Abstract
Background
Understanding the longitudinal trajectory of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies is crucial for diagnosis of prior infection and predicting future immunity.
Methods
We conducted a longitudinal analysis of coronavirus disease 2019 convalescent patients, with neutralizing antibody assays and SARS-CoV-2 serological assay platforms using SARS-CoV-2 spike (S) or nucleocapsid (N) antigens.
Results
Sensitivities of serological assays in diagnosing prior SARS-CoV-2 infection changed with time. One widely used commercial platform that had an initial sensitivity of >95% declined to 71% at 81–100 days after diagnosis. The trajectories of median binding antibody titers measured over approximately 3–4 months were not dependent on the use of SARS-CoV-2 N or S proteins as antigen. The median neutralization titer decreased by approximately 45% per month. Each serological assay gave quantitative antibody titers that were correlated with SARS-CoV-2 neutralization titers, but S-based serological assay measurements better predicted neutralization potency. Correlation between S-binding and neutralization titers deteriorated with time, and decreases in neutralization titers were not predicted by changes in S-binding antibody titers.
Conclusions
Different SARS-CoV-2 serological assays are more or less well suited for surveillance versus prediction of serum neutralization potency. Extended follow-up should facilitate the establishment of appropriate serological correlates of protection against SARS-CoV-2 reinfection.
Keywords: SARS-CoV-2, COVID-19, Serology, Neutralizing antibodies
Analysis of coronavirus disease 2019 convalescent patients reveals that neutralizing antibody levels decline rapidly early after infection. Some clinical serological assay platforms give quantitative outputs that predict neutralizing antibody titer, but in some diagnostic sensitivity deteriorates with time after infection.
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent for coronavirus disease 2019 (COVID-19), has resulted in a devastating global pandemic. Diagnosis of SARS-CoV-2 infection is principally dependent on reverse-transcription polymerase chain reaction (RT-PCR) testing of nasal and throat swab samples, which is not ideally suited to mass population testing. Numbers of RT-PCR–diagnosed cases have therefore underestimated the prevalence of SARS-CoV-2 infection, and serological assays must be deployed to determine the true number of infections using a surveillance approach. Serological assays also have a critical role in screening volunteers for vaccine trials and convalescent plasma donation, as well as in predicting immunity. Although several commercial SARS-CoV-2 immunoassays are in common use, evaluation of their sensitivity has often used samples from hospitalized patients soon after infection. Knowledge of the long-term kinetics of antibody titers and the corresponding effectiveness of commercial assays is sparse [1–3].
Serological assays for SARS-CoV-2 use viral nucleocapsid (N) or spike protein (S) antigens. Because S binds target cells through its receptor binding domain (RBD), it is the target of neutralizing antibodies. Therefore, S-based assays may be preferable to N-based assays for assessing future reinfection risk [4]. This premise is based on the assumptions (1) that neutralizing antibodies constitute a major mechanism of protective immunity and (2) that S-based serological assays accurately predict neutralizing activity.
Outstanding questions about the use of SARS-CoV-2 serological assays include (1) how circulating antibody levels change with time after natural infection and (2) which serological assays best predict protective immunity. The prognostic value of antibody measurements with respect to reinfection has yet to be demonstrated, and it is important to understand postinfection serological findings in order to establish correlates of protection. We present a longitudinal study of mildly symptomatic, nonhospitalized COVID-19–positive patients during the first few months of convalescence. We compared the ability of 4 high-throughput automated serological assays to diagnose prior SARS-CoV-2 infection and predict serum neutralizing activity.
METHODS
Participants
Participants with prior RT-PCR–diagnosed COVID-19 were recruited. Recruits were surveyed to determine the date of the positive PCR test, the date of onset of symptoms, and whether their symptoms required hospitalization. Serum samples were taken at a baseline visit (approximately 3.5–8.5 weeks after the PCR test) and 2 (visit 2), 4 (visit 3) and 8 (visit 4) week later. In total, 97 participants who were not hospitalized during the course of their illness completed ≥3 visits. Their mean age was 44.2 years (range, 21–65 years), and 70 participants (72% of cohort) were female. The average time since the PCR test was 40.8 (range, 24–61) days at visit 1 (baseline), 55.1 (40–79) days at visit 2 (2 weeks after baseline), 69.8 (55–95) days at visit 3 (4 weeks after baseline), and 98.4 (85–110) days at visit 4 (8 weeks after baseline). Ethical approval was obtained for this study to be carried out through the NHS Lothian BioResource. All recruits gave written and informed consent for serial blood sample collection. Deidentified sample were shipped to The Rockefeller University whose institutional review board reviewed and approved the study.
High-Throughput Automated Serological Assays
Four commercial assays, which use either S or N protein antigens and are designed for high throughput in healthcare settings were used. All the assays generate a qualitative positive or negative result based on assay-dependent signal thresholds. The Abbott SARS-CoV-2 immunoglobulin (Ig) G assay detects anti-N IgG using a 2-step chemiluminescent microparticle immunoassay method with acridinium-labeled anti-human IgG. The DiaSorin SARS-CoV-2 IgG assay is also a 2-step chemiluminescent microparticle immunoassay targeting undisclosed epitopes in the SARS-CoV-2 S protein and uses an isoluminol-conjugated anti-human IgG. The Roche Anti-SARS-CoV total antibody assay is a 2-step bridging electrochemiluminesent immunoassay using ruthenium-labeled and biotin-conjugated N protein. The Siemens SARS-CoV-2 total antibody assay is a 1-step bridging CLIA method that detects antibodies against the RBD, using acridinium and biotinylated S1 RBD. Assays were performed on the Abbott Architect and DiaSorin Liason (NHS Lothian), Roche Elecsys (NHS Lanarkshire), and Siemens Atellica (NHS Tayside) platforms. Serum samples, collected and stored according to the manufacturer’s recommendations, were used in all cases.
SARS-CoV-2 Neutralization Assays
To measure neutralizing activity, serum samples, beginning with a 1:12.5 dilution, were serially diluted 5-fold in 96-well plates over 4 dilutions. Thereafter, approximately 5 × 103 infectious units of a human immunodeficiency virus type 1 (HIV-1)NLΔEnv-NanoLuc/SARS-CoV-2 pseudotype virus [5] were mixed with the serum dilutions at a 1:1 ratio and incubated for 1 hour at 37ºC. The mixture was then added to 293T/ACE2cl.22 target cells [5] plated at 1 × 104 cells per well in 100 µL of medium in 96-well plates the previous day. Thus, the final starting serum dilution was 1:50. Cells were cultured for 48 hours and harvested for NanoLuc luciferase assays, as described elsewhere [5].
RESULTS
The cohort consisted of participants who were not hospitalized during the course of their illness and were therefore relatively mildly symptomatic. Approximately 70% of people reported ≥1 of the 3 main World Health Organization–identified symptoms, namely, fever, cough, and anosmia. The most common of symptom was anosmia and the majority of participants reported the presence of 2 of these 3 symptoms (Table 1). Serum samples were collected from 97 participants at 3 visits, at approximately 4, 6, and 8 weeks after diagnosis (by RT-PCR). In addition, serum samples were collected from a subset of participants (28 of 97) at approximately 12 weeks after diagnosis (visit 4).
Table 1.
Proportions of Participants Displaying the 3 Main World Health Organization Symptoms
| Symptoms | Participants, No. (%) (N = 97)a |
|---|---|
| Fever | 65 (67) |
| Cough | 69 (71) |
| Anosmia | 74 (76) |
| No. of symptoms | |
| 0 | 1 (1) |
| 1 | 19 (16) |
| 2 | 42 (43) |
| All 3 | 35 (36) |
| Self-reported recovered (n = 90) | 44 (49) |
aN = 97 for all reported symptoms except “self-reported recovered” (as assessed by the response to the question ‘Do you feel you now fully recovered from COVID-19?’); only 90 individuals responded to this survey question.
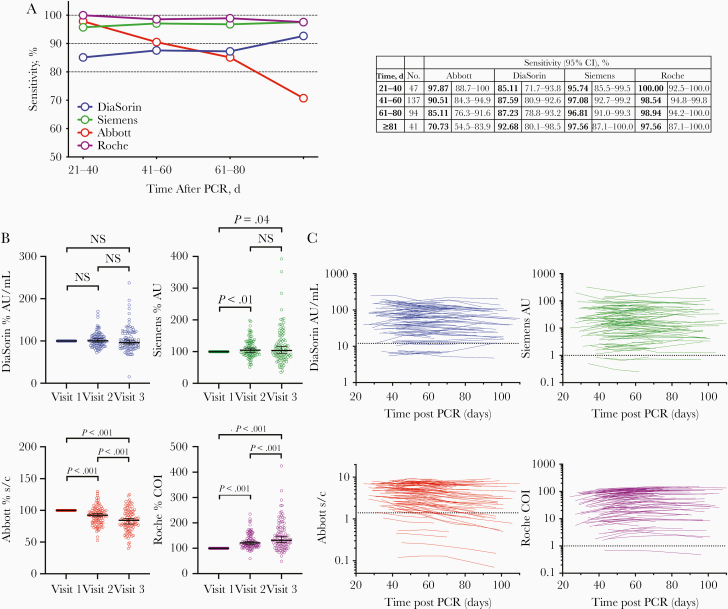
We compared the diagnostic sensitivity of 4 high-throughput SARS-CoV-2 serological assays that are routinely used in hospital settings. Each assay gives a qualitative positive or negative result, based on assay-specific thresholds, and sensitivities were calculated for each assay using these thresholds. Interassay and intra-assay analytical precision for each assay is detailed in Supplementary Table 1. To account for the differences in time after PCR diagnosis that participants made their first visit, sensitivities across a 20-day rolling time window were calculated.
The Abbott, Roche, and Siemens assays all had sensitivities of 95%–100% at 21–40 days after a PCR-positive test result, while the DiaSorin assay had a lower sensitivity of 85% (Figure 1A). However, the relative sensitivities of the assays changed with time. Specifically, the sensitivity of the Abbott assay declined to 85% in the 61–80-day window, and 71% at >81 days after diagnosis (Figure 1A). Conversely, the sensitivities of the other assays were maintained or increased over time (Figure 1A). In terms of intra-individual change, 14 of 91 participants with positive Abbott assay results at visit 1 had negative results by visit 3 or 4, whereas with the other assays none of the participants with a positive result at visit 1 had negative results at visit 3 or 4. For the DiaSorin assay, 2 participants with negative results at visit 1 had positive results at visit 3 (both had an equivocal result at visit 1 and showed a small increase above the assay threshold at visit 3). In the Siemens assay, 3 participants had consistently negative results, and in the Roche assay only a single participant had a negative result at each visit.
Figure 1.
Longitudinal analysis of serum samples from participants with coronavirus disease 2019. A, Sensitivity of the Abbott, DiaSorin, Siemens, and Roche serological assays, measured in samples collected at 4 time points after polymerase chain reaction (PCR) testing, with 95% confidence intervals (CIs). B, Relative antibody titers for the DiaSorin, Siemens, Abbott, and Roche assays at visits 1–3, normalized to visit 1 (serological assay values are reported in the assay-specific units of measurement and are expressed as % of visit 1). Horizontal lines indicates median values with 95% CIs. Statistical significance was assessed with the Wilcoxon test. C, Values for DiaSorin, Siemens, Abbott, and Roche serological assays for each participant plotted over time (each line represents 1 participant) (serological assay values are reported in the assay-specific units of measure). Assay thresholds are indicated by dotted horizontal lines. Abbreviations: AU, arbitrary units; COI, cutoff index; NS, not significant; s/c, sample to calibrator index.
The serological assays give a quantitative assessment of antibody titer as well as a threshold-based positive or negative result. We next analyzed changes in the quantitative results over time for each platform (Figure 1B and 1C). Mean antibody titers decreased in the Abbott assay at visits 2 and 3 compared with visit 1 (Figure 1B) but increased in the DiaSorin and, particularly, the Roche assays and remained approximately constant in the Siemens assay (Figure 1B). Notably, 79 of 97 participants (81%) showed a decrease in antibody titer on the Abbott platform, while 82 of 97 (85%) showed an increase on the Roche assay, despite the fact that both assays detect N-specific antibodies (Figure 1B and 1C). Negative or positive changes were approximately equally likely in the S-based assays; specifically, 57% and 47% of intraindividual changes were negative for the DiaSorin and Siemens assays. respectively (Figure 1B and 1C).
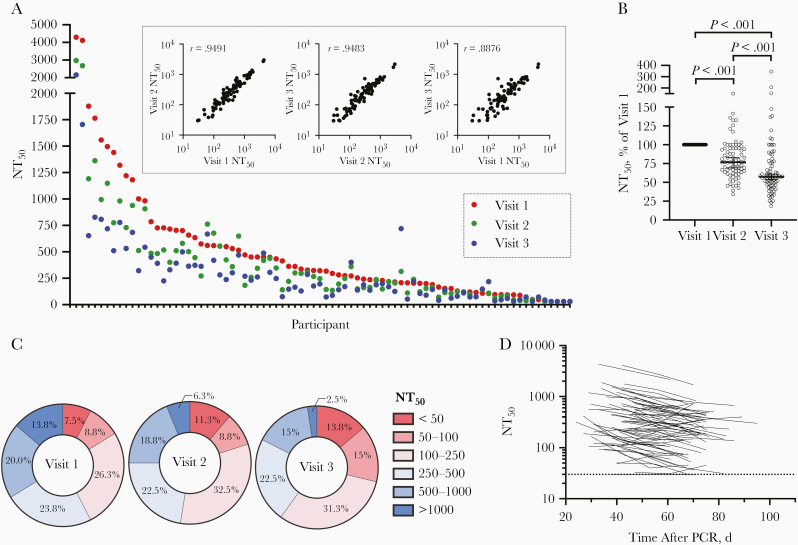
We measured neutralizing activity in serum samples from the first 3 visits for 80 of the 97 participants, using a SARS-CoV-2 pseudotyped virus neutralization assay that is amenable to high throughput and does not require a biosafety level 3 facility. This assay uses HIV-1–based virions carrying a NanoLuc luciferase reporter, pseudotyped with the SARS-CoV-2 spike protein. Neutralization titers obtained using these pseudotyped particles are correlated well with titers obtained using neutralization of authentic SARS-CoV-2 [5]. Moreover, this assay has been successfully applied for analysis of convalescent plasma samples and in a campaign to identify potent human monoclonal antibodies [6, 7]. Consistent with our analyses of other cohorts [6, 7], a broad range of neutralizing titers was evident in serum samples collected from 80 participants at 3 time points (Figure 2A). In samples collected at visit 1, the neutralizing activity, as determined by half-maximal neutralizing titer (NT50), ranged from <30 to 4300, with a geometric mean of 234 (arithmetic mean, 411) (Figure 2A, red symbols). Consistent with other cohorts [6, 7] 34 of 80 (42%) had NT50 values <250, while only 11 of 80 participants (14%) had NT50 values >1000.
Figure 2.
Neutralization activity in serum samples from participants with coronavirus disease 2019. A, Half-maximal neutralization titer (NT50) for each individual participant, measured in serum samples collected at 3 visits. Inset shows correlation of NT50 values for samples collected at each visit, with Spearman r values (P < .001). B, Relative NT50 values in serum samples obtained at visits 1–3, normalized to visit 1. Horizontal line represents median with 95% confidence interval. Statistical significance was assessed with the Wilcoxon test. C, Frequency of serum samples with NT50 values falling to various quantitative categories at each visit. D, NT50 values for each participant plotted over time (each line represents 1 participant). The limit of detection is indicated by a dotted horizontal line. Abbreviation: PCR, polymerase chain reaction.
NT50 values measured at each time point for individual participants were correlated with each other, although they diverged in NT50 values over time (Figure 2A, inset). Notably, neutralizing activity decreased at each time point for the majority of participants (Figure 2A, blue and green symbols). Overall, the decrease in median NT50 was approximately 25% per 2-week sampling interval, resulting in an approximately 45% reduction in NT50 over the 4 weeks between visit 1 and visit 3 (Figure 2B). As a result, the distribution of NT50 values in the cohort differed between visits (Figure 2C). The relative declines in NT50 between visits 1 and 2 versus between visits 2 and 3 did not differ significantly, and the majority of participants exhibited similar relative decreases in neutralizing activity over time, regardless of their initial NT50 values or the days after PCR at visit 1, suggesting exponential decay (Figure 2D).
NT50 values at each sampling time point were poorly correlated with age (Supplementary Figure 1A), and no correlation was observed between age and NT50 decay with time. As has been previously reported, there was a trend toward lower NT50 values in female than in male patients [6, 7], but there was no difference between sexes in NT50 decay with time (Supplementary Figure 1B). Individual clinical parameters, such as gastrointestinal symptoms, fever, or recovery time, did not predict NT50, serological values or decay parameters for any antibody measurement.
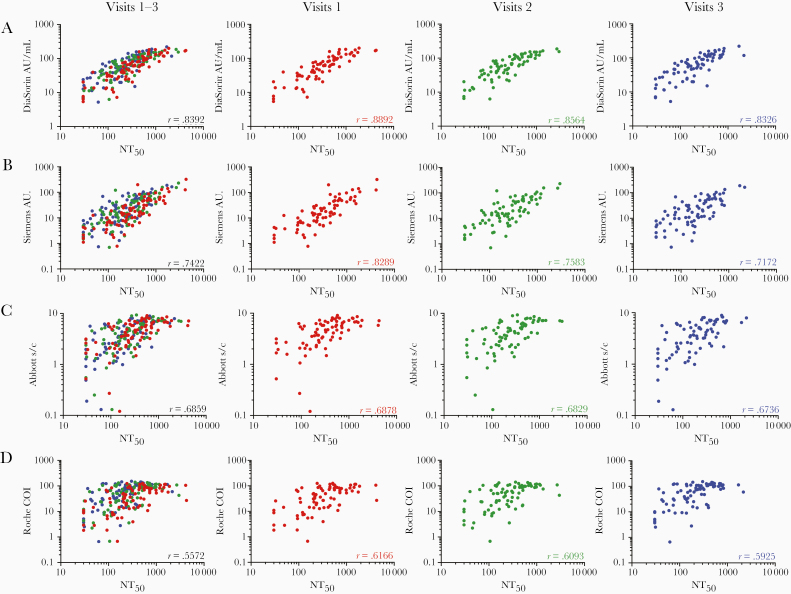
Next, we compared neutralizing activity in serum with quantitative results obtained from the serological assays. Analysis of combined results from the 3 visits by 80 participants revealed a significant correlation between any combination of 2 serological assays (Supplementary Figure 2). However, stronger correlations were observed between the 2 S-based assays, Siemens and DiaSorin (r = 0.92; P < .001) and between the 2 N-based assays Abbott and Roche (r = 0.81; P < .001).The S-based assays were correlated less well, but significantly (P < .001), with the N-based assays (Supplementary Figure 2).
All of the serological assays gave quantitative values that were correlated with NT50 values, but, as expected, the S-based assay measurements were correlated more closely with NT50 values (Figure 3A–3D). The S1/S2-based DiaSorin assay was the best predictor of NT50 (r = 0.84; P < .001) (Figure 3A), followed by the RBD-based Siemens assay (r = 0.74; P < .001) (Figure 3B), the N-based Abbott assay (r = 0.69; P < .001) (Figure 3C) and, finally, the Roche assay (r = 0.56; P = .001) (Figure 3D).
Figure 3.
Correlation of serological results with neutralization titers. A–D, Serological assay values for the DiaSorin (A), Siemens (B), Abbott (C) and Roche (D) assays (serological assay values are reported in the assay-specific units of measure) versus half-maximal neutralizing titer (NT50) values. Samples collected at each visit are indicated by color and are plotted individually as well as on a composite graph. Spearman r values for all visits (black) and for individual visits are indicated (P < .001). Abbreviations: AU, arbitrary units; COI, cutoff index; s/c, sample to calibrator index.
The correlation between NT50 values and individual serological assay results was best at the first visit and deteriorated to some extent thereafter (Figure 3A–3D; see color-coded r values in individual graphs; P < .001 for all correlations), The decrease in the strength of correlation might be attributable in part to the fact that later sampling time points have more samples with lower NT50 values, which may reduce measurement precision. The magnitude of the deterioration in predictive value differed between serological assays, with S-based assays exhibiting larger decreases in correlation coefficients (DiaSorin and Siemens assays, r = 0.89 and 0.83 at visit 1, respectively, and r = 0.83 and 0.71 at visit 3; Figure 3A–3D). Despite the increasing disparity over time, the DiaSorin assay was clearly superior for predicting NT50 at all visits (Figure 3A–3D).
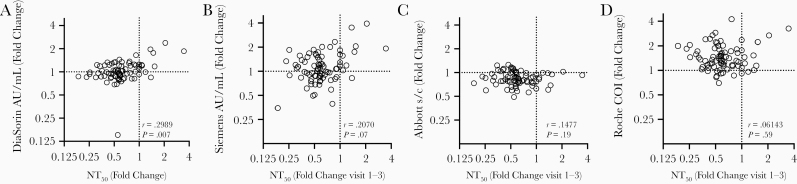
Interestingly, only minimal correlation was revealed by comparing the extent of change in neutralization activity over the 4-week observation interval with the concomitant change in values obtained using serological assays (Figure 4A–4D and Supplementary Figure 3). Notably, in most participants, the decline in serum neutralizing activity was clearly greater than the decline in antibody titer measured using any serological assay (Figure 4A–4D and Supplementary Figure 3). Even for the DiaSorin assay, which gave the best prediction of neutralizing activity at each time point (Figure 4A), declines in neutralizing activity were not well predicted by declines in DiaSorin assay measurements (Figure 4A and Supplementary Figure 3). While both the Abbott assay and the NT50 measurements exhibited declining antibody titers with time, the magnitudes of these declines did not correlate with each other (Figure 4D and Supplementary Figure 3).
Figure 4.
Lack of correlation of changes in serological results with changes in neutralization titers. A–D, Fold change (from visit 1 to visit 3) in serological assay values (reported in the assay-specific units of measure) for the DiaSorin (A), Siemens (B), Abbott (C), and Roche (D) assay versus fold change in half-maximal neutralizing titer (NT50) values. Spearman r and P values are shown. Abbreviations: AU, arbitrary units; COI, cutoff index; s/c, sample to calibrator index.
DISCUSSION
Serological assays for infectious agents have 2 major and distinct uses, namely, (1) to diagnose chronic infections (eg, HIV-1) and (2) to determine past infection or immunization status (eg, for measles or varicella-zoster virus) which may be able to predict immunity from future infection. The use of SARS-CoV-2 serological assays requires understanding of how these assays perform in populations over time. During the current SARS-CoV-2 pandemic it has become clear that the magnitude of serological immune responses is highly variable [6, 7]. Nevertheless, the vast majority of individuals with a PCR-confirmed SARS-CoV-2 infection generate antibodies at a sufficient level for diagnosis of recent infection [8]. A number of high-throughput commercial assays have been deployed for SARS-CoV-2 antibody testing and evaluated mostly using hospitalized participants [9, 10]. Nonhospitalized patients with mild disease typically have lower levels of antibodies than hospitalized patients with severe illness [11–15], Differences in antibody titers between individuals may be driven in part by differences in antigen exposure. However, several factors, including variable viral load trajectories, variable time of diagnosis and sampling relative to infection, variable sampling efficiency using swab samples, and variable relationship between nasal viral load and systemic antigen exposure would make relationships between viral load and antibody responses difficult to establish.
Using our cohort of nonhospitalized participants with mild disease, all 4 assays evaluated herein had sensitivities at visit 1 (an average of 40.8 days after PCR testing) that were comparable to those reported using hospitalized patients [16]. This would therefore make all 4 assays suitable for the detection of COVID-19 antibodies shortly after infection as a confirmatory test for diagnostic purposes, when used in conjunction with RT-PCR assays and clinical history. However, differences in assay diagnostic sensitivity become apparent at later time points. Specifically, the sensitivity of the widely used Abbott assay declined with time, to approximately 70% at >81 days after PCR. Consequently, this assay is not appropriate for seroprevalence studies, for identification of SARS-CoV-2 naive vaccine trial participants, or for investigation of individuals presenting with long-term chronic symptoms. Altering the positive/negative threshold, may mitigate this issue [17] but would not ultimately alter the downward trend in assay signal over time. Notably our study is one of the few that would capture this information, as most other studies have examined seroconversion at early time points [14, 18–20].
Reasons for the differences in assay performance over time are unclear but cannot be attributed solely to the choice of antigen. Although other studies have reported an inherent difference in the dynamics of S versus N antibodies [21] our findings do not support this contention, during the first 100 days or so of convalescence. Both Abbott and Roche assays use the N proteins as an antigen, but Abbott assay titers decline while those in the Roche assay increase during this time period. One possible explanation for this difference is the use of an antigen bridging approach in the Roche assay, where declines in the total amount of antibody might be compensated by increases in affinity or avidity as antibodies mature through somatic hypermutation. Alternatively, it is possible that the range of N epitopes recognized by serum samples might change with time. Whatever the explanation, it is clear that that the trajectories of antibody titers measured using assays based on recognition of the same or related antigens can differ [22–25]. Overall, given their superior sensitivity at each of the time points investigated thus far, the Siemens and Roche assays appear most appropriate for diagnosis of recent SARS-CoV-2 infection and would report a higher population prevalence than Abbott or DiaSorin assays in the 1–4-month postinfection period.
Serological assays that serve a diagnostic function are likely optimized for sensitivity or specificity rather than for dynamic range, Thus, a quantitative signal in a high-throughput serological assay might not correspond linearly with antibody titer. Nevertheless, convalescent plasma is being used therapeutically, with unit selection based on titers measured using serological assays [26].
In addition, because vaccine- or infection-elicited neutralizing antibodies will likely confer protection against SARS-CoV-2 infection, it was of interest to evaluate the potential ability of serological assays to predict neutralizing antibody titer. While the Roche assay exhibited the best diagnostic sensitivity during this time period, it had the lowest ability to predict neutralizing antibody titer. This finding might be expected, because neutralizing antibodies are directed to the S protein, whereas N-specific antibodies are not expected to be neutralizing. The DiaSorin assay best predicted neutralizing titers and marginally outperformed the Siemens assay in this regard, perhaps because the dominant neutralizing and/or S-binding activity in some serum samples is provided by antibodies that recognize epitopes outside the RBD [27, 28]. It is important to recognize, however, that many S-binding antibodies are not neutralizing—measurements of S-binding antibodies remain correlates, and not direct measures, of neutralizing antibodies [7].
Differences in the mechanism of detection likely affects the relationship between antibody titers and assay signal output. Changes in the abundance of different antibody classes over time could also differentially affect readout in serological assays (eg, IgM is polyvalent and short-lived and might give greater signals in bridging assays). Nevertheless, changes in antibody class composition do not easily explain the trends that we observe; for instance, IgG is a longer-lived antibody response and the Abbott assay is IgG specific, but Abbot assay titers were the least stable of those evaluated. Neutralization is not specific for any antibody isotype or subclass, and neutralizing titers will reflect the combined activity of all neutralizing antibodies in a sample, Again the comparative stability of the IgG response and the dominant role of IgG in neutralizing plasma [29] do not comport with the idea that changing antibody class abundance could explain declining neutralizing titers.
Recent reports have similarly indicated that SARS-CoV-2 neutralizing antibody titers decline with time [23, 24], while another study reported that neutralization titers remained stable for ≥3 months after infection [30]. However, in the latter case, neutralization titers were inferred based on a serological enzyme-linked immunosorbent assay measurement that was calibrated using a neutralization assay performed on a small subset of samples. As shown herein, neutralizing antibody levels indeed decline in most patients, even when S-binding or RBD-binding antibody titers measured in serological assays are maintained. Thus, the trajectory of neutralizing antibody levels cannot necessarily be accurately deduced from serological measurements.
Key future questions include to what degree the titers of neutralizing antibodies, or antibodies that simply bind to S or N are correlated with protection from reinfection or severe disease. Many adults possess circulating antibodies to seasonal human coronaviruses (CoVs) OC43 and 229E [31], and children seroconvert to NL63 and 229E before about 3.5 years of age [32]. These baseline levels increased on infection, returning to baseline within 1 year. High levels of circulating neutralizing antibody correlated with protection from reinfection [33, 34]. However, human CoV reinfections occur [34, 35], often within 12 months [35], but with more mild illness.
Thus, these data suggest that seasonal human CoV immunity wanes with time. For SARS-CoV and Middle East respiratory syndrome (MERS) CoV, antibody responses also decline in the majority of infected individuals [36] Indeed, analyses of the decay of SARS-CoV antibodies indicate kinetics consistent with those reported herein [37]. Moreover, recent studies have documented SARS-CoV-2 reinfection [38–40], in one case in the context of waning neutralizing antibodies [41]. Overall, it seems possible that SARS-CoV-2 reinfection might be common. Importantly, however, the magnitude and stability of antibody responses to SARS-CoV-2 vaccines might be quite different from those after natural infection. Specifically, if the great variability in postinfection SARS-CoV-2 antibody levels are indeed a consequence of variable antigen exposure, then differences might be mitigated in the context of vaccination.
If neutralizing antibodies constitute a major protective mechanism against SARS-CoV-2 infection, then serological assays that use S-based antigens and best predict neutralizing titer are the most appropriate for prognostication of immunity. Conversely, if other immune mechanisms, such as long-lived memory T cells play a dominant role in protection [42–45], then the optimal choice of antigen for serological assays might differ. Because detailed analyses of T-cell responses are not currently feasible in a high-throughput clinical setting, future work should examine the frequency of reinfection and clinical outcomes with detailed longitudinal analyses of serum antibodies to both N and S antigens to determine the prognostic value of such measurements.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We acknowledge the support and hard work of the NHS Lothian BioResouce team, including, in particular, David Harrison for his support with initiating this study, Frances Rae and Craig Marshall for their advice, and Linda MacLeod for her advice and hard work. NHS Lothian laboratories and outpatient teams have also worked extremely hard to enable the provision of this research study. In particular, we would like to thank Joan Donnelly for her support and Susan Taylor for her dedication and hard work.
Author contributions. H. W., T. H., S. J., and P. D. B. conceived and designed the study. H. W., B. B., M. S., E. S., C. R., J. M., S. C., E. F., N. G., G. H., K. T., and S. J. acquired and analyzed data using the serological assay platforms. F. M. and J. C. C. L. performed the neutralization assays. F. M. and T. H. did additional data analysis. F. M., H. W., B. B., S. J., and P. D. B. wrote the first draft of the manuscript. F. M., H. W., B. B., T. H., S. J., and P. D. B. critically reviewed and revised the draft. All authors approved the final version of the manuscript for submission. H. W., B. B., and M. S. contributed equally. P. D. B. attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Disclaimer. The funders played no role in the design, analysis, or reporting of this research.
Financial support. This work was supported by the UK National Health Service and the National Institutes of Allergy and infectious Diseases (grants R37AI640003 to P. D. B. and R01AI078788 to T. H.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Andersson M, Low N, French N, et al. Rapid roll out of SARS-CoV-2 antibody testing—a concern. BMJ 2020; 369:m2420. [DOI] [PubMed] [Google Scholar]
- 2. Özçürümez MK, Ambrosch A, Frey O, et al. SARS-CoV-2 antibody testing—questions to be asked. J Allergy Clin Immunol 2020; 146:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. GeurtsvanKessel CH, Okba NMA, Igloi Z, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun 2020; 11:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hess AS, Shardell M, Johnson JK, et al. Methods and recommendations for evaluating and reporting a new diagnostic test. Eur J Clin Microbiol Infect Dis 2012; 31:2111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 2020; 217:e20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luchsinger LL, Ransegnola B, Jin D, et al. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID19 patients. J Clin Microbiol 10.1128/jcm.02005-20. Published 11 September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020; 584:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wajnberg A, Mansour M, Leven E, et al. Humoral immune response and prolonged PCR positivity in a cohort of 1343 SARS-CoV 2 patients in the New York City region. medRxiv [Preprint: not peer reviewed]. 5 May 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.04.30.20085613v1. [Google Scholar]
- 9. The National SARS-CoV-2 Serology Assay Evaluation Group. Head-to-head benchmark evaluation of the sensitivity and specificity of five immunoassays for SARS-CoV-2 serology on >1500 samples. figshare. Collection. 2020. 10.6084/M9.FIGSHARE.C.5046032.V1. [DOI]
- 10. Jääskeläinen A, Kuivanen S, Kekäläinen E, et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 2020; 129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cervia C, Nilsson J, Zurbuchen Y, et al. Systemic and mucosal antibody secretion specific to SARS-CoV-2 during mild versus severe COVID-19. bioRxiv [Preprint: not peer reviewed]. 23 May 2020. Available at: https://www.biorxiv.org/content/10.1101/2020.05.21.108308v1. [Google Scholar]
- 12. Dogan M, Kozhaya L, Placek L, et al. Novel SARS-CoV-2 specific antibody and neutralization assays reveal wide range of humoral immune response during COVID-19. medRxiv [Preprint: not peer reviewed]. 8 July 2020. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7359542/. [Google Scholar]
- 13. Klein S, Pekosz A, Park HS, et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest 2020; 130:6141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 15. Rijkers G, Murk JL, Wintermans B, et al. Differences in antibody kinetics and functionality between severe and mild SARS-CoV-2 infections. medRxiv [Preprint: not peer reviewed]. 12 June 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.06.09.20122036v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perkmann T, Perkmann-Nagele N, Breyer MK, et al. Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. Clin Chem doi:10.1093/clinchem/hvaa198. Published 10 August 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 2020; 58:e00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lou B, Li TD, Zheng SF, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Resp J doi:10.1183/13993003.00763-2020. Published 19 May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pickering S, Betancor G, Pedro Galao R, et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog 2020; 16:e1008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staines HM, Kirwan DE, Clark DJ, et al. Dynamics of IgG seroconversion and pathophysiology of COVID-19 infections. medRxiv [Preprint: not peer reviewed]. 9 June 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.06.07.20124636v2. [Google Scholar]
- 21. Grandjean L, Saso A, Ortiz A, et al. Humoral response dynamics following infection with SARS-CoV-2. medRxiv [Preprint: not peer reviewed]. 22 July 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.07.16.20155663v2. [Google Scholar]
- 22. Perreault J, Tremblay T, Fournier MJ, et al. Longitudinal analysis of the humoral response to SARS-CoV-2 spike RBD in convalescent plasma donors. bioRxiv [Preprint: not peer reviewed]. 17 July 2020. Available at: https://www.biorxiv.org/content/10.1101/2020.07.16.206847v1. [Google Scholar]
- 23. Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild COVID-19. N Engl J Med 2020; 383:1085–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seow J, Graham C, Merrick B, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. Nat Microbiol Dis doi:10.1038/s41564-020-00813-8. Published 26 October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juno JA, Tan HX, Lee WS, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med 2020; 26:1428–34. [DOI] [PubMed] [Google Scholar]
- 26. US Food and Drug Administration. COVID-19 convalescent plasma emergency use authorization 2020. https://www.fda.gov/media/141477/download. Accessed 11 November 2020.
- 27. Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv [Preprint: not peer reviewed]. 22 July 2020. Available at: https://www.biorxiv.org/content/10.1101/2020.07.21.214759v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barnes CO, West AP, Huey-Tubman KE, et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 2020; 182:828–42.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS-CoV-2 neutralization by secretory IgA in vitro. bioRxiv [Preprint: not peer reviewed]. 9 September 2020. Available at: https://www.biorxiv.org/content/10.1101/2020.09.09.288555v1. [Google Scholar]
- 30. Wajnberg A, Amanat F, Firpo A, et al. SARS-CoV-2 infection induces robust, neutralizing antibody responses that are stable for at least three months. medRxiv [Preprint: not peer reviewed]. 22 July 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.07.14.20151126v1. [Google Scholar]
- 31. Macnaughton MR Occurrence and frequency of coronavirus infections in humans as determined by enzyme-linked immunosorbent assay. Infect Immun 1982; 38:419–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dijkman R, Jebbink MF, Gaunt E, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol 2012; 53:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Callow KA Effect of specific humoral immunity and some non-specific factors on resistance of volunteers to respiratory coronavirus infection. J Hygiene 1985; 95:173–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Callow KA, Parry HF, Sergeant M, Tyrrell DAJ. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990; 105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med 2020. doi:10.1038/s41591-020-1083-1. Published 14 September 2020. [DOI] [PubMed] [Google Scholar]
- 36. Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gene Virol 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mo H, Zeng G, Ren X, et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 2006; 11:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 10.1093/cid/ciaa1275. Published 25 August 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Elslande J, Vermeersch P, Vandervoort K, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis 10.1093/cid/ciaa1330. Published 5 September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larson D, Brodniak SL, Voegtly LJ, et al. A case of early reinfection with SARS-CoV-2. Clin Infect Dis 10.1093/cid/ciaa1436. Published 19 September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. To KK, Hung IF, Chan KH, et al. Serum antibody profile of a patient with COVID-19 reinfection. Clin Infect Dis 2020. 10.1093/cid/ciaa1368. Published 23 September 2020. [DOI] [Google Scholar]
- 42. Gallais F, Velay A, Wendling MJ, et al. Intrafamilial exposure to SARS-CoV-2 induces cellular immune response without seroconversion. medRxiv [Preprint: not peer reviewed]. 22 June 2020. Available at: https://www.medrxiv.org/content/10.1101/2020.06.21.20132449v1. [Google Scholar]
- 43. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181:1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020; 584: 457–62. [DOI] [PubMed] [Google Scholar]
- 45. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183:158–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.