Abstract
Interleukin 6 (IL-6) is a pro-inflammatory cytokine that stimulates acute phase responses, hematopoiesis and specific immune reactions. Recently, it was found that the IL-6 plays a vital role in the progression of COVID-19, which is responsible for the high mortality rate. In order to facilitate the scientific community to fight against COVID-19, we have developed a method for predicting IL-6 inducing peptides/epitopes. The models were trained and tested on experimentally validated 365 IL-6 inducing and 2991 non-inducing peptides extracted from the immune epitope database. Initially, 9149 features of each peptide were computed using Pfeature, which were reduced to 186 features using the SVC-L1 technique. These features were ranked based on their classification ability, and the top 10 features were used for developing prediction models. A wide range of machine learning techniques has been deployed to develop models. Random Forest-based model achieves a maximum AUROC of 0.84 and 0.83 on training and independent validation dataset, respectively. We have also identified IL-6 inducing peptides in different proteins of SARS-CoV-2, using our best models to design vaccine against COVID-19. A web server named as IL-6Pred and a standalone package has been developed for predicting, designing and screening of IL-6 inducing peptides (https://webs.iiitd.edu.in/raghava/il6pred/).
Keywords: Interleukin 6 (IL-6), pro-inflammatory cytokine, machine learning, COVID-19, computer-aided prediction
Introduction
The Interleukin 6 gene encodes the pleiotropic cytokine Interleukin 6 (IL-6). It is also known by some alternate names, such as B cell stimulatory factor-2, interferon-β2 (IFN-β2) and plasmacytoma growth factor [1]. It is a multifunctional cytokine that plays a pivotal role in both innate and adaptive immune response [2], numerous inflammatory diseases, rheumatoid arthritis [3], hematopoiesis [4], acute phase responses [5] and organ development [6]. It is mainly produced in response to infections and tissue damage [7, 8]. The production of IL-6 is linked with various cell types, such as macrophages, dendritic cells, mast cells, fibroblasts, endothelial cells, T, and B cells [8, 9]. IL-6 plays a crucial role in regulating many physiological functions, such as the cardiovascular system, central nervous system, immune system, etc. [4] (Figure 1).
Figure 1.
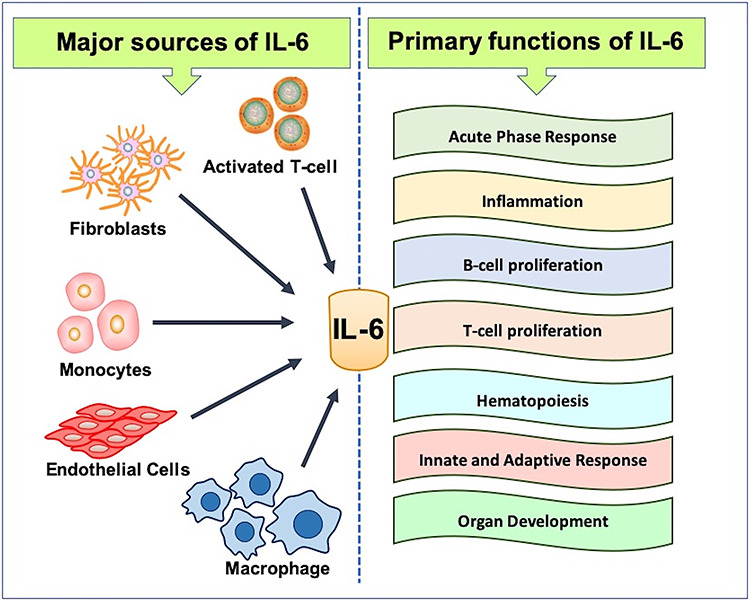
Schematic representation of the mode of IL-6 secretion and its primary functions.
Emerging evidence reveals that the dysregulation of IL-6 leads to several disease states, including various types of cancer development, progression and metastasis [4, 10]. Many studies show that elevated levels of IL-6 are related to a high risk of cancer and other disease conditions such as insulin resistance [11], asthma [12], coronary heart disease [13], advanced-stage cancer and can also work as a prognostic marker for cancer [14, 15]. Previous retrospective studies suggested that the disease progression in the recent outbreak of COVID-19 might be due to the cytokine storm or cytokine release syndrome [16, 17], which is the abnormal release of circulating cytokines [18]. The drastically elevated levels of IL-6 and other pro-inflammatory cytokines (e.g. IL-1, IL-8, IL-12) played a crucial role in deteriorating the health of COVID-19 patients [19, 20]. The increased levels of IL6 in critically ill patients of COVID-19 infection may develop severe pneumonia to acute respiratory distress syndrome, eventually causing multisystem organ failure and leading to high mortality [21, 22]. The elevated concentration of IL-6 constitutes the more massive cytokine storm, which worsens the disease’s consequences. IL-6 might be used as a potential therapeutic target for critical COVID-19 cases [18]. In the past, several methods have been developed to design the subunit vaccines and immuno-therapeutics while focusing on cytokine specific methods [23–25]. CytoPred [26] is a cytokine-specific method that predicts and further classifies the cytokine into its family and sub-family. IFNepitope [27] is a method that was developed to predict and design interferon-gamma (IFN-gamma) inducing peptides. Following methods, IL4Pred [28], IL-10Pred [29] and IL17eScan [30] have been developed for predicting the peptides for inducing IL-4, IL-10 and IL-17, respectively. In addition, methods have been developed for predicting peptides for inducing a group of cytokines, such as ProInflam [31] and PIP-EL [32]. Another tool named AntiInflam, developed by Gupta et al., predicts the peptides or proteins which induce the production of anti-inflammatory cytokines [33]. To the best of our knowledge, there is no method specifically developed for the prediction of IL-6 inducing epitopes. In order to serve the scientific community, an attempt has been made to develop in silico models for predicting peptides that can induce cytokine IL-6 production.
Material and methods
Dataset preparation and pre-processing
We extracted experimentally validated, 583 IL-6 inducing peptides from the immune epitope database (IEDB) [34]. We removed all identical peptides and peptides having a length greater than 25 amino acids. Finally, we got 365 IL-6 inducing peptides, which we called positive dataset in this study. It has been observed that these peptides have been tested either in human or mouse hosts. Due to the lack of sufficient data for humans, we have taken all IL-6 peptides tested in human or mouse hosts. Thus, our method is applicable for both hosts. One of the major challenges is to obtain sufficient experimentally validated data for non-IL6 inducing peptides. In order to generate a negative dataset, we extracted experimentally validated peptides from IEDB that induce cytokines (e.g. IL1α, IL1β, TNFα, IL6, IL8, IL12, IL17, IL18) other than IL-6, called non-IL-6 inducing peptides. Our negative dataset comprises sequences tested either in human or mouse hosts only. We removed all identical peptides and peptides having length above 25 amino acids. Finally, we got 2991 non-IL-6 inducing peptide called negative dataset in this study. Eventually, we got a final dataset of 365 IL-6 inducing and 2991 non-IL-6 inducing unique peptides, known as positive dataset and negative dataset, respectively.
Position conservation analysis
We create a two-sample logo (TSL) [35] to understand the preference of specific amino acids at a particular position. The TSL tool requires a fixed length of input sequence vector criterion. The minimum length of the peptide in both datasets is eight residues. Therefore, we extract eight residues from the N-terminus side and eight residues from the C-terminus side of a peptide. These regions were joined to create a sequence of 16 residues corresponding to each sequence in negative and positive datasets. The following example shows the process of creating a sequence of residues 16 from a peptide of length 10 residues.
Peptide Sequence (original): ARGCGHTRKL.
N-terminus 8-residues (N- > C): ARGCGHTR [1:8].
C-terminus 8-residues (N- > C): GCGHTRKL [3:10].
C-terminus 8-residues (C- > N): LKRTHGCG [10:3].
Peptide of 16-residues: N-terminus(N- > C) + C-terminus residues (C- > N).
Single peptide of 16-residues: ARGCGHTRLKRTHGCG.
Even, we can create a sequence of 16 residues from a peptide of length 8 residues. These sequences of 16-residues created from peptides in our datasets were used for creating TSL. The first eight positions of logo represent N-terminus of peptides, and the last eight residue positions represent the C-terminus of the peptide. We used all IL-6 inducing and non-IL6 inducing peptides to create TSL.
Feature generation
In this study, Pfeature was used to compute a wide range of features from the peptide sequences. It calculates thousands of features/descriptors of protein or peptides sequences. It is useful to annotate different structural and functional properties of peptide sequences [36]. A vector of 9149 features was created from the Pfeature using a composition-based feature module. We computed 15 types of features/descriptors, such as AAC, DPC, TPC, ATC, PCP, RRI, PRI, etc. The complete description of each feature with the length of the feature vector is presented in Table 1.
Table 1.
List of descriptors with brief description and number of features; all features computed using Pfeature
| Name of descriptor | Description of descriptor | Number of features or length of vector |
|---|---|---|
| AAC | Amino acid composition | 20 |
| DPC | Dipeptide composition | 400 |
| TPC | Tripeptide composition | 8000 |
| ABC | Atomic and bond composition | 9 |
| RRI | Residue repeat Information | 20 |
| DDOR | Distance distribution of residue | 20 |
| SE | Shannon-entropy of protein | 1 |
| SER | Shannon entropy of all amino acids | 20 |
| SEP | Shannon entropy of physicochemical property | 25 |
| CTD | Conjoint triad calculation of the descriptors | 343 |
| CeTD | Composition-enhanced transition distribution | 187 |
| PAAC | Pseudo amino acid composition | 23 |
| APAAC | Amphiphilic pseudo amino acid composition | 29 |
| QSO | Quasi-sequence order | 46 |
| SOCN | Sequence order coupling number | 6 |
Machine learning
In this study, several machine learning algorithms have been used to develop models for classifying IL-6 and non-IL-6 inducing peptides. It includes Decision tree (DT), Random Forest (RF), Logistic Regression (LR), XGBoost (XGB), k-nearest neighbors (KNNs) and Gaussian Naive Bayes (GNB); the following is a brief description of these algorithms. Classification using the DT algorithm was based on the non-parametric supervised learning models. The objective was to make a model that can predict the response variable by learning the decision rules from the data features [37]. RF is an ensemble-based method for classification, which fits numerous DTs during the training and predicts the response variable as the individual tree. Averaging DTs improves the prediction accuracy and control on overfitting of the models [38]. The GNB algorithm uses the probabilistic classification approach and builds on the Bayes theorem. It assumes that the continuous variables of each class follow the normal or Gaussian distribution. The fundamental aim was to generate the model that provides the sample/query probabilities to belong to a particular class [39]. LR is a method to obtain the logistic or logit model, which provides a class or event probability. It uses the logistic function to create the model which can predict the class or response variable. This method shares the similarity with the multiple linear regression, with the exception that the dependent variable is binomial [40]. KNN is an instance-based classification technique. It only stores the instances of the training variables and classification is determined from the majority vote of the nearest neighbor of each data point [41]. XGB is a scalable tree boosting classification algorithm which uses an iterative approach for the final prediction. It uses the ensemble method in which the number of models combined to perform the final prediction. We have generated the model by using the parameters tuned on the training dataset, which can predict the response variable or class of the sample [42]. These classification techniques were implemented using python-library scikit-learn [43].
Ranking and selection of features
One of the major challenges in this study is identifying an important set of features from the large dimension of features. There are several methods for feature selection; we have used SVC-L1-based feature selection technique, which implements the support vector classifier (SVC) with linear kernel, penalized with L1 regularization. We used SVC-L1 because it performs several methods to select the best features from a large number feature vector, and it is extremely fast as compared with other techniques [44]. Its primary purpose is to minimize the objective function, which considers the loss function and regularization. SVC-L1 method selects the non-zero coefficients and then applies the L1 penalty to select relevant features to reduce dimensions. The L1 regularization creates the sparse models during the optimization process, and by selecting some of the features out of the model by making the coefficients equal to zero. Using the ‘C’ parameter, it regulates the sparsity, which is directly proportional to the number of selected features; lower the value of the ‘C’, lesser number of features will be selected. We have used the default value of 0.01 for parameter ‘C’ [45]. Based on this technique, 186 important features (Supplementary Table S1) have been identified from the 9149-feature set.
After that, these 186 features were ranked based on their importance in classifying peptides using program feature-selector. The program feature-selector rank features using a DT-based algorithm Light Gradient Boosting Machine, which calculates the rank of feature based on the number of times a feature is used to split the data across all trees [46]. These top-ranked features were examined to understand the nature of IL-6 inducing peptides. Furthermore, we applied machine learning on selected features and computed the performance on top 10, 20, 30 …., and 186 features, respectively.
Cross-validation
We used the 5-fold cross-validation and external validation technique to train, test and evaluate our prediction models. In the past, several studies used an 80:20 proportion for the splitting of the complete dataset into training and validation datasets [47–50]. We also used this standard protocol in this study, where 80% (i.e. 292 IL-6 inducing and 2393 non-IL-6 inducing peptides) of the data was used for training and the remaining 20% (i.e. 73 IL-6 inducing and 598 non-IL-6 inducing peptides) was used for external validation. Then, we implement standard 5-fold cross-validation evaluation techniques, which is frequently used in the previous studies [51, 52]. Firstly, the entire training dataset is divided into five equivalent sets or folds, with all the 5-folds have the same number of positive and negative examples. Then, 4-folds were used for training, while the 5-fold was utilized for testing. This procedure was iterated five times so that each set was used for testing.
Evaluation parameters
In order to evaluate the efficiency of different prediction models, we used well-established evaluation parameters. In this study, we used both threshold-dependent and independent parameters, and we measure threshold-dependent parameters such as sensitivity (Sens), specificity (Spec) and accuracy (Acc) with the help of the following equations. We also used the standard threshold-independent parameter Area Under the Receiver Operating Characteristic (AUROC) curve to measure the performance of the models. AUROC curve is generated by plotting sensitivity against (1-specificity) on various thresholds. These parameters were calculated using the following equations:
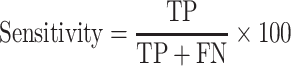 |
(1) |
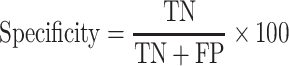 |
(2) |
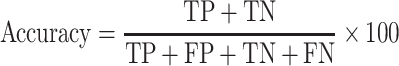 |
(3) |
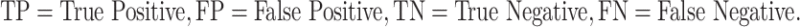 |
Architecture of web server
A web server named as ‘IL6Pred’ (https://webs.iiitd.edu.in/raghava/il6pred) is developed to predict IL-6 inducing and non-inducing peptides. The front end of the web server was developed by using HTML5, JAVA, CSS3 and PHP scripts. It is based on responsive templates which adjust the screen based on the size of the device. It is compatible with almost all modern devices such as mobile, tablet, iMac and desktop. The web server incorporates five major modules, such as Predict, Design, Protein Scan, Motif Scan and Blast Scan.
Results
In this study, we have used 365 peptides as a positive dataset, which can induce IL-6 cytokine. The negative dataset includes 2991 peptides, which do not induce IL-6 cytokine. All the analyses and predictions performed on the IL-6 inducing and non-inducing epitopes or peptides.
Positional analysis
In this analysis, we study the preference of particular amino acid at a specific position in the peptide string; we create a TSL for the IL-6 inducing (positive) and non-inducing (negative) peptides as represented in Figure 2. The most significant amino acid residue represents the relative abundance in the sequence. It is important to note that the first eight positions represent the N-terminal residues of peptides, and the last eight positions represent C-terminus of peptides. We observed that ‘L’ amino acid residue is mostly preferred at 2nd, 4th, 5th, 6th, 7th, 10th, 11th, 12th, 13th, 14th, 15th and 16th positions in the IL-6 inducing peptides. It means that ‘L’ is preferred in N-terminus as well as C-terminus residues. Besides, residue ‘I’ is found to be most abundant at positions 1st, 4th and 7th in IL-6 inducing peptides; it means that ‘I’ is preferred in N-terminus residues. On the other hand, amino acid residue ‘A’ dominates at 4th, 8th, and 16th positions in non-IL-6 inducing peptides.
Figure 2.
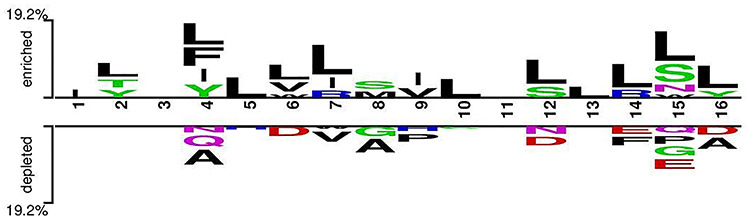
TSL shows the preference of residues in IL-6 and non-IL-16 peptides at different positions. First, eight positions represent the N-terminus of peptides, whereas the last eight positions represent the C-terminus of peptides.
Compositional analysis
In this analysis, we computed amino acid composition (AAC) for both positive and negative datasets. The average composition of IL-6 inducing and non-inducing peptides is shown in Figure 3. The average composition of residues (such as I, L and S) is higher in IL-6 inducing peptides than in non-IL-6 peptides. Besides, the residues (such as A, D and G) are more abundant in non-IL-6 peptides as compared with IL-6 inducing peptides.
Figure 3.
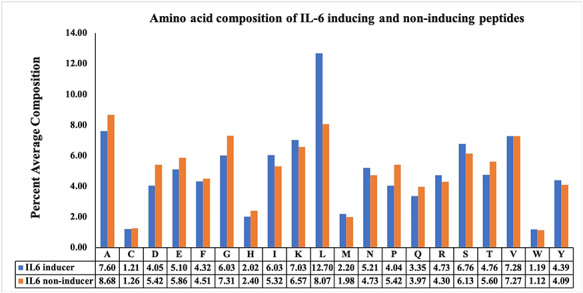
Average AAC of IL-6 inducing and non-inducing peptides.
Prediction models
Machine learning-based prediction models
We develop prediction models using various classifiers such as RF, DT, GNB, XGB and LR. Firstly, we computed the features of the IL-6 inducers and non-inducers from the Pfeature compositional-based module. A total of 9149 features were generated by Pfeature, and then we have implemented the SVC-L1 feature selection technique to select the most relevant features, i.e. 186 features, as shown in Supplementary Table S1. With this feature set, we applied various machine learning models. RF attains maximum performance with AUROC 0.893 and 0.863; accuracy 75.79 and 73.32 on training and validation datasets, respectively, with balanced sensitivity and specificity. XGB also performed well on training and validation datasets with AUROC 0.87 and 0.82, accuracy 86.29 and 84.65, respectively, but there is a considerable difference in sensitivity and specificity. Other classifiers, such as DT, LR, KNN and GNB, perform poorly on the training and validation dataset, as represented in Table 2.
Table 2.
The performance of machine learning-based models developed using 186 selected features on training and validation datasets. It shows the average performance of models with standard deviation on 5-folds
| Classifier | Sensitivity (mean ± SD) | Specificity (mean ± SD) | Accuracy (mean ± SD) | AUROC (mean ± SD) | Sensitivity | Specificity | Accuracy | AUROC |
|---|---|---|---|---|---|---|---|---|
| Training dataset | Validation dataset | |||||||
| DT | 40.07 ± 11.97 | 89.89 ± 03.86 | 84.47 ± 02.77 | 0.66 ± 0.06 | 39.726 | 89.632 | 84.203 | 0.65 |
| RF | 85.96 ± 03.03 | 74.55 ± 03.27 | 75.79 ± 03.24 | 0.89 ± 0.22 | 83.562 | 72.074 | 73.323 | 0.863 |
| LR | 69.18 ± 10.19 | 78.10 ± 02.04 | 77.13 ± 02.40 | 0.80 ± 0.06 | 68.493 | 76.087 | 75.261 | 0.783 |
| KNN | 47.60 ± 03.96 | 59.97 ± 03.32 | 58.62 ± 03.34 | 0.53 ± 0.03 | 52.055 | 56.187 | 55.738 | 0.542 |
| GNB | 57.54 ± 07.55 | 88.88 ± 01.00 | 85.48 ± 01.26 | 0.82 ± 0.03 | 53.425 | 88.294 | 84.501 | 0.782 |
| XGB | 66.10 ± 04.97 | 88.76 ± 02.10 | 86.29 ± 02.10 | 0.87 ± 0.02 | 58.904 | 87.793 | 84.65 | 0.823 |
Sens, Sensitivity; Spec, Specificity; Acc, Accuracy; AUROC, Area Under Receiver Operating Curve.
Performance of top-ranked features
All 186 features were ranked based on their importance according to their normalized and cumulative score, with the help of the feature selector tool. Furthermore, we evaluate the performance of the different feature sets. We identified the feature set with a minimum number of features, which will discriminate between IL-6 inducers and non-inducers with high AUROC and accuracy. Therefore, we build different models on top (10, 20, 30 … … and 186) features, respectively, and evaluate performance on the training and validation dataset. In order to understand the difference between the positive and negative datasets, we computed the average values of the top-10 features of IL-6 inducing and non-inducing peptides, as represented in Table 3.
Table 3.
Brief description of top 10 features and their average values in IL-6 inducing and non-inducing peptides
| Name of descriptors | Description of descriptors | #Average_1 | #Average_2 |
|---|---|---|---|
| hydrogen_bonds | Bond composition of peptide | 157.47 | 151.96 |
| Grantham_gap3 | Quasi sequence order of peptide | 9444.51 | 9614.37 |
| Polarity_2 | Polarity composition of group 2 amino acids in sequence in Composition-enhanced Transition Distribution (CeTD) | 29.18 | 33.14 |
| N_perc | Atomic composition of nitrogen in sequence | 53.16 | 52.59 |
| Charge_gp_3 | Charge composition of group 3 amino acids in Composition-enhanced Transition Distribution (CeTD) | 9.16 | 11.28 |
| NVWV_3 | Normalized Vander Waals Volume of group 3 amino acids in Composition-enhanced Transition Distribution (CeTD) | 26.21 | 25.28 |
| AAC_L | Leucine composition in sequence | 12.7 | 8.07 |
| Hydrophobicity_group_1 | Hydrophobicity of group 3 amino acids in Composition-enhanced Transition Distribution (CeTD) | 29.47 | 30.84 |
| AAC_A | Alanine composition in sequence | 7.6 | 8.68 |
| SS_gp_3 | Secondary structure composition of group 3 amino acids in Composition-enhanced Transition Distribution (CeTD) | 26.09 | 29.01 |
#Average_1: Average values of IL-6 inducing peptides; #Average_2: Average values of IL-6 non-inducing peptides.
The top-10 selected features have reasonable discriminatory power in case of AUROC and accuracy. RF achieves maximum performance with accuracy (77.39 and 73.47), AUROC (0.84 and 0.83) on training and validation dataset with balanced sensitivity and specificity, respectively, as represented in Table 4 and Figure 4. The performance of 10, 20, 30 … …. and 186 selected feature sets is provided in Supplementary Table S2.
Table 4.
The performance of machine learning techniques-based models developed using top-10 features on training and validation datasets. It shows the average performance of models with standard deviations on 5-folds
| Classifier | Sensitivity (mean ± SD) | Specificity (mean ± SD) | Accuracy (mean ± SD) | AUROC (mean ± SD) | Sensitivity | Specificity | Accuracy | AUROC |
|---|---|---|---|---|---|---|---|---|
| Training dataset | Validation dataset | |||||||
| DT | 70.55 ± 06.29 | 69.12 ± 04.44 | 69.27 ± 04.28 | 0.74 ± 0.04 | 69.86 | 68.23 | 68.41 | 0.72 |
| RF | 77.40 ± 08.35 | 77.39 ± 02.45 | 77.39 ± 02.19 | 0.84 ± 0.04 | 75.34 | 73.24 | 73.47 | 0.83 |
| LR | 61.64 ± 06.15 | 58.63 ± 03.27 | 58.96 ± 02.83 | 0.65 ± 0.03 | 64.38 | 57.19 | 57.97 | 0.64 |
| KNN | 58.56 ± 11.21 | 42.29 ± 05.37 | 44.06 ± 04.69 | 0.52 ± 0.06 | 64.38 | 48.16 | 49.93 | 0.58 |
| GNB | 70.21 ± 06.87 | 66.15 ± 05.45 | 66.59 ± 04.36 | 0.74 ± 0.04 | 67.12 | 64.38 | 64.68 | 0.72 |
| XGB | 71.23 ± 06.41 | 72.71 ± 02.41 | 72.55 ± 01.54 | 0.80 ± 0.04 | 71.23 | 67.56 | 67.96 | 0.8 |
Sens, Sensitivity; Spec, Specificity; Acc, Accuracy; AUROC, Area Under Receiver Operating Curve.
Figure 4.
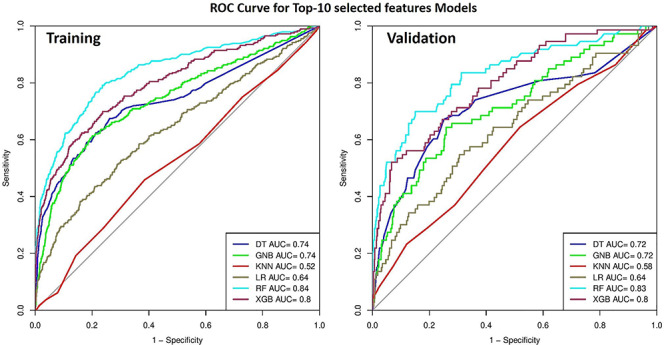
ROC curve shows the performance of models developed using 10 features on training and validation datasets; models were developed using various machine-learning techniques.
Services to the scientific community
In order to serve the scientific community, we develop a user-friendly prediction web server that integrates different modules to predict IL-6 inducing peptides. The prediction models used in the study are implemented in the web server. Users can predict that the given query peptide is IL-6 inducing or non-inducing based on the prediction models score at a different threshold. The web server has five important modules: (i) Predict; (ii) Design; (iii) Protein Scan; (iv) Motif Scan and (v) Blast Scan. The ‘Predict’ module provides the facility to the user to classify IL-6 inducing peptides from non-inducing peptides. The ‘Design’ module allows the user to create all possible analogs of the input sequence and identify the best analog which initiates cytokine, i.e. IL-6 release. The ‘Protein Scan’ module was used to scan IL-6 inducing regions in the given amino-acid sequence. ‘Motif Scan’ module allows the users to map or scan IL-6 motifs in the query sequence. We used MEME/MAST and MERCI software to derive motifs from experimentally validated IL-6 inducing peptides. The ‘Blast Scan’ module is based on a similarity search method, i.e. Basic Local Alignment Search Tool (BLAST). The input query sequence is searched against the database of known IL-6 inducing peptides. A query sequence is predicted as IL-6 inducer if found match or hit in the database; otherwise, it is predicted as non-IL-6 inducer peptide. Users can also download the positive and negative datasets used in this study, and the peptide sequence is available in the FASTA file format. The web server ‘IL-6pred’ was implemented using HTML, JAVA and PHP scripts. The server is user-friendly and compatible with a wide range of devices such as laptops, android mobile phones, iPhone, iPad, etc. The open-source web server is available at ‘https://webs.iiitd.edu.in/raghava/il6pred/’. Additionally, we also develop a standalone package of IL-6Pred in the form of a docker container. This standalone is integrated into the ‘GPSRdocker’ package; the user can download it from our ‘https://webs.iiitd.edu.in/gpsrdocker/’ [53].
Case study: IL-6 inducing peptides in spike proteins of SARS-CoV-2
Recent studies have shown the up-gradation of IL-6 levels in COVID-19 patients. Spike protein of novel coronavirus massively induces the release of proinflammatory cytokine IL-6 [54–57]. To identify the IL-6 inducing peptides in the SARS coronavirus proteins, we used our web server ‘IL-6Pred’ Protein Scan module (https://webs.iiitd.edu.in/raghava/il6pred/scan.php) with the default parameters (i.e. length of peptide 15 and threshold 0.11 with the RF method). We downloaded the SARS-CoV-2 proteins of five different countries, such as India (MT539168), China (NC_004718), USA (MT536976), Germany (MT539726) and Italy (MT528239) from NCBI (https://www.ncbi.nlm.nih.gov/sars-cov-2/). We identify 222 IL-6 inducing peptides out of 1259 peptides from the spike proteins of all the countries, as mentioned above (Supplementary Table S3). Table 5 represented the topmost predicted peptides of the spike protein of USA strain, which can induce IL-6 cytokine release. Furthermore, we identify the IL-6 inducing/non-inducing peptides in other SARS-CoV-2 proteins such as Envelope protein, ORF6, ORF1ab, ORF3a, ORF7a/7b, ORF8, spike protein, nucleocapsid phosphoprotein, ORF10 and Membrane glycoprotein of all the prediction results (USA strain) represented in Supplementary Tables S4–S14. These findings can be further used by the scientific community, working in the field of subunit vaccine designing against deadly coronavirus and other diseases that can be proliferated by the induction of IL-6 level. Researchers can use our web server to identify IL-6 inducing/non-inducing peptides and design a potential vaccine candidate against several diseases.
Table 5.
Potential IL-6 inducing peptides predicted by our method in SARS-CoV-2 spike protein of USA (MT536976) strain, selected based on score
| Sequence | Prediction score |
|---|---|
| YNYLYRLFRKSNLKP | 0.35 |
| NYLYRLFRKSNLKPF | 0.35 |
| NYNYLYRLFRKSNLK | 0.34 |
| QKFNGLTVLPPLLTD | 0.31 |
| FVFLVLLPLVSSQCV | 0.3 |
| CAQKFNGLTVLPPLL | 0.3 |
| AQKFNGLTVLPPLLT | 0.29 |
| MFVFLVLLPLVSSQC | 0.28 |
| ITRFQTLLALHRSYL | 0.27 |
Discussion and conclusion
IL-6 is rapidly produced as an immune response in infection and tissue injuries via strictly controlled transcriptional and post-transcriptional mechanisms [7]. However, dysregulated expression of IL-6 plays a pathological effect on chronic inflammation and autoimmunity [57]. IL-6 stimulates the auto-immune and inflammatory processes in numerous diseases such as alzheimer’s disease [58], atherosclerosis [59], behçet’s disease [60], diabetes [61], depression [62], multiple myeloma [63], prostate cancer [64], rheumatoid arthritis [65] and systemic lupus erythematosus [66]. The elevated level of serum IL-6 has been reported in various COVID-19 confirmed cases [67]. Thus, in various diseases, either anti-IL-6 treatment is essential or the presence of IL-6 inducing entities must be checked.
In this study, we have tried to empathize the nature of IL-6 inducing peptides and developed models to identify the IL-6 inducing potential of peptides. To the best of our knowledge, this is the first attempt to develop the IL-6 inducing peptide prediction tool. The dataset plays a significant role in machine learning; thus, we have constructed the dataset from IEDB. TSL and compositional analytical studies were performed to understand the composition and positional preference, and we observed that IL-6 inducing peptides are enriched in Leucine (L) amino acid. ‘Pfeature’ has been used to compute 9149 features from sequence information. SVC-L1 from the scikit package was used to select relevant features and then ranked by feature selector tools. Our compositional analysis indicates that a certain types of residues (i.e. L, I, S) are preferred in IL-6 peptides, whereas a certain types of other residues (i.e. A, D, G) are not preferred in IL-6 inducing peptides. It is interesting to note that 186 features selected by modern feature selection techniques SVC-L1 also include composition of these residues (i.e. L, I, A, D, G). This indicates that simple compositional-based techniques can identify important features. These 186 features have been used in our study for developing classification models. RF attains maximum performance with AUROC 0.893 and 0.863 on the training and validation datasets, respectively.
Furthermore, various models were developed based on top-ranked features, and a 5-fold cross-validation technique used to validate the performance. To avoid over-optimization of models, we want a minimum set of features with minimum loss in performance (almost equal to the 186 features). We selected top-10 features for the final classification models because the difference in the performance, i.e. AUROC (0.84 and 0.83) on training and validation, is lower in the case of 10-feature in comparison to 186-feature based models.
Additionally, we have predicted 222 IL-6 inducing peptides in the SARS-CoV-2 spike protein. Importantly, six peptide sequences of spike protein are YNYLYRLFRKSNLKP, NYLYRLFRKSNLKPF, NYNYLYRLFRKSNLK, QKFNGLTVLPPLLTD, FVFLVLLPLVSSQCV and CAQKFNGLTVLPPLL, which can induce IL-6 cytokine release with higher prediction score. Thus, these peptides should not be included in the vaccine regions, as they will elicit cytokine storm, especially in the case of IL-6. After that, these IL-6 inducing peptides also mapped on the five SARS-CoV-2 strains collected from the different parts of the globe. In this manner, IL6-Pred will be useful in designing the vaccine region as users can omit or add the IL-6 inducing peptide region as required. In the case of SARS-CoV-2, IL-6 inducing peptide regions will have a negative aspect, so it must be omitted from potential vaccine candidates.
In order to serve the scientific community, we have developed a web server named IL-6Pred (https://webs.iiitd.edu.in/raghava/il6pred/) as well as the standalone version which incorporated our best models. IL-6Pred is freely available and provides numerous facilities to the users. We anticipate that this work will surely benefit the researcher working in vaccine designing and want to include or exclude IL-6 inducing regions. The complete architecture of the IL-6Pred is shown in Figure 5.
Figure 5.
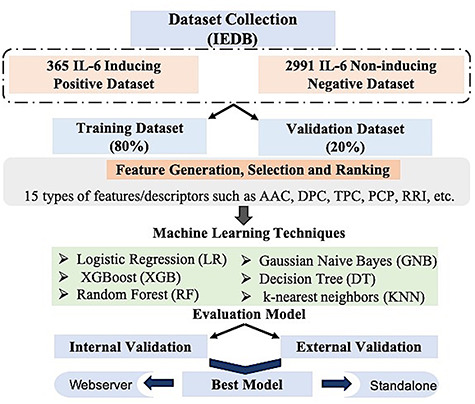
Overall architecture of IL-6Pred includes the creation of datasets, feature creation/selection, machine learning techniques and process of evaluation.
Limitation of the study
In the current study, we have developed a prediction tool for the identification of IL-6 inducing/non-inducing peptides. Due to the limited number of experimentally validated IL-6 inducing peptides, we have considered both human and mouse hosts to develop classification models. Ideally, one should develop host-specific methods for predicting IL-6 inducing peptides. Besides, the negative dataset used in this study is not perfect as it uses peptide inducing other cytokines as non-IL-6 inducing peptides. Ideally, one should have experimentally validated non-IL-6 inducing peptides, which is not available in IEDB. There is a need of perfect data of sufficient size to develop an accurate and reliable method. In this study, a systematic attempt has been made to develop the best possible models in the present conditions.
Key Points
IL-6 plays an important role in the progression of many diseases, including COVID-19.
A method has been developed for predicting IL-6 inducing peptides.
More than 9000 features have been generated for each peptide.
State-of-the-art technique has been used for selecting and ranking features.
It is available as a web server, standalone software and Docker container.
Supplementary Material
Acknowledgement
Authors are thankful to DST-INSPIRE and DBT for fellowships and financial support.
Anjali Dhall is currently working as Ph.D. in bioinformatics from the Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Sumeet Patiyal is currently working as Ph.D. in bioinformatics from the Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Neelam Sharma is currently working as Ph.D. in bioinformatics from the Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Salman Sadullah Usmani completed his Ph.D. in bioinformatics from CSIR-IMTECH, Chandigarh, India, and is currently working as a Research Associate-I in the Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Gajendra P. S. Raghava is currently working as a Professor and the Head of the Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Contributor Information
Anjali Dhall, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Sumeet Patiyal, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Neelam Sharma, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Salman Sadullah Usmani, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Gajendra P S Raghava, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi, India.
Data Availability Statement
All the datasets generated for this study are either included in this article/Supplementary material or available at the ‘IL-6Pred’ https://webs.iiitd.edu.in/raghava/il6pred/dataset.php as mentioned in the Materials and Methods section.
Author Contributions
A.D., S.P., N.S. and S.S.U. collected and processed the datasets. A.D., S.P. and G.P.S.R. implemented the algorithms. S.P. and A.D. developed the prediction models. A.D., S.P. and G.P.S.R. analysed the results. S.P., A.D. and N.S. created the back-end of the web server and front-end user interface. A.D., S.S.U., N.S., S.P., and G.P.S.R. penned the manuscript. G.P.S.R. conceived and coordinated the project and gave overall supervision to the project. All authors have read and approved the final manuscript.
References
- 1. Ataie-Kachoie P, Pourgholami MH, Richardson DR, et al. Gene of the month: interleukin 6 (IL-6). J Clin Pathol 2014;67:932–7. [DOI] [PubMed] [Google Scholar]
- 2. Rose-John S, Winthrop K, Calabrese L. The role of IL-6 in host defence against infections: immunobiology and clinical implications. Nat Rev Rheumatol 2017;13:399–409. [DOI] [PubMed] [Google Scholar]
- 3. Covarrubias AJ, Horng T. IL-6 strikes a balance in metabolic inflammation. Cell Metab 2014;19:898–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for translational therapeutics. Cancer 2007;110:1911–28. [DOI] [PubMed] [Google Scholar]
- 5. Hirano T. Interleukin 6 and its receptor: ten years later. Int Rev Immunol 1998;16:249–84. [DOI] [PubMed] [Google Scholar]
- 6. Su H, Lei CT, Zhang C. Interleukin-6 signaling pathway and its role in kidney disease: an update. Front Immunol 2017;8:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanaka T, Narazaki M, Kishimoto T. Il-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol 2014;6(10):a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, et al. The role of interleukin 6 during viral infections. Front Microbiol 2019;10:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mauer J, Denson JL, Brüning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol 2015;36:92–101. [DOI] [PubMed] [Google Scholar]
- 10. Yu B, Wang Y, Huang SH, et al. Group SM Interleukin-6 as a therapeutic target on human cancer. Targeted Cancer Therapy SM Group. China 2016;1–19. [Google Scholar]
- 11. Mizuhashi S, Nakamura K, Mori Y, et al. Insulin allergy and immunologic insulin resistance caused by interleukin-6 in a patient with lung cancer. Diabetes Care 2006;29:1711–2. [DOI] [PubMed] [Google Scholar]
- 12. Seow A, Ng DP, Choo S, et al. Joint effect of asthma/atopy and an IL-6 gene polymorphism on lung cancer risk among lifetime non-smoking Chinese women. Carcinogenesis 2006;27(6):1240–4. [DOI] [PubMed] [Google Scholar]
- 13. Swerdlow DI, Holmes MV, Kuchenbaecker KB, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 2012;379:1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ujiie H, Tomida M, Akiyama H, et al. Serum hepatocyte growth factor and Interleukin-6 are effective prognostic markers for non-small cell lung cancer. Anticancer Res 2012;32:3251–8. [PubMed] [Google Scholar]
- 15. Zarogoulidis P, Yarmus L, Darwiche K, et al. Interleukin-6 cytokine: a multifunctional glycoprotein for cancer. Immunome Res 2013;9(62):16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen L, Liu HG, Liu W, et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2020;43:E005–5. [DOI] [PubMed] [Google Scholar]
- 17. Zumla A, Hui DS, Azhar EI, et al. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet 2020;395:e35–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Zhao B, Qu Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients [published online ahead of print, 2020 Apr 17] Clin Infect Dis 2020;ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 2020;382(12):1177–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. COVID-19 and the Cytokine Storm the crucial role of IL-6 - Enzo Life Sciences, 2020. https://www.enzolifesciences.com/science-center/technotes/2020/april/covid-19-and-the-cytokine-storm-the-crucial-role-of-il-6/ (1 August 2020, date last accessed). [Google Scholar]
- 21. Mehta P, McAuley DF, Brown M, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 2020;395:1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaim S, Chong JH, Sankaranarayanan V, et al. COVID-19 and multiorgan response. Curr Probl Cardiol 2020;45(8):100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Usmani SS, Kumar R, Bhalla S, et al. In silico tools and databases for designing peptide-based vaccine and drugs. Adv Protein Chem Struct Biol 2018;112:221–63. [DOI] [PubMed] [Google Scholar]
- 24. Nagpal G, Usmani SS, Raghava GPS. A web resource for designing subunit vaccine against major pathogenic species of bacteria. Front Immunol 2018;9:2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar Dhanda S, Sadullah Usmani S, Agrawal P, et al. Novel in silico tools for designing peptide-based subunit vaccines and immunotherapeutics. Brief Bioinform 2017;18(3):467–78. [DOI] [PubMed] [Google Scholar]
- 26. Lata S, Raghava GPS. CytoPred: a server for prediction and classification of cytokines. Protein Eng Des Sel 2008;21(4):279–82. [DOI] [PubMed] [Google Scholar]
- 27. Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct 2013;8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhanda SK, Gupta S, Vir P, et al. Prediction of IL4 inducing peptides. Clin Dev Immunol 2013;2013:263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagpal G, Usmani SS, Dhanda SK, et al. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep 2017;7:42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta S, Mittal P, Madhu MK, et al. IL17eScan: a tool for the identification of peptides inducing IL-17 response. Front Immunol 2017;8:1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta S, Madhu MK, Sharma AK, et al. ProInflam: a webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J Transl Med 2016;14(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manavalan B, Shin TH, Kim MO, et al. PIP-EL: a new ensemble learning method for improved proinflammatory peptide predictions. Front Immunol 2018;9:1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta S, Sharma AK, Shastri V, et al. Prediction of anti-inflammatory proteins/peptides: an insilico approach. J Transl Med 2017;15(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vita R, Mahajan S, Overton JA, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res 2018;47:339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vacic V, Iakoucheva LM, Radivojac P. Two sample logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 2006;22(12):1536–7. [DOI] [PubMed] [Google Scholar]
- 36. Pande A, Patiyal S, Lathwal A, et al Computing wide range of protein/peptide features from their sequence and structure. bioRxiv 2019;599126. [Google Scholar]
- 37. Webb GI, Fürnkranz J, Fürnkranz J, et al. Decision tree. Encycl Mach Learn 2011;63:263–7. [Google Scholar]
- 38. Geurts P, Ernst D, Wehenkel L. Extremely randomized trees. Mach Learn 2006;63:3–42. [Google Scholar]
- 39. Zhang H. Exploring conditions for the optimality of naïve bayes. Int J Pattern Recognit Artif Intell 2005;19:183–98. [Google Scholar]
- 40. Tolles J, Meurer WJ. Logistic regression: relating patient characteristics to outcomes. JAMA 2016;316:533–4. [DOI] [PubMed] [Google Scholar]
- 41. Mucherino A, Papajorgji PJ, Pardalos PM. k-Nearest neighbor classification In: Data Mining in Agriculture. New York, NY: Springer; 2009;83–106. [Google Scholar]
- 42. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System In Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, 2016;785–94.
- 43. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- 44. Tang J, Alelyani S, Liu H. Feature selection for classification: a review. Data Classif Algorithms Appl 2014;37:1871–74. [Google Scholar]
- 45. Fan R-E, Chang K-W, Hsieh C-J, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–4. [Google Scholar]
- 46. Ke G, Meng Q, Finley T, et al. LightGBM: a highly efficient gradient boosting decision tree. Adv Neural Inf Proces Syst 2017;9:3146–54. [Google Scholar]
- 47. Agrawal P, Bhalla S, Chaudhary K, et al. In silico approach for prediction of antifungal peptides. Front Microbiol 2018;9:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagpal G, Chaudhary K, Agrawal P, et al. Computer-aided prediction of antigen presenting cell modulators for designing peptide-based vaccine adjuvants. J Transl Med 2018;16(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qureshi A, Thakur N, Kumar M. VIRsiRNApred: a web server for predicting inhibition efficacy of siRNAs targeting human viruses. J Transl Med 2013;11:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patiyal S, Agrawal P, Kumar V, et al. NAGbinder: an approach for identifying N-acetylglucosamine interacting residues of a protein from its primary sequence. Protein Sci 2020;29(1):201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gautam A, Chaudhary K, Kumar R, et al. In silico approaches for designing highly effective cell penetrating peptides. J Transl Med 2013;11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dhall A, Patiyal S, Kaur H, et al. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front Genet 2020;11:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Agrawal P, Kumar R, Usmani SS, et al. GPSRdocker: a Docker-based resource for genomics, proteomics and systems biology. bioRxiv 2019; 827766. [Google Scholar]
- 54. Wang W, Ye L, Ye L, et al. Up-regulation of IL-6 and TNF-α induced by SARS-coronavirus spike protein in murine macrophages via NF-κB pathway. Virus Res 2007;128(1–2):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feng Z, Diao B, Wang R, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes running title: SARS-CoV-2 infects human spleens and lymph nodes (in press) medRxiv 2020;1:18. [Google Scholar]
- 56. Tay MZ, Poh CM, Rénia L, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020;20(6):363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol 2010;40(7):1830–5. [DOI] [PubMed] [Google Scholar]
- 58. Cojocaru IM, Cojocaru M, Miu G, et al. Study of interleukin-6 production in Alzheimer’s disease. Rom J Intern Med 2011;49(1):55–8. [PubMed] [Google Scholar]
- 59. Hartman J, Frishman WH. Inflammation and atherosclerosis: a review of the role of interleukin-6 in the development of atherosclerosis and the potential for targeted drug therapy. Cardiol Rev 2014;22(3):147–51. [DOI] [PubMed] [Google Scholar]
- 60. Yamakawa Y, Sugita Y, Nagatani T, et al. Interleukin-6 (IL-6) in patients with Behçet’s disease. J Dermatol Sci 1996;11(3):189–95. [DOI] [PubMed] [Google Scholar]
- 61. Akbari M, Hassan-Zadeh V. IL-6 signalling pathways and the development of type 2 diabetes. Inflammopharmacology 2018;26(3):685–98. [DOI] [PubMed] [Google Scholar]
- 62. Hodes GE, Ménard C, Russo SJ. Integrating Interleukin-6 into depression diagnosis and treatment. Neurobiol Stress 2016;4:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stasi R, Brunetti M, Parma A, et al. The prognostic value of soluble interleukin-6 receptor in patients with multiple myeloma. Cancer 1998;82:1860–6. [PubMed] [Google Scholar]
- 64. Culig Z. Proinflammatory cytokine interleukin-6 in prostate carcinogenesis. Am J Clin Exp Urol 2014;2(3):231–8. [PMC free article] [PubMed] [Google Scholar]
- 65. Yoshida Y, Tanaka T. Interleukin 6 and rheumatoid arthritis. Biomed Res Int 2014;2014:698313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tackey E, Lipsky PE, Illei GG. Rationale for interleukin-6 blockade in systemic lupus erythematosus. Lupus 2004;13:339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ulhaq ZS, Soraya GV. Interleukin-6 as a potential biomarker of COVID-19 progression. Med Mal Infect 2020;50(4):382–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated for this study are either included in this article/Supplementary material or available at the ‘IL-6Pred’ https://webs.iiitd.edu.in/raghava/il6pred/dataset.php as mentioned in the Materials and Methods section.


