Abstract
Background
Repeated coronavirus disease 2019 (COVID-19) molecular testing can lead to positive test results after negative results and to multiple positive results over time. The association between positive test results and infectious virus is important to quantify.
Methods
A 2-month cohort of retrospective data and consecutively collected specimens from patients with COVID-19 or patients under investigation were used to understand the correlation between prolonged viral RNA positive test results, cycle threshold (Ct) values and growth of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in cell culture. Whole-genome sequencing was used to confirm virus genotype in patients with prolonged viral RNA detection. Droplet digital polymerase chain reaction was used to assess the rate of false-negative COVID-19 diagnostic test results.
Results
In 2 months, 29 686 specimens were tested and 2194 patients underwent repeated testing. Virus recovery in cell culture was noted in specimens with a mean Ct value of 18.8 (3.4) for SARS-CoV-2 target genes. Prolonged viral RNA shedding was associated with positive virus growth in culture in specimens collected up to 21 days after the first positive result but mostly in individuals symptomatic at the time of sample collection. Whole-genome sequencing provided evidence the same virus was carried over time. Positive test results following negative results had Ct values >29.5 and were not associated with virus culture. Droplet digital polymerase chain reaction results were positive in 5.6% of negative specimens collected from patients with confirmed or clinically suspected COVID-19.
Conclusions
Low Ct values in SARS-CoV-2 diagnostic tests were associated with virus growth in cell culture. Symptomatic patients with prolonged viral RNA shedding can also be infectious.
Keywords: SARS-CoV-2, COVID-19, repeat testing, infectious virus
A correlation between severe acute respiratory syndrome coronavirus 2 diagnostic cycle threshold values and virus growth in cell culture is assessed in the context of prolonged viral RNA shedding and positive after negative results. Droplet digital polymerase chain reaction is used to investigate rates of false-negativity.
Molecular methods for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid detection from nasopharyngeal swabs have been the reference standard for coronavirus disease 2019 (COVID-19) diagnosis. Although diagnostic approaches target different genes within the SARS-CoV-2 genome, they have shown comparable analytical sensitivity and high specificity [1–19]. The accuracy of the assay’s result is associated with the shedding pattern of SARS-CoV-2 RNA and can vary based on the source of respiratory specimens, the sufficiency of specimen collection, and the course of illness [20–25].
Infection control personnel and physicians managing patients with COVID-19 and patients under investigation continue to face several diagnostic dilemmas related to a lack of understanding of the clinical sensitivities of SARS-CoV-2 molecular diagnostics and the correlation between viral RNA detection and shedding of infectious virus. Retesting of patients has become a common practice, especially when there is a strong clinical suspicion or exposure history and an initial negative result [26]. A single positive molecular result should be sufficient for confirming COVID-19 diagnosis; however, repeated testing of hospitalized patients to determine isolation needs and infection control measures has become part of managing this patient population.
Two negative molecular assay results from 2 consecutively collected respiratory specimens >24 hours apart has been the initial strategy used in the United States to enable discontinuation of transmission precautions and return to work [27]. Repeated testing in patients has revealed that SARS-CoV-2 RNA can be detectable for weeks after the onset of symptoms [28]. In general, molecular detection of SARS-CoV-2 RNA does not necessarily denote the presence of recoverable infectious virus. A few studies, as well as data from the Centers for Disease Control and Prevention (CDC), showed that higher viral loads are associated with recovery of infectious virus and that virus recovery is generally not reported >9 days after symptom onset [22, 29, 30]. A case study, in which severe infection was associated with successful recovery of infectious SARS-CoV-2 from stool samples, indicates that the duration of recovery of infectious virus particles might vary based on the severity of disease or the duration of symptoms [31]. A careful interpretation of cell culture results is essential, as variables that include cell lines used for viral isolation and days that cell cultures were held, among other technical factors, might contribute to the success or failure of virus recovery from clinical specimens.
False-negative molecular SARS-CoV-2 results occur, and in some cases a single negative result is not sufficient for excluding a COVID-19 diagnosis. False-negative rates are estimated to range from 5% to 40%, yet a conclusive percentage is currently difficult to determine, owing to the lack of a diagnostic comparator reference standard [32, 33]. Initial false-negative results in the setting of consistent respiratory symptoms have been reported, with some patients having subsequent positive results with serial testing [34]. The Infectious Diseases Society of America recommends repeated testing after initial negative RNA test results in cases with intermediate to high suspicion of COVID-19, but evidence that this practice positively affects outcomes is still lacking [35]. Clinical sensitivity has also been attributed to the specimen type collected and the time of collection in relation to the duration of symptoms [36–46].
In the current study, we analyzed the molecular diagnostics data from Johns Hopkins Hospital in the time frame from 11 March to 11 May 2020. Our study aimed to dissect different diagnostic dilemmas by incorporating statistics of repeated testing, cycle threshold (Ct) values, virus isolation in cell culture, whole-genome sequencing, and droplet digital polymerase chain reaction (ddPCR). We address questions that include the following: How does a positive molecular test correlate with growth in cell culture? Are patients with prolonged viral RNA shedding also shedding infectious virus? Are there changes in viral sequences during prolonged shedding? Does a positive test result after undetectable viral RNA correlate with virus recovery in cell culture? And, finally, can false-negative results due to an assay’s analytical limitation (limit of detection) be detected with ddPCR?
METHODS
Study Site, Ethics, and Safety
This study was performed in the Molecular Virology Laboratory of Johns Hopkins Hospital. Cell culture studies were conducted at the Johns Hopkins Bloomberg School of Public Health. The study was approved by the Johns Hopkins University School of Medicine Institutional Review Board. Specimen handling for clinical diagnostic assays were performed in a biosafety level (BSL) 3 laboratory or a BSL-2 laboratory with BSL-3 personal protective equipment including either a powered air-purifying respirator or an N95 mask with face shield. Cell culture experiments were performed in a BSL-3 laboratory using procedures approved by the Institutional Biosafety Committee.
Clinical Data, Standard-of-Care Assays, and Specimens
Repeated testing was identified by pulling the data of all molecular COVID-19 testing that was conducted in the Johns Hopkins Hospital Microbiology laboratory from 11 March to 11 May 2019. Data were pulled using the SOFT laboratory information system. Specimens used were remnant nasopharyngeal swab specimens collected in viral transport medium (commercially purchased or custom made at Johns Hopkins University based on the CDC recipe [47]) available at the completion of standard-of-care (SOC) testing at the Johns Hopkins Laboratory. SOC testing was performed <24 hours after receipt of specimens in the laboratory, and specimens were refrigerated in the meantime. Leftover original specimens as well as nucleic acid extracts were frozen at −70°C. Cell culture and ddPCR were performed after a single freeze-thaw cycle.
During the time frame reported, several molecular diagnostic assays for SARS-CoV-2 were used, including primarily the RealStar SARS-CoV-2 reverse-transcription (RT) polymerase chain reaction (PCR) Kit 1.0 from Altona Diagnostics [3] and the NeuModx SARS-CoV-2 assay [48]. Additional assays include the CDC COVID-19 RT-PCR panel assay, the GenMark ePlex SARS-CoV-2 test [3, 49] the BD SARS-CoV-2 reagents for the BD MAX system [50], and the Xpert Xpress SARS-CoV-2 assay [51]. The Ct values shown are for specimens diagnosed using either the RealStar or the NeuModx SARS-CoV-2 assay. For simplicity, we show the Ct values of only 1 gene target per assay: the spike (S) gene for the RealStar SARS-CoV-2 and the nonstructural protein 2 gene for the NeuMoDx SARS-CoV-2 assay. Our data indicate that Ct values are comparable for the 2 genes [19]. Specimens were selected for further testing that include cell culture, sequencing, and ddPCR, largely based on the availability of leftover clinical specimens or nucleic acid extracts. Clinical data were extracted by manual chart reviews. Positive serologic results were extracted from patients charts, and the method used for serology at Johns Hopkins Hospital was described in detail elsewhere [52].
Nucleic Acid Extractions
Nucleic acid extractions for the RealStar SARS-CoV-2 assay, the ddPCR assays, and Nanopore whole-genome sequencing were performed as described elsewhere [3]. The NucliSENS easyMag or eMAG instruments (bioMérieux) were used, with software version 2.1.0.1. This extraction method was validated for our clinical diagnostic assays owing to constraints of safety and throughput compared with manual approaches. The volume of the input specimens was 500 µL, and the final elution volume was 50 µL. Specimens for automated systems were processed following each assay’s Food and Drug Administration emergency use authorization (EUA) package insert.
SARS-CoV-2 Virus Isolation
Vero E6 cells (American Type Culture Collection CRL-1586) were cultured at 37°C with 5% carbon dioxide in a humidified chamber, using complete medium (CM) consisting of Dulbecco modified Eagle medium (Sigma Life Sciences; D5796) supplemented with 10% fetal bovine serum (Gibco; sterile filtered), 1-mmol/L glutamine (Invitrogen), 1-mmol/L sodium pyruvate (Invitrogen), 100-µg/mL penicillin (Invitrogen), and 100-µg/mL streptomycin (Invitrogen). Cells were plated in 24-well dishes and grown to 75% confluence. The use of a 24-well plates allowed for more convenient isolation of larger numbers of clinical samples. The CM was removed and replaced with 150 µL of infection medium (IM), which is identical to CM but with the fetal bovine serum reduced to 2.5%. After that, 50–100 µL of the clinical specimen was added to 1 well, and the cells incubated at 37°C for 1 hour. The inoculum was aspirated and replaced with 0.5 mL of IM and the cells cultured at 37°C for 4 days. Inoculum removal minimized nonspecific cytopathic effects associated with viral transport medium.
When a cytopathic effect was visible in most of the cells, the IM was harvested and stored at −70°C. Pilot experiments using 10 infectious units of SARS-CoV-2/USA-WA-1/2020 inoculated into 1 well of a 24-well plate routinely showed nearly complete cytopathic effect within 4 days of culture. The presence of SARS-CoV-2 was verified in 1 of 2 ways. SARS-CoV-2 viral RNA was extracted using the Qiagen Viral RNA extraction kit (Qiagen), and viral RNA detected using quantitative RT-PCR, as described elsewhere [53]. Alternatively, SARS-CoV-2 viral antigen was detected by infecting Vero E6 cells grown on 4 chamber LabTek slides (Sigma Aldrich), with 50 µL of the Vero E6 virus isolate diluted in 150 µL of IM for 1 hour at 37°C. The inoculum was replaced with IM, and the culture incubated at 37°C for 12–18 hours. The cultures were fixed with 4% paraformaldehyde for 20 minutes at room temperature and processed for indirect immunofluorescence microscopy, as described elsewhere [54]. The humanized monoclonal antibody D-006 (Sino Biological) was used as the primary antibody to detect spike or S protein, followed by Alexa Fluor 488–conjugated goat anti-human immunoglobulin G. The cells were mounted on ProLong Antifade mounting medium and imaged at ×40 on a Zeiss Axio Imager M2 wide-field fluorescence microscope [55].
Oxford Nanopore Whole-Genome Sequencing
Whole-genome sequencing was conducted using the Oxford Nanopore platform following the ARTIC protocol for SARS-CoV-2 sequencing with the V3 primer set [56]. Eleven indexed samples (and 1 negative control) were pooled for each sequencing run, and 20 ng of the final pooled library was run on the Oxford Nanopore GridION instrument with R9.4.1 flow cells. Base calling and demultiplexing was performed with Guppy software, version 3.5.2, and reads were assembled using a custom pipeline modified from the ARTIC network bioinformatics pipeline (https://artic.network/ncov-2019). As part of this custom pipeline, reads were mapped to a SARS-CoV-2 reference genome (GenBank MN908947.3) using minimap2 software [57]. Coverage was normalized across the genome and variant calling was performed with Nanopolish software, version 0.13.2 [58]. Sites with low coverage (based on the negative control coverage) were marked as N. Variant calls were also independently validated with 2 other variant callers—medaka (https://nanoporetech.github.io/medaka/snp.html) and samtools (https://wikis.utexas.edu/display/bioiteam/Variant+calling+using+SAMtools)—and all sites with disagreements or allele frequency <75% were manually inspected using Integrated Genome Viewer [59]. Sites with minor allele frequency 25%–75% were replaced with International Union of Pure and Applied Chemistry ambiguity codes. Details about our SARS-CoV-2 sequencing protocols and analysis pipeline validation are available elsewhere [52].
RT-ddPCR Procedure
The ddPCR procedure followed the assay’s EUA package insert [60]. Briefly, RNA isolated from nasopharyngeal specimens (5.5 µL) was added to the master mix, comprising 1.1 µL from the CDC ddPCR triplex assay for SARS-CoV-2, 2.2 µL of proprietary reverse-transcriptase, 5.5 µL of SuperMix (ThermoFisher), 1.1 µL of dithiothreitol, and 6.6 µL of nuclease-free water. Twenty-two microliters from these samples and master mix RT-ddPCR mixtures were loaded into the wells of a 96-well PCR plate (Bio-Rad). The mixtures were then fractionated in up to 20 000 nanoliter-sized droplets in the form of a water-in-oil emulsion in the Automated Droplet Generator (Bio-Rad). The 96-well RT-ddPCR ready plate containing droplets was sealed with foil using a plate sealer (Bio-Rad) and thermocycled to achieve RT of RNA, followed by PCR amplification of complementary DNA in a C1000 Touch thermocycler (Bio-Rad). After PCR, the plate was loaded into the QX200 Droplet Reader (Bio-Rad); the droplets in each well were singulated and flowed past a 2-color fluorescence detector. The FAM and HEX fluorescence intensity of each droplet was measured and droplets were determined to be positive or negative for each target within the Bio-Rad SARS-CoV-2 ddPCR test: N1, N2, and Rnase P. The fluorescence data were then analyzed using QuantaSoft 1.7 and QuantaSoft Analysis Pro 1.0 Software to determine the presence of SARS-CoV-2 N1 and N2 in the specimen.
Statistical Analysis
Paired t tests were used to determine the mean difference in Ct values between groups, which showed successful virus growth on cell culture versus no growth.
RESULTS
COVID-19 Testing in the Johns Hopkins Hospital Network
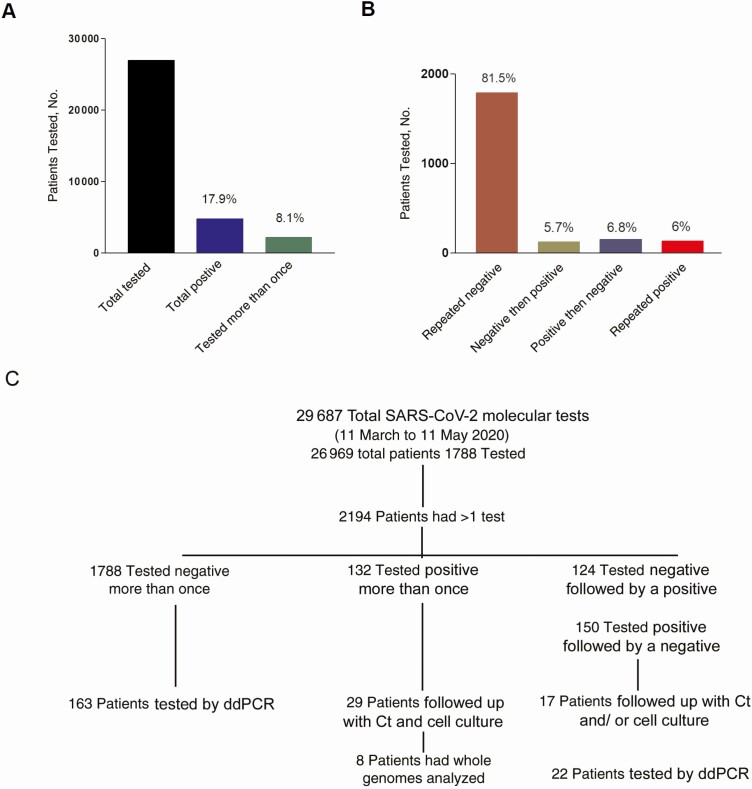
The Johns Hopkins molecular virology laboratory processed a total of 29 686 COVID-19 molecular diagnostic tests from 26 969 patients (or patients under investigation), from 11 March (first day of in-house testing) to 11 May 2020. There were 2194 patients tested more than once. Of these 1788 patients repeatedly tested negative, 132 continued to test positive at all time points, 124 had an initial negative result followed by a positive result, and 150 had an initial positive result followed by a negative result (Figure 1A and 1B). Our data indicate that of all the patients with repeated testing, 81.5% continued to have negative results, 5.7% had an initially negative followed by a positive result, and 6.8% had a final negative result after an initial positive result (Figure 1B). Figure 1C provides a layout for the subset of patients selected for subsequent testing, as discussed below and Figure 2 provides cumulative metadata for patients 1–57 (Figures 3–5).
Figure 1.
Coronavirus disease 2019 molecular testing at the Johns Hopkins Hospital. A, Total number of patients tested from 11 March through 11 May 2020, patients with positive results, and patients tested more than once. B, Total number of patients with repeated testing, assay results. (Percentages in [A] and [B] represent proportion of total.) C, Flow chart showing how patients were selected for additional analyses. Abbreviations: Ct, cycle threshold; ddPCR, droplet digital polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
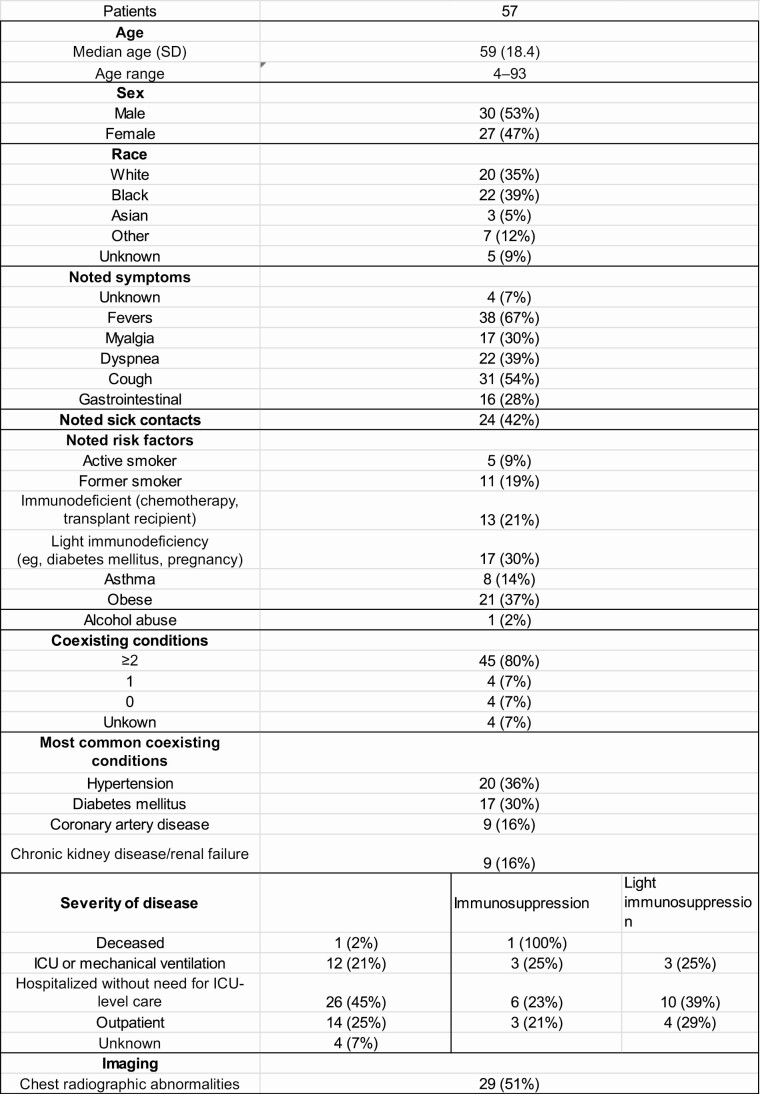
Figure 2.
Details of study patient population. Abbreviations: ICU, intensive care unit; SD, standard deviation.
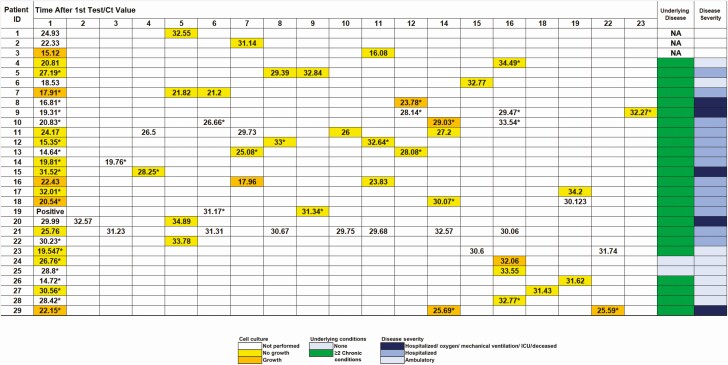
Figure 3.
Findings in patients with multiple positive molecular results over time and correlation between time of testing, isolation of infectious virus in cell culture, and cycle threshold (Ct) value of the diagnostic assay. Asterisks indicate presence of symptoms at the time of specimen collection. Abbreviations: ICU, intensive care unit; ID, identifier; NA, clinical information not available.
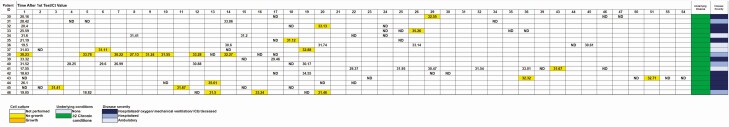
Figure 4.
Patients with positive molecular results after ≥1 negative result and correlation with time of testing, isolation of infectious virus in cell cultures, and cycle threshold (Ct) value of the diagnostic assay. Abbreviations: ID, identifier; ND, target not detected.
Figure 5.
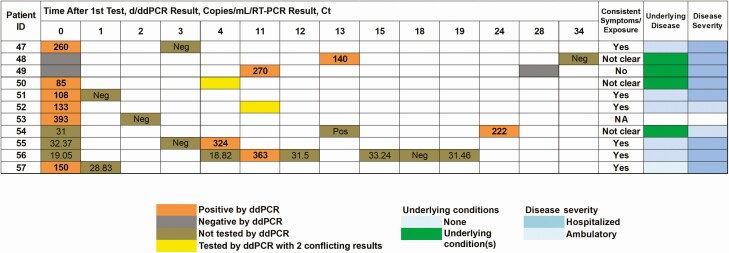
Droplet digital polymerase chain reaction (ddPCR) sensitivity of detection in patients with consecutive negative results (patients 47–53) and negative specimens collected from known positive patients (patients 54–57). ddPCR copies are shown for the N1 target. Note that a sputum sample was used in patient 51. Abbreviations: Ct, cycle threshold; ID, identifier; NA, clinical information not available; Neg, negative result with standard-of-care reverse-transcription polymerase chain reaction (RT-PCR); Pos, positive result with standard-of-care RT-PCR, with no available Ct value.
Infectious Virus Isolation and Viral RNA Load
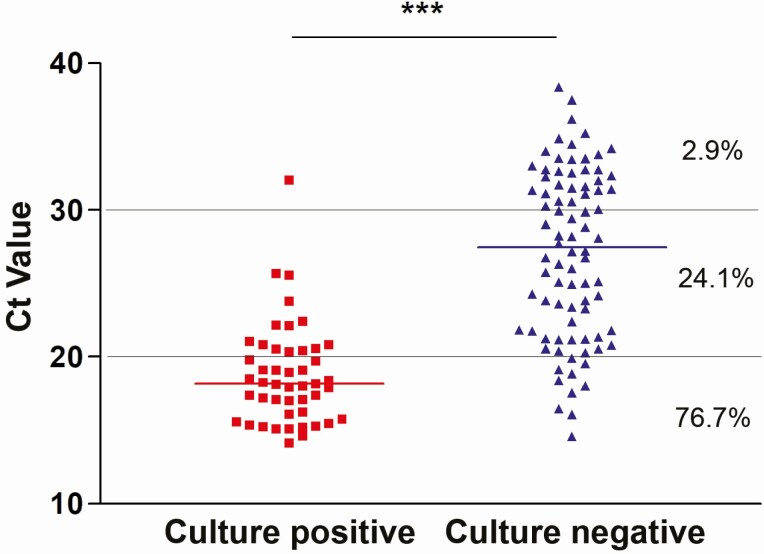
To understand the correlation between a positive molecular result and growth in cell culture, 131 patients’ specimens that were positive by molecular testing were cultured on Vero E6 cells. The cultured specimens spanned a wide range of Ct values reflecting different viral loads. The development of cytopathic effect was monitored for up to 4 days after infection of Vero E6 cells. The mean and median Ct values associated with recoverable virus were 18.8 (3.4) and 18.17, respectively, which was significantly lower than the mean and median Ct values that were not correlated with virus growth in cell culture (27.1 [5.7] and 27.5, respectively) (P < .001; paired t test) (Figure 6). Virus growth in cell culture was highly efficient in specimens with Ct values between 10 and 20 (76.7% positive isolation rate), and dropped to 24.1% for values between 20–30 and 2.9% for values between 30–40 (Figure 6). Of note, virus isolates of all SARS-CoV-2 genetic lineages circulating in the US National Capital Region were obtained [52] indicating our virus isolation method was not biased for specific virus genotypes.
Figure 6.
Correlation between severe acute respiratory syndrome coronavirus 2 growth in cell cultures and cycle threshold (Ct) values. Nasopharyngeal specimens were cultured on Vero E6 cells, and the recovery of virus and development of cytopathic effect were monitored for up to 4 days after infection. Percentages of viral growth–positive samples with given Ct values are shown on the right. Viral growth was confirmed by means of antigen staining or polymerase chain reaction. ***P < .001 (paired t test).
Prolonged Viral RNA Detection and Infectious Viral Load
Patients who received repeated testing with longitudinal positive results were tested within a time frame ranging from <1 day to >45 days. To assess the correlation between repeated positivity, viral loads, and virus growth in cell culture, we evaluated a randomly selected subset of 29 patients. We examined the Ct values of all test results, days between testing, as well as viral growth on cell culture (if performed) (Figure 3). Except for 2 patients (patients 24 and 25) (and the first 3 whose clinical information was not accessible), this cohort of patients had chronic underlying conditions.
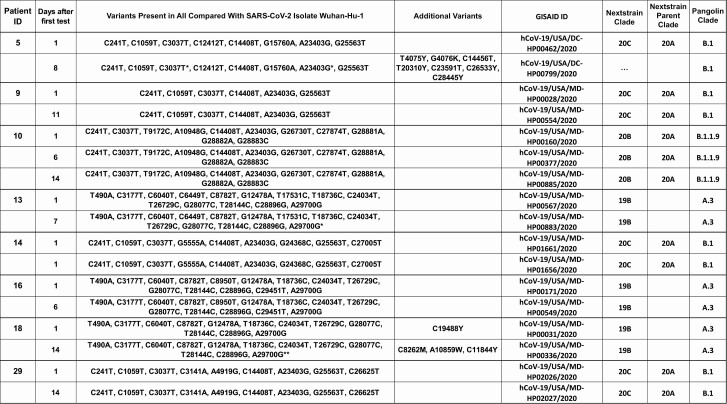
The observed general trend was an increase in the Ct values over time, indicating a reduction in the viral RNA load, and further correlated, in the majority of the patients, with failure to recover infectious virus on cell culture. Interestingly, 4 patients had infectious virus recovered from specimens collected up to 22 days after the first positive result; however, infectious virus shedding was not associated with a specific outcome, as 1 patient was never admitted (patient 24), 1 was hospitalized with no oxygen requirements (patient 10), and 2 had more severe disease (patients 8 and 29). Positive virus growth in cell culture was associated with persistence of symptoms in all but 1 patient (patient 24). Longitudinal specimens of patients were sequenced to assess any changes in the viral genome that could have resulted in prolonged shedding or could possibly suggest a reinfection. The successful recovery of complete viral genome sequences at multiple time points from 7 patients provided evidence that these patients were carrying the same virus over time; however, in 1 case the sample from the second time point had additional variants, and in 2 cases minor variants appeared in the later sample (denoted as IUPAC ambiguity codes, because 2 alleles are present in the sequencing reads) (Figure 7). Of note, 2 isolates collected from patient 14 on the same day were included in this analysis to validate our sequencing reproducibility.
Figure 7.
Sequence comparison of whole viral genomes from consecutive positive nasopharyngeal samples (subset of patients from Figure 3). Single asterisks indicate that limited read data are consistent with specified mutation (>75% of reads support variant), but position is ambiguous (N) owing to low coverage. Double asterisks indicate that limited read data provide some evidence for possible mutation or mixture (<75% of reads support variant), but position is ambiguous (N) owing to low coverage. Abbreviations: ID, identifier; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Testing-Based Discontinuation of Transmission Precautions for Patients With COVID-19
One hundred twenty-four patients who tested negative for SARS-COV-2 showed a subsequent positive result. A subset of patients (n = 17) who received repeated testing and had mixed negative and positive results were examined for the Ct values of the positive results that followed negative results, as well as the recovery of virus in cell culture. The follow-up positive testing in patients who previously tested negative produced Ct values >29.5 (Figure 4). Attempted virus culture from these specimens was negative.
Repeated Negative Results in Patients With Clinical Disease or History of Exposure to COVID-19
A total of 1788 patients were tested more than once between 11 March and 11 May 2020, without any positive result. To examine the possibility of false-negative results of the SOC molecular SARS-CoV-2 diagnostic assay due to limitations in the analytical sensitivity, we used SARS-CoV-2 ddPCR. We selected 198 negative specimens from 185 patients who underwent repeated testing over time, of whom 163 patients had at least 2 and up to 5 negative results. We selected 15 who had positive SARS-CoV-2 serologic along with multiple negative RT-PCR results. We included 22 specimens from patients who had an initial positive result but turned negative on a repeated test or the reverse. Of the total 198 tested, 11 specimens were positive by ddPCR (Figure 5). Only 1 patient with positive serologic result (patient 51) had a positive ddPCR result, and 4 of the 11 patients had positive specimens by RT-PCR collected on other days (patients 54–57).
DISCUSSION
The molecular detection of the SARS-CoV-2 genome has been valuable not only in diagnosis, but also in making infection control–related decisions. Several outcomes were observed with repeated molecular testing, including (1) prolonged viral RNA shedding, (2) alternating negative and positive results, and (3) false-negative results. Our data show that prolonged positivity could be associated with growth of virus in cell culture, especially when symptoms persist. Our data also show that RNA-positive specimens after a negative result are not associated with viral culture growth. In general, we believe our data support the current CDC guidelines updated to discourage the use of testing-based methods for return to work. In addition, our data indicate that although Ct values might be used to determine whether a patient is likely infectious, caution is warranted, as higher Ct values were occasionally associated with viral growth on cell culture.
The ddPCR assay detected a few positive specimens at very low viral loads that were missed by our SOC testing in the subset of patients who were highly suspected of infection. Overall, our data confirm that SARS-CoV-2 RNA is detectable for a prolonged time and show that the analytical sensitivities of SOC molecular diagnostics are largely influenced by variables other than the assay’s performance.
The use of a diagnostic test’s Ct values as an indicator of the presence of infectious virus has been proposed. One report suggested that a Ct value above 33–34 is not associated with cell culture viral recovery [61], and another concluded that cell culture infectivity is observed when the Ct values were <24 and within 8 days after symptom onset [29]. Our data show that the average Ct value associated with cell culture growth is 18.8. Virus growth in cell culture was possible from some specimens with Ct values as high as 32.1 and in others collected up to 22 days after the first positive result, especially in patients symptomatic at the time of sample collection. One report noted successful recovery of virus in culture for a prolonged time in severely ill patients with COVID-19, which could be correlated with high Ct values [62]. This indicates that the interpretation of Ct values and cell culture results should be used to guide clinical or infection control decisions with caution, owing to the lack of sufficient clinical outcome studies. This is especially important because of variability in specimen collection, the assays used for diagnosis, the lack of a standardized quantification assays, and inconsistencies in cell culture protocols between different laboratories.
A significant number of our cultured specimens that yielded no infectious virus had low Ct values (28.6% with Ct <23; Figure 1), indicating that variables other than the viral genome copies play a role in isolating infectious virus on cell culture. The integrity of the viral genome and variables related to sampling and storage of specimens have been proposed to affect virus recovery in cell culture [63]. Virus particles may be bound to neutralizing antibodies and therefore unable to initiate infection [64]. Generally, prolonged shedding of viral RNA was previously noted for many other viruses, including SARS-CoV, Middle Eastern respiratory syndrome coronavirus, influenza, and measles viruses [65–69]. Subgenomic SARS-CoV-2 RNA was assessed, as compared with cell culture, as a surrogate of infectiousness, with good agreement [22, 70]. Validating this approach will be valuable, because it might overcome variability in recovering the virus in cell culture.
Positive molecular results after negative tests were noticed in patients with COVID-19, and it is not certain whether that indicates a relapsed infection or reinfection. Our data showed that RNA detection after RNA-negative tests was not associated with positive viral growth in cell culture. It is likely that detectable viral RNA in convalescence is associated with prolonged viral RNA shedding, especially since the viral loads are usually lower than what is detectable during the early stages of infection. In addition, positive test results after negative molecular RNA tests that are associated with new symptoms are more perplexing, and reinfection has not been ruled out. Our ddPCR data also show that some of these negatives are associated with viral loads below the assays analytical sensitivities. Comprehensive studies that combine understanding the development of protective immunity and compare isolated viral genomes will help in understanding the enigma of reinfection by SARS-CoV-2. Our group previously showed that different viral clades circulate in the Baltimore/Washington, DC, metropolitan area [52]. Dissecting the differences between viral clades in the efficiency of growth in cell culture, development of immune responses, and prolonged shedding or reinfection is under investigation by our group.
Compared with the standard diagnostic molecular techniques, ddPCR offers an absolute quantification of targets after partitioning the specimen into thousands of droplets, which increases the accuracy of detection [71–73]. ddPCR showed a slightly higher sensitivity in detecting SARS-CoV-2 RNA in a subset of specimens from patients with high suspicion of COVID-19 and negative RT-PCR results. Our data are consistent with published reports of studies comparing ddPCR with RT-PCR [37, 74]. It is important to note that the analytical sensitivity of the ddPCR assay as reported in the EUA package insert (645 copies/mL) is comparable to that of SOC RT-PCR methods used for diagnosis, including the CDC panel assay, among others [3]. All the positive results detected with the ddPCR assay in this study were below the ddPCR assay’s analytical limit of detection, which explains a few conflicting results in a few specimens with repeated testing (Figure 5). The Bio-Rad ddPCR assay uses primers and probes that are the same as those used in the CDC assay, including the human RNase P gene as an internal control. Including this control is very valuable for excluding insufficient sampling as a cause of false-negative results [75]. Only a few samples that tested negative with the standard PCR methods were later positive by ddPCR (5.6%), even in a cohort with a high suspicion of COVID-19. Overall, this suggests that false-negative results in some cases are secondary to low viral loads, likely associated with temporal aspects of viral shedding.
Our study findings indicate that prolonged viral RNA shedding is associated with growth of the virus in cell culture in a subset of patients and seems to be correlated with persistence of symptoms. Higher Ct values and positive RNA test results detected after viral RNA clearance were not associated with successful recovery of virus in culture in our tested cohort. ddPCR can add increased sensitivity in detecting viral RNA. Our study was limited by relatively small number of patients analyzed and the overrepresentation of hospitalized patients with various comorbid conditions as well as the retrospective and single-center nature of the study. Additional studies are required to inform the use of Ct values and cell culture in making clinical decisions, and diagnostic strategies that can differentiate shedding from active replication will be very valuable for infection control.
Notes
Acknowledgments. The authors thank the entire clinical microbiology laboratory for the rapid response to the pandemic and for offering a unique testing capacity. Droplet digital polymerase chain reaction was performed in collaboration with Bio-Rad Laboratories. They thank Winston Temp, PhD, and Stuart C. Ray, MD, for their valuable contribution to the severe acute respiratory syndrome coronavirus 2 genomic analysis pipeline.
Financial support. This work was supported by the Department of Pathology, Johns Hopkins School of Medicine, the National Institutes of Health (NIH) Johns Hopkins Center of Excellence in Influenza Research and Surveillance (grant HHSN272201400007C to A. P. and H. H. M.), and the Molecular and Cellular Basis of Infectious Diseases program, NIH (grant T32A1007417 to H. P. and B. S.).
Potential conflicts of interest. K. C. C. reports grants to her institution from MeMed, BD Diagnostics, and LBT Innovations and scientific advisory board fees from Scanogen and Pattern Diagnostics. outside the submitted work. H. H. M. reports research collaboration and contribution with equipment and reagents from Bio-Rad Laboratories and DiaSorin Molecular, during the conduct of the study. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Zhen W, Manji R, Smith E, Berry GJ. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol 2020; 58:e00743– 20. doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol 2020; 58:e00760– 20. doi: 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhteg K, Jarrett J, Richards M, et al. . Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol 2020; 127:104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordi L, Piralla A, Lalle E, et al. . Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa™ COVID-19 direct assay. J Clin Virol 2020; 128:104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman JA, Pepper G, Naccache SN, Huang ML, Jerome KR, Greninger AL. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol 2020; 58:e00821– 20. doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00783– 20. doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craney AR, Velu P, Satlin MJ, et al. . Comparison of two high-throughput reverse transcription-polymerase chain reaction systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol 2020; 58:e00890– 20. doi: 10.1128/JCM.00890-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan CA, Sahoo MK, Huang C, et al. . Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J Clin Virol 2020; 127:104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington A, Cox B, Snowdon J, et al. . Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol 2020; 58:e00798– 20. doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu A, Zinger T, Inglima K, et al. . Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J Clin Microbiol 2020; 58:e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore NM, Li H, Schejbal D, Lindsley J, Hayden MK. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV RT-PCR assay for the detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00798– 20. doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smithgall MC, Scherberkova I, Whittier S, Green DA. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol 2020; 128:104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan CA, Garamani N, Lee AS, et al. . Comparison of the Accula SARS-CoV-2 test with a laboratory-developed assay for detection of SARS-CoV-2 RNA in clinical nasopharyngeal specimens. J Clin Microbiol 2020; 58:e01072– 20. doi: 10.1128/JCM.01072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran A, Beavis KG, Matushek SM, et al. . The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J Clin Microbiol 2020; 58:e00772– 20. doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pujadas E, Ibeh N, Hernandez MM, et al. . Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol 2020; 92:1695–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poljak M, Korva M, Knap Gašper N, et al. . Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol 2020; 58:e00599–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avaniss-Aghajani E, Sarkissian A, Fernando F, Avaniss-Aghajani A. Validation of the Hologic’s Aptima Unisex and Multitest specimen collection kits used for endocervical and male urethral swab specimen (Aptima Swab) for sample collection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00753– 20. doi: 10.1128/JCM.00753-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostafa HH, Hardick J, Morehead E, Miller JA, Gaydos CA, Manabe YC. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J Clin Virol 2020; 130:104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mostafa HH, Lamson DM, Uhteg K, et al. . Multicenter evaluation of the NeuMoDx™ SARS-CoV-2 test. J Clin Virol 2020; 130:104583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol 2020; 58:e00512– 20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.To KK, Tsang OT, Leung WS, et al. . Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wölfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 23.Ai T, Yang Z, Hou H, et al. . Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020; 296:E32–E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCulloch DJ, Kim AE, Wilcox NC, et al. . Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 2020; 3:e2016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu YP, Jennings R, Hart B, et al. . Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med 2020; 383:494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doll ME, Pryor R, Mackey D, et al. . Utility of retesting for diagnosis of SARS-CoV-2/COVID-19 in hospitalized patients: impact of the interval between tests. Infect Control Hosp Epidemiol 2020:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centers for Disease Control and Prevention. Discontinuation of transmission-based precautions and disposition of patients with COVID-19 in healthcare settings (interim guidance). Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html.
- 28.He X, Lau EHY, Wu P, et al. . Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26:672–5. [DOI] [PubMed] [Google Scholar]
- 29.Bullard J, Dust K, Funk D, et al. . Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020; 71:2663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu WD, Chang SY, Wang JT, et al. . Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect 2020; 81:318–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao F, Sun J, Xu Y, et al. . Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis 2020; 26:1920–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissleder R, Lee H, Ko J, Pittet MJ. COVID-19 diagnostics in context. Sci Transl Med 2020; 12:eabc1931. doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 33.Long DR, Gombar S, Hogan CA, et al. . Occurrence and timing of subsequent SARS-CoV-2 RT-PCR positivity among initially negative patients. Clin Infect Dis 2021; 72:323– 6. doi: 10.1093/cid/ciaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang Y, Zhang H, Xie J, et al. . Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 2020:200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Infectious Diseases Society of America. Guidelines on the diagnosis of COVID-19. 2020. [DOI] [PMC free article] [PubMed]
- 36.Wang W, Xu Y, Gao R, et al. . Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843– 4. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu F, Yan L, Wang N, et al. . Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis 2020; 71:793– 8. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.COVID-19 Investigation Team. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020; 26:861–8. [DOI] [PubMed] [Google Scholar]
- 39.Tu YP, Jennings R, Hart B, et al. . Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med 2020; 383:494– 6. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheuk S, Wong Y, Tse H, et al. . Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis 2020; 71:2939– 46. doi: 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamal AJ, Mozafarihashjin M, Coomes E, et al. . Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2021; 72:1064– 6. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasomsub E, Watcharananan SP, Boonyawat K, et al. . Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect 2021; 27:285.e1– 285.e4. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00776– 20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020; 173:262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Ren L, Yang S, et al. . Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 2020; 71:778– 85. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furukawa NW, Brooks JT, Sobel J. Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg Infect Dis 2020; 26:e201595. doi: 10.3201/eid2607.201595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith KP, Cheng A, Chopelas A, et al. . Large-scale, in-house production of viral transport media to support SARS-CoV-2 PCR testing in a multihospital health care network during the COVID-19 pandemic. J Clin Microbiol 2020; 58:e00913– 20. doi: 10.1128/JCM.00913-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Food and Drug Administration. NeuMoDx™ SARS-CoV-2 assay. Available at: https://www.fda.gov/media/136565/download. 2020. Accessed 28 July 2020.
- 49.Food and Drug Administration. ePlex®SARS-CoV-2 test. Available at: https://www.fda.gov/media/136282/download. 2020. Accessed 28 July 2020.
- 50.Food and Drug Administration. BD SARS-CoV-2 reagents for BD MAX™ system. Available at: https://www.fda.gov/media/136816/download. 2020. Accessed 28 July 2020.
- 51.Food and Drug Administration. Xpert® Xpress SARS-CoV-2. Available at: https://www.fda.gov/media/136314/download. 2020. Accessed 28 July 2020.
- 52.Thielen PM, Wohl S, Mehoke T, et al. . Genomic diversity of SARS-CoV-2 during early introduction into the United States National Capital Region. medRxiv 2020. [Preprint: not peer reviewed]. 23 August 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.08.13.20174136v2.
- 53.Waggoner JJ, Stittleburg V, Pond R, et al. . Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020; 26:1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaecher SR, Diamond MS, Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J Virol 2008; 82:9477–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, Grantham ML, Pekosz A. Mutations in the influenza A virus M1 protein enhance virus budding to complement lethal mutations in the M2 cytoplasmic tail. J Virol 2017; 92:e00858– 17. doi: 10.1128/JVI.00858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyson JR, James P, Stoddart D, et al. . 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. doi: 10.1101/2020.09.04.283077. [DOI] [Google Scholar]
- 57.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018; 34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 2015; 12:733–5. [DOI] [PubMed] [Google Scholar]
- 59.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. . Integrative genomics viewer. Nat Biotechnol 2011; 29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Food and Drug Administration. Bio-Rad SARS-CoV-2 ddPCR test. Available at: https://www.fda.gov/media/137579/download. 2020. Accessed 28 July 2020.
- 61.La Scola B, Le Bideau M, Andreani J, et al. . Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 2020; 39:1059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Folgueira MD, Luczkowiak J, Lasala F, Perez-Rivilla A, Delgado R. Persistent SARS-CoV-2 replication in severe COVID-19.medRxiv [Preprint: not peer reviewed]. 12 June 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.06.10.20127837v1. [Google Scholar]
- 63.Huang CG, Lee KM, Hsiao MJ, et al. . Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol 2020; 58:e01068– 20. doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atkinson B, Petersen E. SARS-CoV-2 shedding and infectivity. Lancet 2020; 395:1339–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peiris JS, Chu CM, Cheng VC, et al. ; HKU/UCH SARS Study Group . Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003; 361:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan KH, Poon LL, Cheng VC, et al. . Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis 2004; 10:294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh MD, Park WB, Choe PG, et al. . Viral load kinetics of MERS coronavirus infection. N Engl J Med 2016; 375:1303–5. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Guo Q, Yan Z, et al. ; CAP-China Network . Factors associated with prolonged viral shedding in patients with avian influenza A(H7N9) virus infection. J Infect Dis 2018; 217:1708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin WH, Kouyos RD, Adams RJ, Grenfell BT, Griffin DE. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc Natl Acad Sci U S A 2012; 109:14989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perera RAPM, Tso E, Tsang OTY, et al. . SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 2020; 26:2701–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hindson BJ, Ness KD, Masquelier DA, et al. . High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011; 83:8604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hindson CM, Chevillet JR, Briggs HA, et al. . Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013; 10:1003–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor SC, Carbonneau J, Shelton DN, Boivin G. Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for quantification of oseltamivir-resistant subpopulations. J Virol Methods 2015; 224:58–66. [DOI] [PubMed] [Google Scholar]
- 74.Falzone L, Musso N, Gattuso G, et al. . Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med 2020; 46:957–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kinloch NN, Ritchie G, Brumme CJ, et al. . Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis 2020; 222:899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]