ABSTRACT
Coronaviruses are a group of viruses causing disease in a wide range of animals, and humans. Since 2002, the successive emergence of bat-borne severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), swine acute diarrhea syndrome coronavirus (SADS-CoV) and SARS-CoV-2 has reinforced efforts in uncovering the molecular and evolutionary mechanisms governing coronavirus cell tropism and interspecies transmission. Decades of studies have led to the discovery of a broad set of carbohydrate and protein receptors for many animal and human coronaviruses. As the main determinant of coronavirus entry, the spike protein binds to these receptors and mediates membrane fusion. Prone to mutations and recombination, spike evolution has been studied extensively. The interactions between spike proteins and their receptors are often complex and despite many advances in the field, there remains many unresolved questions concerning coronavirus tropism modification and cross-species transmission, potentially leading to delays in outbreak responses. The emergence of SARS-CoV-2 underscores the need to address these outstanding issues in order to better anticipate new outbreaks. In this review, we discuss the latest advances in the field of coronavirus receptors emphasizing on the molecular and evolutionary processes that underlie coronavirus receptor usage and host range expansion.
Keywords: coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), host cell receptor, virus entry, host tropism, pathogenesis
This review reframes and puts into perspective SARS-CoV-2 receptor usage in the broader landscape of coronavirus spike interactions with animal host cell receptors.
INTRODUCTION
Coronaviruses are a large group of enveloped, positive-strand RNA viruses of rich diversity with complex evolutionary histories belonging to the Nidovirales order (de Groot et al. 2012). These viral pathogens infect a wide range of wild and domesticated birds and mammals, including humans (Woo et al. 2009). It is well recognized that bat species in particular constitute one of the most important animal reservoirs for emerging coronaviruses (Li et al. 2005b; Ithete et al. 2013; Corman et al. 2014; Drexler, Corman and Drosten 2014; Corman et al. 2015; Ge et al. 2016; Huang et al. 2016; Hu et al. 2017; Tao et al. 2017; Zhou et al. 2018; Fan et al. 2019). Coronaviruses possess the longest known viral RNA genome at around 30 kb in length and while the coronavirus replicase complex is endowed with proof-reading activity thanks to its non-structural protein 14 (nsp14) exoribonuclease, the genomic RNA is still subject to frequent recombination and mutation (Gorbalenya et al. 2006; Minskaia et al. 2006; Woo et al. 2009). Coronaviruses are classified into four distinct genera: Alpha-, Beta-, Gamma- and Deltacoronavirus.
Well known for their agility to cross host species barriers, coronaviruses have repeatedly manifested their ability for zoonotic and anthroponotic transmission, as well as their involvement in spillover events. This has been exemplified most strikingly during the zoonotic emergence of three highly pathogenic human coronaviruses in the first two decades of this century, all of which are thought to originate from bat species and intermediate mammalian hosts: severe acute respiratory syndrome coronavirus (SARS-CoV) during the 2002–2003 outbreak that originated in Guangdong province, China, with additional laboratory-acquired and sporadic cases reported after the initial outbreak (Fouchier et al. 2003; Kuiken et al. 2003; Peiris et al. 2003; Lim et al. 2004; Normile 2004; Richardson et al. 2004); the Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak that began in 2012 and is still ongoing (Corman et al. 2012; van Boheemen et al. 2012; Zaki et al. 2012); more recently the current coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2, a virus which is related to the 2002–2003 SARS-CoV and which originated in Wuhan, China (Chan et al. 2020; Coronaviridae Study Group of the International Committee on Taxonomy of 2020; Li et al. 2020; Zhou et al. 2020b; Zhu et al. 2020).
Thanks to unprecedented research efforts and the decades of accumulated knowledge gained through studies on related coronaviruses, in particular SARS-CoV, a wealth of information and insights have been gained on SARS-CoV-2 in terms of its basic biology, epidemiology and phylogeny. Notably, its genome became available in a short period of time (Zhou et al. 2020b). Structural and functional analyses of its spike protein and its receptor, angiotensin converting enzyme 2 (ACE2), have been obtained within a few months (Lan et al. 2020; Shang et al. 2020b; Walls et al. 2020; Wrapp et al. 2020; Yan et al. 2020). Despite these impressive achievements, there is still much to learn about this fast spreading and deadly coronavirus, in particular the details of its host cell and species tropism, receptor usage and how that relates to its pathogenicity and evolutionary history. This review aims at reframing SARS-CoV-2 receptor recognition in the broader context of coronavirus spike–receptor interactions.
In various mammalian hosts, many instances of coronavirus interspecies jumping and co-infections have been reported. This is particularly well characterized for viruses of veterinary importance in the Alphacoronavirus genus, where canine coronavirus (CCoV), feline coronavirus (FCoV) and transmissible gastroenteritis virus of swine (TGEV) are considered to form a single prototype species, Alphacoronavirus-1. Co-infections and recombination events among these viruses have been well characterized (Herrewegh et al. 1998; Regan et al. 2012; Terada et al. 2014; Guo et al. 2020). Recently, the emergence of a large-scale and deadly outbreak of swine acute diarrhea syndrome (SADS) was shown to be caused by SADS-CoV, a member of the Alphacoronavirus genus originating from the bat-derived coronavirus HKU2 (BatCoV-HKU2) (Zhou et al. 2018). Another example is the alpaca coronavirus (ACoV), also belonging to the Alphacoronavirus genus that was found to be closely related to the human HCoV-229E virus, which suggests possible viral transmission, either zoonotic or anthroponotic, occurring between humans and alpacas (Crossley et al. 2012).
In the betacoronaviruses, HCoV-OC43 is thought to have originated from bovine coronavirus (BCoV) with molecular clock analysis revealing a zoonotic transmission event estimated to have occurred in the 1890s (Vijgen et al. 2005). Retrospective analyses of sera demonstrated that MERS-CoV has been circulating in dromedary camel populations for around 30 years prior to the first known human cases (Müller et al. 2014). For SARS-CoV, reports during the 2002–2003 epidemic have shown that cats and ferrets could be experimentally infected with the virus (Martina et al. 2003; van den Brand et al. 2008). Similarly, SARS-CoV-2 infections in cats, ferrets, dogs and other mammals have been reported (Leroy, Gouilh and Brugere-Picoux 2020; Sit et al. 2020; Stout et al. 2020). Phylogenetic analyses of fully sequenced genomes of SARS-CoV-2 have shown that, much like SARS-CoV, the novel human coronavirus very likely originated from a bat host (Zhou et al. 2020b). For SARS-CoV, it was shown that masked palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) served as intermediate hosts (Guan et al. 2003).
It is thought that the origins of most human coronaviruses (HCoV), such as the alphacoronaviruses HCoV-229E (Corman et al. 2015), HCoV-NL63, and the betacoronaviruses MERS-CoV, SARS-CoV and SARS-CoV-2 can ultimately be traced back to viruses infecting bat species (Cui, Li and Shi 2019; Fan et al. 2019). This notion is based on the richness of the diversity of coronaviruses detected in bat species, which was found to far exceed that of other mammalian hosts as well as the identification of bat coronaviruses very closely related to human viruses, such as Bat-229E-like, Bat-NL63-like and Bat-SARS-like viruses (Drexler, Corman and Drosten 2014; Cui, Li and Shi 2019).
The quantity and diversity of SARS-like coronavirus species that have been identified from various bat species in China during the past 15 years is a matter of concern, highlighting the pressing need for accrued surveillance of bat coronavirus strains currently circulating and to better discern those that have gained enhanced capability to cross host species barriers. This is especially concerning in the context of the COVID-19 pandemic where the proximal origin of SARS-CoV-2 is still unknown and the fact that several bat-derived SARS-related coronaviruses have been demonstrated to be capable of using human receptors for entry (Ge et al. 2013; Yang et al. 2015).
A question that arises from the above assessment is to ask what makes coronaviruses, and bat coronaviruses in particular, especially prone to crossing host species barriers. Several criteria govern the propensity for a given virus to successfully transmit to a new host species, including susceptibility (receptor compatibility), permissiveness, accessibility of susceptible cells and ability to evade host immune responses (Flint et al. 2015). For coronaviruses, the ability to cross the species barrier can be attributed most prominently thanks to their large and distinctive spike envelope glycoprotein (S).
The coronavirus spike is typically 1200–1400 amino acids in length and is encoded by the S gene located downstream of ORF 1ab encoding the replicase polyprotein (Fig. 1A). In addition, spike is a class I fusion protein that forms homotrimers, with each monomer composed of a globular S1 subunit responsible for engagement with host cell receptor(s) and the S2 subunit folded in a metastable spring-loaded conformation and tasked with membrane fusion (Fig. 1A and B). The S1 subunit can be further subdivided into functional and structural domains: the N-terminal domain (NTD, also named domain A or S1A), the C-terminal domain (CTD, also named C-domain or domain B or S1B), as well as the SD1 (domain C) and SD2 (domain D) subdomains that are located upstream of the S1/S2 boundary. NTD and CTD domains have the potential to bind to distinct receptors, notably carbohydrates or transmembrane proteins (Fig. 1C).
Figure 1.
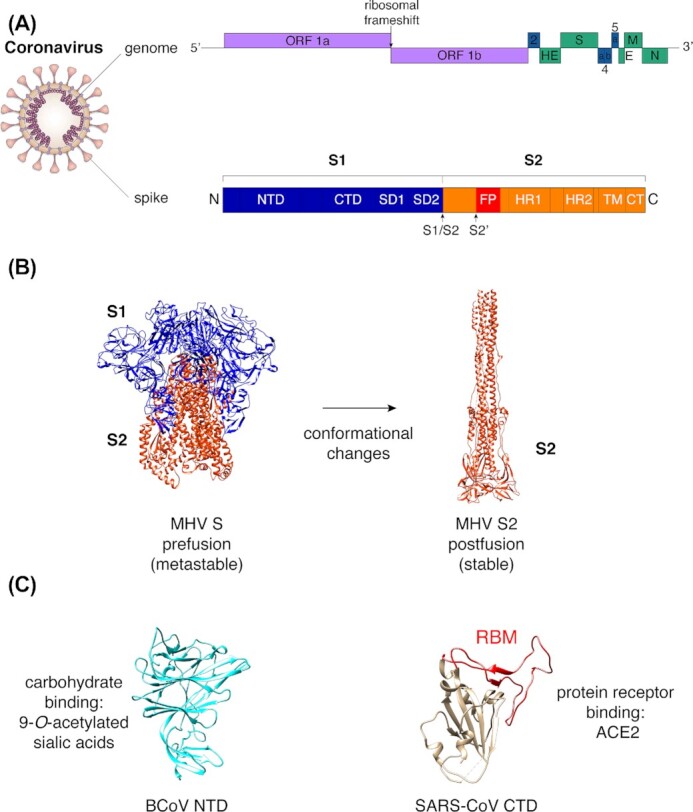
General features of the coronavirus spike protein. (A) Diagram of coronavirus virion with schematic of genome organization (based on MHV) and domains of the coronavirus spike protein. For the genome schematic, purple boxes represent ORF 1a and ORF 1b, which encode the replicase polyproteins pp1a and pp1ab. Blue boxes indicate genes encoding accessory proteins (ns2, ns4a, ns4b and ns5a), while green boxes indicate structural proteins. Abbreviations used: HE, hemagglutinin esterase; S, spike protein; E, envelope protein; M, membrane protein; N, nucleoprotein; NTD, N-terminal domain; CTD, C-terminal domain; SD1, subdomain 1; SD2, subdomain 2; S1/S2, S1/S2 cleavage site; S2′, S2′ cleavage site; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; and CT, C-terminal domain. (B) Representative structure of coronavirus S protein. MHV S protein in pre-fusion (S1 and S2 subunits, PDB 3JCL) and post-fusion (S2 subunit, PDB 6B3O) conformations. (C) Representative structures of coronavirus NTD subdomain (BCoV NTD, PDB 4H14) and CTD subdomain (SARS-CoV CTD, PDB 2AJF) with receptor-binding motif (RBM) highlighted in red.
For each genus, the receptor-binding domain within the CTD is relatively well conserved structurally, but contains variable extended loops that contain specific receptor-binding motifs (RBMs), which are prone to mutations and are responsible for receptor specificity (Li 2015). The different receptors of coronaviruses will be covered and discussed in this review.
An interesting property of coronavirus spike proteins is that the subdomain responsible for binding to the receptor, the receptor-binding domain or RBD, can be found at different locations within the S1 subdomain (Hulswit, de Haan and Bosch 2016), and in some cases several functional RBDs can be found, allowing binding to different receptors.
The coronavirus field has benefited greatly from structural studies of the spike protein especially by cryo-electron microscopy (cryo-EM) analyses. These studies considerably advanced our mechanistic understanding of how spike proteins function, in particular the various structural conformation changes that spike proteins undergo during virus entry, from a pre-fusion to a post-fusion conformation (Fig. 1B). Also, the structure of the spike RBD in contact with host receptors has been characterized at atomic resolution, capturing in exquisite detail the receptor-binding interface. This has been exemplified with the SARS-CoV spike RBD bound to the ACE2 receptor, revealing receptor- and virus-binding motifs (RBM and VBM) within spike and ACE2, respectively (Li et al. 2005a; Li 2015). More recently, cryo-EM studies of nearly complete ectodomains of pre-fusion S trimers have provided groundbreaking insights into the mechanisms by which coronaviruses bind to host cell receptors and mediate membrane fusion (Walls et al. 2017; Tortorici et al. 2019; Wrapp et al. 2020; Yan et al. 2020), as well as glycan shielding strategies coronaviruses employ to evade host immune recognition by antibodies (Walls et al. 2016b; Xiong et al. 2018; Watanabe et al. 2020; Yang et al. 2020).
The list of spike structures keeps on growing and includes representatives of the Alphacoronavirus genus, for example FCoV and HCoV-NL63 (Walls et al. 2016b; Yang et al. 2020), several Betacoronavirus genus representatives such as MHV (Walls et al. 2016a), HCoV-HKU1 (Kirchdoerfer et al. 2016), SARS-CoV and MERS-CoV (Gui et al. 2017; Yuan et al. 2017; Heise et al. 2018), HCoV-OC43 (Tortorici et al. 2019), and more recently SARS-CoV-2 (Walls et al. 2020; Wrapp et al. 2020). The structures of the ectodomains of the spike glycoprotein representatives of Gammacoronavirus and Deltacoronavirus genera have also been obtained for infectious bronchitis virus (IBV) and porcine deltacoronavirus (PDCoV), respectively (Shang et al. 2018b; Xiong et al. 2018).
Coronavirus spike proteins are prone to accumulate mutations and can easily recombine. Some regions (e.g. the RBD) have been identified as mutational and/or recombination hotspots. Such propensity for mutations and recombination has led to the designation of the coronavirus spike protein as being modular (Graham and Baric 2010). In the case of SARS-CoV-2, the RBD region within the spike gene is believed to have undergone recombination (Zhou et al. 2020a).
Coronaviruses offer unique examples of virus–receptor interactions underscoring the modular nature of the spike glycoprotein and possible mechanisms explaining the propensity for coronavirus interspecies jumping, offering some clues to better anticipate their zoonotic potential. While far from being exhaustive, this review highlights the decades-long efforts in understanding coronavirus–receptor interactions and their consequences in host cell entry, tropism and interspecies jumping. Importantly, many studies reviewed here have greatly facilitated the rapid and insightful achievements in understanding how SARS-CoV-2 recognizes the ACE2 protein receptor. The review also emphasizes on important areas in SARS-CoV-2 and other emerging coronavirus research that have so far been underappreciated and await further investigation, in particular the potential for carbohydrate binding and the functional role played by such interactions.
PROTEIN RECEPTORS OF CORONAVIRUSES
To initiate successful infection, virus binding to the host cell surface is a prerequisite and critical first step that governs to a large extent host cell susceptibility to infection and capacity for host tropism change and interspecies transmission. Coronavirus spike proteins are capable of mediating attachment to carbohydrates and protein receptors found at the cell surface (Fig. 2), with RBDs located in either the NTD or the CTD within the S1 subunit of the spike proteins. In most studied cases, protein receptor engagement is carried out by the CTD.
Figure 2.
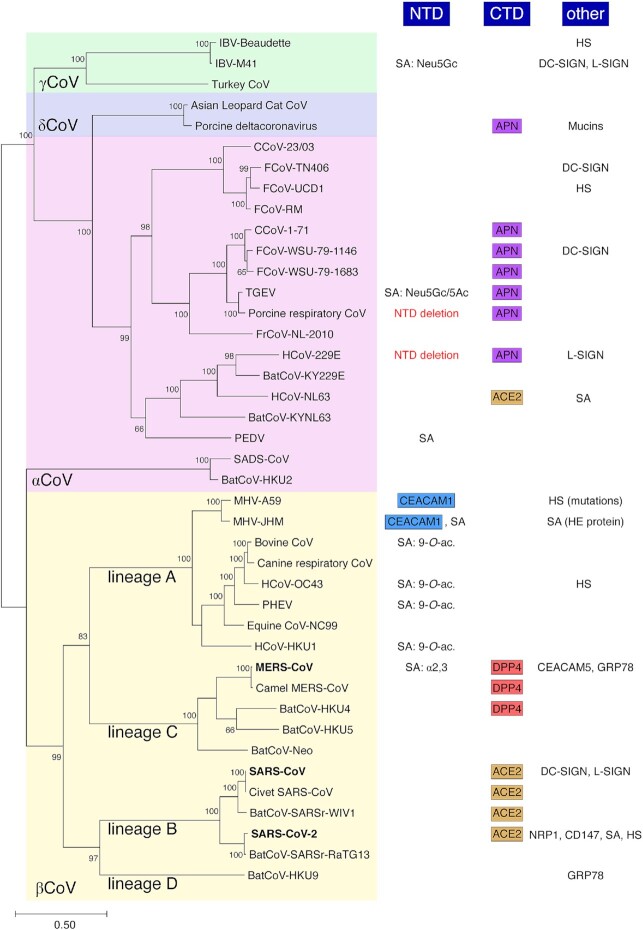
Coronavirus carbohydrate and protein receptor usage. A phylogenetic analysis of coronavirus spike proteins from representatives of all four genera was performed with corresponding carbohydrate and protein receptors shown along with receptor-binding domains involved. Selected coronavirus spike protein sequences were aligned and a maximum-likelihood (ML) phylogenetic tree was generated. Bootstrap values shown at nodes were calculated from 1000 replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Abbreviations used: NTD, N-terminal domain; CTD, C-terminal domain; SA, sialic acid; Neu5Gc, N-glycolylneuraminic acid; Neu5Ac, N-acetylneuraminic acid; CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1; 9-O-ac., 9-O-acetylated sialic acids; α2,3, α2,3-linked sialic acids; APN, aminopeptidase N; ACE2, angiotensin converting enzyme 2; DPP4, dipeptidyl peptidase 4; HS, heparan sulfate; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin; L-SIGN, liver/lymph node-specific intracellular adhesion molecules-3 grabbing non-integrin; HE, hemagglutinin esterase; CEACAM5, carcinoembryonic antigen-related cell adhesion molecule 5; GRP78, 78-kDa glucose-regulated protein; NRP1, neuropilin-1; and CD147, basigin.
To date, four protein receptors have been characterized as main receptor for coronavirus binding to host cells: several members of the Alphacoronavirus genus are known to bind to amino peptidase N (APN, CD13) of their respective host species, using an RBD located in the CTD of the S protein; murine hepatitis virus (MHV) of the Betacoronavirus genus utilizes murine carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), the first coronavirus protein receptor identified (Fig. 2). Interestingly, MHV uses its spike NTD as RBD, which is unusual as NTDs are more typically known to bind carbohydrates; SARS-CoV, SARS-CoV-2 and certain SARS-related viruses of bats have been shown to use angiotensin-converting enzyme 2 (ACE2), using their S1 CTD as RBD; MERS-CoV, camel-derived MERS-CoV and the related BatCoV-HKU4 are known to utilize the protein receptor named dipeptidyl peptidase 4 (DPP4), which involves the S1 CTD as RBD (Fig. 2).
Among the known protein receptors of coronaviruses, it is interesting to point out that three (APN, ACE2 and DPP4) have peptidase activity. However, studies have shown that their proteolytic activity is not required for successful binding and entry of the respective coronaviruses (Raj et al. 2013; Bosch, Smits and Haagmans 2014). It was shown that coronavirus spike proteins bind to outer regions distinct from the catalytic sites of these receptors. Related to this, spike protein proteolytic activation by transmembrane protease/serine subfamily member 2 (TMPRSS2) has been shown to be an important step in the viral entry process of SARS-CoV, SARS-CoV-2, as well as other coronaviruses such as MERS-CoV (Shulla et al. 2011; Shirato, Kawase and Matsuyama 2013; Heurich et al. 2014; Hoffmann et al. 2020). Remarkably, in the context of SARS-CoV entry, TMPRSS2 and ACE2 were observed to coprecipitate, with TMPRSS2 even found to proteolytically process ACE2 receptor (Shulla et al. 2011). In addition to the direct role played by TMPRSS2 in activating SARS-CoV spike protein, the TMPRSS2-mediated cleavage event on ACE2 was found to have an enhancing effect on SARS-CoV S-mediated virus entry (Heurich et al. 2014).
Aminopeptidase N (APN, CD13)
APN has been identified as the main receptor for several species within the Alphacoronavirus genus, including human HCoV-229E (Yeager et al. 1992), and members of the Alphacoronavirus-1 species, such as TGEV of swine (Delmas et al. 1992), the related porcine respiratory coronavirus (PRCV)—a deletion mutant of TGEV that possesses a spike protein with a deleted NTD (see below), as well as type II (clade B) feline and canine coronaviruses (FCoV and CCoV) (Tresnan, Levis and Holmes 1996; Tresnan and Holmes 1998) (Fig. 2). APN is a type II transmembrane protein and a zinc aminopeptidase. It has a wide tissue distribution and is found in epithelial cells of the intestines, in the nervous system, as well as immune cells such as monocytes and dendritic cells (Mina-Osorio 2008; Mesel-Lemoine et al. 2012). Interestingly, in the brush border membrane of the small intestine APN was found to be associated with broad neutral (0) amino acid transporter 1 (B0AT1) also named solute carrier family 6 member 19 (SLC6A19) (Fairweather et al. 2012). Whether B0AT1 plays a role in regulating virus entry and tropism of APN-dependent coronaviruses remains to be investigated.
Alphacoronaviruses bind to their respective host species APN using their spike protein S1 CTD domain. However, while HCoV-229E and Alphacoronavirus-1 members use the same host receptor, the molecular details of the S-APN binding interface differ. In particular, the mode by which the spike CTD RBD interacts with APN in the VBM region of the receptor appears distinct in different alphacoronaviruses (Chen et al. 2012). A structural investigation showed that porcine alphacoronaviruses PRCV and TGEV differ in the length of RBD loops that contact APN compared with human HCoV-229E. These differences, along with differential glycosylation patterns, suggest distinct virus-APN residue contacts in humans (Reguera et al. 2012). Interestingly, it was previously demonstrated that feline APN could serve as a functional receptor of type II CCoV, TGEV and human coronavirus HCoV-229E (Tresnan, Levis and Holmes 1996). This important finding suggested the possibility that co-infections in cats could lead to generation of novel recombinant strains (Tresnan and Holmes 1998). Interestingly, it was recently shown that APN usage was not restricted to alphacoronaviruses as it was demonstrated that PDCoV binds to and utilizes APN as its main host receptor via an interaction mediated by the spike CTD subdomain (Li et al. 2018) (Fig. 2). The common usage of APN from members of two distinct coronavirus genera can be explained by the relatively close evolutionary relationships between the spike proteins of alphacoronaviruses and deltacoronaviruses (Fig. 2). However, SARS-CoV-2, a member of the Betacoronavirus genus, was shown to be unable to use APN (Zhou et al. 2020b).
Protein receptor usage for the porcine pathogen SADS-CoV was assessed using pseudotyped particle infectivity assays and it was shown that APN was not a functional receptor for the virus (neither were ACE2 and DPP4) (Zhou et al. 2018). Although classified within the Alphacoronavirus genus based on replicase domain conservation, SADS-CoV and the closely related BatCoV-HKU2 possess comparatively very short spike proteins (1130 and 1128 amino acids, respectively), which are more closely related to that of the Betacoronavirus genus (Lau et al. 2007; Yu et al. 2020) (Fig. 2). SADS-CoV and BatCoV-HKU2 exemplify the complex evolutionary patterns of coronaviruses.
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1)
Murine carcinoembryonic antigen-related cell adhesion molecule 1 (mCEACAM1), which belongs to the immunoglobulin superfamily, was demonstrated to be the main receptor of Betacoronavirus genus prototype species murine hepatitis virus (MHV) strain A59 (Dveksler et al. 1991) (Fig. 2). Members of the carcinoembryonic antigen (CEA) family are glycosyl phosphatidyl inositol (GPI)-anchored glycoproteins involved in cellular adhesion, development and modulation of immune responses.
Uniquely among coronaviruses, the RBD of MHV-A59 is located within the spike NTD, which usually functions as a lectin for other coronaviruses. However, structural analysis revealed that MHV-A59 NTD conserved protein folds of human galectins, a finding that has led to the hypothesis that an ancestral coronavirus acquired the NTD from a host galectin and co-opted its carbohydrate-binding function (Peng et al. 2011). This lectin function has been retained in many other coronavirus species, as detailed below. Among the various strains of MHV, the JHM strain stands out for its neurotropic and neurovirulent phenotype compared with the hepatotropic MHV-A59 strain. Interestingly, in a phenomenon described as ‘receptor-independent spread’, it was shown that the JHM strain, but not the MHV-A59 strain, can spread from infected mouse cells to cells lacking mCEACAM1a (Gallagher, Buchmeier and Perlman 1992). In addition, it has been demonstrated that MHV-JHM is able to infect and kill Ceacam1-deficient mice (Miura et al. 2008). While the JHM strain of MHV can initiate brain infection in Ceacam1-deficient mice, expression of mCEACAM1a potentiates susceptibility to infection. These findings suggested that the JHM strain can use an alternative, less effective receptor to initiate infection. Once primary infection is established in the murine host glial cells, the JHM strain was shown to rapidly spread via cell–cell fusion and syncytia formation in a receptor-independent manner (Nakagaki and Taguchi 2005).
Angiotensin-converting enzyme 2 (ACE2)
SARS-CoV, which belongs to the Betacoronavirus genus and is the etiological agent of the 2002–2003 SARS outbreak associated with 8000 confirmed infections and a mortality rate of around 10%, was found to utilize human ACE2 as host cell receptor (Li et al. 2003) (Fig. 2). ACE2 is a type I transmembrane protein and a metalloenzyme whose primary function is to convert angiotensin II into angiotensin 1–7. It is expressed in type II pneumocytes, enterocytes of small intestines, vascular endothelial cells and cortical neurons and glial cells. Similarly to APN, ACE2 was found to interact with the neutral amino acid transporter B0AT1 in intestinal epithelial cells (Fairweather et al. 2012; Jando et al. 2017). Recently, structural determination of the full-length ACE2 protein was obtained using the ACE2-B0AT1 complex, which assembles in a dimer of heterodimers, providing further evidence of the importance of the interaction between the peptidase and the amino acid transporter for their overall structural integrity (Yan et al. 2020). The study also suggested that B0AT1 may play a regulatory role in coronavirus enteric tract infections.
From a pathogenesis standpoint, it has been demonstrated that ACE2 expression in lungs of mice has a protective effect on experimentally induced lung injury (Imai et al. 2005). Interestingly, SARS-CoV infection and the spike protein itself were shown to reduce cell surface expression of ACE2 resulting in an exacerbation of lung injury (Kuba et al. 2005). This led to the interesting view of the dual role of ACE2 in SARS-CoV pathogenesis as both main receptor for the virus and also as a protective factor for the host preventing acute lung injury (Zhang et al. 2020).
Following the landmark discovery of ACE2 as SARS-CoV receptor, many subsequent studies have investigated the molecular details of the SARS-CoV spike–ACE2 interaction, in particular the residues in both spike and ACE2 involved in the spike–receptor interaction at the binding interface (Li et al. 2005a), as well as adaptive mutations that occurred during emergence of the virus from bats to mask palmed civet to humans. Comprehensive reviews on these adaptive mutations can be found in Graham and Baric (2010) and Li (2015).
On a structural level, SARS-CoV CTD was shown to adopt a core structure formed by a single beta-sheet with an extended loop that contains all the residues contacting the receptor. In the case of SARS-CoV, this single loop is designated as the receptor-binding motif or RBM (Fig. 1C). Subsequently, closely related bat coronaviruses, such as BatCoV-SARS-related strains WIV1 and WIV16, were also found to be able to use ACE2. Notably, both bat coronaviruses WIV1 and WIV16 were able to use bat, civet and human ACE2 as functional receptor (Ge et al. 2013; Yang et al. 2015).
Rather unexpectedly, ACE2 was shown to also be the main receptor of the distantly related HCoV-NL63, a virus belonging to the Alphacoronavirus genus (Hofmann et al. 2005) (Fig. 2). In both SARS-CoV and HCoV-NL63, the CTDs of the respective spike proteins are implicated in the binding to ACE2. However, the receptor-binding interfaces differ greatly as HCoV-NL63’s core CTD structure is formed by a beta-sandwich composed of two stacked beta-sheets and with three short loops that function as three discrete RBMs contacting the receptor rather than the single RBM loop identified for SARS-CoV. While HCoV-NL63 and SARS-CoV show differences in the structure of their CTDs, they both bind within the same region on ACE2, with several key residues in the VBM shared between the two viruses. Within the alphacoronaviruses, the CTD core structure is conserved; however, differences in the extended loop structures allow for different receptor specificity, such as in the case of HCoV-NL63 and PRCV, which bind ACE2 and APN, respectively (Hulswit, de Haan and Bosch 2016).
SARS-CoV-2 was also found to bind to and use ACE2 as main receptor (Hoffmann et al. 2020; Letko et al. 2020; Walls et al. 2020; Wrapp et al. 2020; Zhou et al. 2020b). Surface plasmon resonance analysis revealed that the SARS-CoV-2 spike ectodomain could bind ACE2 with a measurably higher affinity than SARS-CoV spike (Wrapp et al. 2020). It has also been suggested that while the RBD of SARS-CoV-2 has a higher binding affinity, it is less exposed than the RBD of SARS-CoV (Shang et al. 2020a). The cryo-electron microscopy study by Yan and colleagues revealed the overall structure formed by the ternary complex composed of SARS-CoV-2 RBD-ACE2-B0AT1 (Yan et al. 2020). Crystal structure analyses of the SARS-CoV-2 RBD-ACE2 binding interface revealed that although the overall binding mode was similar to that of SARS-CoV, some key residue changes in SARS-CoV-2 RBD were responsible for the increase in binding affinity to the receptor (Lan et al. 2020; Shang et al. 2020b). A landmark study by Hou and colleagues established a reverse genetics system for SARS-CoV-2 and revealed by highly sensitive RNA in situ mapping that ACE2 expression was most abundant in the nasal cavity with a decreasing gradient of expression along the respiratory tract (Hou et al. 2020). This finding mirrored the gradient of susceptibility to SARS-CoV-2 infection observed from proximal to distal pulmonary epithelial cell cultures.
The bat coronavirus strain RaTG13, which was found to be the closest known relative to SARS-CoV-2 to date with over 96% genomic sequence identity, was found to have an RBD 89% identical to SARS-CoV-2 RBD and pseudotyped viruses harboring RaTG13 spike were able to use human ACE2 for cellular entry, albeit with much less efficiency than SARS-CoV-2 pseudovirions (Shang et al. 2020b). A more recent comparative analysis has shown that SARS-CoV-2 spike binding to ACE2 was 1000-fold tighter than that of RaTG13 spike (Wrobel et al. 2020). Furthermore, the bat coronavirus strain RmYN02 identified in Rhinolophus malayanus fecal samples from Yunnan Province, China, was found to have a more divergent RBD with only 62% identity to SARS-CoV-2 RBD (Zhou et al. 2020a). Based on structural modeling considerations, it was suggested that bat coronavirus RmYN02 uses a different receptor than ACE2. Malayan pangolins (Manis javanica) illegally smuggled into Southern China in 2017 and 2019 were found to harbor SARS-CoV-2-related viruses (Lam et al. 2020). Interestingly, one strain obtained from a pangolin in Guangdong Province in 2019 was found to possess an RBD sequence almost identical to SARS-CoV-2 with 97% sequence identity at amino acid level (Jaimes et al. 2020a), which suggested a likely recombination event (Zhou et al. 2020a).
Dipeptidyl peptidase 4 (DPP4, CD26)
DPP4 is the main receptor for MERS-CoV and was identified soon after the discovery and characterization of the first isolates of the virus (Raj et al. 2013) (Fig. 2). DPP4 is a type II transmembrane protein that forms a homodimer with each monomer composed of a membrane-proximal α/β hydrolase domain and a membrane-distal β-propeller domain.
Functionally, DPP4 is a serine exopeptidase known to cleave a broad range of cellular proteins. Human DPP4 carries important metabolic functions as it is a major participant in glucose regulation, thanks to its ability to inactivate incretins and because it binds to extracellular adenosine deaminase allowing local regulation of adenosine levels. DPP4 is expressed in many tissues and cell types, including type I and II pneumocytes, alveolar macrophages, vascular endothelial cells, as well as thymocytes and in intestine, liver and kidney cells (Meyerholz et al. 2016).
Shortly after the discovery of the MERS-CoV receptor, the molecular details of the interaction between MERS-CoV S and DPP4 were revealed at atomic resolution by X-ray crystallography analyses (Lu et al. 2013; Wang et al. 2013). The RBD of MERS-CoV, located in the CTD, binds to the β-propeller domain of DPP4 and recognizes blades 4 and 5 of this domain. Subsequently, functional studies based on chimeric DPP4 receptors with the swapping of blades 4 and 5 regions of various mammalian species demonstrated that MERS-CoV is capable of recognizing DPP4 from different mammals, including camel and horse DPP4, and to a lesser extent goat and bat DPP4 (Barlan et al. 2014). MERS-CoV is not able to bind to murine DPP4 because of a divergence identified in the blade 4 and 5 region.
Investigation of receptor usage in closely related bat coronavirus species BatCoV-HKU4 and BatCoV-HKU5 found that only the former was capable of DPP4 usage, with a preference for bat DPP4 over human DPP4 (Yang et al. 2014). The more distantly related SARS-CoV-2, a lineage B Betacoronavirus genus member, was shown to be unable to use DPP4 as host receptor (Zhou et al. 2020b). Similarly to what is observed in alphacoronaviruses, the betacoronaviruses SARS-CoV and MERS-CoV share common structural features in their CTD cores, but differences in the extended loops allow each virus to bind different receptors, in this case ACE2 and DPP4, respectively (Li 2015).
Other host proteins as attachment factors and other considerations
While this review has so far covered the four main protein receptors of coronaviruses, it is important to consider that other cell surface membrane proteins have been shown to be attachment factors and/or co-receptors. These include proteins such as C-type lectins dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) and liver/lymph node-specific intracellular adhesion molecule-3-grabbing non-integrin (L-SIGN) (Belouzard et al. 2012). These lectins have been well described for enhancing entry of HIV-1 in trans whereby dendritic cells expressing DC-SIGN capture HIV-1 virions and enable transport to lymphoid tissues where efficient transmission of virions to T cells can occur (Geijtenbeek and van Kooyk 2003). DC-SIGN and/or L-SIGN were found to be used during cell entry by coronaviruses such as SARS-CoV, HCoV-229E, FCoV and IBV (Jeffers et al. 2004; Yang et al. 2004; Jeffers, Hemmila and Holmes 2006; Regan and Whittaker 2008; Regan, Ousterout and Whittaker 2010; Zhang et al. 2012). In addition, MERS-CoV entry was found to be facilitated by CEACAM5 attachment factor (Chan et al. 2016). Further, both MERS-CoV and the lineage D betacoronavirus BatCoV-HKU9 were found capable of using 78-kDa glucose-regulated protein (GRP78) as a host cell binding factor (Chu et al. 2018).
CARBOHYDRATE RECEPTORS OF CORONAVIRUSES
In addition to the well-characterized capacity to bind protein receptors, many coronavirus spike proteins can also bind carbohydrates. This additional ability for cell attachment has been generally underappreciated, perhaps because it is considered to be ancillary to the protein binding function of most coronavirus spike proteins. However, carbohydrate binding can play important roles in coronavirus tropism modification and pathogenicity, as discussed below. This function is typically carried out by the N-terminal NTD subdomain, suggested to have been co-opted from a cellular galectin gene by a hypothetical ancestral coronavirus that harbored a spike protein with only a CTD present in its S1 subunit (Peng et al. 2011; Li 2015). Depending on the coronavirus considered, spike proteins can either bind carbohydrate moieties, a proteinaceous receptor or both.
Sialic acids or sialosides
Different members of the four coronavirus genera have the capacity to bind carbohydrates, including the PDCoV, which was shown to have a spike NTD that could bind to mucins, a family of heavily glycosylated proteins produced by animal epithelial tissues (Shang et al. 2018a) (Fig. 2).
The most well-characterized types of carbohydrates recognized by coronavirus NTDs on both structural and functional levels are sialic acids or sialosides. Sialic acids are acidic carbohydrates with a nine-carbon backbone derived from neuraminic acid, which are terminally linked to oligosaccharides on glycoproteins or gangliosides and are used by a wide variety of enveloped and non-enveloped viruses for attachment and entry, such as species of parainfluenza viruses, rotaviruses, adenoviruses and polyomaviruses (Neu, Bauer and Stehle 2011). They are perhaps best known for being the main receptors of influenza viruses (Stencel-Baerenwald et al. 2014).
Sialic acid binding activity has been confirmed for the alphacoronaviruses TGEV and PEDV (Schultze et al. 1996; Liu et al. 2015) (Fig. 2). Very interestingly for TGEV, point mutations or a short deletion near the N-terminus of spike was shown to abrogate sialic acid binding and such variations were associated with markedly lower viral pathogenicity (Krempl et al. 1997). Many betacoronaviruses from lineage A such as members of the Betacoronavirus-1 species BCoV, PHEV, HCoV-OC43, as well as HCoV-HKU1, the JHM strain of MHV and also lineage C MERS-CoV were shown to bind sialosides (Peng et al. 2012; Li et al. 2017; Hulswit et al. 2019; Park et al. 2019; Tortorici et al. 2019; Qing et al. 2020) (Fig. 2). IBV, a representative of the Gammacoronavirus genus, is another example of the spike protein's ability to recognize such carbohydrate moieties (Winter et al. 2006; Promkuntod et al. 2014).
9-O-acetylated sialic acids (9-O-Ac-Sias) were identified to be the main ligand of the NTDs of lineage A betacoronaviruses as shown for the prototype BCoV virus and related human virus HCoV-OC43, as well as HCoV-HKU1 and porcine hemagglutinating encephalitis virus PHEV (Figs 2 and 3). Attempts by crystallization to pinpoint the exact location of the 9-O-Ac-Sias binding site within BCoV S has proven difficult (Peng et al. 2012; Hulswit et al. 2019); however, structural analysis of the related PHEV S revealed a site within the NTD formed by two hydrophobic pockets compatible with 9-O-Ac-Sias binding and which is conserved for other betacoronaviruses (Hulswit et al. 2019). Further, a recent cryo-electron microscopy analysis of HCoV-OC43 S revealed in unprecedented detail the binding pocket in the NTD that allows interaction with 9-O-Ac-Sias (Tortorici et al. 2019). The identified receptor recognition site was confirmed to be conserved among betacoronaviruses known to bind 9-O-Ac-Sias, such as members of the Betacoronavirus-1 species HCoV-OC43, BCoV and PHEV, as well as HCoV-HKU1. Interestingly, the 9-O-Ac-Sias binding region shares architectural features with coronavirus hemagglutinin esterase (HE) and influenza C hemagglutinin esterase fusion (HEF) protein. Indeed, lineage A betacoronaviruses such as Betacoronavirus-1 species members and MHV are known to harbor a HE protein (Zeng et al. 2008). Structural analysis revealed details of the intriguing evolutionary relationship of coronavirus HE with influenza C HEF protein. The coronavirus HE protein is a dimer and functions both as a lectin and a receptor destroying enzyme (RDE), thanks to its sialate-9-O-acetylesterase activity (Zeng et al. 2008). Notably, for MHV, binding to O-acetylated sialic acids was shown to be mediated solely by its HE protein and not S (Langereis et al. 2010).
Figure 3.
Structures of the NTD of members of the Betacoronavirus genus. Displayed are the structures of BCoV NTD (PDB 4H14) known to bind to 9-O-acetylated sialic acids, MHV NTD, which recognizes the proteinaceous receptor mCEACAM1a but conserves structural features of the galectin fold (PDB 3JCL), MERS-CoV NTD (PDB 6Q06), which binds sialosides with a preference for α2,3-linked sialic acids, and SARS-CoV-2 NTD (PDB 6ZGE), which was suggested to bind to α,N-acetyl neuraminic acid (see text for details).
Other examples of sialic acids known to be recognized by other coronaviruses include N-glycolylneuraminic acid (Neu5Gc), N-acetylneuraminic acid (Neu5Ac) for TGEV and only Neu5Gc for IBV (Li 2015). MERS-CoV was shown to bind to sialic acids, with a selective preference for binding to α2,3-linked sialosides receptors over α2,6-linked sialic acid receptors, a finding which correlates with the major sites of replication in the upper and lower respiratory tracts of dromedary camels and humans, respectively (Li et al. 2017). Detailed cryo-electron microscopy analysis of MERS-CoV S revealed the conserved groove within the NTD that mediates interaction with sialic acids and the molecular determinants for the observed selectivity for α2,3-linked sialic acid receptors (Park et al. 2019).
To date, coronavirus CTDs appear specialized in binding to protein receptors and coronavirus binding to carbohydrates is mainly attributed to the NTD, with the clear exception of MHV-A59 NTD that binds mCEACAM1a protein (Fig. 3). From an evolutionary standpoint, it appears likely that ancestral coronavirus spike proteins first evolved the capacity to bind to protein receptors, thanks to their CTD domain, which forms a basic foundation for coronavirus receptor recognition and binding. Later acquisition by an ancestral coronavirus, possibly by integration of a host galectin gene at the N-terminus of spike allowed coronaviruses to gain the ability to bind to carbohydrates (Peng et al. 2011; Li 2015). Such co-opted lectin function underscores the modular nature of the coronavirus spike proteins and likely considerably expanded coronavirus tropism and host range (Hulswit, de Haan and Bosch 2016).
This additional binding ability to carbohydrates appears to be dispensable for entry of some coronaviruses. This notion is supported by the fact that several coronaviruses harbor S proteins with deletions of the entire NTD such as PRCV and HCoV-229E (Fig. 2). The presence of two distinct NTDs in a 229E-like bat coronavirus spike protein adds further evidence to the notion that NTD domains were added at a later evolutionary stage (Corman et al. 2015). In this context, it is interesting to consider the consequences of deletions of NTD in contemporary coronaviruses. Perhaps the most well-described example can be found in PRCV, which is very closely related to TGEV but has lost its ability for carbohydrate binding due to a deletion in its NTD (Rasschaert et al. 1990). This NTD deletion and the accompanied loss in the ability to bind sialic acids led to a switch in tropism and pathogenicity from TGEV, which can infect both the respiratory and enteric tracts to PRCV, which is predominantly respiratory tract-tropic (Krempl et al. 1997).
For alphacoronaviruses, the above examples have led to the notion that NTD binding to sialic acids is an important adaptation for enteric tropism, but is dispensable for respiratory tract infections (Hulswit, de Haan and Bosch 2016). It has been suggested that sialic acid binding allows enteric coronaviruses to bind to soluble sialoglycoconjugates effectively shielding virions from the harsh environment of the stomach. Also, binding to mucins would allow coronaviruses to navigate through thick mucus barriers and gain access to intestinal epithelial cells in order to initiate infection (Schwegmann-Wessels et al. 2003).
A recent study by Qing and colleagues has provided important mechanistic insights in the role played by sialic acid binding for betacoronaviruses and sheds new light on the receptor-independent spread phenomenon described previously for MHV-JHM (Qing et al. 2020). In this study, contrary to the A59 strain of MHV, the neurovirulent strain JHM was found to bind to sialic acids. This sialoside binding activity was mapped to the NTD (S1A) domain of MHV-JHM, which can still bind to the proteinaceous mCEACAM1a receptor. This remarkable finding illustrates the flexible nature of coronavirus spike proteins with an NTD capable of dual-binding modalities enabling attachment to both carbohydrate (sialic acids) and protein (mCEACAM1a) receptors. The study suggests that MHV-JHM attachment likely occurs in a two-step fashion with low affinity binding to sialic acids followed by higher affinity binding to mCEACAM1a protein receptor. In addition, sialic acid binding was shown to allow cell–cell fusion and spread even in the absence of the protein receptor. The authors have analyzed the functional role of sialic acid binding during MERS-CoV infection. It was also shown that entry likely occurred in a similar two-step manner, with cell–cell spread facilitated by MERS-CoV S NTD–sialic acid binding.
Other studies on MERS-CoV have demonstrated that neutralizing antibodies that block NTD–sialic acid interactions could be proposed as complementary or alternatives to the more conventional approaches targeting spike CTD–DPP4 interaction since NTD-targeting approaches were found to reduce infection and pathogenesis in animal models (Chen et al. 2017; Widjaja et al. 2019; Zhou et al. 2019).
In light of what is currently known regarding coronavirus NTD–sialic acid binding, the question whether SARS-CoV-2 can bind to and functionally use sialic acids for entry still remains to be more fully investigated (Ou et al. 2020). An analysis with the related SARS-CoV did not find evidence for binding to mucins (Peng et al. 2011). A recent cryo-EM study of SARS-CoV-2 spike has revealed much more extended loops as previously reported, particularly in the NTD region (Wrobel et al. 2020). This allows comparison of the overall structure of SARS-CoV-2 NTD with that of BCoV NTD, which binds 9-O-acetylated sialic acids (Fig. 3). Intriguingly, SARS-CoV-2 NTD appears to retain many core structural features of the NTD of BCoV and other members of the Betacoronavirus genus, with a ceiling featuring enrichment of beta strands (Fig. 3). A recent report based on the use of polymer-stabilized, multivalent gold nanoparticles bearing sialic acid derivatives suggests that SARS-CoV-2 spike can bind to α,N-acetyl neuraminic acid (Baker et al. 2020). Since binding to carbohydrates and sialic acids in particular has been shown to play an important role in regulating coronavirus tropism and pathogenicity, particularly in the context of enteric tract infections, it is apparent that this is an area that calls for further investigation. If these findings are confirmed, they could pave the way to the identification of novel neutralization-vulnerable sites within the NTD of SARS-CoV-2 spike.
Heparan sulfate (HS)
Under certain conditions, such as during serial passaging or through persistent infection, some coronaviruses are known to have acquired mutations in the spike protein enabling novel binding capacity to heparan sulfate (HS), a distinct type of linear polysaccharide occurring as a proteoglycan and found in most animal tissues. Binding to HS involves multi-basic motifs typically XBBXBX or XBXXBBBX (where B is a basic residue, lysine K, arginine R or histidine H) as described by Cardin and Weintraub (Cardin and Weintraub 1989; Liu and Thorp 2002).
HS binding has been clearly demonstrated for MHV, where persistent infection by MHV-A59 virus, which is solely dependent on mCEACAM1a binding for entry, led to the emergence of variant strains, in particular the MHV/BHK strain, with mutations and even a short 7 amino acid insertion in the S1 subunit (492TQTTRTKKVPKPKS505, insertion underlined) that introduces multi-basic sites at different locations within the spike protein (Schickli et al. 2004; de Haan et al. 2005). These modifications allow entry into cells in a heparan sulfate-dependent manner. It was found that the mutations and insertions were responsible for the extended host cell range of this particular strain of MHV (Schickli et al. 2004). Intriguingly, the 7 amino acid insert that was identified in MHV/BHK, located in the CTD, was found to allow for dual-binding competency to HS and mCEACAM1a as well as dependency of both factors for host cell entry (de Haan et al. 2006). Moreover, the added mutations introducing a multi-basic HS-binding site identified in the S2 subunit was found to obviate the requirement for mCEACAM1a binding with virus entry shown to be solely dependent on HS. These mutations are specifically located at the S2′ cleavage site. While HS-binding may allow facilitated entry into host cells in vitro, it is important to bear in mind that this could well be a product of cell culture adaptation. In vivo, HS binding could potentially be deleterious for virus entry into host cells as HS can also likely act as decoy receptor.
Another remarkable instance of HS binding was identified in FCoV. Type I (Clade A) FCoVs and CCoVs harbor a distinct spike protein that contains an insertion introducing a multi-basic motif at the S1/S2 junction that can be cleaved by furin (Whittaker, André and Millet 2018; Jaimes et al. 2020b). Such site is absent in type II (clade B) FCoV. Intriguingly, it has been demonstrated that a point mutation in the S1/S2 cleavage site that arose during cell culture adaptation of a type I FCoV strain abrogates furin cleavage (UCD1 variant strain 782NHTHSRRSRGSTSTSV797, with the mutation from parental UCD strain underlined) (de Haan et al. 2008) (Fig. 2). At the same time, the R791G mutation allows preservation of a multi-basic HS-binding motif establishing a novel dependency for HS during viral entry (de Haan et al. 2008). Similarly, the study by de Haan and colleagues has shown that a gain in HS dependency during cell culture adaptation was also identified for HCoV-OC43, a human virus of the Betacoronavirus genus. In addition, the alphacoronavirus HCoV-NL63 was found to functionally use HS proteoglycans for host cell attachment (Milewska et al. 2014).
As mentioned, several type I FCoV and CCoV spike proteins are predicted to be cleaved by furin at the S1/S2 multi-basic site (Jaimes et al. 2020b). In the case of the FCoV strain UCD, this site is also predicted to bind HS (de Haan et al. 2008). However, cleavage of the spike protein appears to prevent the binding to HS in this strain, suggesting that an intact spike is required to allow binding. This finding agrees with a previous report by the same group, where MHV/BHK was shown to have introduced a mutation at the S1/S2 site that also prevented furin cleavage (de Haan et al. 2005). The introduction of this mutation also resulted in a predicted HS multi-basic binding site. However, further analysis of this finding showed that from the three predicted HS-binding sites in MHV/BHK, the one located at the S1/S2 junction revealed less biological relevance in the adaptation of the strain to use HS as a receptor for cell entry (de Haan et al. 2006). According to that study, the two additional genetic modifications in the MHV/BHK spike gene that were described previously in this review (a 7 amino acid insert at the CTD and mutations at the S2 subunit) are considered determinant for the loss of the mCEACAM1 dependency, and the transition to a HS-dependent entry pathway. At the time when those studies were performed, the structure of the MHV spike was not yet solved. Modeling of MHV/BHK based on the known cryo-EM structure of MHV-A59 (PDB 3JCL), as performed previously with FCoV (Jaimes and Whittaker 2018), suggests that the basic residues at the HS-binding sites located at the CTD and S1/S2 junction are not well exposed, indicating less possible interaction with HS. In contrast, the basic residues at the HS-binding site located in the S2 subunit (S2′ cleavage site) are well exposed, which could facilitate interaction with HS. This last observation supports the suggestion that the multi-basic HS-binding site at the S2 subunit is determinant for HS binding, therefore regulating receptor adaptation, as was put forward previously (de Haan et al. 2006).
The Beaudette strain of IBV, a representative of the Gammacoronavirus genus, is yet another example of a coronavirus spike harboring a heparan sulfate-binding motif 685SSRRKRSL693 at the S2′ cleavage site, similar to what was described for MHV/BHK (Fig. 2). It was revealed that HS binding to the spike allows IBV-Beaudette to attach to cell surfaces (Madu et al. 2007). It was suggested that this HS binding capacity, which is not found in other IBV strains, could explain in part the broadened tropism of the Beaudette strain. IBV Beaudette harbors an additional multi-basic site at the S1/S2 position. However, it does not fulfill the requirements for a HS-binding site XBBXBX or XBXXBBBX. Interestingly, other IBV strains such as California 99, 065846/10 and AL/4614/98 harbor HS-binding sequences at their S1/S2 site. In fact, multi-basic sequences are commonly found in IBV at the S1/S2 junction. However, furin cleavage is predicted to occur at this site, which would prevent binding to HS, based on previous studies with FCoV UCD. Nevertheless, the role of the multi-basic sequences in HS binding in other IBV strains is yet to be studied.
An enigmatic feature of SARS-CoV-2 spike protein is the presence of a 4 amino acid insertion introducing a multi-basic site at the S1/S2 junction (676TQTNSPRRARSVAS689, insertion underline). This insert has attracted intense scrutiny as it appears to be evolutionary distinct with structural modeling suggesting it forms an extended and proteolytically sensitive activation loop (Andersen et al. 2020; Coutard et al. 2020; Hoffmann et al. 2020; Jaimes et al. 2020a; Walls et al. 2020; Wrapp et al. 2020; Zhou et al. 2020a). Biochemical analyses performed using fluorogenic peptide mimetics have revealed that such site could be recognized and cleaved by a wide range of host cell proteolytic enzymes including trypsin-like proteases, cathepsins and proprotein convertases such as furin (Jaimes, Millet and Whittaker 2020c). Other analyses performed on full-length spike protein expressed in mammalian cells have demonstrated that SARS-CoV-2 spike could be cleaved by furin-like proteases during maturation of the protein, however cleavage always appears partial with a substantial proportion of the S protein remaining uncleaved, as shown in various western blot analyses performed by several independent groups (Hoffmann et al. 2020; Letko et al. 2020; Ou et al. 2020; Shang et al. 2020a). Intriguingly, while the SARS-CoV-2 insert contains a minimal furin cleavage site (682RRXR685, containing the minimal P1 position arginine, R685 and the P4 position arginine, R682), it lacks a more basic character that is found in the hemagglutinin (HA) proteins of highly pathogenic avian influenza strains (Kawaoka and Webster 1988). The HA of highly pathogenic H5 and H7 strains typically contain longer stretches of basic K and R residues, a feature that defines their pathogenicity (Nao et al. 2017). Importantly, the SARS-CoV-2 S1/S2 insert lacks a basic residue at the P2 position of the cleavage site. In the uncleaved protein, the insert contains a consensus motif XBBXBX for heparan sulfate binding.
In view of the precedents for coronaviruses gaining HS binding via insertion of multi-basic sites, such as for MHV and FCoV detailed earlier, it would be of interest to study whether the SARS-CoV-2 S1/S2 loop could allow the virus to bind to HS. In fact, a recent study by Kim and colleagues suggests that SARS-CoV-2 can indeed bind HS via the S1/S2 site (Kim et al. 2020). The preliminary findings will require confirmation whether this binding allows entry into host cells, and functional validation of the consequence of this on host cell and tissue tropism, transmission and pathogenicity. More recent work confirmed that SARS-CoV-2 spike does interact with heparan sulfate, although binding was shown to occur via its receptor binding domain rather than the S1/S2 site (Clausen et al. 2020).
Interestingly, a study reported that SARS-CoV-2 virus growth in Vero-E6 cell culture and plaque purification quickly resulted in the isolation of deletion variants of varying sizes at the S1/S2 site (Lau et al. 2020). The deletions of the S1/S2 cleavage site is likely an adaptive response of the virus to cell culture and/or a new host cell proteolytic environment. However, in light of the previous considerations regarding HS binding, it would be interesting to study whether potentially negative effects of heparan sulfate binding to cell entry could be at play in this context.
CONCLUDING REMARKS
The dynamic nature and diverse array of strategies for coronaviruses have evolved to recognize various host cell receptors are remarkable. This mirrors their complex evolutionary histories and their propensity to adapt to new environments as well as new hosts and offer insights into how novel coronaviruses from bats, such as SARS-CoV-2, emerge, cross species barriers and conquer entirely new host populations. The body of work obtained by studying coronavirus–receptor interactions conducted over several decades by generations of research groups provides a solid foundation from which principles of zoonotic potential and pandemic preparedness can be drawn.
For instance, it is noteworthy to highlight the fact that many coronaviruses can bind to both carbohydrates through their spike NTD and protein receptors through their CTD. While the carbohydrate-binding NTD is in some cases dispensable, it may be considered as a lower affinity, non-specialized binding module that many coronaviruses conserve. The CTD on the other hand is highly specialized and binds specifically and with high affinity to protein receptors, often in a host species-dependent manner. The high specificity is often the result of adaptive mutations, which have been characterized extensively in the case of SARS-CoV during its emergence in different hosts (Fig. 4). Perhaps the NTD may play a critical role during species barrier crossing events by allowing an emerging coronavirus to adapt to a new host environment and maintain a minimal level of binding that would allow infection of new host cells via sialic acids, while the CTD readjusts and gains adaptive mutations for optimizing binding to a new host protein receptor. In some ways, it can be considered that the coronavirus S1 has evolved the NTD and CTD binding modules as a means to allow broadening of host cell tropism within a host as well as between differing host species. It would be of interest to investigate further the potential for evolutionary cooperation between the NTD and CTD binding modules (Fig. 4).
Figure 4.
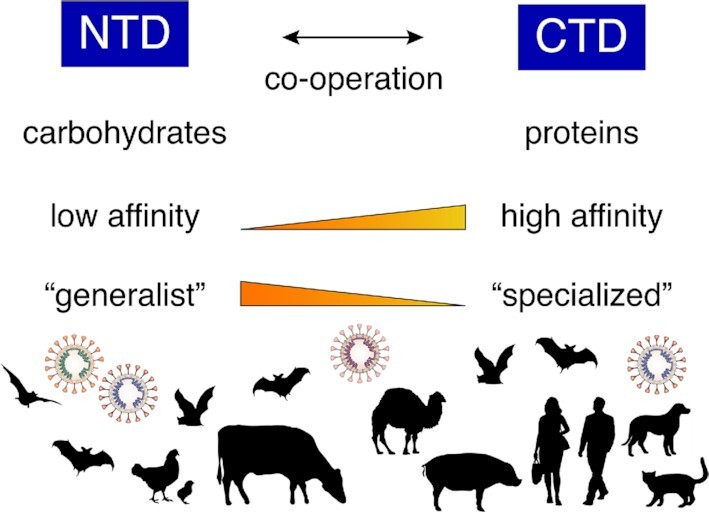
Co-operative roles of the spike protein NTD and CTD receptor binding modules in coronavirus ecology. Schematic of coronavirus NTD and CTD subdomains with their complementary characteristics and roles in coronavirus tropism and host range.
In the case of SARS-CoV-2, binding of its NTD to sialic acids and/or other carbohydrates requires further investigation. In that regard, it appears important to study whether its NTD played a role during its emergence and zoonotic transmission from bats to humans.
Regarding the role of the NTD in enteric tract infection and pathogenicity, it is interesting to point out that sequences of BatCoVs often originate from anal swabs or fecal samples (Li et al. 2005b; Poon et al. 2005; Tang et al. 2006; Samuel et al. 2007; Ge et al. 2013; Yang et al. 2015; Ge et al. 2016; Hu et al. 2017). For example, the SARS-CoV-2-related virus BatCoV-RaTG13 was obtained through a fecal swab. This would suggest that in many cases BatCoVs can productively infect the enteric tracts of bats, resulting in viral shedding. A basic question that arises from this observation is to ask what is the role played by the NTD of the spike proteins of these enteric tract tropic bat coronaviruses. Related to this, it was recently demonstrated that SARS-CoV-2 can infect human gut enterocytes (Lamers et al. 2020). Although ACE2 is known to be highly expressed in the brush border of intestinal enterocytes and likely plays a major role for attachment and infection of these cells (Qi et al. 2020), it would be of interest to probe whether SARS-CoV-2 NTD can recognize sialic acids in this context as such binding could be of importance for protection during transit within the enteric tract and infection of target enteric cells, as described above.
Two recent reports on SARS-CoV-2 provide fascinating insights on an intriguing link between furin-mediated cleavage of the coronavirus spike S1/S2 site and expansion of protein receptor usage as well as extension of cellular and tissue tropism (Cantuti-Castelvetri et al. 2020; Daly et al. 2020). Daly and colleagues demonstrated that proteolytic processing by furin of the SARS-CoV-2 S1/S2 site generates an exposed C-terminal motif 682RRAR685 at the end of S1 that complies with the so-called C-end rule (CendR) allowing for binding to neuropilin receptors, in particular neuropilin-1 or NRP1. The S1–NRP1 interaction was confirmed biochemically and NRP1 expression in ACE2-expressing cells was shown to potentiate SARS-CoV-2 infection in cell culture (Daly et al. 2020). It was further shown that a monoclonal antibody that recognizes the b1b2 extracellular domain of NRP1 inhibits SARS-CoV-2 infectivity (Cantuti-Castelvetri et al. 2020). Analysis of NRP1 expression patterns revealed that it is expressed abundantly in cells of the respiratory and olfactory epithelium, with the highest levels of expression observed in endothelial and epithelial cells that face the nasal cavity. Histological analyses of human COVID-19 samples demonstrated that SARS-CoV-2 infected cells that were positive for NRP1 staining from the olfactory bulb and epithelium, most notably endothelial cells. Mouse studies conclusively showed that NRP1 allowed transport of virus-sized nanoparticles into the central nervous system after intranasal administration. These findings can explain the extended tropism and spread observed in COVID-19 patients, in particular those suffering from anosmia and neurological symptoms (Cantuti-Castelvetri et al. 2020).
It would be of interest to investigate whether NRP-1 usage is limited to SARS-CoV-2 or is a more general phenomenon for betacoronaviruses that have a spike protein cleaved by furin at the S1/S2 site, such as HCoV-OC43, which is known for its neurotropism (Le Coupanec et al. 2015), as well as BCoV, strains of MHV, MERS-CoV or HCoV-HKU1 (Millet and Whittaker 2015). Besides NRP1, CD147 (basigin) has been proposed as being a potential receptor for SARS-CoV-2 (Wang et al. 2020). Further studies are needed to confirm whether these newly identified host factors act as bona fide receptors or play a co-receptor function in concert with the main ACE2 receptor, akin to the situation during HIV-1 infections.
As exemplified by the recent NRP1 discoveries for SARS-CoV-2 and despite extensive studies on the known coronavirus protein receptors, there are likely many more to be discovered. In particular, very little is known about protein receptors used by bat coronaviruses, despite their huge diversity. In the case of ACE2, it can be argued that the relatively high degree of conservation in various mammalian species could enable ACE2-dependent coronaviruses to gain access to a broad range of host species. In the context of the COVID-19 pandemic, a recent analysis of cross-species conservation of ACE2 based on a vast dataset of 410 vertebrates has shown a high degree of likelihood that many mammalian species, including endangered ones, have the potential to being infected by SARS-CoV-2 and could become new reservoirs and/or intermediate host species (Damas et al. 2020).
When considering protein receptors of coronaviruses, it is important to highlight that they often operate with other host cell factors such as proteases (e.g. ACE2-TMPRSS2 association) or other membrane proteins (e.g. APN-B0AT1 and ACE2-B0AT1). Furthermore, for some coronaviruses such as MERS-CoV, binding and proteolytic priming events occur in membrane microdomains enriched in tetraspanins such as CD9 (Earnest et al. 2015, 2017; Hantak et al. 2018). These tetraspanin-enriched microdomains offer a scaffold enabling clustering of the virus receptor, DPP4, and activating protease, TMPRSS2, allowing for efficient membrane fusion and virus entry.
As described in this review, the identification, characterization and study of coronavirus receptors have allowed to gain deep insights into the tropism, host range, interspecies transmission and pathogenesis of this highly successful and diverse group of viruses. These studies have been particularly invaluable to better understand emerging coronaviruses such as SARS-CoV-2. Many questions remain however, particularly regarding bat coronaviruses and other emerging coronaviruses and it is likely that further studies on coronavirus–receptor interactions will continue to shed new light on the biology of these viruses.
ACKNOWLEDGMENTS
We thank members of the Whittaker and Daniel labs at Cornell University for helpful comments and discussion.
Contributor Information
Jean K Millet, Université Paris-Saclay, INRAE, UVSQ, Virologie et Immunologie Moléculaires, 78352 Jouy-en-Josas, France.
Javier A Jaimes, Department of Microbiology and Immunology, Cornell University, Ithaca, NY 14853, USA.
Gary R Whittaker, Department of Microbiology and Immunology, Cornell University, Ithaca, NY 14853, USA; Master of Public Health Program, Cornell University, Ithaca, NY 14853, USA; Cornell Feline Health Center, Ithaca, NY 14853, USA.
FUNDING
This work was supported by the National Institutes of Health (research grant R01AI35270).
Conflict of interest
None declared.
REFERENCES
- Andersen KG, Rambaut A, Lipkin WIet al. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AN, Richards S-J, Guy CSet al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent Sci. 2020, DOI: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan A, Zhao J, Sarkar MKet al. Receptor variation and susceptibility to MERS coronavirus infection. J Virol. 2014;88:4953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BNet al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, Smits SL, Haagmans BL. Membrane ectopeptidases targeted by human coronaviruses. Curr Opin Virol. 2014;6C:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L, Ojha R, Pedro LDet al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. bioRxiv. 2020. [Google Scholar]
- Cardin AD, Weintraub HJ. Molecular modeling of protein–glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. [DOI] [PubMed] [Google Scholar]
- Chan CM, Chu H, Wang Yet al. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of middle east respiratory syndrome coronavirus. J Virol. 2016;90:9114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF-W, Yuan S, Kok K-Het al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet North Am Ed. 2020;395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lin YL, Peng Get al. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc Natl Acad Sci USA. 2012;109:17966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lu S, Jia Het al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect. 2017;6:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Chan CM, Zhang Xet al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem. 2018;293:11709–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TM, Sandoval DR, Spliid CBet al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Baldwin HJ, Tateno AFet al. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 2015;89:11858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Eckerle I, Bleicker Tet al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. [DOI] [PubMed] [Google Scholar]
- Corman VM, Ithete NL, Richards LRet al. Rooting the phylogenetic tree of MERS-Coronavirus by characterization of a conspecific virus from an African Bat. J Virol. 2014;88:11297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Valle C, de Lamballerie Xet al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley BM, Mock RE, Callison SAet al. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses. 2012;4:3689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JL, Simonetti B, Antón-Plágaro Cet al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. bioRxiv. 2020, DOI: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Hughes GM, Keough KCet al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA. 2020;117:22311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot RJ, Cowley JA, Enjuanes Let al. Order Nidovirales. In: King AMQ, J. AM, B. CE, J. LE (eds). Virus Taxonomy. San Diego: Elsevier, 2012, 784–94. [Google Scholar]
- de Haan CA, Li Z, te Lintelo Eet al. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J Virol. 2005;79:14451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CA, Te Lintelo E, Li Zet al. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J Virol. 2006;80:10909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CAM, Haijema BJ, Schellen Pet al. Cleavage of Group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J Virol. 2008;82:6078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B, Gelfi J, L'Haridon Ret al. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler GS, Pensiero MN, Cardellichio CBet al. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest JT, Hantak MP, Li Ket al. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017;13:e1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest JT, Hantak MP, Park J-Eet al. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J Virol. 2015;89:6093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairweather SJ, Broer A, O'Mara MLet al. Intestinal peptidases form functional complexes with the neutral amino acid transporter B(0)AT1. Biochem J. 2012;446:135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Zhao K, Shi ZLet al. Bat coronaviruses in China. Viruses. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint SJ, Racaniello VR, Rall GFet al. In: Principles of Virology. Washington, DC: ASM Press, 2015. [Google Scholar]
- Fouchier RA, Kuiken T, Schutten Met al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher TM, Buchmeier MJ, Perlman S. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology. 1992;191:517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TBH, van Kooyk Y. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. In: Steinkasserer A (ed). Dendritic Cells and Virus Infection. Berlin, Heidelberg: Springer Berlin Heidelberg, 2003, 31–54. [DOI] [PubMed] [Google Scholar]
- Ge X-Y, Li J-L, Yang X-Let al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XY, Wang N, Zhang Wet al. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol Sin. 2016;31:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Enjuanes L, Ziebuhr Jet al. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng BJ, He YQet al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. [DOI] [PubMed] [Google Scholar]
- Gui M, Song W, Zhou Het al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Fan B, Chang Xet al. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology. 2020;545:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantak MP, Qing E, Earnest JTet al. Tetraspanins: architects of viral entry and exit platforms. J Virol. 2018;93:e01429–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise MT, Song W, Gui Met al. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh AA, Smeenk I, Horzinek MCet al. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72:4508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A, Hofmann-Winkler H, Gierer Set al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder Set al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Pyrc K, van der Hoek Let al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. PNAS. 2005;102:7988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Okuda K, Edwards CEet al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–46.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu WJ, Xu Wet al. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 2016;12:e1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zeng LP, Yang XLet al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit RJG, Lang Y, Bakkers MJGet al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci USA. 2019;116:2681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Rao Set al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete NL, Stoffberg S, Corman VMet al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19:1697–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Andre NM, Chappie JSet al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020a;432:3309–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Millet JK, Stout EAet al. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses. 2020b;12:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1. /S2 site. iScience. 2020c;23:101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Whittaker GR. Feline coronavirus: insights into viral pathogenesis based on the spike protein structure and function. Virology. 2018;517:108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jando J, Camargo SMR, Herzog Bet al. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLoS One. 2017;12:e0184845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers SA, Hemmila EM, Holmes KV. Human coronavirus 229E can use CD209L (L-SIGN) to enter cells. Adv Exp Med Biol. 2006;581:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers SA, Tusell SM, Gillim-Ross Let al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Webster RG. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci USA. 1988;85:324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jin W, Sood Aet al. Glycosaminoglycan binding motif at S1/S2 proteolytic cleavage site on spike glycoprotein may facilitate novel coronavirus (SARS-CoV-2) host cell entry. bioRxiv. 2020, DOI: 10.1016/j.antiviral.2020.104873. [DOI] [Google Scholar]
- Kirchdoerfer RN, Cottrell CA, Wang Net al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C, Schultze B, Laude Het al. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol. 1997;71:3285–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao Set al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Fouchier RA, Schutten Met al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart Jet al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Shum MH-H, Zhu H-Cet al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020. [DOI] [PubMed] [Google Scholar]
- Langereis MA, van Vliet ALW, Boot Wet al. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by the hemagglutinin-esterase and not by the spike protein. J Virol. 2010;84:8970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu Jet al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KSet al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SY, Wang P, Mok BWet al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9:837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Coupanec A, Desforges M, Meessen-Pinard Met al. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11:e1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Gouilh MAR, Brugere-Picoux J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a One-Health strategy to control the COVID-19 pandemic. One Health. 2020;10:100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li W, Farzan Met al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005a;309:1864–8. [DOI] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PL, Kurup A, Gopalakrishna Get al. Laboratory-acquired severe acute respiratory syndrome. N Engl J Med. 2004;350:1740–5. [DOI] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu Pet al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Tang J, Ma Yet al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol. 2015;89:6121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Thorp SC. Cell surface heparan sulfate and its roles in assisting viral infections. Med Res Rev. 2002;22:1–25. [DOI] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Kenney SPet al. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 2018;115:E5135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Widjaja Iet al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci USA. 2017;114:E8508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva Net al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu Met al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science. 2005b;310:676–9. [DOI] [PubMed] [Google Scholar]
- Lu G, Hu Y, Wang Qet al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu IG, Chu VC, Lee Het al. Heparan sulfate is a selective attachment factor for the avian coronavirus infectious bronchitis virus Beaudette. Avian Dis. 2007;51:45–51. [DOI] [PubMed] [Google Scholar]
- Martina BE, Haagmans BL, Kuiken Tet al. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M, Millet J, Vidalain P-Oet al. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol. 2012;86:7577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz DK, Lambertz AM, McCray PB Jr. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol. 2016;186:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A, Zarebski M, Nowak Pet al. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med. 2008;14:361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E, Hertzig T, Gorbalenya AEet al. Discovery of an RNA virus 3'→5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura TA, Travanty EA, Oko Let al. The spike glycoprotein of murine coronavirus MHV-JHM mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a-/- Mice. J Virol. 2008;82:755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MA, Corman VM, Jores Jet al. MERS Coronavirus Neutralizing Antibodies in Camels, Eastern Africa, 1983–1997. Emerg Infect Dis. 2014;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagaki K, Taguchi F.. Receptor-independent spread of a highly neurotropic murine coronavirus JHMV strain from initially infected microglial cells in mixed neural cultures. J Virol. 2005;79:6102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nao N, Yamagishi J, Miyamoto Het al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio. 2017;8:e02298–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu U, Bauer J, Stehle T. Viruses and sialic acids: rules of engagement. Curr Opin Struct Biol. 2011;21:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Second lab accident fuels fears about SARS. Science. 2004;303:26. [DOI] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei Xet al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Walls AC, Wang Zet al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26:1151–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Lai ST, Poon LLet al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Sun D, Rajashankar KRet al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci USA. 2011;108:10696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Xu L, Lin YLet al. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. 2012;287:41931–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LL, Chu DK, Chan KHet al. Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promkuntod N, van Eijndhoven REW, de Vrieze Get al. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Qian S, Zhang Set al. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E, Hantak M, Perlman Set al. Distinct roles for sialoside and protein receptors in coronavirus infection. mBio. 2020;11:e02764–02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, Smits SLet al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasschaert D, Duarte M, Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J Gen Virol. 1990;71:2599–607. [DOI] [PubMed] [Google Scholar]
- Regan AD, Millet JK, Tse LPet al. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan AD, Ousterout DG, Whittaker GR. Feline lectin activity is critical for the cellular entry of feline infectious peritonitis virus. J Virol. 2010;84:7917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan AD, Whittaker GR.. Utilization of DC-SIGN for entry of feline coronaviruses into host cells. J Virol. 2008;82:11992–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Santiago C, Mudgal Get al. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SE, Tellier R, Mahony J. The laboratory diagnosis of severe acute respiratory syndrome: emerging laboratory tests for an emerging pathogen. Clin Biochem Rev. 2004;25:133–41. [PMC free article] [PubMed] [Google Scholar]
- Samuel RD, Thomas JOS, Lauren MOet al. Detection of Group 1 coronaviruses in bats in North America. Emerg Infect Dis. 2007;13:1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickli JH, Thackray LB, Sawicki SGet al. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J Virol. 2004;78:9073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Krempl C, Ballesteros MLet al. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J Virol. 1996;70:5634–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C, Zimmer G, Schroder Bet al. Binding of transmissible gastroenteritis coronavirus to brush border membrane sialoglycoproteins. J Virol. 2003;77:11846–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo Cet al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020a;117:11727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi Ket al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020b;581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Zheng Y, Yang Yet al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol. 2018a;92:e01556–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Zheng Y, Yang Yet al. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018b;14:e1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus (MERS-CoV) infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87:12552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A, Heald-Sargent T, Subramanya Get al. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit THC, Brackman CJ, Ip SMet al. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald JE, Reiss K, Reiter DMet al. The sweet spot: defining virus–sialic acid interactions. Nat Rev Microbiol. 2014;12:739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout AE, André NM, Jaimes JAet al. Coronaviruses in cats and other companion animals: where does SARS-CoV-2/COVID-19 fit? Vet Microbiol. 2020;247:108777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XC, Zhang JX, Zhang SYet al. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006;80:7481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Shi M, Chommanard Cet al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and Their recombination history. J Virol. 2017;91:e01953–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Matsui N, Noguchi Ket al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One. 2014;9:e106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici MA, Walls AC, Lang Yet al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan DB, Holmes KV. Feline aminopeptidase N is a receptor for all Group I coronaviruses. In: Enjuanes L, Siddell SG, Spaan W, (eds). Coronaviruses and Arteriviruses. Boston, MA: Springer US, 1998, 69–75. [DOI] [PubMed] [Google Scholar]
- Tresnan DB, Levis R, Holmes KV. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol. 1996;70:8669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S, de Graaf M, Lauber Cet al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00473–00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand JM, Haagmans BL, Leijten Let al. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet Pathol. 2008;45:551–62. [DOI] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moes Eet al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici MAet al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–92.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Bosch B-Jet al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016a;531:114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Frenz Bet al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016b;23:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Snijder Jet al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017;114:11157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Chen W, Zhou Y-Set al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020. [Google Scholar]
- Wang N, Shi X, Jiang Let al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Allen JD, Wrapp Det al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker GR, André NM, Millet JK. Improving virus taxonomy by recontextualizing sequence-based classification with biologically relevant data: the case of the alphacoronavirus 1 species. mSphere. 2018;3:e00463–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja I, Wang C, van Haperen Ret al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg Microbes Infect. 2019;8:516–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, Schwegmann-Wessels C, Cavanagh Det al. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol. 2006;87:1209–16. [DOI] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Huang Yet al. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood). 2009;234:1117–27. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KSet al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel AG, Benton DJ, Xu Pet al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020;27:763–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Tortorici MA, Snijder Jet al. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J Virol. 2018;92:e01628–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Chang YC, Ko TPet al. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc Natl Acad Sci USA. 2020;117:1438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Hu B, Wang Bet al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol. 2015;90:3253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Du L, Liu Cet al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA. 2014;111:12516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z-Y, Huang Y, Ganesh Let al. pH-Dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Yet al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager CL, Ashmun RA, Williams RKet al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Cao D, Zhang Yet al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Qiao S, Guo Ret al. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat Commun. 2020;11:3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TMet al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Langereis MA, van Vliet ALWet al. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. 2008;105:9065–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Penninger JM, Li Yet al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buckles E, Whittaker GR. Expression of the C-type lectins DC-SIGN or L-SIGN alters host cell susceptibility for the avian coronavirus, infectious bronchitis virus. Vet Microbiol. 2012;157:285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu Tet al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020a;30:2196–203.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen Y, Zhang Set al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat Commun. 2019;10:3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fan H, Lan Tet al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XGet al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020b;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang Wet al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]