; Table 1). Of the 12 blood samples tested, one was collected the day after molecular testing; the remaining 11 were collected 5 to 28 days after molecular testing, allowing for assessment of cross-reactivity in the convalescent window. No cross-reactivity was exhibited except in 2 cases using the Euroimmun IgA assay. We then assessed 33 samples from patients with respiratory symptoms that warranted an ePlex respiratory pathogen panel. These specimens were drawn from 0 to 22 days after molecular testing, allowing for assessment of acute and convalescent cross-reactivities (Table 1). One specimen exhibited cross-reactivity in the Beckman Coulter and both Euroimmun assays. This patient was critically ill with idiopathic pulmonary fibrosis and serial lung transplants, in addition to stage III acute kidney injury. One RVPE specimen had insufficient volume to be measured on the Roche assay but was measured on all other platforms and demonstrated no cross-reactivity.
Table 1.
Assay Specificity
| Specimen Description | No. | Beckman Coulter IgG | Siemens Centaur Total | Siemens Vista Total | Roche Total | Euroimmun IgA | EuroimmunIgA |
|---|---|---|---|---|---|---|---|
| ANA | 18 | 0 | 0 | 0 | 0 | 2 | 0 |
| CMV (IgG) | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| CoV | 12 | 0 | 0 | 0 | 0 | 2 | 0 |
| EBV | 11 | 0 | 0 | 0 | 0 | 1 | 0 |
| HBV | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCV | 10 | 0 | 0 | 0/9a | 0 | 0 | 0 |
| Healthy volunteers | 24 | 0 | 0 | 0 | 0 | 1 | 0 |
| HIV | 19 | 0 | 1 | 0 | 0 | 1 | 0 |
| HSV1/2 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| RPR/SYPH | 21 | 0 | 0 | 0 | 0 | 2 | 0 |
| Rubella | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| RVPE | 33 | 1 | 0 | 0 | 0/32a | 1 | 1 |
| TOXG | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| VZV | 8 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 184 | 1 | 1 | 0 | 0 | 10 | 2 |
| % Specificity | 99.46 | 99.46 | 100.00 | 100.00 | 94.57 | 98.91 |
ANA, antinuclear antibodies; CMV, cytomegalovirus; CoV, coronavirus; EBV, Epstein-Barr virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HSV1/2, herpes simplex virus 1 or 2; RPR, rapid plasma reagin; SYPH, syphilis; TOXG, toxoplasmosis; VZV, varicella zoster virus.
aOne specimen did not have sufficient quantity for this assay; total sample size for these tests is denoted in the denominator.
We also assessed samples that were positive for other infectious disease serologies, autoimmunity markers, and healthy volunteers (Table 1). The Euroimmun IgA demonstrated several cross-reactivities (10/184) in samples that tested positive for antinuclear antibodies, Epstein-Barr virus, HIV, syphilis, coronaviruses, and RVPE. The Euroimmun IgG exhibited cross-reactivity to a varicella-zoster–reactive specimen, and the Siemens-C had 1 cross-reaction on an HIV-reactive specimen. Specificity was greater than 99% for the Beckman Coulter, Roche, Siemens-C, and Siemens-V assays. The Euroimmun IgG assay had specificity of 98.9%; however, cross-reactivity was noted for the Euroimmun IgA assay, with specificity of 94.6%. No single specimen was shown to have cross-reactivity in a majority of the assays.
Seroconversion
Samples from acutely infected and critically ill patients with SARS-CoV-2 with serial remnant specimens that initially tested negative for either IgG or IgA antibodies by the Euroimmun ELISA were assayed by other tested methods (n = 11). These were sequential patients and represented all patients who met these inclusion criteria at the time of cohort consolidation. Day of symptom onset was determined by chart review. It is notable that symptom onset can be vague in chart reviews (eg, about a week ago), but we found that the date from PCR positivity was less reliable because early in the pandemic, patients often were not tested until they were critically ill. For these 11 patients, symptom onset was well defined in the patient chart. Known viral exposure was often significantly earlier than symptom onset.
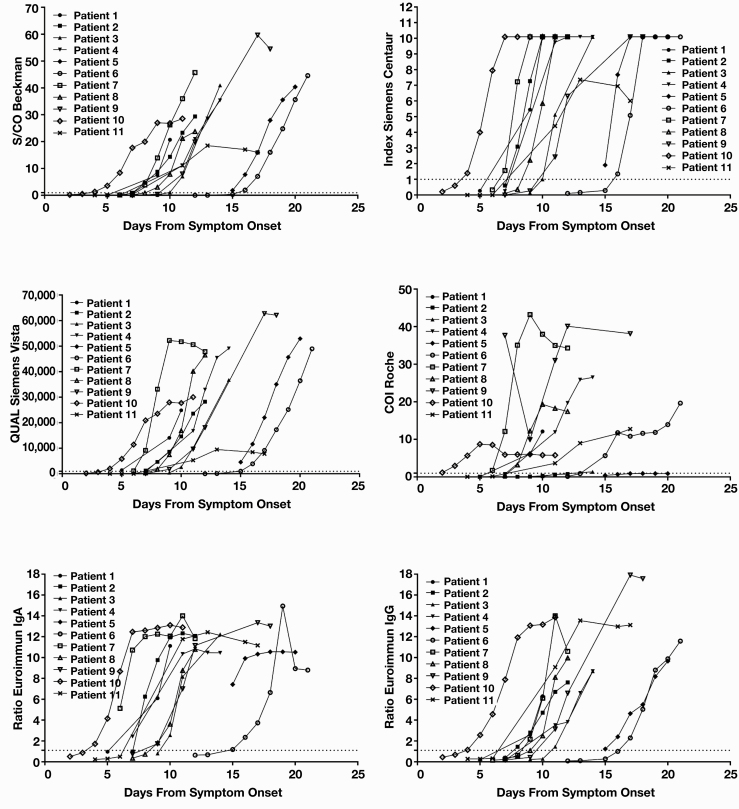
Index values were graphed against days from symptom onset for each methodology to visualize seroconversion by method Figure 1. The Euroimmun assays were outside a linear measurement range at a ratio greater than approximately 10. The Siemens-C assay has an upper limit of 10.0 index, and samples greater than 10 are graphed as 10.1. Examination of average day to seroconversion between assays Table 2 demonstrated that the Roche and Siemens-V assays detected seroconversions earliest, followed by the Euroimmun IgA, Siemens-C, Beckman Coulter, and Euroimmun IgG assays. In all patients without exogenous immunosuppressants, seroconversion occurred in less than 2 weeks. Assays had a difference in day of seroconversion that ranged from 1 to more than 5 days between assays for each patient (Table 2).
Figure 1.
Seroconversion by assay. Serial remnant samples from 11 patients hospitalized for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection were assayed for presence of antibodies against SARS-CoV-2 spike protein by the Euroimmun IgG and IgA assays. The first 11 patients with a first sample negative for either IgG or IgA and at least 2 subsequent samples were included in this cohort. Antibody results for these patients separated by assay are plotted vs days from symptom onset. Dotted line represents assay cutoff for positivity. The units of measurement vary by assay and are indicated on their respective y-axes.
Table 2.
Day of Seroconversion
| Patient | Beckman Coulter IgG | Siemens Centaur Total | Siemens Vista Total | Roche Total | Euroimmun IgA | Euroimmun IgG |
|---|---|---|---|---|---|---|
| 1 | 7 | 7 | 5 | 7 | 7 | 7 |
| 2 | 8 | 8 | 7 | >12 | 8 | 8 |
| 3 | 10 | 10 | 10 | 13 | 10 | 11 |
| 4 | 9 | 9 | 9 | 9 | 7 | 9 |
| 5 | 15 | 15 | 15 | >20 | 15 | 15 |
| 6 | 16 | 16 | 15 | 14 | 15 | 17 |
| 7 | 8 | 7 | 6 | 6 | 6 | 9 |
| 8 | 9 | 9 | 8 | 8 | 9 | 10 |
| 9 | 10 | 10 | 8 | 7 | 9 | 10 |
| 10 | 4 | 4 | 4 | 2 | 4 | 4 |
| 11 | 8.5 | 8.5 | 8.5 | 8.5 | 8.5 | 8.5 |
| Average | 9.5 | 9.4 | 8.7 | >9.7 | 9.0 | 9.9 |
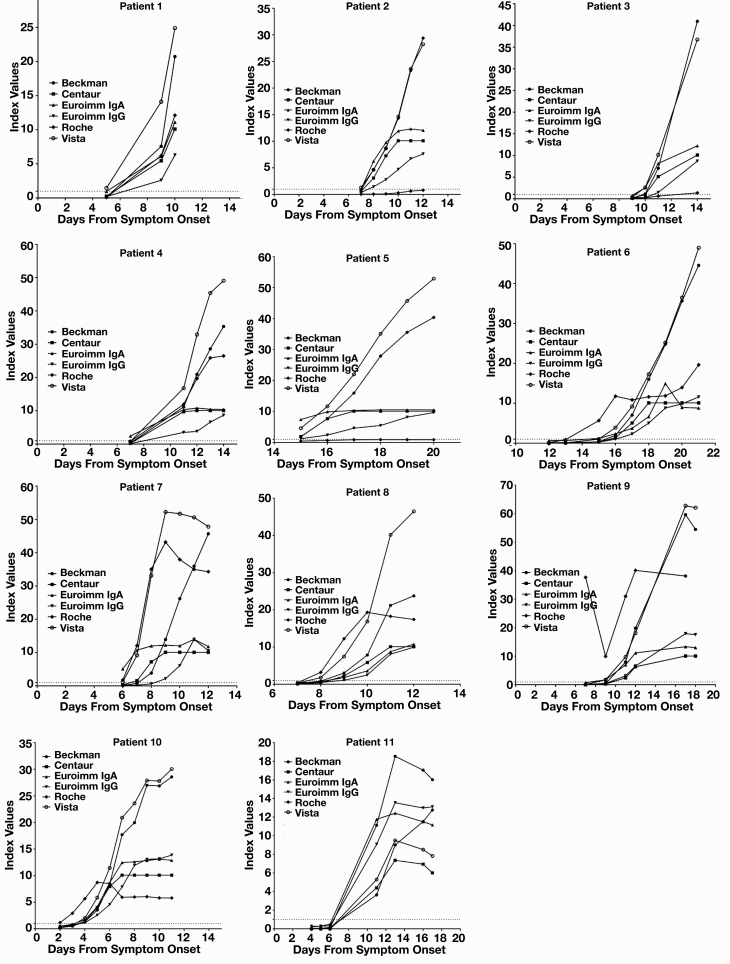
Method differences are more apparent when index values are graphed by patient against days from symptom onset Figure 2. Assays have different measurement ranges, and truncated measurement is visible in the Euroimmun IgG and IgA assays and the Siemens-C assay. Patients 2 and 5 did not produce a detectable nucleocapsid antibody response, and patient 3 appears to have an attenuated antibody response to nucleocapsid antigen.
Figure 2.
Seroconversion by patient. Serial remnant samples from 11 patients hospitalized for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection were assayed for presence of antibodies against SARS-CoV-2 spike protein by the Euroimmun IgG and IgA assays. The first 11 patients with a first sample negative for either IgG or IgA and at least 2 subsequent samples were included in this cohort. Antibody results for each assay separated by patient are plotted vs days from symptom onset. Dotted line at 1.0 represents assay cutoff for positivity for Beckman Coulter, Siemens, and Roche. The cutoff value for Euroimmun is 1.1, which is approximated by the dotted line. Vista QUAL units have been brought within the range of other assays by dividing by 1,000, with resultant assay cutoff for positivity of 1.0.
Assay Sensitivity
Serologic assay sensitivity should be assessed, if possible, by examining antibody levels as a function of time from infection. The samples available to us did not include many from patients who were symptomatic but confirmed those who were PCR negative; therefore, our assessment of assay sensitivity has inherent biases. All samples used in the seroconversion assessment were also included in the sensitivity assessment (n = 69). Additional samples that had SARS-CoV-2 IgG and IgA testing performed as part of their clinical care (n = 15), presurgical bloodwork (n = 12), or for donation of convalescent plasma (n = 58) were also included for a total of 154 specimens that were tested in all assays. Chart review was performed for all specimens to retrieve available molecular SARS-CoV-2 testing, days from symptom onset, and clinical COVID-19 diagnosis if possible. If chart information was not available for patients donating convalescent plasma, they were assigned days from symptom onset of more than 30 days (graphed as 30 days) because this was a prerequisite for enrolling patients for screening. Percentage agreement was assessed among all samples, samples that were confirmed as PCR positive for SARS-CoV-2, samples that were presumed molecularly positive (patients donating convalescent plasma), and samples at varying days from symptom onset that were both PCR confirmed and presumed positive. Agreement for each assay was assessed between the assay and the consensus of all assays, and agreement for each assay was compared with the clinical diagnosis Table 3. Assay consensus was assigned the value split when the assays were evenly divided and both reactive and nonreactive were considered to be in agreement. The clinical diagnosis was determined by PCR confirmation, and chart review for physician-assigned or -excluded diagnosis of SARS-CoV-2 infection. If chart review was unclear, the clinical diagnosis was assessed as “equivocal” and either R or NR were considered in agreement. For specimens with either molecular SARS-CoV-2 positivity or clinical diagnosis for convalescent plasma donation at more than 14 days after symptom onset, the sensitivity compared with clinical status was 86%, 95%, 78%, 90%, 73%, and 96% for Beckman Coulter, Siemens-C, Siemens-V, Roche, Euroimmun IgA, and Euroimmun IgG, respectively. The concordance between assays was 97%, 97%, 86%, 85%, 82%, and 99% for Beckman Coulter, Siemens-C, Siemens-V, Roche, Euroimmun IgA, and Euroimmun IgG, respectively.
Table 3.
Qualitative Positive Agreement Between Assays (%)
| No. | Beckman Coulter | Siemens Centaur | Siemens Vista | Roche | Euroimmun IgA | EuroimmunIgG | |
|---|---|---|---|---|---|---|---|
| Assay vs assay consensus | |||||||
| All specimens | 154a | 96 | 97 | 90 | 83 | 84 | 94 |
| PCR positive | 100a | 96 | 98 | 90 | 78 | 85 | 95 |
| PCR and presumptive positive | 132a | 96 | 98 | 89 | 81 | 86 | 95 |
| >7 d | 114a | 97 | 97 | 89 | 81 | 86 | 96 |
| >14 d | 74a | 97 | 97 | 86 | 85 | 82 | 99 |
| >21 d | 57 | 96 | 96 | 84 | 93 | 77 | 98 |
| Assay vs clinical status | |||||||
| All specimens | 151a | 81 | 84 | 79 | 81 | 72 | 81 |
| PCR positive | 100a | 78 | 78 | 79 | 75 | 68 | 77 |
| PCR and presumptive positive | 132a | 80 | 83 | 78 | 80 | 70 | 81 |
| >7 d | 114a | 87 | 91 | 83 | 84 | 75 | 89 |
| >14 d | 74a | 86 | 95 | 78 | 90 | 73 | 96 |
| >21 d | 57 | 84 | 95 | 72 | 98 | 67 | 96 |
PCR, polymerase chain reaction.
aRoche is n-1.
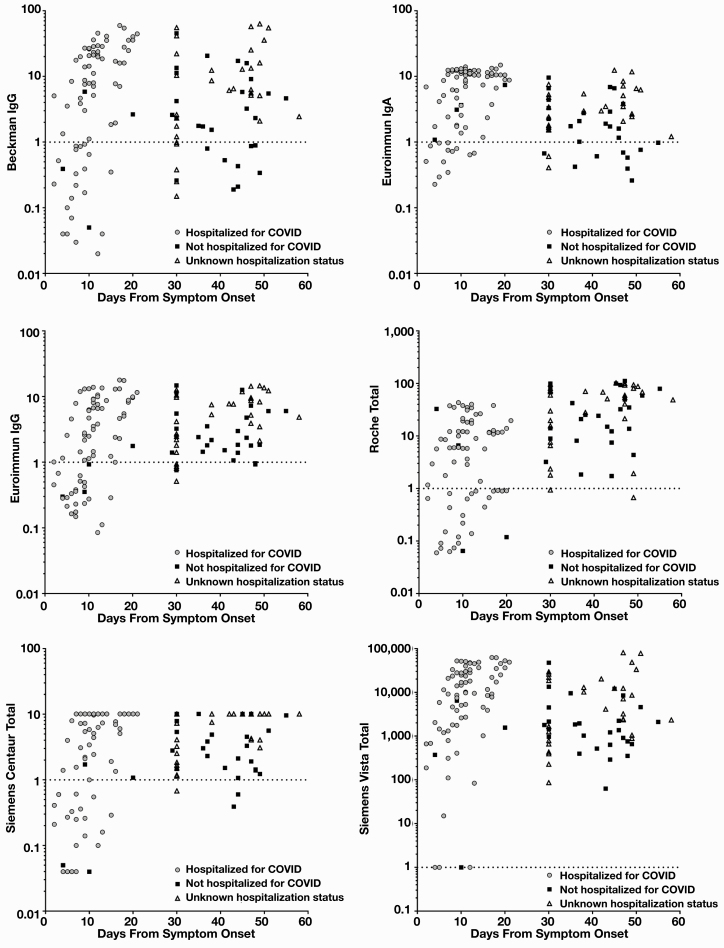
To allow for improved data visualization, days from symptom onset were graphed against the log index value to understand general SARS-CoV-2 antibody detection over time between assays, all 135 specimens with documented molecular SARS-CoV-2 testing, and clinical diagnosis for convalescent plasma donation Figure 3. At both earlier and later time points, antibody detection varies between assays. At more than 30 days, the Beckman Coulter, Euroimmun IgA, and Siemens-V assays detected fewer prior infections (46, 48, and 44 of 76, respectively; 6 and 7 equivocal by Beckman Coulter and Euroimmun IgA, respectively) than the Roche, Siemens-C, and Euroimmun IgG assays (60, 59, and 58 of 76 respectively; 11 equivocal by Euroimmun IgG).
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody reactivity compared with symptom onset. Remnant samples from patients hospitalized for SARS-CoV-2, having SARS-CoV-2 antibody testing performed as part of their medical care, or being assessed for convalescent plasma donation were assessed (n = 135). Patients being screened for convalescent plasma donation without chart notes with symptom onset information or polymerase chain reaction testing information were assigned a value of 30 days after symptom onset, as this was the minimum amount of time required before screening for plasma donation. Patients were classified as hospitalized for coronavirus disease (COVID), not hospitalized for COVID, or unknown status based on chart review. Results for this cohort are plotted separated by assay vs days from symptom onset. Dotted line represents assay cutoff for positivity.
Discussion
Serologic identification of past exposure to SARS-CoV-2 is currently of clinical and epidemiologic interest, with a future interest in assessing antibody response in vaccine trials.18,19 Specific, sensitive, and reliable assays are required to meet these needs. Working in a large academic medical center, we sought to validate and compare the new assays that would be implemented across our hospital sites or that had been initially assessed early in the pandemic.
Specificity testing indicated, as expected, that detection of anti–SARS-CoV-2 IgA lacked sufficient specificity as a stand-alone screening test. All other tests demonstrated sufficient specificity of approximately 99% (Table 1), although one critically ill patient with idiopathic pulmonary fibrosis, serial lung transplants, and stage III acute kidney injury demonstrated cross-reactivity on both the Beckman Coulter and Euroimmun IgG platforms but not in any of the total antibody platforms. The specificity cohort was chosen to include specimen types known to have higher cross-reactivity with serologic testing than the general population. Because of RNA testing limitations early in the pandemic, there were an insufficient number of molecularly tested negative samples to provide the basis for adequate specificity assessment. The samples we used were presumed negative, given their collection before known SARS-CoV-2 presence in our area, and assay concordance appears to confirm that status. It is a limitation that molecular SARS-CoV-2 testing was not performed to confirm negativity in these samples. In addition, we were able to address the concerns about cross-reactivity with the common coronaviruses by assessing patient specimens from early convalescent infection that would be expected to have higher antibody titers, as these titers may wane quickly. We found no evidence of cross-reactivity except in the Euroimmun IgA assay.
A limitation of our specificity studies is the small study size. Further studies are needed, particularly for use of these assays in large-scale serosurveys, in which even the minor cross-reactivities noted in this article will lead to large inaccuracies in prevalence estimates due to low pretest probabilities. Consequently, to determine true positives, a validation assay is needed. Because we found that the individual false positives (or cross-reactivities) did not align across platforms, we suggest that any of the IgG or total Ig platforms can be considered orthogonal and thus used to confirm specificity, in accordance with the current CDC interim testing guidelines for SARS-CoV-2 antibody testing.20
Initial assessment of seroconversion was markedly different between patients based on reported time from symptom onset but similar between assays with only minor average differences overall (Table 2). Patient 10 appears to have had early seroconversion, but this patient was transferred to the hospital from an assisted care facility with other positive cases of SARS-CoV-2 where this patient’s initial mild symptoms may have also been overlooked as they presented with significant hypoxia (Figure 1, Table 2). Patients 5 and 6 had later seroconversion than other patients (Figure 1, Table 2). Patient 5 was immunosuppressed with liver and renal transplantation history, likely accounting for a delayed seroconversion. Patient 6 did not have a history of immunosuppression but was recently diagnosed with an adenocarcinoma of the colon, which may have contributed to either secondary immune suppression or unclear symptom onset due to comorbid conditions. All of the 11 patients proceeded to critical illness, which likely accounts for the early trending seroconversion overall and is in keeping with other studies that demonstrate earlier seroconversion for severe disease but later seroconversion for mild and asymptomatic disease.3,21-23
The assays generally demonstrate high index levels of antibody response in these critically ill patients, which is in keeping with current findings that severe illness tends to correlate with high antibody titers.3,21-23 Although some assays had reasonably low upper ranges for antibody index levels, the Beckman Coulter, Siemens-V, and Roche assays all demonstrated that antibody titers for several patients continued to rise through the course of their in-patient stay, whereas other patients appeared to plateau (Figure 2). We did not find any correlations with disease course and continued antibody rise or plateau, although the number of patients was small and thus could miss a small but significant correlation. Interestingly, critically ill patients did not seem to have a uniform antibody response to nucleocapsid protein, as detected by the Roche assay. Unfortunately, we had only 1 nucleocapsid antibody assay; consequently, between-assay comparisons could not be made. Two patients (patients 2 and 5) failed to generate detectable nucleocapsid antibodies despite robust antibodies against the receptor-binding or spike protein domains. Patient 3 barely produced detectable nucleocapsid antibody at day 14 after symptom onset, despite seroconversion for antibodies against spike protein from around day 10 for most assays. This is curious because one would expect that patients who are severely ill would have more replication, leading to greater nucleocapsid presentation. Differential antibody responses have been noted in patients infected with SARS-CoV-2; however, T-cell responses to spike, N, and M appear to be codominant.24 It is unclear if the differences between immune responses are temporal, intrapatient, or method dependent, and further larger scale studies are needed to tease out these differences.
Our assessment of overall assay sensitivity found significant differences, particularly for samples more than 21 days from symptom onset (Table 3, Figure 3). Euroimmun IgA detection appeared to begin to wane in many patients after 30 days, which is not unexpected and may serve to aid in identification of more recent viral exposure. This identification could be of use as we progress to subsequent waves of SARS-CoV-2 outbreaks and may have an epidemiologic need to differentiate exposure in prior vs current outbreaks. The Roche antinucleocapsid assay appeared to continue to robustly detect prior SARS-CoV-2 infection at later time points. Our time points are likely insufficient to detect significant titer waning in nucleocapsid, as the recently reported half-life of these titers is estimated to be approximately 2 months.12 For the subgroup at more than 21 days, there was heterogeneity among the anti–spike protein assays, despite close agreement between assays when earlier time points were included (Table 3). The Euroimmun IgG and Siemens-C assays both detected antibody response in most specimens at more than 21 days (96% and 95%, respectively), but the Beckman Coulter and Siemens-V assays missed a larger percentage of cases (sensitivity of 84% and 72%, respectively). This result raises concerns that seroprevalence studies may vary significantly based on the serologic assay utilized, even when the assays are from reliable manufacturers with proven methodologies and have similar targets and initial specificity and sensitivity measures. The Siemens-C and Siemens-V assays, despite both being total SARS-CoV2 antibody assays against the same antigen, provided different responses at more than 21 days. This difference may be due to method differences between the platforms and highlights the need to robustly test assays in the appropriate population for intended use. It is a significant limitation that we do not have patients that span the time period from acute seroconversion to late convalescence or pairwise comparison. In addition, patients with symptom onset at less than 21 days were predominantly hospitalized for COVID-19, whereas patients with symptom onset at more than 21 days were predominantly unhospitalized with a mild disease course. A mild disease course often produces a lower titer antibody response than severe illness from SARS-CoV-2,12,23 and that may bias these data.
Overall, we have shown that all manufacturers’ IgG or total anti–SARS-CoV2 assays have acceptable specificity. The serologic detection of SARS-CoV-2 spike protein antibodies in critical illness can be detected in under 2 weeks for patients without immune complications, although nucleocapsid antibodies may be less reliably produced early in disease. Assessment of convalescent plasma donors at more than 30 days from symptom onset should be undertaken on assays that have been correlated with plaque reduction neutralization assays; it is unclear if the differences noted at later time points in this study may reflect loss of neutralizing antibodies or limitations of the assays. Seroprevalence studies should likewise ensure that the sensitivity of the assay used is known or calculated for the time from infection being assessed in the study. These findings should be further corroborated by larger studies or by meta-analysis as additional information becomes available.
Acknowledgments
The authors thank the following technologists who were involved in running specimens and who provided critical administrative support: Chad Chilleo, Darla Lower, David Jordan, Diane DeGrave, Gina Pillage, Greg Acero, Jayne Rasmussen, Jayne Zona, Jeff Tischler, Jim Bird, John Mahon, Katie Mulvey, Mary Jane Horenzy, Mary Yost, Spiros Giannoutsos, Stephanie Phillips, Tammy Garret, and Vicki Hoover.
References
- 1. World Health Organization. WHO coronavirus disease (COVID-19) dashboard https://covid19.who.int/. Accessed July 29, 2020.
- 2. Centers for Disease Control and Prevention. CDC COVID data tracker: United States COVID-19 cases and deaths by state. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html. Accessed July 29, 2020.
- 3. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020. DOI: 10.1093/cid/ciaa344/5812996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qu J, Wu C, Li X, et al. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. DOI: 10.1093/cid/ciaa489/5825506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kai-Wang To K, Tak-Yin Tsang O, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020. DOI: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lou B, Li T-D, Zheng S-F, et al. Early view serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020. DOI: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71:778-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020:2020.03.24.20042382. DOI: 10.1101/2020.03.24.20042382. [DOI] [Google Scholar]
- 9. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845-848. [DOI] [PubMed] [Google Scholar]
- 10. Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. SSRN Electronic J. 2020. DOI: 10.2139/ssrn.3566211. [DOI] [Google Scholar]
- 11. Klimstra WB, Tilston-Lunel NL, Nambulli S, et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected, hospitalized COVID-19 patients. J Gen Virol. 2020. DOI: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grandjean L, Mbbs AS, Torres A, et al. Humoral response dynamics following infection with SARS-CoV-2. DOI: 10.1101/2020.07.16.20155663. [DOI]
- 13. Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye M, Fu D, Ren Y, et al. Treatment with convalescent plasma for COVID‐19 patients in Wuhan, China. J Med Virol. 2020;92:1890-1901. DOI: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weidner L, Gänsdorfer S, Unterweger S, et al. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J Clin Virol. 2020;129:104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Theel ES, Harring J, Hilgart H, et al. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2 2020. https://jcm.asm.org/content/58/8/e01243-20. Accessed July 29, 2020.
- 17. Tang MS, Hock KG, Logsdon NM, et al. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem. 2020;66:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Theel ES, Slev P, Wheeler S, et al. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol. 2020;58 DOI: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deeks JJ, Dinnes J, Takwoingi Y, et al. ; Cochrane COVID-19 Diagnostic Test Accuracy Group Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Centers for Disease Control and Prevention. Interim guidelines for COVID-19 antibody testing https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html. Accessed July 31, 2020.
- 21. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58 DOI: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489-1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]