Abstract
Estimates of seroprevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies have been hampered by inadequate assay sensitivity and specificity. Using an enzyme-linked immunosorbent assay–based approach that combines data about immunoglobulin G responses to both the nucleocapsid and spike receptor binding domain antigens, we show that excellent sensitivity and specificity can be achieved. We used this assay to assess the frequency of virus-specific antibodies in a cohort of elective surgery patients in Australia and estimated seroprevalence in Australia to be 0.28% (95% Confidence Interval, 0–1.15%). These data confirm the low level of transmission of SARS-CoV-2 in Australia before July 2020 and validate the specificity of our assay.
Keywords: SARS-CoV-2, COVID-19, seroprevalence, ELISA, antibodies
The determination of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody seroprevalence in low-transmission settings requires highly sensitive and specific assays. We combined data about antibody responses to the spike receptor binding domain and nucleocapsid to confirm the low prevalence of SARS-CoV-2 in Australia.
Reported cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are likely to represent only a fraction of actual SARS-CoV-2 infections, as approximately 40% of cases are mild or asymptomatic, or otherwise undiagnosed [1]. Detection of antibodies that recognize viral antigens specific for SARS-CoV-2 has become an important molecular sentinel of current or prior exposure to SARS-CoV-2 [2]. Since a significant number of people either present with mild symptoms of coronavirus disease 2019 (COVID-19) infection or are asymptomatic, serological measurements will have ongoing utility in gauging exposure and prevalence in the community [3]. Such studies will provide valuable information on the time course and longevity of antibody responses to SARS-CoV-2 [4]. Further, serological testing is likely to be valuable in the assessment of vaccine efficacy. However, analyses of seroprevalence, especially in low-prevalence settings, are hampered by assays with inadequate sensitivity and specificity [5].
Australia has reported low case numbers of COVID-19 per head of population compared to other developed Westernized countries, especially before the July–August 2020 outbreak in Melbourne, Victoria (Australian Department of Health). However, nucleic acid testing generally only reveals a fraction of the total numbers of infections; thus, the overall number of previous infections is unknown [3, 6]. Nonetheless, it is likely that the total of previously infected individuals is low as a proportion of the population (<1%). Thus, to assess the seroprevalence of SARS-CoV-2 in Australia, we developed a dual-antigen enzyme-linked immunosorbent assay (ELISA), which gave high sensitivity and specificity suitable for this setting.
MATERIALS AND METHODS
Samples and Ethics Statement
Collection of blood from individuals pre-2020 (n = 184) was carried out after provision of informed consent, using procedures approved by the Human Research Ethics Committees (HRECs) of the Australian National University (2016/317) and Australian Capital Territory (ACT) Health (1.16.011 and 1.15.015). Samples from SARS-CoV-2–positive individuals (n = 43) were collected after consent under the following protocols: Alfred Hospital HREC (280/14); James Cook University HREC (H7886); ACT Health HREC (1.16.011); Charité Ethics Committee (EA2/066/20) [7]. Approval for the elective surgery study was given by HRECs at the Alfred Hospital (339/20) and The Australian National University (2020/379). Whole blood was collected by venipuncture into a syringe containing 3.2% (w/v) trisodium citrate (healthy donors) or a red-capped serum Vacutainer tube (patients), rested for 1 hour, then centrifuged (1000g, 10 minutes, 4°C) and the upper plasma or serum phase removed by aspiration to a new tube and immediately frozen. All samples were heated to 56°C for 1 hour prior to analysis. Due to limited sample availability, some samples were not used for all tests; the sample number is given in the relevant figure legends.
ELISA Protocol
Our ELISA protocol was based on previously published methodologies with modifications [8]. In brief, white 96-well Maxisorp microtiter plates (Nunc 436110) were coated overnight at 4°C with 100 µL of 500 ng/mL S1 spike domain (GenScript Biotech, Piscataway, New Jersey; Z03501) spike receptor binding domain (RBD; GenScript Biotech, Z03483) or nucleocapsid (GenScript Biotech, Z03480) protein in 1× Dulbecco’s phosphate-buffered saline (PBS) pH 7.4 (Sigma D1408). Wells were washed 3 times with PBS containing 0.2% (v/v) Tween-20 (PBS-T), blocked with 100 µL 3% (w/v) bovine serum albumin (BSA) in PBS with 0.1% (v/v) Tween-20 for 1 hour at room temperature, then washed once with PBS-T, before addition of 50 µL serum diluted to 1:100 in 1% (w/v) BSA in PBS with 0.1% (v/v) Tween-20. Plate washing was performed by repeated plunging of plates into a bucket filled with PBS-T and flicking of well contents into a sink. After 1 hour of incubation at room temperature, wells were washed 5 times with PBS-T and incubated with 100 µL of horseradish peroxidase (HRP)–conjugated anti-human immunoglobulin G (IgG), immunoglobulin M (IgM), or immunoglobulin A (IgA) antibodies diluted to the optimal concentration in 1% BSA (w/v) in PBS with 0.1% Tween-20 for 1 hour at room temperature. Wells were washed 5 times with PBS-T, then 100 μL of Super Signal ELISA Pico enhanced chemiluminescent (ECL) HRP substrate (Pierce, Rockford, Illinois) was added and light emission (stable after 1 minute) was measured using a Victor-Nivo luminescence plate reader. In some assays, 100 µL 0.4 mg/mL o-phenylene diamine (OPD; Sigma, St Louis, Missouri) stopped with 50 µL 10% sodium dodecyl sulfate was used as the detection reagent and absorbance read at 450 nm. For high-throughput screening of samples, steps downstream of sample addition were automated as outlined in the Supplementary Materials.
Statistical Analysis
ELISA data were expressed as the normalized log10 emission at 700 nm. Receiver operating characteristic analysis and cutoffs were determined using GraphPad Prism 8 software. Estimates of seroprevalence were calculated using R with 95% confidence calculated by bootstrapping. Bayesian analysis to determine the probability of positivity for each sample was determined using R based on the distributions of the positive and negative values described as mixed distributions. Full details of statistical analysis, code, and data are given in the Supplementary Materials.
RESULTS
Optimization of Manual and Automated ELISA Protocol Conditions
To optimize our assay, we used a library of 184 plasma samples collected pre-2020 as negative controls, and a panel of 43 sera from individuals infected with SARS-CoV-2 as positive controls. Initial optimization of assay conditions was carried out with defined pools of sera from 5 positive donors and 5 negative donors. Noting that even small gains in specificity can substantially reduce the number of false positives in large serosurveys, we optimized the concentration and amount of antigen used for coating, blocking, and washing conditions. Overall, we found that the principal factors affecting assay performance were the coating conditions and the necessity of stringent washing (Supplementary Table 1 and Supplementary Figure 1).
To handle large numbers of samples, we optimized our ELISA assay for automation. We investigated the use of an ECL substrate as these substrates have superior sensitivity compared to traditional colorimetric absorbance substrates such as OPD [9] and do not require a stopping step facilitating automation. Comparing different protocols to distinguish our responses to the N antigen in positive and negative donors, we determined that ECL was marginally superior to OPD with a larger separation between positive and negative control values (Supplementary Figure 2A and 2B). Importantly, conducting the analysis on a robotic platform did not compromise assay sensitivity or specificity (Supplementary Figure 2C and 2D).
Combining IgG Responses to Multiple Antigens Gives Optimal Sensitivity and Specificity
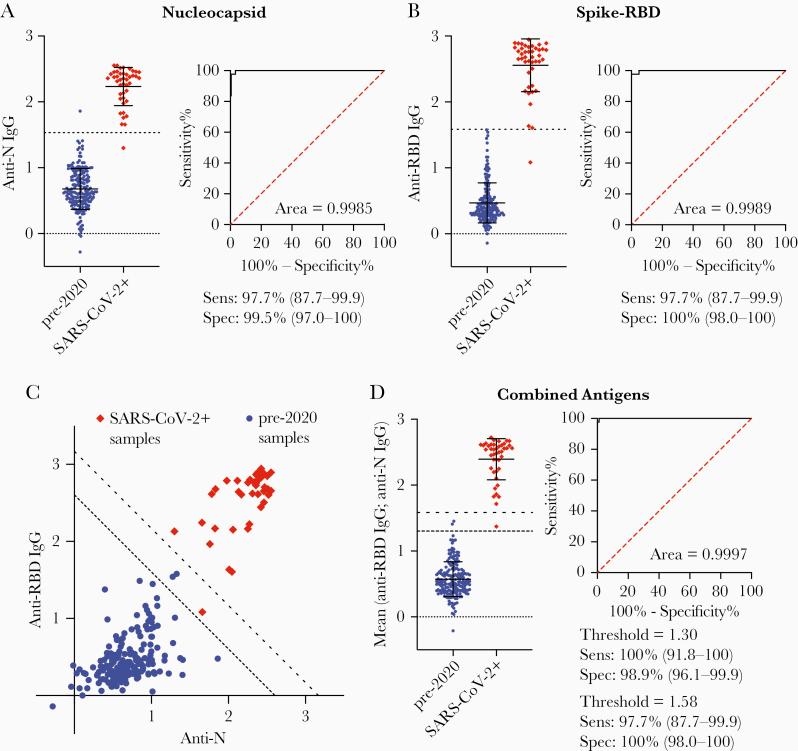
Having established optimal ELISA conditions, we wanted to determine the optimal antigen, or combination of antigens for seroprevalence surveys. We therefore compared responses to the S1 domain of the spike protein (S1), nucleocapsid protein (N), and the RBD of the spike protein. Overall, the sensitivity and specificity of responses to the N and RBD were comparable, with the N protein being slightly superior (Figure 1A and 1B). Surprisingly, the S1 protein gave poor sensitivity and specificity with many negative samples giving high values (Supplementary Figure 3). Plotting responses to the RBD and N antigens revealed that even the less responsive positive control samples generally had at least elevated responses to both antigens (Figure 1C). We can therefore present the data as the mean of the responses to the RBD and N responses (Figure 1D). Analyzed this way, we found that a cutoff of 1.30 gave 100% (95% Confidence Interval [CI], 91.8%–100%) sensitivity and 98.91% (95% CI, 96.1%–99.9%) specificity, whereas a more stringent cutoff of 1.58 gave 97.7% (95% CI, 87.7%–99.9%) sensitivity but 100% (95% CI, 98.0%–100%) specificity (Figure 1D). Neither IgA nor IgM responses distinguished positive and negative donors as well as IgG, and averaging IgA or IgM responses to both antigens did not substantively improve the assay (Supplementary Figure 4).
Figure 1.
Combining immunoglobulin G (IgG) responses to different antigens improves sensitivity and specificity. IgG responses to the nucleocapsid (N) antigen (A) and spike receptor binding domain (RBD) antigen (B) among positive (n = 43) and negative (n = 184) control samples and corresponding receiver operating characteristic (ROC) curve used to determine the 100% sensitivity and specificity cutoffs for enzyme-linked immunosorbent assays using that antigen (dashed black lines on graph); individual data and mean ± standard deviation are shown. C, Relationship between responses to the N and RBD antigens among positive and negative control samples. Dashed lines represent the 100% specificity and sensitivity cutoffs derived from the mean of the IgG responses to the N and RBD antigens. D, Mean responses to the N and RBD antigens among positive and negative control samples and corresponding ROC curve.
Seroprevalence of SARS-CoV-2 Is Low in Australia
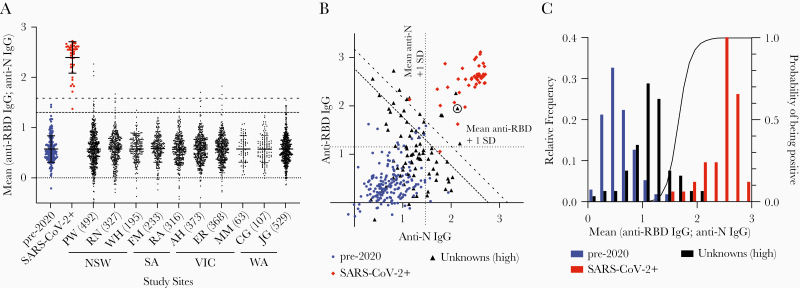
We next used our dual-antigen IgG ELISA to assess the seroprevalence of SARS-CoV-2 infection among 2991 individuals, comprising 1531 women and 1460 men with mean age 54 years (range, 15–95 years) providing blood samples at 10 hospital sites across 4 states in Australia in June–July 2020. These individuals were enrolled in a prospective cohort study to determine the prevalence of asymptomatic SARS-CoV-2 infection in individuals undergoing elective surgery in Australia; full demographic information is given in a separate manuscript describing this study (Coatsworth et al, unpublished data). In our initial screen, 41 of 2991 were above our lower cutoff of 1.302 (Figure 2A); correcting for the specificity of our assay, we calculated the seroprevalence to be 0.28% (0–1.15%). Performing a similar analysis but using the more stringent cutoff of 1.58 to minimize the number of false positives gave 7 of 2991 above the cutoff, leading to a slightly lower estimate of seroprevalence but with a smaller CI: 0.24% (.07%–.44%). To confirm our positive results, we retested the top 2.7% of samples from each site in parallel with our complete set of positive and negative control samples. In this analysis, 15 individuals remained above the 100% specificity cutoff (Figure 2B); however, plotting the RBD and N values showed that only 5 samples were strongly positive for both antigens, clustering with our positive controls. In contrast, the remaining 10 putative positives were close to the cutoff and were in many cases strongly positive for only 1 or other antigen; thus, we reasoned these might be false positives. Of note, 1 of 5 (20%) of our high-confidence positive samples was a contact of a known SARS-CoV-2–positive individual, compared to 14 of 2986 (0.47%) in the remainder of the cohort (P = .0248 by 2-tailed Fisher exact test; odds ratio = 53.1 [95% CI, 4.07–357]), giving us confidence that our assay was detecting true positive individuals.
Figure 2.
Estimation of seroprevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Australia. A, Normalized averaged responses to the spike receptor binding domain (RBD) and N antigens for each of the 2991 individuals in the study separated by study site and state. B, Anti-N and anti-RBD responses for the top 2.7% samples from each site (n = 80) compared to the positive and negative controls; the circled unknown sample was a contact of a SARS-CoV-2–positive individual. C, Frequency distribution of the negative, positive, and unknown samples (bars) plotted against the calculated probability of positivity in a Bayesian model based on the distributions of the positive and negative samples. Abbreviations: AH, Alfred Hospital; CG, Sir Charles Gairdner Hospital; ER, Epworth Richmond; FM, Flinders Medical Centre; IgG, immunoglobulin G; JG, St John of God Hospital; MM, Monash Medical Centre; NSW, New South Wales; PW, Check gamma symbol roman; RA, Royal Adelaide Hospital; RBD, receptor binding domain; RN, Royal North Shore Hospital; SA, South Australia; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; VIC, Victoria; WA, Western Australia; WH, Westmead Hospital.
To avoid biases associated with the use of cutoffs, we also calculated the probability of each of our 80 retested samples being positive based on the known distributions of the positive and negative results (Figure 2C). This analysis determined that the top 6 samples each had a >50% (range, 58%–99%) probability of being positive, whereas the remaining 9 potentially positive samples had individual probabilities of being positive of 10%–47%. By summing the probabilities of positivity among these samples, we can estimate that approximately 8 (0.27%) individuals in our cohort would be positive, which is similar to our original estimates of seroprevalence.
DISCUSSION
Here we report results from the first large-scale seroprevalence survey in Australia and estimate a seroprevalence of 0.28% (95% CI, 0–1.15%). This would equate to a point estimate of 71 400 infections nationwide; however, our cohort may not reflect the general population of Australia, with older individuals in particular being overrepresented. At the start of sample collection (2 June 2020), 7387 cases and 102 deaths had been reported in Australia, rising to 11 190 cases and 116 deaths by 17 July 2020 when sample collection finished, suggesting that testing was capturing 10%–15% of cases, similar to other jurisdictions with high testing rates [10]. A further caveat of our study is that the positive controls used for assay validation are skewed to hospitalized individuals, and thus we do not know with certainty the performance characteristics of the assays for asymptomatic cases that are known to have lower antibody levels [4, 11]. Moreover, a recent study has suggested that asymptomatic cases may not always seroconvert, though the assays used there had lower sensitivity than we report for our assay [5, 11]. Overall, however, these data suggest that the low case number seen in Australia was reflective of low community transmission, not inadequate testing. This is supported by the fact that the subsequent outbreak in Melbourne in July–August 2020 emerged from breaches of hotel quarantine of overseas travelers rather than undetected community transmission.
A variety of assays have been put forward for the assessment of seroprevalence of antibodies to SARS-CoV-2. Lateral flow devices were used in early studies, but these devices have insufficient sensitivity and specificity for use in low-prevalence settings [12]. However, more recent studies using ELISA and electrochemiluminescence-based assays with greater statistical rigor have overcome some of these issues and have given reliable estimates of seroprevalence in higher-transmission areas such as the United States [3, 13]. Of note, reports on important differences in seropositivity between different SARS-CoV-2 antigens and the effectiveness of measuring dual antigens in low-prevalence populations have emerged [14, 15]. By combining results from responses to antigens and using convergent statistical approaches, we show how an assay that can be established in ordinarily equipped laboratories can obtain credible estimates of SARS-CoV-2 seroprevalence, even in low-transmission settings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors acknowledge the contributions of all participants, study sites, local investigators, surgical and anesthesia teams, and study coordinators Sophie Wallace and Lucy Morris.
Financial support. This work was sponsored by the Australian Government Department of Health; and by Medibank Better Health Foundation.
Potential conflicts of interest. N. C. is an employee of the Australian Government Department of Health. N. C. contributed to study design, interpretation of results, and approval of the manuscript. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med 2020; 173:362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 3. Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May 12, 2020 [manuscript published online ahead of print 21 July 2020]. JAMA Intern Med 2020. doi:10.1001/jamainternmed.2020.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 5. GeurtsvanKessel CH, Okba NMA, Igloi Z, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun 2020; 11:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pollán M, Pérez-Gómez B, Pastor-Barriuso R, et al. ; ENE-COVID Study Group Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet 2020; 396:535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurth F, Roennefarth M, Thibeault C, et al. Studying the pathophysiology of coronavirus disease 2019: a protocol for the Berlin prospective COVID-19 patient cohort (Pa-COVID-19). Infection 2020; 48:619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samineni S, Parvataneni S, Kelly C, Gangur V, Karmaus W, Brooks K. Optimization, comparison, and application of colorimetric vs. chemiluminescence based indirect sandwich ELISA for measurement of human IL-23. J Immunoassay Immunochem 2006; 27:183–93. [DOI] [PubMed] [Google Scholar]
- 10. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland [manuscript published online ahead of print 1 September 2020]. N Engl J Med 2020. doi:10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183:158–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bond K, Nicholson S, Lim SM, et al. Evaluation of serological tests for SARS-CoV-2: implications for serology testing in a low-prevalence setting. J Infect Dis 2020; 222:1280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flannery DD, Gouma S, Dhudasia MB, et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci Immunol 2020; 5:eabd5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burbelo PD, Riedo FX, Morishima C, et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis 2020; 222: 206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dingens AS, Crawford KHD, Adler A, et al. Serological identification of SARS-CoV-2 infections among children visiting a hospital during the initial Seattle outbreak. Nat Commun 2020; 11:4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.