Abstract
Background
It is unknown whether a positive serology result correlates with protective immunity against SARS-CoV-2. There are also concerns regarding the low positive predictive value of SARS-CoV-2 serology tests, especially when testing populations with low disease prevalence.
Methods
A neutralization assay was validated in a set of PCR-confirmed positive specimens and in a negative cohort. In addition, 9530 specimens were screened using the Diazyme SARS-CoV-2 IgG serology assay and all positive results (N = 164 individuals) were reanalyzed using the neutralization assay, the Roche total immunoglobin assay, and the Abbott IgG assay. The relationship between the magnitude of a positive SARS-CoV-2 serology result and neutralizing activity was determined. Neutralizing antibody titers (50% inhibitory dilution, ID50) were also longitudinally monitored in patients confirmed to have SARS-CoV-2 by PCR.
Results
The SARS-CoV-2 neutralization assay had a positive percentage agreement (PPA) of 96.6% with a SARS-CoV-2 PCR test and a negative percentage agreement (NPA) of 98.0% across 100 negative control individuals. ID50 neutralization titers positively correlated with all 3 clinical serology platforms. Longitudinal monitoring of hospitalized PCR-confirmed patients with COVID-19 demonstrated they made high neutralization titers against SARS-CoV-2. PPA between the Diazyme IgG assay alone and the neutralization assay was 50.6%, while combining the Diazyme IgG assay with either the Roche or Abbott platforms increased the PPA to 79.2 and 78.4%, respectively.
Conclusions
These 3 clinical serology assays positively correlate with SARS-CoV-2 neutralization activity observed in patients with COVID-19. All patients confirmed SARS-CoV-2 positive by PCR develop neutralizing antibodies.
Keywords: neutralizing antibodies, COVID-19, serology, immunity, SARS-CoV-2
Introduction
The 2019 coronavirus pandemic is caused by the highly pathogenic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that was first discovered in Wuhan, China (1, 2). As governments across the world struggle to contain the spread of the virus, the worldwide economy has been impacted by the resulting shutdown and social distancing protocols that have been implemented (3–7). As nations begin to “reopen their economies” in stages, a major question that surrounds COVID-19 antibody testing is whether SARS-CoV-2 serology tests can be used to identify individuals with protective immunity against the virus (8). Neutralizing antibodies play a key role in the quest for protective immunity against SARS-CoV-2 (9), and 50% inhibitory dilution (ID50) neutralization titers have been the reference method to assess protective immunity against smallpox, polio, and influenza viruses following vaccine administration (10–12). A number of reports have characterized the clinical performance of commercially available SARS-CoV-2 serology assays (13–20). However, whether commercial SARS-CoV-2 serology platforms correlate with the presence of neutralizing antibodies and protective immunity against COVID-19 still needs further exploration (21–27).
A recent comparison of 6 SARS-CoV-2 immunoassays and a microneutralization assay against SARS-CoV-2 (28) demonstrated that 41 of 62 patients with COVID-19 showed neutralizing antibodies. Here, we evaluated the clinical sensitivity and specificity of a pseudovirus-based neutralization assay and investigated the correlation between the neutralization assay (29, 30) and SARS-CoV-2 serology platforms in a large cohort of clinical specimens. We determined the orthogonality of the Diazyme, Roche, and Abbott assays to initiate a dual immunoassay approach for confirming positive SARS-CoV-2 serology results, as suggested by the Centers for Disease Control and Prevention (31). This screen and confirm approach was applied retrospectively to 9530 SARS-CoV-2 tests, demonstrating that combining 2 orthogonal serology assays substantially improved the predictive value for identifying neutralizing antibodies.
Materials and Methods
Study Design and Patient Cohort
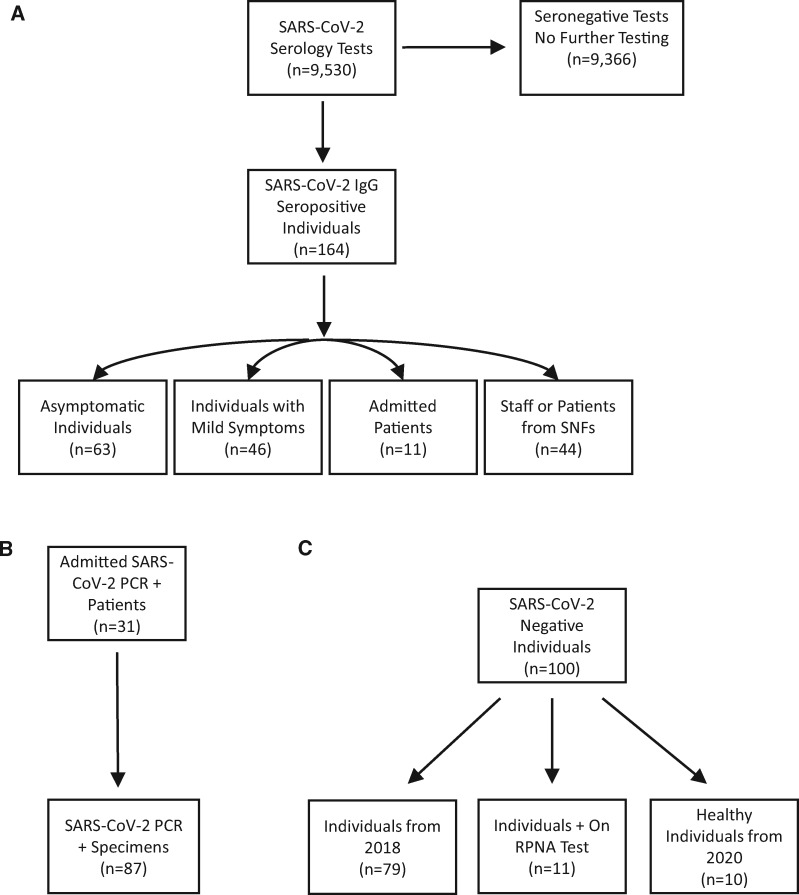
The patient cohorts used in this study are illustrated in Fig. 1. Briefly, the main cohort (Fig. 1, A) consisted of 9530 consecutive specimens (K-EDTA, lithium-heparin plasma separator tubes, and/or serum separator tubes) that were screened at UC San Diego Health using the Diazyme IgG assay from April 14 until May 12, 2020. The specimens that were positive using the Diazyme IgG assay (N = 164 specimens, N = 164 individuals) were separated into subgroups depending on symptoms, if they were admitted, or if they were from a skilled nursing facility. All of the specimens that screened positive with the Diazyme assay were retrospectively tested using the neutralization, Roche, and Abbott assays. A separate cohort (Fig. 1, B) was used to evaluate the relationship between hospitalized patients confirmed SARS-CoV-2 positive by PCR (31 patients, 87 specimens) and the neutralization assay. The last cohort (Fig. 1, C) was used to evaluate the relationship between SARS-CoV-2 negative specimens (100 specimens from 100 individuals) and the neutralization assay. The negative cohort consisted of 7 patients positive for other coronaviruses (229E, HKU1, NL63, or OC42), 4 patients positive for rhinovirus , 10 apparently healthy individuals, and 79 COVID-19 naïve specimens (individual patients collected in 2018, stored −20 °C).
Fig. 1.
Flowcharts of the cohorts used for A) Main cohort, B) SARS-CoV-2 PCR positive cohort, and C) SARS-CoV-2 negative cohort. SNF, skilled nursing facility, RPNA, respiratory pathogen nucleic acid.
The group (N = 251 specimens, N = 195 individuals) used to evaluate a cutoff for neutralization activity on each of the commercial serology platforms, shown in (Fig. 5), included the 164 specimens from 164 individuals that were seropositive for IgG on the Diazyme platform and the 87 specimens from the 31 patients who were SARS-CoV-2 PCR confirmed (that were also positive on the Diazyme IgG assay). The median number of days post PCR positivity for these 87 specimens from 31 patients was 9 days, with an interquartile range of 5–15 days. The median number of days postsymptom onset for these specimens was 13 days, with an interquartile range of 9–18 days. Specimens were split into 2 groups (negative and positive) based on the absence or presence of SARS-CoV-2 neutralization activity.
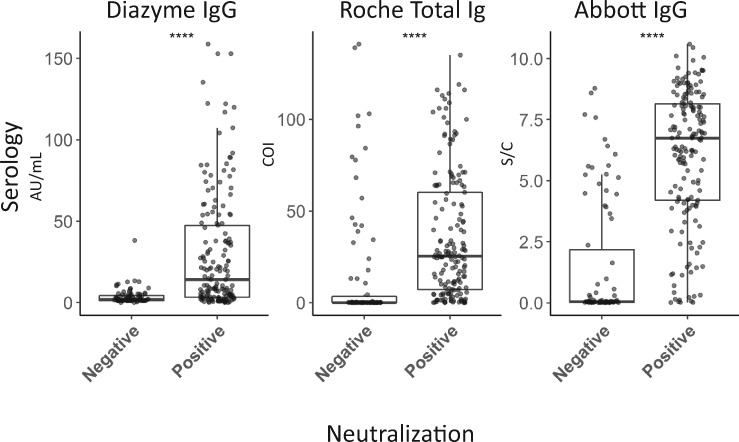
Fig. 5.
Distribution of commercial serology platform results in specimens negative and positive for SARS-CoV-2 neutralization activity that initially screened positive by the Diazyme IgG assay. Median AU/mL, COI, and S/C values (Y-axis) on the Diazyme IgG, Roche total Ig, and Abbott IgG platforms, respectively, are shown in boxplots (whiskers are up to but no greater than 1.5 times the IQR) for specimens without (negative) or with (positive) ID50 neutralization titers against SARS-CoV-2 (X-axis). **** indicates significant difference between positive and negative group at a p value of <0.00001 by Mann-Whitney.
All patient specimens were collected under UCSD IRB protocol 181656.
Classification of Symptoms
Individuals were classified into 3 groups based on symptom severity. Asymptomatic individuals had no COVID-19 related symptoms (shortness of breath, fever, cough, loss of taste, diarrhea, etc.). Mildly symptomatic individuals had mild COVID-19 related symptoms (as above), and did not require hospital admission. Admitted patients had COVID-19 related symptoms and were admitted to the hospital due to the severity of their symptoms on presentation.
Confirmation of Patients who were SARS-CoV-2 Positive
All 31 patients with SARS-CoV-2 were positively confirmed for COVID-19 by an emergency use authorized nucleic acid test that had also been validated in our laboratory (Abbott ID NOW, GenMark ePlex, Abbott RealTime, Roche Cobas 6800) (13, 20). The Abbott RealTime and Cobas 6800 are RT-PCR based tests, while the Abbott ID Now and GenMark ePlex are nucleic acid amplification-based tests. For simplicity, these patients will be referred to as SARS-CoV-2 PCR-confirmed patients.
Serology
Serology was performed on the Diazyme DZ-Lite 3000 plus clinical analyzer as previously described for IgG (13). Serology was performed on the Roche Cobas 8000 e801 analyzer (Roche Elecsys Anti-SARS-CoV-2 total Ig) and the Abbott ARCHITECT i1000SR analyzer (Abbott SARS-CoV-2 IgG) according to the manufacturer’s instructions. Plasma (Li-Heparin or K-EDTA) and serum specimens were analyzed in a manner consistent with the package inserts. The Diazyme platform targets antibodies produced against SARS-CoV-2 nucelocapsid (N) and spike (S) proteins, while the Roche and Abbott platforms target only the N protein. The Diazyme platform reports results as absorbance units per mL (AU/mL); values ≥1.00 AU/mL are considered reactive. The Roche platform reports results in the form of a cutoff index (COI; signal of sample/cutoff); values ≥1.00 COI are considered reactive. The Abbott platform reports results in the form of an Index value (S/C); Index values ≥1.4 S/C are considered reactive. For consistency, we refer to reactive and nonreactive to mean the same as positive and negative throughout this report.
SARS-CoV-2 and SARS-CoV-1 Pseudovirus Neutralization Assays
The neutralization assays were performed as previously described using a pseudovirus (PSV) (30). The PSV assay was established for both SARS-CoV-1 and SARS-CoV-2 using murine leukemia virus-based PSV. The assay used single cycle infectious viral particles containing firefly luciferase. The amount of luminescence in HeLa cells that stably expressed the cell surface receptor angiotensin converting enzyme 2 were measured after viral infection. Titers of 50% inhibitory dilution (ID50) were determined. An ID50 titer of greater than or equal to 50 was considered a positive neutralization result. The PSV assay was compared with a live replication-competent virus and neutralizing antibodies identified with the assay provided protection against high dose SARS-CoV-2 infection in a hamster animal model (29). All neutralization studies were conducted in a blinded manner.
GenMark ePlex Respiratory Pathogen Nucleic Acid Test
The respiratory pathogen nucleic acid test was performed as previously described to confirm patients with other respiratory pathogens (13).
Calculation of Positive Percentage Agreement and Negative Percentage Agreement
Positive percentage agreement (PPA) between the Diazyme IgG and Roche assay was calculated by taking the 164 Diazyme IgG screened positives and subsequently testing them on the Roche platform. Specimens that were positive on the Roche were treated as in agreement and specimens that were negative on the Roche were treated as not in agreement. The PPA was defined as:
This was done in the same manner for calculating PPA between the Diazyme IgG and Abbott assays.
PPA between the neutralization assays and the Diazyme IgG assay was calculated by taking the 164 Diazyme IgG screened positives and testing them for the presence of neutralizing antibodies. Specimens with ID50 neutralization titers >50 were treated as in agreement and specimens with ID50 neutralization titers <50 were treated as not in agreement, and the PPA is defined as above.
Negative percentage agreement (NPA) between the neutralization assays and the Diazyme IgG assay was calculated by testing specimens from 100 SARS-CoV-2 PCR negative individuals (Fig. 1, C) for the presence of neutralizing antibodies. Specimens with ID50 neutralization titers <50 were treated as in agreement and specimens with ID50 neutralization titers >50 were treated as not in agreement. The NPA was then defined as:
Statistical Analyses
Data was analyzed using R in Rstudio and linear regression analysis for all figures were performed in excel or Rstudio. Box and whisker plots were generated in Rstudio. Mann–Whitney and Fishers exact test were performed in R for the box and whisker plots and demographics table, respectively.
Results
Cohort Demographics
Demographics of the Diazyme IgG seropositive individuals, which were divided based on symptom severity, are shown for age, gender, body mass index, ethnicity, underlying medical conditions, PCR positivity for COVID-19, and positivity for neutralization activity against SARS-CoV-2 in Table 1. Median [interquartile (IQR)] age was significantly different among all 3 symptom severity groups using the Fishers exact test (P value <0.0001) with asymptomatic patients being younger. However, no significant difference was observed for body mass index or gender. Significant differences in ethnicity were observed across all 3 groups (P = 0.005), with 40% of the admitted group being Hispanic and 63% of the mildly symptomatic group being white. A significant difference was observed for underlying medical conditions (P = 0.018), 60% of admitted patients had underlying conditions vs 32% in the asymptomatic group. A significant difference (P < 0.0001) was observed in the percentage of individuals that were positive for COVID-19 by PCR, with 88% of admitted patients having a positive PCR test for SARS-CoV-2 vs 2% testing in the asymptomatic group. A significant difference (P < 0.0001) was observed across all 3 groups for the presence of neutralization activity against SARS-CoV-2, with 98% of admitted patients having neutralization activity vs 19% in the asymptomatic group. Demographics for the 44 Diazyme IgG seropositive patients and staff from skilled nursing facilities could not be retrieved because they were deidentified prior to analysis.
Table 1.
Demographics of seropositive individuals.
| Asymptomatic N = 63 |
Mild symptoms N = 46 |
Admitted N = 42 |
P | |
|---|---|---|---|---|
| Age (years) | 43 (33–49) | 49 (36–64) | 52 (43–68) | <0.0001 |
| Gender | 0.21 | |||
| Male | 33 (52%) | 25 (54%) | 29 (69%) | |
| Female | 30 (48%) | 21 (46%) | 13 (31%) | |
| Body Mass Index | 0.23 | |||
| Median (IQR) | 25 (23–29) | 26 (24–28) | 28 (24–30) | |
| Not available | 15 (24%) | 1 (2%) | 0 (0%) | |
| Ethnicity | 0.005 | |||
| Asian | 9 (14%) | 1 (2%) | 4 (10%) | |
| Black | 5 (8%) | 0 (0%) | 3 (7%) | |
| Filipino | 7 (11%) | 7 (15%) | 2 (5%) | |
| Hispanic | 14 (22%) | 8 (17%) | 17 (40%) | |
| Mixed | 3 (5%) | 0 (0%) | 1 (2%) | |
| White | 20 (32%) | 29 (63%) | 15 (36%) | |
| Not available | 5 (8%) | 1 (2%) | 0 (0%) | |
| Underlying medical conditions | 0.018 | |||
| Yes | 20 (32%) | 19 (41%) | 25 (60%) | |
| COVID-19 PCR | <0.0001 | |||
| Negative | 54 (86%) | 17 (37%) | 5 (12%) | |
| Positive | 1 (2%) | 15 (33%) | 37 (88%) | |
| Not tested | 8 (13%) | 14 (30%) | 0 (0%) | |
| Neutralization activity (SARS-CoV-2) | <0.0001 | |||
| Negative | 51 (81%) | 25 (54%) | 1 (2%) | |
| Positive | 12 (19%) | 21 (46%) | 41 (98%) | |
IQR, interquartile range.
Clinical Evaluation of a SARS-CoV-2 Neutralization Assay
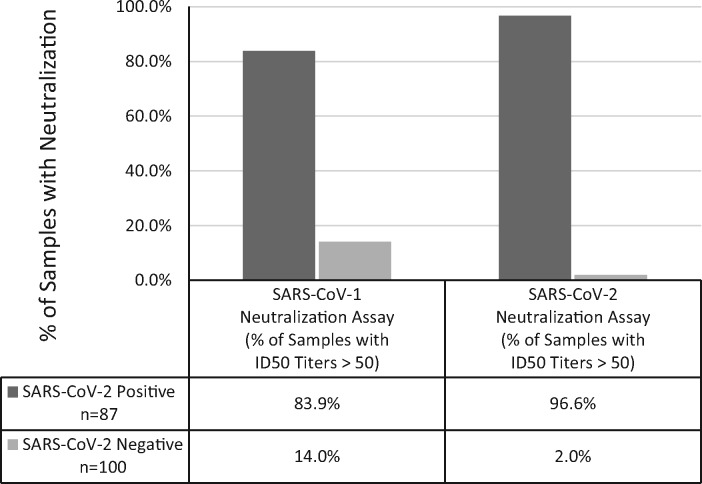
The SARS-CoV-2 neutralization assay detected neutralization activity in 96.6% of specimens from SARS-CoV-2 PCR-confirmed patients, and in 2.0% of the SARS-CoV-2 negative specimens (Fig. 2). 83.9% of the specimens from SARS-CoV-2 PCR-confirmed patients and 14.0% of the SARS-CoV-2 negative specimens had SARS-CoV-1 neutralization activity (Fig. 2) . SARS-CoV-1 or SARS-CoV-2 neutralization was not detected in 7 patients infected with non-COVID-19 coronaviruses and 4 patients infected with rhinovirus (Table 1 in the online Data Supplement).
Fig. 2.
COVID-19 patients produce neutralization activity against SARS-CoV-2 and SARS-CoV-1. Percentage of specimens (N=87) from SARS-CoV-2 PCR confirmed patients (N=31, dark grey) and a SARSCoV-2 negative patient cohort (N=100 specimens from 100 individuals, light grey) that had ID50 titers >50 against SARS-CoV-2 or SARS-CoV-1.
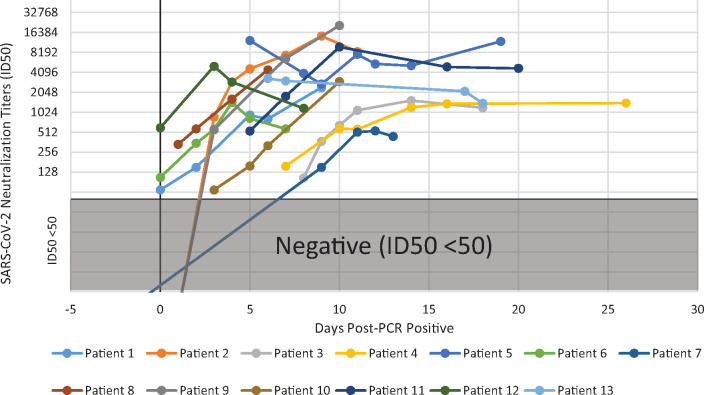
Fig. 3.
Longitudinal monitoring of SARS-CoV-2 neutralization titers in COVID-19 patients. The ID50 titersof 13 SARS-CoV-2 PCR confirmed patients are plotted on a semi-log scale (Y-axis) with the number of days following a positive PCR result indicated for each sample (X-axis). ID50 values of <50 are considered negative for neutralization activity and represented by the greyed-out area.
Correlation of ID50 Neutralization Titers to Commercial SARS-CoV-2 Serology Assays
Linear regression analysis was performed to evaluate the relationship between ID50 neutralization titers and 3 commercially available SARS-CoV-2 serology platforms for PCR-confirmed patients (Supplemental Fig. 1). All 3 platforms showed positive correlation to SARS-CoV-2 ID50 titers.
Longitudinal Monitoring of ID50 Neutralization Titers in Patients with COVID-19
SARS-CoV-2 ID50 neutralization titers were longitudinally monitored in 13 PCR-confirmed SARS-CoV-2 patients. Serial sampling revealed that SARS-CoV-2 ID50 neutralization titers increased with disease progression in 11 of the 13 patients, while the remaining 2 patients maintained ID50 neutralization titers of ≥1000 (Fig. 3). Once elevated, neutralization titers appeared to reach a plateau that stayed elevated up to 25 days (longest time tested). Notably, all patients developed high titers by 2 weeks after becoming PCR positive.
Retrospective Analysis of Diazyme Screened Seropositive Patients
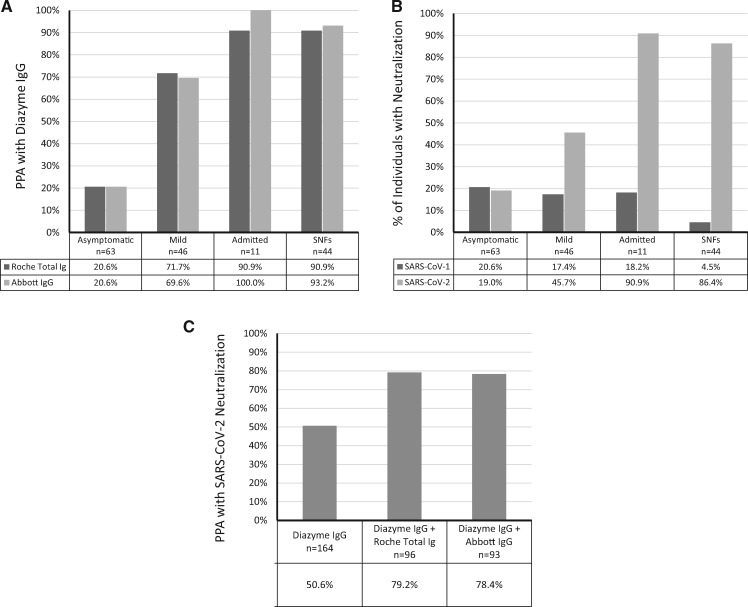
The 164 Diazyme screened IgG seropositive patient specimens were also analyzed on the Roche Total Ig and Abbott IgG SARS-CoV-2 serology platforms. The overall PPA between the Diazyme IgG assay and the Roche total Ig or Abbott IgG assays were 58.5 and 59.1%, respectively (Supplemental Fig. 2). The PPA between Diazyme and Roche was 20.6% for asymptomatic individuals, 71.7% for patients with mild symptoms, 90.9% for admitted patients, and 90.9% for patients/staff from skilled nursing facilities (Fig. 4, A). The PPAs between Diazyme and Abbott were similar to what was observed with the Roche assay (Fig. 4, A). The percentage of individuals with detectable levels of SARS-CoV-2 neutralization activity in the asymptomatic, mild, admitted, and skilled nursing facility groups (Fig. 4, B) closely mirror the observed PPAs between the Diazyme IgG and Roche/Abbott platforms for each respective group (Fig. 4, A). In contrast, the percentage of individuals with detectable levels of SARS-CoV-1 neutralization activity decreased with increasing PPA between serology platforms (Fig. 4, B). Overall, 50.6% of Diazyme IgG seropositive individuals (N = 164) had detectable neutralization activity, from which a total of 96 and 93 of these individuals were also seropositive on the Roche and Abbott platforms, respectively (Fig. 4, C). The percentage of specimens that were positive on 2 serology platforms that had neutralization activity was 79.2 and 78.4% when using the Diazyme + Roche and Diazyme + Abbott, respectively (Fig. 4, C). Confidence intervals for SARS-CoV-2 ID50 titers across the 4 different populations that were tested demonstrated significant overlap (Supplemental Table 2). All specimens from PCR-confirmed positive patients with a positive serology result by either the Diazyme IgG (N = 71), Roche total Ig (N = 76), and Abbott IgG (N = 75) assays had neutralization activity (>ID50).
Fig. 4.
Interplatform agreement between serology and neutralization activity. A) The PPA between the Diazyme IgG and Roche assay and the Diazyme IgG and Abbott assays are shown for 4 different populations of individuals. B) The percentage of individuals with neutralization activity against SARS-CoV-2 or SARS-CoV-1 are shown for 4 different populations of individuals. C) The overall PPA between the Diazyme IgG assay alone and in combination with the Roche or Abbott serology platforms with SARS-CoV-2 neutralization activity are indicated for 164 seropositive individuals identified on the Diazyme IgG platform.
Multiplatform Cutoff Evaluation for SARS-CoV-2 Neutralization Activity
Boxplots of the observed responses (AU/mL, COI, S/C) on the Diazyme, Roche, and Abbott platforms, from Diazyme IgG seropositives, which were split based on negative or positive SARS-CoV-2 neutralization activity, are shown in (Fig. 5). The medians (IQR) for the Diazyme IgG assay were 1.9 (1.2–4.3) AU/mL and 14.1 (3.3–47.4) AU/mL for the negative and positive groups, respectively. The medians (IQR) for the Roche total Ig assay were 0.1 (0.09–3.4) COI and 25.5 (7.5–63.6) COI for the negative and positive groups, respectively. The medians (IQR) for the Abbott IgG assay was 0.04 (0.02–1.1) S/C and 6.8 (4.2–8.2) S/C for the negative and positive groups, respectively. For the group that was positive, the median response value was explored as a predictive cutoff for neutralization activity (Diazyme, 14.1 AU/mL; Abbott, 6.8 S/C; and Roche, 25.5 COI). The percentage of seropositive specimens above these cutoff values that had detectable levels of SARS-CoV-2 neutralization activity (>ID50) were 98.8% for Diazyme, 95.1% for Abbott, and 83.5% for Roche.
Discussion
The SARS-CoV-2 neutralization assay had a PPA of 96.6% with a SARS-CoV-2 PCR test (N = 87 specimens, N = 31 patients) and an NPA of 98.0% across 100 negative control subjects. Interestingly, 83.9% of specimens from SARS-CoV-2 PCR-confirmed patients had neutralizing activity for SARS-CoV-1, which was responsible for the original 2002–2004 SARS epidemic (32–35). This supports the finding that SARS-CoV-2 infected individuals produce neutralizing antibodies that cross-react with SARS-CoV-1 (36). Likewise, neutralization activity against SARS-CoV-1 was observed in 14.0% of COVID-19 negative individuals and could be a result of past exposure to SARS-CoV-1, lower specificity of the SARS-CoV-1 neutralization assay, or greater cross-reactivity to other common coronaviruses. The high agreement between a positive SARS-CoV-2 PCR result and the SARS-CoV-2 neutralization assay suggests that neutralizing antibodies are made quickly in response to viral infection. Overall, the SARS-CoV-2 neutralization assay had PPA and NPA similar to published results for clinically validated SARS-CoV-2 serology assays (13) and provides a direct approach to assess protective immunity against SARS-CoV-2.
Regression analysis of SARS-CoV-2 neutralization titers in SARS-CoV-2 PCR-confirmed positive patients correlated with the corresponding values on the Diazyme, Roche, and Abbott platforms. The Diazyme IgG assay had the strongest correlation with SARS-CoV-2 neutralization titers, and could be a result of the linear characteristics of this platform (13). Furthermore, all platforms were highly sensitive for the detection of ID50 neutralization titers greater than 1000 (Supplemental Fig. 1). At ID50 titers >1000, 98.9% of the specimens were seropositive on the Diazyme IgG assay, and 100% of the specimens were seropositive for both the Roche and Abbott platforms. Longitudinal monitoring of SARS-CoV-2 ID50 neutralization titers in PCR-confirmed SARS-CoV-2 PCR positive hospitalized patients revealed that all patients developed robust neutralization titers during the course of infection (Fig. 3).
Using the Roche or Abbott SARS-CoV-2 serology platforms to confirm specimens initially screened positive on the Diazyme IgG assay demonstrated that nearly 40% of the 164 IgG seropositive individuals would have been classified as false positives by the confirmatory assay. As predicted previously (13), the percentage of individuals that were retrospectively identified as falsely positive depended greatly on the population of individuals being tested. The retrospective false positive rate was as high as 79.4% in asymptomatic individuals, approximately 30% in individuals with mild symptoms, and potentially 0% in admitted patients. One limitation with using a secondary platform to confirm SARS-CoV-2 seropositive specimens, is that there is no reference assay for COVID-19 serology at this time. Based on prevalence and specificity of this assay it is likely that the discordant specimens observed are false positives, but prudent practice would be to call these specimens indeterminate and ask for repeat testing. Altogether, this emphasizes the effect of disease prevalence on the positive predictive values, and highlights the danger of screening low prevalence populations even when using a highly specific SARS-CoV-2 serology assay.
One hundred percent of specimens that tested positive on all 3 commercially available serology platforms had detectable ID50 titers (>50) against SARS-CoV-2. In contrast, only 50.6% of the 164 seropositive individuals identified on the Diazyme platform had neutralization titers (Fig. 4, C), a percentage similar to the PPA between Diazyme and the Roche and Abbott platforms (Fig. 4, A). However, using the Roche or Abbott platforms to confirm IgG seropositive individuals, would have resulted in nearly 80% of reported seropositive individuals also having neutralization activity against SARS-CoV-2 (Fig. 4, C). Confidence intervals for SARS-CoV-2 ID50 titers across the 4 different populations that were tested demonstrated significant overlap (Supplemental Table 2), suggesting that individuals make comparable levels of neutralizing antibodies regardless of symptoms. One limitation of our cohort is that the length of symptom duration for most of these 164 individuals was not available, which could affect the neutralizing antibody response that an individual generates. The finding that 80% of two-platform confirmed positive serology results have neutralizing activity is an important result, addressing a major concern when using COVID-19 serology to screen low disease prevalence populations. However, roughly 20% of the two-platform confirmed seropositive individuals did not have SARS-CoV-2 neutralization activity suggesting that a clinically validated SARS-CoV-2 neutralization assay is required for understanding if patients have protective immunity against COVID-19.
In principle, not all patients with COVID-19 should have neutralizing antibodies, as these take time to develop after infection, and neutralizing antibodies represent a small subset of antibodies produced (30). It is therefore encouraging that 84 of 87 specimens (96.6%) from SARS-CoV-2 PCR-confirmed positive individuals also had neutralizing antibodies, and may encourage more aggressive steroid treatment in patients with symptoms of cytokine storm as production of neutralizing antibodies occurs shortly after infection. We note that the 3 specimens that did not have neutralizing antibodies were from different patients and these specimens were collected early in the disease process. One was collected 2 days before the patient became PCR positive and the other 2 were collected one day after the patient was confirmed positive using PCR. All 3 patients had neutralizing antibodies on the next specimen that was collected (day 3, 3, and 9). It is therefore important to consider symptom duration when testing for neutralizing antibodies as testing too early in the course of disease could lead to false negative results.
We explored using the median positive serology result as the cutoff value for determining the presence of neutralizing antibodies (Fig. 5). Specimens with serology results above the median were highly associated with neutralizing antibodies. 98.9, 95.1, and 83.5% of specimens above the median serology value contained neutralizing antibodies with ID50 titers greater than 50 when using the Diazyme, Abbott, and Roche assays, respectively.
We observed significant differences (P < 0.005) in the demographics of 151 Diazyme seropositive individuals that were asymptomatic, mildly symptomatic, and hospital admitted. In particular, although the percentage of Hispanic individuals comprised 25.8% of this population, they accounted for 40% of the hospital admitted cases; of which 98% were PCR positive for COVID-19. This is similar to another recent report which observed that 60% of all pregnant women hospitalized for COVID-19 were of Hispanic ethnicity (37).
The retrospective analysis of only the 164 seropositive SARS-CoV-2 individuals and not the population of 9530 individuals is a limitation of the study, preventing a fair comparison of the Diazyme platform with the Roche and Abbott SARS-CoV-2 serology platforms. This is because these individuals were effectively prescreened for SARS-CoV-2 seropositivity before analysis on either the Roche or Abbott platforms. Moreover, out of the 9530 tests performed, the estimated overall false positive rate of the Diazyme IgG assay was between 0.7 and 0.9%, and is equal to or lower than its published value of 0.9% (13). This suggests that our findings would have been similar if either the Roche total Ig or Abbott IgG assays were used as the screening platform and not the confirmatory platform, as both assays have been reported to have specificity that exceeds 99% (14–16); emphasizing the dangers of using any one single serology platform to screen low prevalence populations for SARS-CoV-2 serology. Another limitation is that we did not use live virus to perform the neutralization assays, however, Rogers et al. (30) showed a high correlation between the PSV and live virus, indicating that the use of a more hazardous live virus assay may be unnecessary. A final limitation is that the 7 patients with other coronaviruses were not serologically tested for the presence of antibodies against these viruses and although they were PCR positive, it is not certain that a humoral response had occurred in these patients.
We report a comprehensive retrospective study of 164 SARS-CoV-2 seropositive specimens from 9530 SARS-CoV-2 serology tests and demonstrate that serology results correlate with neutralization. Our study demonstrates the risk of using a single serology platform to identify SARS-CoV-2 seropositive individuals in low prevalence populations and highlights the benefits of a two-platform approach. Finally, the inclusion of serology and neutralization activity found in mild and asymptomatic cases of COVID-19 are also unique, as cohorts used for other reports are generally of hospitalized patients.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Author Declaration
A version of this article was previously posted as a preprint on medRxiv as https://www.medrxiv.org/content/10.1101/2020.07.10.20150946v1.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
R.T. Suhandynata, statistical analysis; M.A. Hoffman, statistical analysis; D. Huang, statistical analysis; R.W. McLawhon, financial support, administrative support, provision of study material or patients; D. Nemazee, financial support, statistical analysis, provision of study material or patients.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
None declared.
Consultant or Advisory Role
None declared.
Stock Ownership
None declared.
Honoraria
None declared.
Research Funding
D. Nemazee, NIH.
Expert Testimony
None declared.
Patents
None declared.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments
We thank all of the staff in the UC San Diego Health clinical laboratories for their help identifying specimens for sensitivity and specificity testing. We also thank Amy Rockefeller and Ernestine Ferrer for valuable technical expertise. We would like to thank Waters Corporation (RTS) and Roche Diagnostics (MAH) for supporting the clinical chemistry fellowship at the University of California-San Diego.
References
- 1. Ren L-L, Wang Y-M, Wu Z-Q, Xiang Z-C, Guo L, Xu T, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J 2020;133:1015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg 2020;78:185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCall B. Shut down and reboot—preparing to minimise infection in a post-COVID-19 era. Lancet Digit Health 2020;2:e293–e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilder-Smith A, Freedman DO.. Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak. J Travel Med 2020;27:taaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pak A, Adegboye OA, Adekunle AI, Rahman KM, McBryde ES, Eisen DP.. Economic consequences of the COVID-19 outbreak: the need for epidemic preparedness. Front Public Health 2020;8:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arshad Ali S, Baloch M, Ahmed N, Arshad Ali A, Iqbal A.. The outbreak of coronavirus disease 2019 (COVID-19)—an emerging global health threat. J Infect Public Health 2020;13:644–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weinstein MC, Freedberg KA, Hyle EP, Paltiel AD.. Waiting for certainty on Covid-19 antibody tests—at what cost? N Engl J Med 2020;383:e37. [DOI] [PubMed] [Google Scholar]
- 9. Jiang S, Hillyer C, Du L.. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 2020;41:355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zinkernagel RM. On natural and artificial vaccinations. Annu Rev Immunol 2003;21:515–46. [DOI] [PubMed] [Google Scholar]
- 11. Fenner F. Mouse-pox (infectious ectromelia of mice): a review. J Immunol 1949;63:341–73. [PubMed] [Google Scholar]
- 12. Sabin AB. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J Infect Dis 1985;151:420–36. [DOI] [PubMed] [Google Scholar]
- 13. Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL.. Longitudinal monitoring of SARS-CoV-2 IgM and IgG seropositivity to detect COVID-19. J Appl Lab Med 2020;5:908–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Favresse J, Eucher C, Elsen M, Marie T-H, Dogné J-M, Douxfils J.. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies. Clin Chem 2020;66:1104–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, et al. Clinical performance of the Roche SARS-CoV-2 serologic assay. Clin Chem 2020;66:1107–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 2020;58:e00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonelli F, Sarasini A, Zierold C, Calleri M, Bonetti A, Vismara C, et al. Clinical and analytical performance of an automated serological test that identifies S1/S2 neutralizing IgG In COVID-19 patients semiquantitatively. J Clin Microbiol 2020;58:e01224–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chew KL, Tan SS, Saw S, Pajarillaga A, Zaine S, Khoo C, et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect 2020;26:1256.e9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicol T, Lefeuvre C, Serri O, Pivert A, Joubaud F, Dubée V, et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol 2020;129:104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL.. Multi-platform comparison of SARS-CoV-2 serology assays for the detection of COVID-19. J Appl Lab Med 2020;5:1324–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. “Immunity Passports” in the context of COVID-19. https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19 (Accessed June 2020).
- 22. Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 2020;26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease. Clin Infect Dis 2020;ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 2020;71:778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol 2020;92:1518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020;368:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 28. Jääskeläinen A, Kuivanen S, Kekäläinen E, Ahava M, Loginov R, Kallio-Kokko H, et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020;360:731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020;369:956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CDC. Information for Laboratories about Coronavirus (COVID-19). 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html (Accessed June 2020).
- 32. Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003;348:1977–85. [DOI] [PubMed] [Google Scholar]
- 33. Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967–76. [DOI] [PubMed] [Google Scholar]
- 34. Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003;348:1986–94. [DOI] [PubMed] [Google Scholar]
- 35. Demmler GJ, Ligon BL.. Severe acute respiratory syndrome (SARS): a review of the history, epidemiology, prevention, and concerns for the future. Semin Pediatr Infect Dis 2003;14:240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lv H, Wu NC, Tsang OT-Y, Yuan M, Perera RAPM, Leung WS, et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep 2020;31:107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldfarb IT, Clapp MA, Soffer MD, Shook LL, Rushfirth K, Edlow AG, et al. Prevalence and severity of coronavirus disease 2019 (COVID-19) illness in symptomatic pregnant and postpartum women stratified by Hispanic ethnicity. Obstet Gynecol 2020;136:300–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.