Abstract
Background
Rapid, reliable, and widespread testing is required to curtail the ongoing COVID-19 pandemic. Current gold-standard nucleic acid tests are hampered by supply shortages in critical reagents including nasal swabs, RNA extraction kits, personal protective equipment, instrumentation, and labor.
Methods
To overcome these challenges, we developed a rapid colorimetric assay using reverse-transcription loop-mediated isothermal amplification (RT-LAMP) optimized on human saliva samples without an RNA purification step. We describe the optimization of saliva pretreatment protocols to enable analytically sensitive viral detection by RT-LAMP. We optimized the RT-LAMP reaction conditions and implemented high-throughput unbiased methods for assay interpretation. We tested whether saliva pretreatment could also enable viral detection by conventional reverse-transcription quantitative polymerase chain reaction (RT-qPCR). Finally, we validated these assays on clinical samples.
Results
The optimized saliva pretreatment protocol enabled analytically sensitive extraction-free detection of SARS-CoV-2 from saliva by colorimetric RT-LAMP or RT-qPCR. In simulated samples, the optimized RT-LAMP assay had a limit of detection of 59 (95% confidence interval: 44–104) particle copies per reaction. We highlighted the flexibility of LAMP assay implementation using 3 readouts: naked-eye colorimetry, spectrophotometry, and real-time fluorescence. In a set of 30 clinical saliva samples, colorimetric RT-LAMP and RT-qPCR assays performed directly on pretreated saliva samples without RNA extraction had accuracies greater than 90%.
Conclusions
Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric RT-LAMP is a simple, sensitive, and cost-effective approach with broad potential to expand diagnostic testing for the virus causing COVID-19.
Keywords: SARS-CoV-2, COVID-19, acute respiratory syndrome, LAMP, RT-LAMP, saliva, rapid diagnosis, point-of-care testing, high-throughput testing, cost-effective, preventive medicine
Introduction
Establishing rapid and widespread testing for coronavirus disease 2019 (COVID-19) is essential to containing the pandemic and safely reopen society. The current gold-standard test measures viral nucleic acids extracted from clinical swabs by reverse-transcription quantitative polymerase chain reaction (RT-qPCR). This assay requires trained medical personnel, specialized instrumentation, supply-limited reagents, and substantial technical labor. Isothermal nucleic acid amplification tests are an alternative to conventional PCR methods that do not require expensive instruments to perform the reaction or interpret the results. Specifically, loop-mediated isothermal amplification with simultaneous reverse-transcription (RT-LAMP) allows for rapid and analytically sensitive detection of nucleic acids within 1 hour in an easily interpretable colorimetric assay that requires only a heat source (1, 2).
Several groups are currently developing LAMP-based protocols for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing COVID-19 (3–13). The analytical sensitivity of LAMP on purified RNA compares well to RT-qPCR, and LAMP may achieve higher sensitivity on crude clinical samples (5). Robustness of LAMP to PCR inhibitors makes it especially well-suited and widely used for pathogen detection in unpurified samples (14). This confers a major potential advantage over current testing protocols as it enables skipping the cost-, labor-, time-, and reagent-consuming RNA extraction step.
Saliva is a promising sample for expanding and facilitating testing due to the ease, safety, and noninvasive nature of its collection and its relatively high viral load (15, 16). Direct comparison of saliva to nasopharyngeal (NP) swabs from the same individuals revealed that saliva samples provided more consistent and clinically sensitive results for SARS-CoV-2 detection (17). Here, we sought to establish and optimize a simple RT-LAMP assay for the qualitative detection of SARS-CoV-2 directly from saliva without an RNA extraction step.
Materials and Methods
LAMP Reactions
The 20-µL LAMP reactions containing 3 µL of sample were performed following New England Biolabs’ recommended protocol using WarmStart Colorimetric LAMP 2X Master Mix (NEB, M1800L). Primer sequences are provided in Table 1 in the online Data Supplement.
Table 1.
Accuracy of LAMP and RT-qPCR assays on pretreated clinical saliva samples.
| True positive | False negative | True negative | False positive | Clinical sensitivity (%, 95% CI) | Clinical specificity (%, 95% CI) | Accuracy (%, 95% CI) | |
|---|---|---|---|---|---|---|---|
| Colorimetric LAMP | 17 | 3 | 10a | 0a | 85 (62.1–96.8) | 100 (69.1–100) | 90 (73.5–97.9) |
| RT-qPCR | 19 | 1 | 10 | 0 | 95 (75.1–99.9) | 100 (69.1–100) | 96.7 (82.8–99.9) |
After retesting.
SARS-CoV-2 Standards and Controls
In vitro transcribed RNA standards were prepared as described (18). Individual aliquots (10 µL of aliquots were frozen at −80 ˚C in 8-tube strips to prevent multiple freeze-thaws). Heat-inactivated SARS-CoV-2 particles were acquired from the US Centers for Disease Control and Prevention (CDC) through BEI Resources. DNA plasmid coronavirus controls corresponding to SARS-CoV-2 and MERS were obtained from IDT as plasmid DNA solutions.
Saliva Pretreatment Protocols
We initially implemented a heat treatment of 55 ˚C for 15 min followed by 95 ˚C for 5 min. We later optimized heat treatment to 65 ˚C for 15 min followed by 95 ˚C for 5 min. Samples were cooled to 4 ˚C for 5 min before being assayed. In initial experiments, proteinase K from NEB (no. P8107S) was added to undiluted saliva at 1/10 volume (5 µL in 50 µL of saliva). Later, proteinase K was used at 100× in samples diluted 1:1 in TE buffer (10 mM Tris, 0.1 mM EDTA) or phosphate buffered saline (PBS). Optimized experiments included 1× RNAsecure (25×, ThermoFisher, AM7006). We tested the HUDSON method (19), and other pretreatment protocols (Supplemental Data).
RT-qPCR
RT-qPCR reactions were performed according to CDC Emergency Use Authorization guidelines using TaqPath 1-Step RT-qPCR Master Mix GC (ThermoFisher, A15300) and the nCoV-N1 probe from the 2019-nCoV RUO Kit (IDT). Reactions were performed on Quantstudio 3 and 6 Real-Time PCR systems (ThermoFisher). A volume of 3 µL of sample was used as input to match the LAMP protocol.
High-Throughput Colorimetric Assay
Assay scale-up was performed in a 96-well plate format (BioRad) with minor modifications to the LAMP reaction. Here, 4 µL of saliva samples were used in 25 µL total volume reactions and pretreated with the original heat treatment, proteinase K, and RNasin (Promega). Samples were run in technical triplicate at each dilution. Samples were analyzed using a BioTek Epoch microplate spectrophotometer measuring absorbances at 430 and 560 nM wavelengths (Supplemental Data).
Quantitative Real-Time LAMP (qLAMP)
qLAMP was performed by adding the DNA-binding dye SYTO 9 (ThermoFisher) to the colorimetric LAMP reaction (1 µM) and performing the reaction on a QuantStudio 3 or 6 RT-PCR system. Machines were programmed to run 90 or 120 isothermal cycles of 30 s at 65 ˚C, then slowly ramped up to 95 ˚C for inactivation and melt-curve analysis.
Data Analysis
Data were analyzed and plotted in R (v.3.5.1) using ggplot2. qLAMP/RT-qPCR experiments were analyzed using Quantstudio Design & Analysis Software (v.2.3.3, ThermoFisher), or exported for analysis in R. For sensitivity analysis, we fit a probit regression model to estimate the limit of detection (LOD).
Clinical Samples
Saliva collection was approved by the institutional review board at Washington University School of Medicine (WU350, IRB no. 202003085). Informed consent was obtained for all participant samples. Saliva samples from individuals who were COVID-19 positive and negative were diluted 1:1 in PBS to facilitate pipetting and then frozen. Some samples were heat-treated at 56 ˚C for 30 minutes for viral inactivation. Most samples underwent several freeze-thaw cycles prior to assaying.
Results
LAMP Primer Screening
We compared the performance of 5 sets of recently developed LAMP primer sets targeting different regions of the SARS-CoV-2 genome (3–6). Of these, the NEB Gene N-A (3) and Lamb et al. (4) primers targeting the nucleocapsid (Gene N) and Orf1ab regions, respectively, had the highest analytical sensitivity, lowest rates of false positives in water-only controls, and no cross-reactivity with MERS coronavirus controls (Supplemental Fig. 1).
RT-LAMP Reaction Optimization in Simulated Samples
Next, we validated these primers on both RNA standards and heat-inactivated viral particles spiked into water or human saliva to simulate clinical samples (Supplemental Fig. 2, A). Saliva strongly inhibited LAMP detection of SARS-CoV-2 compared to water (Supplemental Fig. 2, B). Particles were weakly detected in saliva whereas their detection in water was on par with detection of RNA (Supplemental Fig. 2, C) indicating the presence of an inhibitor in saliva that impaired the assay. Colorimetric interpretation was time-sensitive with many samples, including negative controls, turned yellow in LAMP reactions longer than 40 minutes due to nonspecific amplification (20, 21). A 30-minute incubation provided a reliable readout.
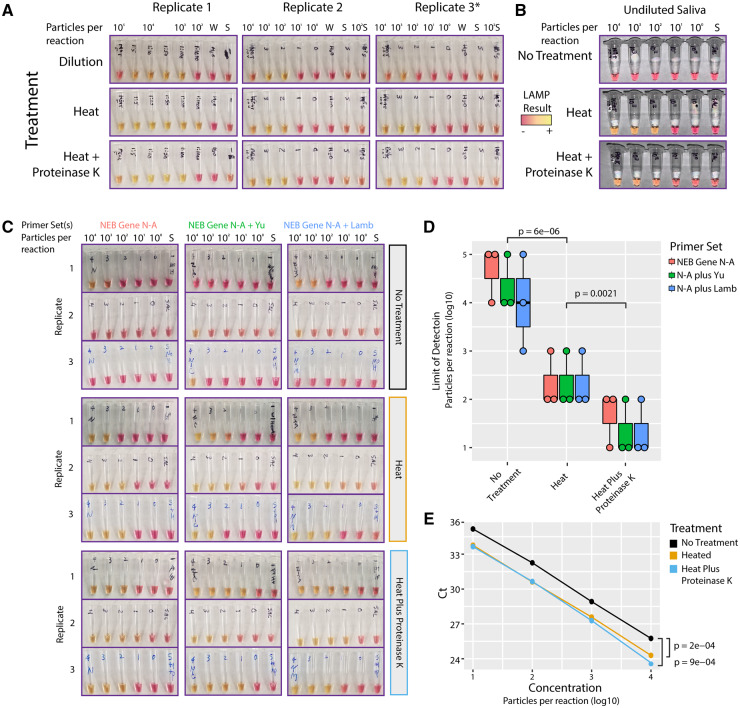
To neutralize or otherwise reduce inhibitors in human saliva, we tested several approaches that have been demonstrated to improve viral RNA detection in crude samples (19, 22–25). First, we found that simple dilution of saliva into water enabled sensitive detection of SARS-CoV-2 particles using LAMP (Fig. 1, A, top). Heat treatment with or without proteinase K further improved LAMP assay sensitivity (Fig. 1, A) and enabled SARS-CoV-2 particle detection in undiluted human saliva samples (Fig. 1, B). This simple pretreatment conferred a consistent LOD on the order of 102 particles per reaction, representing a 10 000-fold improvement in sensitivity over assays on untreated saliva.
Fig. 1.
Dilution, heat, and proteinase K treatments improve SARS-CoV-2 detection from saliva. (A), Dilution of particle-containing saliva into water improved LAMP detection by at least two orders of magnitude from undetectable to ∼103 particles per reaction. Heat treatment and heat treatment plus proteinase K further increased LAMP sensitivity to ∼102 viral genome equivalents per reaction. *Replicate 3 used Lamb et al. primers but gave nearly identical results to NEB Gene N-A primers. (B), Heat treatment with or without proteinase K increased LAMP sensitivity from 106 to ∼102 viral genome equivalents in undiluted saliva. (C), Multiplexed primers improved LAMP sensitivity. LAMP reactions using NEB Gene N-A primers alone or in combination with Yu et al. or Lamb et al. primers are shown. S = negative control saliva. Viral particles per reaction are indicated. (D), Saliva pretreatments significantly improved LAMP sensitivity. Heat treatment improved limit of detection (P = 6e-6, t-test, two-tailed vs ‘No Treatment’). Proteinase K treatment further improved heat treatment (P = 0.002, t-test, two-tailed vs ‘Heat’). Multiplexed primers increased the frequency of detection at ∼101 particles/reaction. N = NEB Gene N-A. (E), RT-qPCR on crude saliva using the CDC N1 probe showed increased sensitivity with either heat or proteinase K treatment (P < 1e-3 for either treatment, two-tailed paired t-test).
We experimented with additional heat and chemical pretreatments including the HUDSON protocol (19) and various detergents, but each of these conditions decreased assay sensitivity or interfered with colorimetry (Supplemental Fig. 3, A–C). Varying the amount of crude sample input to the LAMP reaction, we found that adding up to 8 µL of direct saliva was compatible with the assay (Supplemental Fig. 3, D).
Multiplexing LAMP Primer Sets
To further improve assay accuracy, we sought to multiplex LAMP primer sets in a single reaction. Combining primers can potentially increase sensitivity through additive signals of simultaneous amplification reactions (8). Nonspecific primer interactions, however, could result in increased rates of false positives. We compared pairwise combinations of NEB Gene N-A primers with the other 4 primer sets targeting various regions across the SARS-CoV-2 genome. All pairs of primer sets outperformed the NEB Gene N-A primer set alone, with no apparent increase in spurious background amplification (Supplemental Fig. 4).
We next tested whether multiplexing primer sets could improve signal detection in untreated and heat and chemical treated particle-containing saliva (Fig. 1, C). Heat treatment alone gave a marked improvement in SARS-CoV-2 particle detection from saliva (Fig. 1, D, P < 1e-5, two-sided t-test). Heat treatment plus proteinase K further improved assay sensitivity compared to heat alone (P < 0.003, two-sided t-test). Multiplexed primer sets slightly improved the sensitivity of the assay, increasing the frequency of detection in samples with ∼101 particles per reaction. At this sensitivity, the multiplexed LAMP assay would detect the vast majority of COVID-19 positive samples based on reported saliva viral loads (median ∼102–103 viral copies per µL) (16, 17), and virtually all infectious individuals (26). As viral loads and contagiousness peak around the time of symptom onset, LAMP would have the highest accuracy at this critical timepoint for isolating infectious carriers (27).
To determine whether our extraction-free protocol also improved the sensitivity of SARS-CoV-2 detection by conventional RT-qPCR, we performed RT-qPCR using the CDC Gene N1 hydrolysis probe set directly on untreated and treated simulated saliva samples. We found that RT-qPCR had similar analytical sensitivity to LAMP on crude samples, reliably detecting SARS-CoV-2 in all samples down to ∼101 particles per reaction (Fig. 1, E). We observed strong improvements in cycle thresholds (Ct) using either heat alone or heat plus proteinase K (P < 1e-3, two-tailed paired t-tests), increasing the sensitivity of viral RNA detection by RT-qPCR by 3 to 4-fold. Taken together, our results show that a simple, extraction-free pretreatment protocol can significantly improve the LOD of downstream nucleic acid-based assays.
Initial Assay Validation on Clinical Samples
We validated the initial colorimetric RT-LAMP assay on 5 COVID-19 positive samples. Four of 5 samples pretreated with heat plus proteinase K tested positive after a 30-minute RT-LAMP reaction (Supplemental Fig. 5, A). We performed RT-qPCR directly on the untreated and treated samples. Results were qualitatively concordant with the RT-LAMP results, identifying the same 4 positives (Supplemental Fig. 5, B). Pretreatment significantly improved viral detection by RT-qPCR (Supplemental Fig. 5, C). Together, these results demonstrated the feasibility of our assay on actual clinical samples.
Establishing a High-Throughput Quantitative Assay
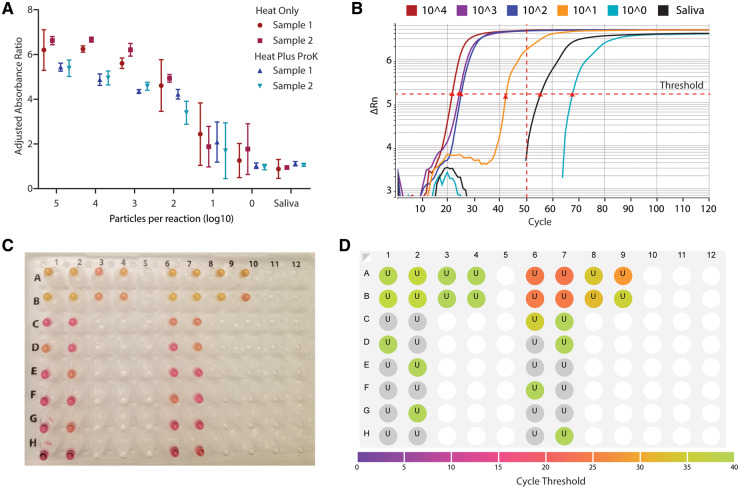
To enable substantial scale-up of testing capacity using RT-LAMP, we adapted our protocol to a 96-well plate format measuring absorbance. Spectrophotometric plate scanning before and after the assay provided an unbiased, quantitative interpretation. Heat treatment with and without proteinase K enabled unbiased and sensitive detection of viral particles in saliva samples down to 102 particles per reaction (Fig. 2, A).
Fig. 2.
Establishing high-throughput LAMP assays with quantitative readouts. (A), RT-LAMP assay was adapted to a high-throughput 96-well plate format with a quantitative absorbance readout, achieving a limit of detection <102 particles per reaction from saliva samples. Absorbance for 430 nM (yellow) and 560 nM (red) wavelengths was measured before and after the LAMP reaction and normalized to negative controls. Heat indicates 55 ˚C for 15 minutes, 95 ˚C for 3 minutes, with or without proteinase K (ProK). Two biological replicates were each run in triplicate. (B), Real-time quantitative fluorescent LAMP results are shown for a dilution series of particles in saliva. Change in fluorescence (delta Rn) is monitored over 120 ‘cycles’ of 30-second incubations at 65 ˚C. Cycle thresholds (Cts), indicated by red triangles, represent the time at which total fluorescence reaches a given level. Samples with higher viral loads reach this threshold earlier. Nonspecific amplification may arise after 50 cycles, corresponding to 25 minutes. (C), Colorimetric and (D), fluorescent results for the same reaction show that the fluorescent dye does not interfere with colorimetric interpretation. Results are concordant with colorimetric LAMP, and fluorescent results are more quantitative.
We next sought to establish a quantitative real-time fluorescent-based LAMP assay (qLAMP) using the DNA intercalating dye SYTO 9 (28). qLAMP offers several potential advantages over colorimetric LAMP including real-time reaction monitoring and melt-curve analysis to discriminate false positives. We benchmarked qLAMP using contrived samples of known amounts of viral particles in diluted saliva, and we determined that a Ct of 50 (25 minutes) reliably discriminated positive reactions from nonspecific amplifications (Fig. 2, B). SYTO 9 did not interfere with colorimetric RT-LAMP allowing assay interpretation by colorimetry or fluorimetry (Fig. 2, C) with qualitatively concordant results (Fig. 2, D). Spectrophotometry and real-time LAMP therefore represent 2 alternative modalities for high-throughput, unbiased LAMP implementation.
Improving Compatibility with Point-of-Care Testing
We sought to develop a saliva pretreatment compatible with a single isothermal heat source to reduce equipment requirements and facilitate point-of-care testing (Supplemental Data). Pretreatment at 65 ˚C for 15 min of diluted saliva improved detection (Supplemental Fig. 6). RNAsecure, guanidine hydrochloride (40 mM), and primer multiplexing further enhanced assay sensitivity. Pulse spinning samples in a microfuge prior to the LAMP reaction improved assay reliability. While not quite as sensitive as pretreatments including a 95 ˚C heat step, these optimizations enable assaying saliva by colorimetric RT-LAMP using a single heat source, simplifying point-of-care testing.
Reoptimizing the RT-LAMP Assay
Given the improvements we observed in assay sensitivity using RNAsecure, guanidine, and sample dilution into TE, we incorporated these into an optimized protocol. We increased the 55 ˚C stage of the original heat treatment to 65 ˚C for better inactivation of virus, RNases, and reaction inhibitors. In these conditions, we observed a low rate of nonspecific amplification arising in experiments with multiplexed primers (20) (Supplemental Fig. 7). Switching to primer sequences redesigned by NEB targeting the nucleocapsid and envelope small membrane protein (E) genes (NEB-N2 and NEB-E1 primers) (13) solved this issue.
We validated the performance of the new primers with both colorimetric RT-LAMP and qLAMP. These experiments indicated the new NEB-N2 primer set outperformed the previous primers in both sensitivity and time to threshold (Supplemental Fig. 8, A and B). RNAsecure improved analytical sensitivity across all primer sets (Supplemental Fig. 8, C). Multiplexing NEB-N2 with NEB-E1 gave consistent results with no false positives in saliva-only controls (Supplemental Fig. 8, D). Guanidine improved both the speed and sensitivity of the LAMP reactions (P < 0.001, Supplemental Fig. 8, D and E) (13).
LOD Analysis of Optimized Assay
We assayed serial dilutions of viral particles to estimate the concentration of target viral particles that could be detected with a probability of 0.95 (LOD95). To account for variation across donors, we spiked viral particles into saliva from at least 3 donors. In parallel, we tested whether proteinase K inclusion was beneficial. Our optimized assay was highly sensitive across all donors, with a LOD95 of 59 (44–104) particle copies per reaction and 100% (86–100) specificity (Supplemental Fig. 9, A).
Proteinase K significantly improved SARS-CoV-2 detection, especially in samples with the lowest viral amounts (P = 0.006, Supplemental Fig. 9, B). For samples that included proteinase K, LOD95 was estimated to be 27 (22–47) particles per reaction (Supplemental Fig. 10, A), a major improvement compared to assays without proteinase K treatment [LOD95: 79 (55–175), Supplemental Fig. 10, B].
We retested the optimal amount of saliva that should be added to the reaction and found that increasing amounts of pretreated saliva impeded reaction times (Supplemental Fig. 9, E). Based on this observed reaction inhibition at higher levels of input saliva, we recommend adding 1–4 µL of pretreated saliva per 20-µL LAMP reaction. Reaction volumes and saliva amounts can be scaled up proportionally to increase assay sensitivity but at higher costs per reaction.
Finally, our optimized saliva pretreatment protocol again markedly improved the detection of SARS-CoV-2 by RT-qPCR (Supplemental Fig. 9, F). Compared to untreated saliva, heat, and proteinase K pretreatment improved viral detection by an average 3.4-fold (P < 3e-9, t-test). RT-qPCR on direct saliva without RNA extraction is thus another viable option to overcome bottlenecks limiting widespread testing.
Clinical Validation of Optimized Assay
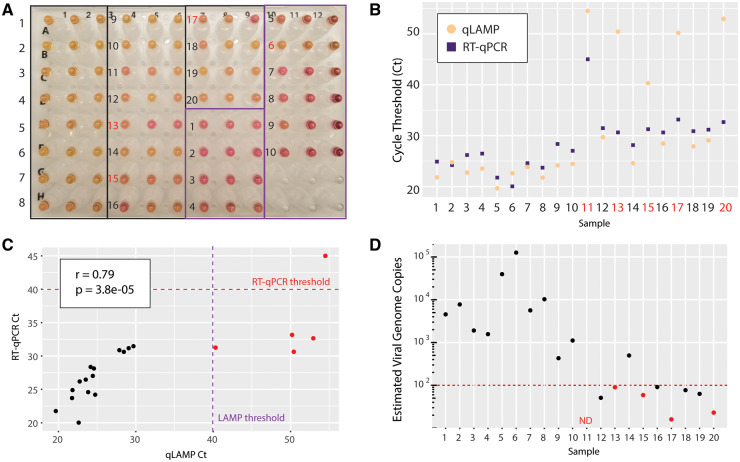
We tested our optimized colorimetric RT-LAMP and RT-qPCR protocols on 30 additional clinical samples (20 positive and 10 negative samples). By naked-eye interpretation of colorimetric RT-LAMP, 17/20 positive samples were correctly called positive and 9/10 negative samples were called negative (Fig. 3, A). The false positive result was called negative when retested with an alternate set of LAMP primers, indicating possible carryover contamination of LAMP amplicons (29). This can be prevented by including uracil (dUTP) and uracil glycosylase in the reactions (30), as implemented in clinical RT-qPCR. qLAMP Cts had strong overall agreement with colorimetric results. qLAMP Cts for 5 samples were too high to distinguish between weak positive signal or nonspecific amplification (Fig. 3, B). RT-qPCR correctly called 19/20 positive samples (Fig. 3, B, purple squares), and 10/10 negative samples (Table 1). RT-qPCR Cts were well-correlated with LAMP Cts (Fig. 3, C). One positive sample was undetected by either method, suggesting sample degradation or very low viral levels.
Fig. 3.
Clinical validation of optimized colorimetric LAMP assay and RT-qPCR. (A), Colorimetric LAMP results on 30 clinical samples (20 positives, 10 negatives). Correct calls are numbered in black. Incorrect calls are numbered in red. (B), qLAMP and RT-qPCR results on pretreated samples are shown for each sample. Red numbers indicate false negative calls by qLAMP. (C), qLAMP and RT-qPCR Ct values were highly correlated. Thresholds of 40 cycles were used for determining positivity for LAMP and RT-qPCR as indicated. Samples called negative by either of these methods are colored red. (D), Estimates of viral genome copy number were made for each sample using a quantitative standard curve by RT-qPCR. Samples called negative by LAMP are colored red. Red-dotted line indicates 100 viral genomes. ND = not detected.ry
We included a quantitative dilution series of particles in the RT-qPCR assay to estimate viral loads in clinical samples. The median estimated viral copy number in positive samples was ∼500 copies per µL (range 16–126 000). All false negatives in the LAMP assay had fewer than 100 estimated viral copies per µL, which is likely below the threshold for viral transmission (26). Nevertheless, among 8 samples below this level, LAMP still detected virus in half the samples. Above this level, the assay achieved 100% (71.5 to 100) sensitivity. Therefore, colorimetric RT-LAMP on pretreated saliva samples without RNA extraction is a cheap, fast, and accurate method for SARS-CoV-2 testing.
Discussion
Our proposed approach combines 3 promising avenues to enable rapid and widespread SARS-CoV-2 testing: (a) colorimetric RT-LAMP, (b) self-collected saliva specimens, and (c) direct testing on crude saliva samples without RNA extraction. This approach solves 2 major bottlenecks in massively scaling up COVID-19 nucleic acid testing: sample collection and RNA extraction, and it enables test result turnaround times of <1 hour. Using both colorimetric RT-LAMP and RT-qPCR directly on treated samples without RNA extraction, we demonstrated high accuracy in simulated and actual clinical saliva samples.
Due to its ease of use, rapid amplification of nucleic acids, and high specificity RT-LAMP has been widely used for pathogen detection. Sensitive diagnostic assays have been developed for viruses including Zika (31), and such assays are being developed for SARS-CoV-2 by several groups including ours (3–13). Its low cost, fast turnaround time, and simple colorimetric readout make RT-LAMP an effective solution for ramping up global testing capacity. Further, because it does not require specialized equipment or training for performing or interpreting the assay, colorimetric RT-LAMP is especially well-suited for point-of-care detection.
NP swabs are uncomfortable and must be carefully performed by a trained health-care worker using personal protective equipment. Mid-nasal swabs are a promising alternative to NP swabs because they can be self-administered and contain high viral loads (32–34). We instead focused on saliva due to its ease of collection, high viral load, and potential swab shortages (17). Saliva is a challenging clinical matrix due to variability across individuals in pH and viscosity and the presence of reaction inhibitors (35). Here, we have overcome these challenges and developed a saliva pretreatment protocol that enables sensitive detection of SARS-CoV-2 by both RT-LAMP and RT-qPCR without an RNA extraction step.
While RNA extraction methods improve sample purity and increase viral concentration, they are cost-, labor-, and time-consuming, and some reagents are still in short supply. Several groups are optimizing workarounds to avoid the RNA extraction step for nucleic acid-based SARS-CoV-2 testing (22, 23, 25). We achieved a careful balance between the inactivation of reaction inhibitors and the preservation of viral RNA with heat and chemical pretreatment. A 1:1 dilution of saliva followed by treatment with RNAsecure and 65 ˚C incubation potently reduces reaction inhibitors. This can be implemented with a single heat source isothermal with the LAMP reaction in a point-of-care setting. Proteinase K addition and a brief incubation at 95 ˚C further reduce inhibitors, ease saliva handling, and improve assay sensitivity. For high-throughput testing in a centralized laboratory, this pretreatment protocol can be coupled with colorimetric RT-LAMP using a spectrophotometric or fluorescent readout, or with RT-qPCR.
Our protocol has several potential limitations for clinical implementation. While many studies have shown that saliva samples have higher and more stable viral loads than found in NP swabs (17), NP and other clinical specimens may provide higher sensitivity for viral detection (34). The saliva collection protocol is limited to individuals who can produce enough saliva. Contaminants in saliva, such as food or mucus, might influence downstream assays, although our protocol and additives should mitigate these factors. Carryover contamination of LAMP amplicons is a major concern for clinical implementation (29), but this can be prevented by including dUTP and a uracil-DNA glycosylase in the reaction (30). While qLAMP facilitated the comparison of multiple conditions, future implementation of digital LAMP could enable more exhaustive optimization of reaction conditions (21). For clinical assay implementation, samples should be tested in duplicate across multiple primer sets targeting the SARS-CoV-2 genome and an internal human RNA control. Finally, the complexity of the optimized two-step protocol of saliva pretreatment and downstream assay still requires some training and equipment that may preclude at-home implementation.
Direct colorimetric RT-LAMP on saliva has broad potential to increase COVID-19 screening speed and capacity, and has high flexibility in implementation depending on equipment availability. Extension of testing to asymptomatic individuals and increased test frequency would promote the application of predictive, preventive, and personalized medicine (36). Expanded testing would improve the predictive reliability of modeling disease spread, inform better containment policies, and identify and protect vulnerable populations (37). Integrating test results into contact tracing tools would help provide personalized risk assessments, enabling the self-isolation of exposed individuals and the avoidance of high-risk areas by healthy individuals (38). Expanded testing and contact tracing are essential for successful management of the pandemic.
In summary, we have developed a saliva pretreatment protocol which enables sensitive SARS-CoV-2 detection from unpurified samples in an optimized colorimetric RT-LAMP assay and RT-qPCR. This optimization overcomes the burdensome step of RNA extraction, and alleviates some of the time, labor, and reagent bottlenecks of the current gold-standard nucleic acid-based tests. Our extensive optimizations have enabled reliable detection below ∼102 viral genomes per reaction from saliva samples. Individuals who are COVID-19 positive with viral loads below this level are likely not infectious (26). Colorimetric RT-LAMP and RT-qPCR assays achieved high accuracy on clinical saliva specimens without RNA extraction. Because of the flexibility of implementation and readout, our assay can be deployed as a point-of-care test or in a centralized laboratory facility and has broad potential to expand diagnostic testing for the virus causing COVID-19.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Nonstandard Abbreviations
- COVID-19
Coronavirus disease 2019
- RT
reverse-transcription
- LAMP
loop-mediated isothermal amplification
- qPCR
quantitative polymerase chain reaction
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- NP
nasopharyngeal
- TE
Tris EDTA
- qLAMP
quantitative real-time fluorescent-based LAMP assay
- Ct
cycle threshold
- LOD
limit of detection
- N
nucleocapsid gene
- E
envelope small membrane protein gene
Contributor Information
Matthew A Lalli, Email: mlalli@wustl.edu.
Robi D Mitra, Email: rmitra@wustl.edu.
Jeffrey Milbrandt, Email: jmilbrandt@wustl.edu.
Author Declaration
A version of this article was previously posted as a preprint on medRxiv as https://www.medrxiv.org/content/10.1101/2020.05.07.20093542v2.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
M.A. Lalli, X. Chen, S.J. Langmade, C.S. Sawyer, L.C. Burcea, M.N. Wilkinson, and W.J. Buchser developed and optimized the LAMP assay with significant intellectual contribution from M. Heinz, R.D. Mitra, R.S. Fulton, R.D. Head, J. Milbrandt. Clinical samples were obtained, prepared, and tested by M.A. Lalli, C.C. Fronick, S.J. Langmade, and R.S. Fulton. Spectrophotometric assay scale-up and quantitative readout were implemented and analyzed by C.S. Sawyer, L.C. Burcea, M. Heinz, and R.D. Head. Fluorescent assay scale-up was implemented and analyzed by M.A. Lalli, X. Chen, S.J. Langmade, M.N. Wilkinson, and W.J. Buchser. M.A. Lalli analyzed the data and prepared the manuscript, with editing and revision by all authors.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
None declared.
Consultant or Advisory Role
J. Milbrandt, Disarm Therapeutics.
Stock Ownership
J. Milbrandt, Disarm Therapeutics.
Honoraria
None declared.
Research Funding
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, Heat Inactivated, NR-52286. These studies were supported by donations to the WUSM COVID Research fund, the Department of Genetics and the McDonnell Genome Institute, and NIH grants P30 CA91842 and UL1 TR000448. The Alvin J. Siteman Cancer Center at Washington University School of Medicine (WUSM) and Barnes-Jewish Hospital, the Institute of Clinical and Translational Sciences (ICTS), and the Tissue Procurement Core provided saliva samples. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant no. P30 CA091842 and the ICTS is funded by NCATS Clinical and Translational Science Award (CTSA) program grant no. UL1 TR002345.
Expert Testimony
None declared.
Patents
M.A. Lalli, provisional; W.J. Buchser, pending patent around the implementation of this; R.D. Head, provisional patent; R.D. Mitra, patent application filed related to this work.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine (WUSM) and Barnes-Jewish Hospital, the Institute of Clinical and Translational Sciences (ICTS), and the Tissue Procurement Core, which provided saliva samples. We thank Feiming Chen for the R code used to fit a probit curve and calculate LOD95.
References
- 1. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000;28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanner NA, Zhang Y, Evans TC.. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 2015;58:59–68. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. Preprint at https://www.medrxiv.org/content/10.1101/2020.02.26.20028373v1 (2020).
- 4. Lamb LE, Bartolone SN, Ward E, Chancellor MB.. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020;15:e0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El-Tholoth M, Bau HH, Song J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. A version of this paper is available as a preprint at https://chemrxiv.org/articles/A_Single_and_Two-Stage_Closed-Tube_Molecular_Test_for_the_2019_Novel_Coronavirus_COVID-19_at_Home_Clinic_and_Points_of_Entry/11860137 (2020).
- 6. Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem 2020;66:975–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci USA2020;117:24450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhadra S, Riedel TE, Lakhotia S, Tran ND, Ellington AD. High-surety isothermal amplification and detection of SARS-CoV-2, including with crude enzymes. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.13.039941v3 (2020). [DOI] [PMC free article] [PubMed]
- 9. Osterdahl MF, Lee KA, Lochlainn MN, Wilson S, Douthwaite S, Horsfall R, et al. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis 2020;20:783. [DOI] [PMC free article] [PubMed]
- 10. Schmid-Burgk JL, Li D, Feldman D, Slabicki M, Borrajo J, Strecker J, et al. LAMP-Seq: population-scale COVID-19 diagnostics using a compressed barcode space. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.06.025635v2 (2020).
- 11. Butler DJ, Mozsary C, Meydan C, Danko DC, Foox J, Rosiene J, et al. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.20.048066v5 (2020). [DOI] [PMC free article] [PubMed]
- 12. Ben-Assa N, Naddaf R, Gefen T, Capucha T, Hajjo H, Mandelbaum N, et al. Direct on-the-spot detection of SARS-CoV-2 in patients. Exp Biol Med (Maywood) 2020;245:1187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, Tanner NA.. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques 2020;69:178–85. [DOI] [PubMed] [Google Scholar]
- 14. Francois P, Tangomo M, Hibbs J, Bonetti E-J, Boehme CC, Notomi T, et al. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 2011;62:41–8. [DOI] [PubMed] [Google Scholar]
- 15. To KK-W, Tsang OT-Y, Yip CC-Y, Chan K-H, Wu T-C, Chan JM-C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 2020;71:841–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020;20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 2020;383:1283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol 2020;5:1299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018;360:444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meagher RJ, Priye A, Light YK, Huang C, Wang E.. Impact of primer dimers and self-amplifying hairpins on reverse transcription loop-mediated isothermal amplification detection of viral RNA. Analyst 2018;143:1924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rolando JC, Jue E, Schoepp NG, Ismagilov RF.. Real-time, digital LAMP with commercial microfluidic chips reveals the interplay of efficiency, speed, and background amplification as a function of reaction temperature and time. Anal Chem 2019;91:1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marzinotto S, Mio C, Cifu’ A, Verardo R, Pipan C, Schneider C, et al. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. pPreprint at https://www.medrxiv.org/content/10.1101/2020.04.06.20054114v1 (2020). [DOI] [PMC free article] [PubMed]
- 23. Bruce EA, Huang M-L, Perchetti GA, Tighe S, Laaguiby P, Hoffman JJ, et al. Direct RT-qPCR detection of Sars-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol 2020;18:e3000896. [DOI] [PMC free article] [PubMed]
- 24. Li L, He J, Wang W, Xia Y, Song L, Chen Z, et al. Development of a direct reverse-transcription quantitative PCR (dirRT-qPCR) assay for clinical Zika diagnosis. Int J Infect Dis 2019;85:167–74. [DOI] [PubMed] [Google Scholar]
- 25. Smyrlaki I, Ekman M, Lentini A, Rufino de Sousa N, Papanicolaou N, Vondracek M, et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun 2020;11:4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. Preprint at https://www.medrxiv.org/content/10.1101/2020.06.22.20136309v3 (2020). [DOI] [PMC free article] [PubMed]
- 27. He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020;26:672–5. [DOI] [PubMed] [Google Scholar]
- 28. Oscorbin IP, Belousova EA, Zakabunin AI, Boyarskikh UA, Filipenko ML.. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP). Biotechniques 2016;61:20–5. [DOI] [PubMed] [Google Scholar]
- 29. Tomita N, Mori Y, Kanda H, Notomi T.. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 2008;3:877–82. [DOI] [PubMed] [Google Scholar]
- 30. Hsieh K, Mage PL, Csordas AT, Eisenstein M, Soh HT.. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem Commun 2014;50:3747–9. [DOI] [PubMed] [Google Scholar]
- 31. Bhadra S, Saldaña MA, Han HG, Hughes GL, Ellington AD.. Simultaneous detection of different Zika virus lineages via molecular computation in a point-of-care assay. Viruses 2018;10:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivatsan S, Han PDRKvan Wolf CR, McCulloch DJ, Kim AE, et al. Preliminary support for a “dry swab, extraction free” protocol for SARS-CoV-2 testing via RT-qPCR. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.22.056283v1 (2020).
- 33. Tu Y-P, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med 2020;383:494-6. [DOI] [PMC free article] [PubMed]
- 34. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ochert AS, Boulter AW, Birnbaum W, Johnson NW, Teo CG.. Inhibitory effect of salivary fluids on PCR: potency and removal. Genome Res 1994;3:365–8. [DOI] [PubMed] [Google Scholar]
- 36. Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J 2016;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaari L, Golubnitschaja O.. Covid-19 pandemic by the “real-time” monitoring: the Tunisian case and lessons for global epidemics in the context of 3PM strategies. EPMA J 2020;11:133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radanliev P, De Roure D, Walton R, Van Kleek M, Montalvo RM, Santos O, et al. COVID-19 what have we learned? The rise of social machines and connected devices in pandemic management following the concepts of predictive, preventive and personalized medicine. EPMA J 2020;11:311–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.