Abstract
Novel coronaviruses, including SARS-CoV-2, SARS, and MERS, often originate from recombination events. The mechanism of recombination in RNA viruses is template switching. Coronavirus transcription also involves template switching at specific regions, called transcriptional regulatory sequences (TRS). It is hypothesized but not yet verified that TRS sites are prone to recombination events. Here, we developed a tool called SuPER to systematically identify TRS in coronavirus genomes and then investigated whether recombination is more common at TRS. We ran SuPER on 506 coronavirus genomes and identified 465 TRS-L and 3,509 TRS-B. We found that the TRS-L core sequence (CS) and the secondary structure of the leader sequence are generally conserved within coronavirus genera but different between genera. By examining the location of recombination breakpoints with respect to TRS-B CS, we observed that recombination hotspots are more frequently colocated with TRS-B sites than expected.
Keywords: Key words: coronavirus, transcriptional regulatory sequences (TRS), recombination
Introduction
In the past two decades, at least three novel coronaviruses have spilled over from animals to humans, SARS, MERS, and SARS-CoV-2 (Cui et al. 2019; Guarner 2020). Coronaviruses are positive-sense, single-stranded RNA viruses with large genomes and are grouped into four genera: Alphacoronaviruses, Betacoronaviruses, Gammacoronaviruses, and Deltacoronaviruses (Cui et al. 2019). Recombination between coronaviruses plays an important role in coronavirus evolution and can alter host range, pathogenicity, and transmission patterns (Lai et al. 1985; Keck et al. 1988; Wang et al. 1993; Zhang et al. 2005; Lau et al. 2010; Tian et al. 2014; Xiao et al. 2016; Wang et al. 2017; Bentley and Evans 2018; Graham et al. 2018). Inter-coronavirus recombination requires two different but related coronaviruses to coinfect a cell (Graham and Baric 2010; Simon-Loriere and Holmes 2011; Graham et al. 2018). Recombination in coronaviruses occurs during genome replication when the RNA-dependent RNA polymerase (RdRp) replicating the genome dissociates from one viral genome currently serving as the template and reassociates with a different viral genome while retaining the nascent RNA in a process called template switching (Sawicki and Sawicki 1998; Simon-Loriere and Holmes 2011). Therefore, template switching generates a recombinant RNA originating from the genomes of two coronaviruses (Simon-Loriere and Holmes 2011; Bentley and Evans 2018).
Coronavirus transcription also involves template switching (Sawicki and Sawicki 1998). After a coronavirus infects a cell, it replicates its positive-strand RNA genome into a negative-strand genome with RdRp (Sawicki and Sawicki 1998). The negative-strand genome subsequently serves as a template for the production of positive-strand genomes and subgenomic messenger RNAs (sgmRNAs), a set of 3′ coterminal RNAs encoding structural genes (Sawicki and Sawicki 1998). The sgmRNAs share a common 5′ sequence, called a leader sequence, which is located at the beginning of the coronavirus genome (Zhang et al. 1994). The leader sequence is added to the 5′ end of all sgmRNAs through RdRp template switching (Sawicki and Sawicki 1998, 2005). Template switching occurs as RdRp is transcribing the negative strand and encounters transcriptional regulatory sequences (TRS) preceding each gene called the body TRS (TRS-B) (Sola et al. 2015). The TRS-B site has a 7–8-nt conserved core sequence (CS) which is thought to enhance the likelihood of RdRp template switching by hybridizing with an identical or nearly identical CS in the leader TRS (TRS-L) (Zúñiga et al. 2004; Sola et al. 2015). The occurrence of this programmed template switching leads to the generation of sgmRNAs with identical 5′ and 3′ sequences, but alternative central regions corresponding to the beginning of each structural ORF (Sawicki and Sawicki 1998, 2005; Sawicki et al. 2007; Wu and Brian 2007; Sola et al. 2015).
Because TRS-B is a signal for RdRp to switch templates, it is reasonable to hypothesize that recombination events are more likely to occur at or near TRS-B sites (Graham et al. 2018). Once RdRp has dissociated from the original template after encountering TRS-B, it could reassociate with a different genome, leading to recombination. Identical or nearly identical TRS sites between different coronaviruses could hybridize promoting recombination at TRS-B sites. Therefore, in this study, we investigate the relationship between genome recombination in coronaviruses and template switching at TRS-B sites. TRS-B sites are not annotated in every coronavirus genome because there is not a systematic tool to identify them, nor has there been a project focused on TRS-B annotation. Therefore, there has not been a systematic study of whether recombination events are more common around TRS-B sites. Thus, we developed a tool called SuPER (Subgenomic mRNA Position Exploration with RNA-sequencing) to systematically identify TRS sites within coronavirus genomes to better understand the relationship between coronavirus recombination and template switching. After systematically identifying TRS sites and recombination events in coronaviruses, we found that 8 of 91 (8.7%) of TRS-B sites are within breakpoint hotspots and that recombination hotspots are more frequently colocated with TRS-B sites than expected.
Results
Systematic Identification of TRS in Coronavirus Genomes
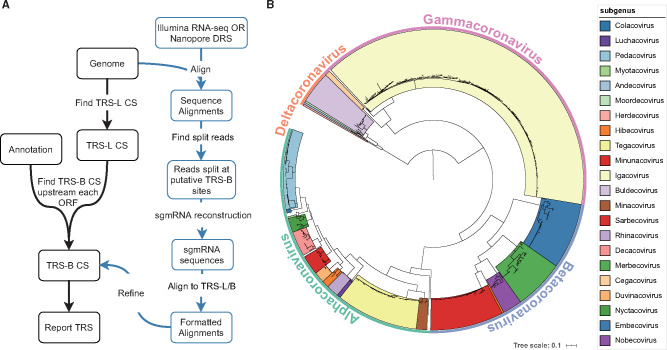
To examine how template switching at TRS-B sites contributes to genome recombination in coronaviruses, we needed to systematically identify TRS-B sites. Therefore, we developed the tool SuPER to identify TRS-B sites in coronavirus genomes (fig. 1A). SuPER first uses a covariance model derived from Rfam (Kalvari, Argasinska, et al. 2018; Kalvari, Nawrocki, et al. 2018) to identify TRS-L via profile-based sequence and structure scoring. Then, SuPER identifies TRS-B sites either by identifying template switching junctions with RNA-seq (RNA-sequencing) or in the absence of RNA-seq by identifying sequences preceding genes that are similar to the TRS-L CS as putative TRS-B CS.
Fig. 1.
(A) SuPER workflow. The main pipeline is shown with black text boxes and arrows. Analysis procedures specific to sequencing data are shown with blue text and arrows. (B) The phylogenetic tree built from RdRp sequences from the 506 representative coronavirus genomes used in the study.
To validate SuPER, we ran it on the SARS and SARS-CoV-2 genomes. The TRS-L/B of SARS and SARS-CoV-2 can be identified or inferred from previous research (Rota et al. 2003; Kim et al. 2020). We found that when using SuPER with RNA-seq data, we can correctly identify 94% (16/17) known TRS-B sites from these two genomes with zero false positives (supplementary table S1, Supplementary Material online). The only TRS-B CS not being captured by SuPER is associated with ORF7b in SARS-CoV-2, which was either reported with low abundance (Kim et al. 2020) or even undetectable (Taiaroa et al. 2020) in previous studies. When running SuPER without RNA-seq data, if only high confidence TRS-B sites are reported, SuPER can also predict 16 (out of 17) known TRS-B CS accurately and precisely. SuPER also reports extra TRS-B but labels them as “not recommended” due to the fact that their hamming distance to TRS-L CS is more than 1 bp, with hamming distance indicating the number of positions between two strings of equal length at which the corresponding symbols are different. The reason we still report low confidence TRS-B is due to the concern of missing potential noncanonical TRS-B CS.
To systematically identify TRS-B in coronaviruses, we used a set of 506 nonredundant representative genomes from the family Coronaviridae (supplementary table S2, Supplementary Material online; see Materials and Methods). Not all coronavirus genomes were assigned to a subgenus in the NCBI Virus database. To place the unassigned genomes into genera and subgenera, we built a phylogenetic tree based on the RdRp protein sequence (Wolf et al. 2018) from all representative genomes (fig. 1B). In total, we assigned 498 genomes into 23 subgenera (supplementary table S2, Supplementary Material online). We identified putative TRS-L and TRS-B CS in the 506 coronavirus genomes with SuPER. Of those, SuPER was run on 11 genomes with RNA-seq data and the remaining genomes without RNA-seq data (supplementary tables S3 and S4, Supplementary Material online). In total, SuPER identified 465 TRS-L and 3509 TRS-B.
TRS Characterization in Coronavirus Genomes
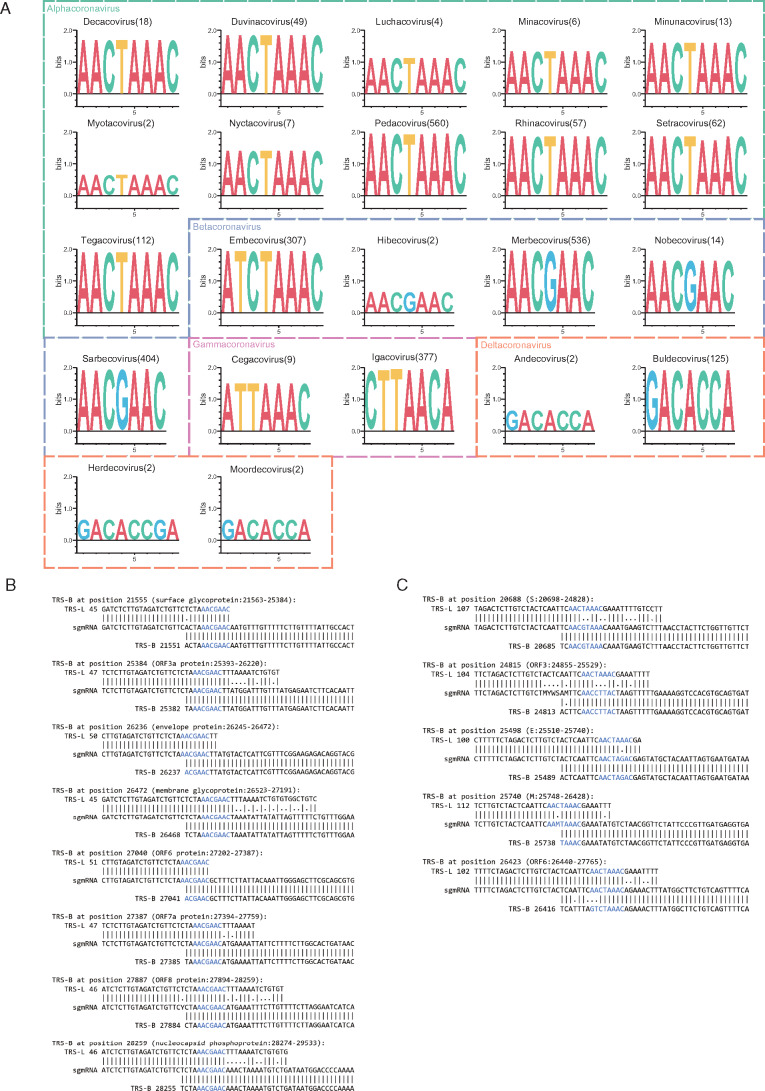
Using the results from SuPER, we examined the conservation of TRS-L CS (fig. 2A) and its position in the secondary structure in all coronavirus subgenera (supplementary fig. S1, Supplementary Material online). In general, we found that the TRS-L CS is conserved within genera but differs between genera, with the exception of Embecoviruses where the TRS-L CS is similar to Alphacoronaviruses. The secondary structures of the leader sequences were visualized by VARNA (Darty et al. 2009). We observed that the secondary structure of the leader sequence is relatively conserved within subgenera, but different between subgenera.
Fig. 2.
(A) Sequence logo of the TRS-L CS from all coronavirus subgenera. The number following the subgenus name indicates the number of sequences used to generate the sequence logo. X-axis shows the relative positions of the sequence letters; y-axis showing the total height of the letters depicts the information content of the position (in bits). (B, C) Alignments of TRS-L, sgmRNA (subgenomic messenger RNA), and TRS-B to illustrate the template switching pattern in SARS-CoV-2 (B) and Porcine epidemic diarrhea virus (C). CSs in TRS-B, TRS-L, and sgmRNA are colored blue.
We examined the TRS-B CS detected from coronavirus genomes where RNA-seq was available using SuPER. RNA-seq data can help to identify the template switching patterns and accurately detect the TRS-B CS. We found that TRS-B CS were either identical to TRS-L CS from the same genome, or differed by only one base pair in most genomes including SARS-CoV-2, SARS, MERS, SADS-CoV, and HCoV-HKU1 (fig. 2B). However, in some cases, there is limited similarity between TRS-L and TRS-B, making it difficult to identify TRS-B without RNA-seq data. For example, in Porcine Epidemic Diarrhea Virus, the TRS-B CS can differ by more than two base pairs from the TRS-L CS (fig. 2C). We also observed similarity in a few base pairs upstream and downstream of the CS, which could be critical for mediating base pairing during template switching in cases where the TRS-L and TRS-B CS differ (fig. 2B) (Sola et al. 2005).
Not all annotated ORFs preceded by TRS-B sites are supported by RNA-seq data. This could be due to the lack of enough coverage to detect the template switching events occurring at that position or there is little or no template switching. For example, we did not find RNA-seq data supporting the TRS-B site preceding ORF7b and ORF10 in SARS-CoV-2 with SuPER. The existence of TRS-B site associated with ORF7b remains controversial in previous studies and it was only found in Kim et al.’s work with low amounts of supported reads relative to other TRS-B sites (Kim et al. 2020). On the other hand, the existence of a sgmRNA corresponding to ORF10 were unanimously unvalidated in this and previous studies (Kim et al. 2020; Taiaroa et al. 2020).
Detecting Recombination in Coronaviruses
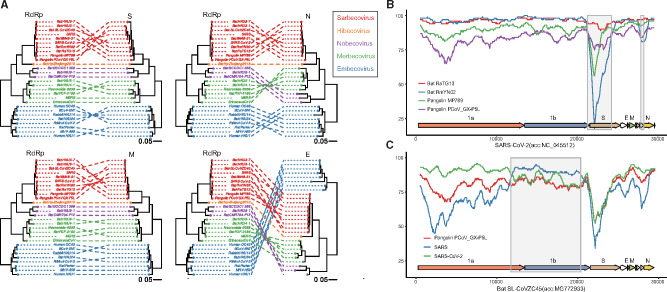
We performed recombination analysis on the genomes for each subgenus using RDP4 (Martin et al. 2015). We chose the subgenus level due to the fact that genomes from different subgenera often share less than 50% nucleotide identity leading to poor alignments, causing issues for recombination detection. By building phylogenetic trees from RdRp and the structural genes S, E, M, and N from Betacoronaviruses, we found that each subgenus formed a distinct clade for RdRp and the structural genes, providing no evidence of recent inter-subgenus recombination (fig. 3A). In contrast, we found conflicting branching orders and incongruent topology of the subgenera phylogenies suggesting recombination events within subgenera. In total, we detected 973 recombination events in 16 subgenera with RDP4 (supplementary data S1, Supplementary Material online). The number of recombination events detected is largely dependent on the number and diversity of representative genomes available for each subgenus.
Fig. 3.
(A) Tanglegrams illustrate phylogenetic incongruence in Betacoronaviruses. The phylogenetic tree built from the RdRp (RNA-dependent RNA polymerase) of Betacoronaviruses is on the left and the phylogenetic tree built from the S, E, M, and N genes is on the right for each tanglegram plot. A line connects genes from the same species. Species from the same subgenus are colored the same. Red: Sarbecovirus; orange: Hibecovirus; purple: Nobecovirus; green: Merbecovirus; blue: Embecovirus. (B, C) Simplot analysis performed with Kimura (two-parameter), a window size of 1000 nt and a step size of 50 nt. The query genome for panel B is SARS-CoV-2 (accession number: NC_045512) and the query genome for panel C is Bat SL-CoVZC45 (accession number: MG772933). Recombinant regions detected by RDP4 are highlighted light gray.
Analyzing Recombination in SARS-CoV-2 and Its Close Relatives
Due to the critical importance of understanding SARS-CoV-2 genome evolution, we performed a focused analysis on recombination in SARS-CoV-2 and its close relatives with SIMPLOT (Lole et al. 1999). The bat-derived coronavirus RmYN02 sequence identity is higher to SARS-CoV-2 than RATG13 over the majority of its length except for the region around S and ORF8 where the identity is lower. The sequence identity of RmYN02 across S and ORF8 is even lower than found in the pangolin coronaviruses MP789 and PCoV-GX_PL5 (fig. 3B). ORF8 of RmYN02 (relative to NC_045512:27882-28259) was identified by RDP4 as a recombinant region derived from an ancestor of the bat coronavirus, BtRs-BetaCoV/GX2013 (KJ473815). The low-identity region between SARS-CoV-2 and RmYN02 detected in the S gene (relative to NC_045512:21259-24264) is likely due to the acquisition of this region by RmYN02 from an unsampled lineage via a recombination event (supplementary data S1, Supplementary Material online) and this region in SARS-CoV-2 appears to be ancestral (MacLean et al. 2020; Zhou et al. 2020). In fact, based on the Sarbecovirus genomes used in this study, RDP4 did not detect any recent recombination events that signify that the S gene in SARS-CoV-2 is recombinant, which is consistent with recent publications (Boni et al. 2020). The coronavirus CoVZC45, which was isolated from a bat in 2017, has a higher sequence identity to SARS-CoV-2 across its genome compared with SARS, except in the region corresponding to the majority of ORF1b (relative to NC_045512:11726-20372) (fig. 3C). Based on the RDP4 inference, the parent sequence of this ORF1b recombinant region should be the ancestor of bat-SL-CoVZC45 (MG772933), a close relative to SARS for this region, whereas the major parental sequence is likely the ancestor of RaTG13 and SARS-CoV-2 (Boni et al. 2020; Cagliani et al. 2020; Lam et al. 2020). These results suggest recombination occurred between the ancestors of SARS and SARS-CoV-2, implying the potential for recombination between SARS and SARS-CoV-2.
TRS-B CS is more likely to be located in a breakpoint hotspot than in a random position in the genome.
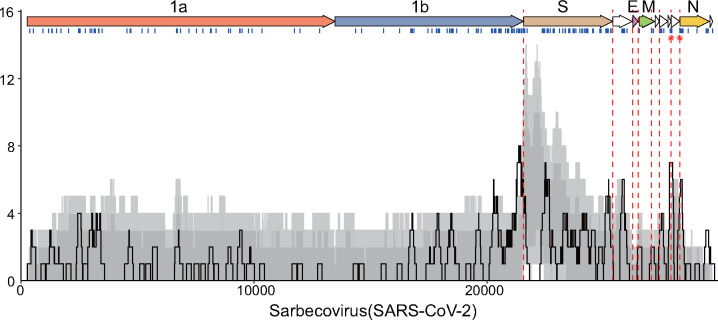
After locating TRS-B sites with SuPER, and systematically identifying recombination with RDP4, we investigated whether recombination is common at TRS-B sites. We determined whether TRS-B sites are located within a breakpoint hotspot, which is defined here as a region with breakpoint frequency higher than the 99th percentile of expected recombination breakpoint clustering assuming random recombination. We identified TRS-B sites from five subgenera located within recombination hotspots (fig. 4 and supplementary fig. S2, Supplementary Material online). In total, 8 out of 91 identified TRS-B sites were located within breakpoint hotspots. The recombination hotspots observed are more frequently colocated with TRS-B sites than expected (two-sample test for equality of proportions without continuity correction: P = 2.2e-07). Interestingly, four of the eight TRS-B sites located within breakpoint hotspots precede gene S, an important determinant of host range. Two of the eight TRS-B sites located within breakpoint hotspots are found within Sarbecoviruses, one before ORF8 and one before gene N.
Fig. 4.
Recombination breakpoint density plot in Sarbecoviruses. Recombination breakpoint density plot illustrating breakpoint positions detected across 123 detectable events in Sarbecovirus using SARS-CoV-2 as the reference. Genes were plotted at the top of the panel as arrows. All detectable unique breakpoint positions are indicated by small vertical blue lines at the top of the graph. The number of breakpoints detected within the 200-nt region was counted and plotted as a solid black line. Light and dark gray areas respectively indicate the 95% and 99% intervals of expected breakpoint events assuming random recombination. Dashed red lines indicate the positions of TRS-B and red stars plotted on the red line to indicate that the TRS-B are located within breakpoint hotspots.
Discussion
Here, we sought to test the hypothesis that template switching at TRS-B sites during genome replication contributes to coronavirus genome recombination. We used SuPER, a tool we developed for identifying TRS sites in coronavirus genomes, to test this hypothesis. It should be noted that SuPER performs best given high-coverage RNA-seq data. Although it still can predict TRS sites without RNA-seq, its performance declines and there are some instances, such as in Pedacovirus, where TRS sites cannot be reliably identified.
Although the association on TRS-B sites on recombination breakpoint sites is not obvious, we found that 8 of the 91 TRS-B sites fell within recombination breakpoint hotspots. However, this is likely an underestimate because our analysis is dependent on the number and diversity of available coronavirus genomes, and as of now, many coronavirus subgenera have only a handful of genomes available. We must also take into account the role of selection, in that only recombination events that produce viable viral genomes will survive and therefore be sequenced. For example, TRS-B sites tend to be intergenic, so recombination events that happen there are more likely to be viable, which could be an alternate explanation to the pattern we observed. As more coronavirus genomes are sequenced in the wake of the COVID-19 pandemic, the contribution of TRS-B sites to coronavirus genome recombination can be revisited. Experiments that examine recombination due to template switching at TRS-B sites that lessen the influence of selection pressure, such as direct RNA sequencing with Oxford Nanopore on cells co-infected with two coronaviruses, could further test this hypothesis. Overall, these results are consistent with, but do not definitively support, the hypothesis that recombinations at TRS-B due to template switching contribute to coronavirus genome recombination.
The worldwide pandemic caused by SARS-CoV-2 will likely lead to a global reservoir of SARS-CoV-2, as transmission to animals such as cats (Halfmann et al. 2020) and minks (Oreshkova et al. 2020) has been documented. A global reservoir of SARS-CoV-2 dramatically increases the chances that SARS-CoV-2 could recombine with other coronaviruses leading to recombinant viruses to which SARS-CoV-2 vaccines do not confer immunity. Furthermore, attenuated coronaviruses used for vaccines could revert to a virulent phenotype through recombination (Almazán et al. 2013; Graham et al. 2018; Pascual-Iglesias et al. 2019). Therefore, it is of critical importance to understand factors and mechanisms that contribute to and underlie coronavirus genome recombination. Our results contribute to this through the systematic annotation of TRS sites in coronaviruses, which helps to inform which coronaviruses are at the highest risk of recombining with SARS-CoV-2 through TRS-B mediated template switching. Though we are limited by the number of available coronavirus genomes, we did not observe evidence for recombination between species in the subgenus Sarbecovirus and species in other Betacoronavirus subgenera or Alpha, Gamma, or Deltacoronaviruses. These results support that recombination between SARS-CoV-2 and other Sarbecoviruses is more likely and therefore, Sarbecoviruses should be the subject of intense surveillance. Overall, this work helps to inform and predict coronavirus genome recombination and could play a small but important role in preventing future coronavirus outbreaks.
Materials and Methods
Identifying Nonredundant Coronavirus Genomes
We downloaded 5,517 complete genomes from the family of Coronaviridae from NCBI Virus (2020-05-19) and added one additional genome (EPI_ISL_412977 RmYN02) (Zhou et al. 2020) from GISAID. To remove the redundancy of the genomes, we used Mash (Ondov et al. 2016) to compute pairwise distance between the genomes (-d 0.01) and clustered them with MCL. We identified 506 clusters and selected one genome to represent each cluster (Shu and McCauley 2017).
Phylogenetic Analysis
Protein sequences of RdRp, S, M, E, and N genes were retrieved and from each of the 506 representative genomes. Multiple sequence alignment was performed using Muscle (Edgar 2004) with default parameters and phylogenetic trees built by FastTree from the alignment (Price et al. 2009). The phylogenetic tree based on RdRp was built to assign the taxonomy for those genomes without genus and subgenus assignments in NCBI Virus.
Putative Recombination Events Detection and Analysis
Representative genomes from the same subgenera were aligned with muscle (Edgar 2004) and then used for RDP4 (Martin et al. 2015) to detect recombination events. The GENECONV, MAXCHI, CHIMAERA, BOOTSCAN, SISCAN, and 3SEQ methods implemented in the RDP4 package were used. Default RDP4 settings were used throughout the analysis. Events detected by four or more of the above methods were accepted and reported. Recombination density plots and hot–cold spots were identified using the RDP4 package. SIMPLOT (Lole et al. 1999) was employed to manually detect recombination events in the Sarbecovirus using a “query versus reference sequence” approach.
Subgenomic mRNA Position Exploration with RNA-Seq
SuPER (Subgenomic mRNA Position Exploration with RNA-seq) is designed to detect TRS sites in coronavirus genomes allowing for the delineation of sgmRNAs start sites. SuPER can be downloaded from https://github.com/ncbi/SuPER. SuPER can use RNA-seq data to precisely delineate sgmRNA start sites and in the absence of RNA-seq, it uses the TRS-L site to predict TRS-B sites. The workflow of SuPER is divided into seven steps: 1) infer TRS-L in the 5′ UTR of reference genomes; 2) find split reads in the alignment of the RNA-seq to the coronavirus genome; 3) detect split sites supported by split reads as potential sgmRNA start sites; 4) refine assigned positions by identifying TRS-B in the reference genome; 5) associate the refined positions with possible downstream ORFs (<100 nt) if the genome annotation file is provided; 6) reconstruct site-specific 5′ end sgmRNA consensus sequences with split reads; and 7) report the alignment of TRS-L, TRS-B and the 5′ sgmRNA sequences.
Detection of TRS-L in Reference Genomes
The curated Stockholm files containing 5′ UTR alignment and consensus RNA secondary structure of major genera of Coronaviridae (namely Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus) were downloaded from the Rfam database (http://rfam.xfam.org/covid-19), from which the CM files were generated by Infernal 1.1.3 (Nawrocki and Eddy 2013) with commands “cmbuild” and “cmcalibrate.” Given a reference genome and genus label, the first 150 nt of the genome was aligned using the corresponding genus CM file with command “cmalign” in Infernal. According to the consensus motif previously marked between SL2 and SL4 (often on SL3 if available) in the secondary structure, the counterpart sequence in the genome was eventually determined as its TRS-L.
Identifying sgmRNA Start Sites Using RNA-Seq
SuPER can use RNA-seq data to precisely detect TRS-B sites, which occur at the beginning of sgmRNAs. If the sequence mapping was obtained by a program using a similar algorithm to BWA (Li and Durbin 2009), SuPER will find the split reads mapped both onto the leader sequence (within the first 10–150 nt in genome) and the sgmRNA 5′ end (between the coordinates 20000 and the end of the genome) on the same strand. On the other hand, if the sequence alignment is derived from reads mapped by a program considering RNA splicing such as HISAT2 (Kim et al. 2015), SuPER will use junction reads instead. The split or junction reads then define a putative sgmRNA start site. The putative sgmRNA start site is further refined by searching for the TRS-B sequence. The region spanning 30 nt upstream and downstream of the putative sgmRNA start site is searched by window sliding with window size of the same length as TRS-L. The sequence with minimal hamming distance from TRS-L is assigned as the TRS-B site. In addition, if a genome annotation file is provided, SuPER will try to connect the sgmRNA start site to the nearest downstream ORF within 170 nt. Results of SuPER on coronavirus genomes with available RNA-seq data in this study can be found in supplementary data S2, Supplementary Material online.
Identifying sgmRNA Start Sites without RNA-Seq
Although the best results are obtained when using RNA-seq, SuPER still functions when only a reference genome and annotation are provided. After inferring the TRS-L site in the reference genome, SuPER is capable of finding possible TRS-B sites throughout the whole genome with a hamming distance from TRS-L less than 1 or less than 2 if a downstream ORF exists. In the situation of multiple positions associated with the same ORF, the position with minimal hamming distance and closest to the ORF is assigned as the TRS-B site.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The work of Y.Y., W.Y., and X.J. was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. The work of A.B.H. was supported by the University of Maryland Department of Cell Biology and Molecular Genetics and the Center for Bioinformatics and Computational Biology. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov).
References
- Almazán F, DeDiego ML, Sola I, Zuñiga S, Nieto-Torres JL, Marquez-Jurado S, Andrés G, Enjuanes L.. 2013. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4(5):e00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K, Evans DJ.. 2018. Mechanisms and consequences of positive-strand RNA virus recombination. J Gen Virol. 99(10):1345–1356. [DOI] [PubMed] [Google Scholar]
- Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, Rambaut A, Robertson DL.. 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 5(11):1408–1417. [DOI] [PubMed] [Google Scholar]
- Cagliani R, Forni D, Clerici M, Sironi M.. 2020. Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. J Virol. 94:e00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L.. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y.. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25(15):1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Baric RS.. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 84(7):3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Deming DJ, Deming ME, Yount BL, Baric RS.. 2018. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun Biol. 1(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J. 2020. Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19. Am J Clin Pathol. 153(4):420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann PJ, Hatta M, Chiba S, Maemura T, Fan S, Takeda M, Kinoshita N, Hattori S-I, Sakai-Tagawa Y, Iwatsuki-Horimoto K, et al. 2020. Transmission of SARS-CoV-2 in domestic cats. N Engl J Med. 383(6):592–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI.. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46(D1):D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI.. 2018. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics. 62(1):e51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J, Matsushima G, Makino S, Fleming J, Vannier D, Stohlman S, Lai M.. 1988. In vivo RNA-RNA recombination of coronavirus in mouse brain. J Virol. 62(5):1810–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee J-Y, Yang J-S, Kim JW, Kim VN, Chang H.. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181(4):914–921.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Baric R, Makino S, Keck J, Egbert J, Leibowitz J, Stohlman S.. 1985. Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol. 56(2):449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Jia N, Zhang Y-W, Shum MH-H, Jiang J-F, Zhu H-C, Tong Y-G, Shi Y-X, Ni X-B, Liao Y-S, et al. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285. [DOI] [PubMed] [Google Scholar]
- Lau SKP, Li KSM, Huang Y, Shek C-T, Tse H, Wang M, Choi GKY, Xu H, Lam CSF, Guo R, et al. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 84(6):2808–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC.. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 73(1):152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean OA, Lytras S, Weaver S, Singer JB, Boni MF, Lemey P, Kosakovsky Pond SL, Robertson DL.. 2020. Natural selection in the evolution of SARS-CoV-2 in bats, not humans, created a highly capable human pathogen. BioRxiv. doi:10.1101/2020.05.28.122366 [DOI] [PMC free article] [PubMed]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B.. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1(1):vev003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM.. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17(1):132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N, Molenaar R-J, Vreman S, Harders F, Munnink BBO, Hakze R, Gerhards N, Tolsma P, Bouwstra R, Sikkema R, et al. 2020. SARS-CoV2 infection in farmed mink, Netherlands, April and May 2020. Eurosurveillance 25(23):2001005. [DOI] [PMC free article] [PubMed]
- Pascual-Iglesias A, Sanchez CM, Penzes Z, Sola I, Enjuanes L, Zuñiga S.. 2019. Recombinant chimeric transmissible gastroenteritis virus (TGEV)—Porcine epidemic diarrhea virus (PEDV) virus provides protection against virulent PEDV. Viruses 11:682.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen M-h.. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624):1394–1399. [DOI] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL.. 1998. A new model for coronavirus transcription. In: Enjuanes L, Siddell SG, Spaan W, editors. Coronaviruses and arteriviruses. Boston: Springer US. p. 215–219. [Google Scholar]
- Sawicki SG, Sawicki DL.. 2005. Coronavirus transcription: a perspective. In: Enjuanes L, editor. Coronavirus replication and reverse genetics. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 31–55. [Google Scholar]
- Sawicki SG, Sawicki DL, Siddell SG.. 2007. A contemporary view of coronavirus transcription. J Virol. 81(1):20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, McCauley J.. 2017. GISAID: global initiative on sharing all influenza data–from vision to reality. Euro Surveill. 22(13):30494.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E, Holmes EC.. 2011. Why do RNA viruses recombine? Nat Rev Microbiol. 9(8):617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazan F, Zuniga S, Enjuanes L.. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2(1):265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Moreno JL, Zúñiga S, Alonso S, Enjuanes L, 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J Virol. 79(4):2506–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiaroa G, Rawlinson D, Featherstone L, Pitt M, Caly L, Druce J, Purcell D, Harty L, Tran T, Roberts J.. 2020. Direct RNA sequencing and early evolution of SARS-CoV-2. BioRxiv. doi:10.1101/2020.03.05.976167
- Tian P-F, Jin Y-L, Xing G, Qv L-L, Huang Y-W, Zhou J-Y.. 2014. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis. 20(10):1735–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fu S, Cao Y, Zhang H, Feng Y, Yang W, Nie K, Ma X, Liang G.. 2017. Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China. Emerg. Microbes Infect. 6(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Junker D, Collisson EW.. 1993. Evidence of natural recombination within the S1 gens of infectious bronchitis virus. Virology 192(2):710–716. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV.. 2018. Origins and evolution of the global RNA virome. mBio 9(6):e02329.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-Y, Brian DA.. 2007. 5′-proximal hot spot for an inducible positive-to-negative-strand template switch by coronavirus RNA-dependent RNA polymerase. J Virol. 81(7):3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, Andino R.. 2016. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe. 19(4):493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liao C-L, Lai M.. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol. 68(8):4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yap Y, Danchin A.. 2005. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch Virol. 150(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, et al. 2020. A Novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 30(11):2196–2203.e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S, Sola I, Alonso S, Enjuanes L, 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol. 78(2):980–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.