Abstract
Background
Pandemic coronavirus disease 2019 (COVID-19) disease represents a challenge for healthcare structures. The molecular confirmation of samples from infected individuals is crucial and therefore guides public health decision making. Clusters and possibly increased diffuse transmission could occur in the context of the next influenza season. For this reason, a diagnostic test able to discriminate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from influenza viruses is urgently needed.
Methods
A multiplex real-time reverse-transcription polymerase chain reaction (PCR) assay was assessed using 1 laboratory protocol with different real-time PCR instruments. Overall, 1000 clinical samples (600 from samples SARS-CoV-2–infected patients, 200 samples from influenza-infected patients, and 200 negative samples) were analyzed.
Results
The assay developed was able to detect and discriminate each virus target and to intercept coinfections. The limit of quantification of each assay ranged between 5 and 10 genomic copy numbers, with a cutoff value of 37.7 and 37.8 for influenza and SARS-CoV-2 viruses, respectively. Only 2 influenza coinfections were detected in COVID-19 samples.
Conclusions
This study suggests that multiplex assay is a rapid, valid, and accurate method for the detection of SARS-CoV-2 and influenza viruses in clinical samples. The test may be an important diagnostic tool for both diagnostic and surveillance purposes during the seasonal influenza activity period.
Keywords: COVID-19, SARS-CoV-2, influenza viruses, multiplex real-time PCR, differential diagnosis
A new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wave may occur in fall/winter 2020 when the competitive influenza driver will be largely present. We report the development of a multiplex real-time polymerase chain reaction assay for detection of SARS-CoV-2 and seasonal influenza viruses.
After the first reported outbreak in Wuhan, China, in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), has swept most countries in 5 continents. In February and March 2020, the novel coronavirus rapidly spread in Europe, where affected countries introduced stringent lockdown measures to contain the epidemic [1, 2]. With >35 million confirmed cases and 1 million deaths reported to the World Health Organization (WHO) as of 5 of October 2020 (https://covid19.who.int), the SARS-CoV-2 pandemic is causing a global health emergency, which is expected to continue in the coming months pending the development of effective pharmacological measures.
Early detection of cases, through rapid diagnosis and reporting, isolation and treatment of cases, contact tracing and individual quarantine, reduction of human mobility, and promotion of social distancing measures were implemented in several countries. These interventions contributed to slow down the spread of the disease, suggesting that draconian and nonpharmacological control measures may contain or mitigate the course of the epidemic [3, 4].
However, due to the relaxation of the lockdown rules, interaction between individuals increased during the summer months. Virus circulation continues also in countries with decreasing incidence rates, while a resurgence of COVID-19 has been documented in Europe (https://www.ecdc.europa.eu/en/cases-2019-ncov-eueea). This trend could worsen in the autumn and winter, when changing human behavior might favor and increase transmission of respiratory viruses, such as influenza viruses. Influenza virus and SARS-CoV-2 may have similar clinical presentations (often characterized by fever and dry cough) and transmission modalities [5]. Therefore, co-circulation of these pathogens may complicate the early detection of SARS-CoV-2.
The WHO and the United States Centers for Disease Control and Prevention (CDC) encourage testing of samples for influenza from COVID-19 patients, inviting countries to report and register data to regional and global platforms [6, 7]. Additionally, Chinese authorities have also reported the enhancement of infectious disease surveillance systems for both viruses [8].
In this view, monitoring the spread of SARS-CoV-2 and influenza viruses, including the detection of possible coinfections in the community, through the improvement of rapid molecular diagnostic assays, becomes extremely important.
To accomplish this specific goal, filling current gaps, we prioritized the development and optimization of 2 different mix reactions (mix 1 and mix 2) for a multiplex 1-step real-time reverse-transcription polymerase chain reaction (RT-PCR) assay for the detection of SARS-CoV-2 and human seasonal influenza viruses. Three gene targets were used for each mix reaction and probes were labeled with common fluorophores compatible with several real-time PCR instruments. A panel of clinical samples previously examined for influenza or COVID-19 infections were used for the evaluation of the method. Hereby, we report the results of this study.
MATERIALS AND METHODS
Study Design and Assessment of the Multiplex Assay
Primers and probes selected for the detection of influenza A/B and SARS-CoV-2 (N2 and E genes) viruses are reported in Table 1; each probe was labeled with a different fluorescent reporter dye (FAM, HEX, or Cy5). The human ribonuclease P gene (RP constitutive gene) was also included as an internal control. The primer and probe concentrations for all mix reactions were singleton optimized and then combined in the multiplex assay. Protocols were set up using the SensiFAST Probe No-ROX One-Step Kit (Bioline) for Roche Real-Time PCR (LC480 II), BioRad Real-Time PCR detection systems (CFX96), Stratagene qPCR instrument (MX3000), Rotor Gene Q (Qiagen), and Applied Biosystems real-time PCR systems (7500 Fast, ViiA7). The SensiFASTProbe Low-ROX One-Step Kit (Bioline) can be also used for Applied Biosystems apparatus. For singleplex and multiplex assays, primers and probes were used at 400 nM and 100 nM, respectively, whereas for RP reaction, primers and probe were employed in a lower concentration (100 nM and 25 nM) as shown in Table 1.
Table 1.
Primers and Probes for Multiplex Real-Time Reverse-Transcription Polymerase Chain Reaction
| Mix Reaction | Primer/Probe | Sequence 5′ > 3′ | Concentrationa | Reference |
|---|---|---|---|---|
| 1 | InfluA-F | GACCRATCCTGTCACCTCTGAC | 400 nM | [9, 11] |
| 1 | InfluA-R | AGGGCATTYTGGACAAAKCGTCTA | 400 nM | [9, 11] |
| 1 | InfluA-P | FAM-TGCAGTCCTCGCTCACTGGGCACG-BHQ1 | 100 nM | [9, 11] |
| 1 | InfluB-F | TCCTCAAYTCACTCTTCGAGCG | 400 nM | [10, 11] |
| 1 | InfluB-R | CGGTGCTCTTGACCAAATTGG | 400 nM | [10, 11] |
| 1 | InfluB-P | HEX-CCAATTCGAGCAGCTGAAACTGCGGTG-BHQ1 | 100 nM | [10, 11] |
| 1 | E-F1 | ACAGGTACGTTAATAGTTAATAGCGT | 400 nM | [13] |
| 1 | E-R2 | ATATTGCAGCAGTACGCACACA | 400 nM | [13] |
| 1 | E-Pcy5 | CY5-ACACTAGCCATCCTTACTGCGCTTCG-BHQ2 | 100 nM | [13] |
| 2 | N2-F | TTA CAA ACA TTG GCC GCA AA | 400 nM | [12] |
| 2 | N2-R | GCG CGA CAT TCC GAA GAA | 400 nM | [12] |
| 2 | N2-P | FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1 | 100 nM | [12] |
| 2 | E-F1 | ACAGGTACGTTAATAGTTAATAGCGT | 400 nM | [13] |
| 2 | E-R2 | ATATTGCAGCAGTACGCACACA | 400 nM | [13] |
| 2 | E-Phex | HEX-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1 | 100 nM | [13] |
| 2 | RP-F | AGATTTGG CCTGCGAGCG | 100 nM | [12] |
| 2 | RP-R | GAGCGGCTGTCTCCACAAGT | 100 nM | [12] |
| 2 | RP-P | CY5-TTCTGACCTGAAGGCTCTGCGCG-BHQ2 | 25 nM | [12] |
aOptimized concentrations are expressed as nanomol per liter (nM) based on the final reaction in the polymerase chain reaction mixture.
Reaction mixture (20 μL) contained final concentrations corresponding to 10 μL of 2X SensiFAST Probe One-Step Mix, each primer and probe at specific concentration, 0.2 μL of reverse transcriptase, 0.4 μL of RiboSafe RNase Inhibitor, and 5 μL of nucleic acid. Amplification was performed on different real-time PCR systems with the following cycling conditions: 45°C for 20 minutes followed by 95°C for 10 seconds and 60°C for 30 seconds (45 cycles). Each specimen was also tested for the RP constitutive gene as an internal control (Table 1). For SARS-CoV-2, an additional mix reaction was performed using N1, N2, and RP gene targets with the identical primers and probe concentrations and thermal cycling protocol (Supplementary Table 1).
Sensitivity, Specificity, and Reproducibility of Single Assays Compared to the Multiplex Assay
The multiplex real-time RT-PCR assay included 2 different reaction mixes, namely mix 1 (InfluA, InfluB, and E) and mix 2 (N2, E, and RP) and were set up with different fluorescent dyes (Table 1).
Both mixes ran simultaneously. Mix 1 and mix 2 were conceived for influenza/SARS-CoV-2 screening and for SARS-CoV-2 confirmation, respectively. The mix 2 is required to monitor nucleotide mutations that may occur during the evolving outbreak and that could compromise the sensitivity and specificity of RT-PCR detection.
The European synthetic single stranded RNA (ssRNA) standard EURM-019 (https://crm.jrc.ec.europa.eu/p/EURM-019) was employed to test the sensitivity of each SARS-CoV-2 target, while purified and quantified RNA from type A/FriuliVeneziaGiulia/228/2019 (belonging to the A/H3N2 subtype–3C.2a1b genetic subgroup and representing A/La Rioja/2202/2018 reference strain) and type B/Parma/4/2019 (belonging to the B/Victoria lineage–1A[∆3]B genetic subgroup and representing B/Washington/02/2019 reference strain) influenza viruses were used as positive controls.
To establish the limit of quantification (LOQ) of each assay, serial dilution containing from 107 to 100 copies were tested in triplicate over 10 runs. Each singleplex was used as baseline control for the development of the multiplex assay. To assess the specificity of the mix real-time PCRs, the cross-reactivity of the primer-probe pairs was examined (5 replicates over 10 runs) by combined equal amount of controls ranging from 104 to 100 copies, and with the only reaction mixture (water, no template control). The endpoint LOQ of each component in the multiplex was directly compared to the single assays using a dilution series of standard and positive controls for each target. Dilutions were tested in triplicate over 10 different runs, and were carried out to guarantee that multiplexing assay did not result in a loss of sensitivity at the endpoint of LOQ, to determine the linearity of the method and for the standard curve production. The cycle threshold number (Ct value) was calculated and each primers-probe set in the single assay was tested, and then combined into mix reactions for multiplex real-time RT-PCR assay.
Inter- and Intra-assay Evaluation and Detection of Mixed Samples
The inter- and intra-assay variability of the multiplex was also assessed to determine the repeatability and the reproducibility of the assay. Each target was tested 10 times at different concentrations (105, 104, and 103 copies) in 1 assay (intra-assay) over 5 separate runs with different users (interassay). The potential cross-detection between the viral pathogens was first measured in order to evaluate the assay specificity. Positive controls and synthetic RNA were combined in equal amounts (from 103 to 100 copy numbers) and used for specificity analysis as the target pool. Groups of mixed pools were run in parallel in all real-time instruments and assessed in multiplex detection.
Specimens
Clinical samples were collected from December 2019 to May 2020 at the COVID-19 National Reference Laboratory and National WHO Influenza Center of the Istituto Superiore di Sanità (ISS), Rome, Italy. A panel of 1000 randomly selected RNA samples purified by the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) from nasopharyngeal swabs (600 SARS-CoV-2, 200 influenza A/B, and 200 negative samples) were employed to evaluate the clinical application of the multiplex real-time PCR assay. All samples were previously tested for the presence/absence of SARS-CoV-2 and/or influenza A/B viruses using the reference methods [9–13]. All positive samples were recorded for Ct values.
A specimen was considered adequate if the internal control (RP) did not cross the threshold line at 35 (Ct ≤35). For samples that exhibited a Ct RP value >35, a retesting of the sample through a new RNA extraction or a resampling was performed.
The use of samples for diagnostic workflow was agreed under the medical ethical rules for the 2019 COVID-19 public health emergency.
Statistical Analyses
To determine the efficiency of the multiplex real-time PCR assay, the Ct values obtained from a series of template RNA dilutions were graphed on the y-axis vs the log of the dilution on the x-axis. The slope of this line was used to determine the efficiency (E) according to the equation E = 10(–1/slope). To evaluate the assay precision, we calculated the coefficient of variation (CV) as follows: CV% = (standard deviation / mean) × 100. All tests were 2-sided and statistically significant differences were assumed when P < .05.
The sensitivity, specificity, positivity, and LOQ were calculated as previously described in the international literature [14, 15]. The optimal cutoff points of the multiplex assay, corresponding to the limit of detection (LOD) of swab samples were statistically established based on receiver operating characteristic (ROC) curve analysis [16]. The ROC curve analysis was evaluated with a LOD selectivity of 0.95, consistent with the Ct at which at most 5% of true-positive samples scored negative. Accuracy estimation was expressed as agreement percentage (%) defined as true-positive or true-negative predictive values (PPV and NPV, respectively) vs the singleplex assays known as the “gold standard” [9–13].
RESULTS
Analytical Sensitivity, Efficiency, and Linearity of Singleplex Assays
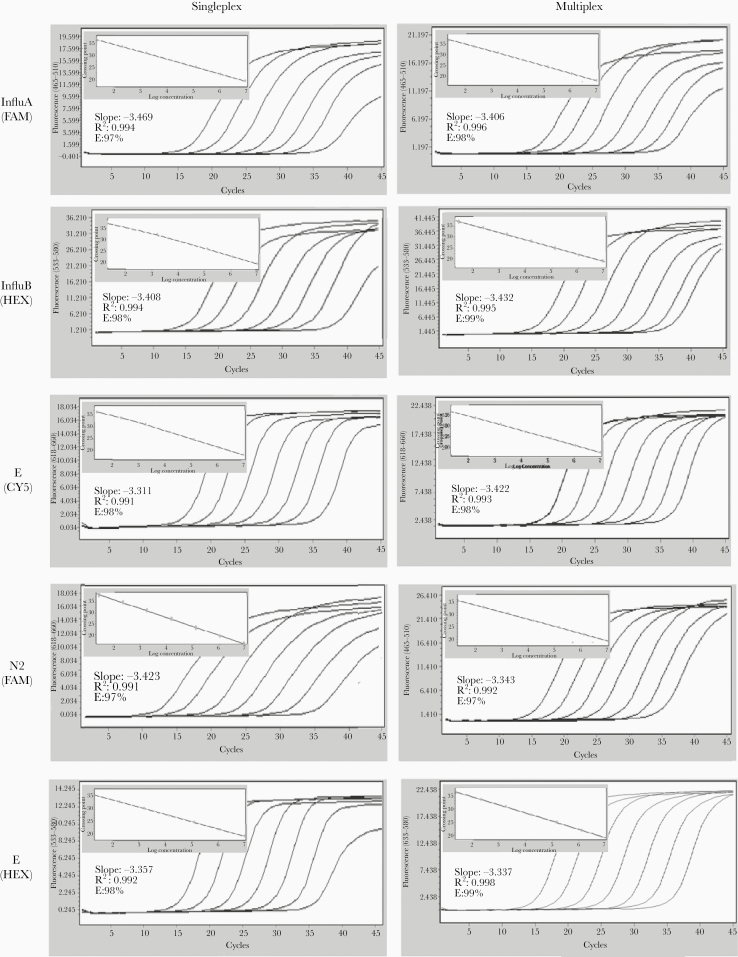
Each target (InfluA, InfluB, E, and N2) was tested using standards to determine the sensitivity and the linearity of the method, while the RP gene was included as internal control. Singleplex assays were performed on LC480II, CFX96, MX3000, Rotor Gene Q, ABI 7500 Fast, and ABI ViiA7 real-time PCR platforms. All instruments showed similar Ct values for all target genes (overall range, 0.2–0.8 ± standard deviation [SD] 0.31; P value < .05) and the results for the LC480 II are shown in Figure 1, with marked slope, R2, and E values. All the slope values were between −3.311 and −3.469 whereas the R2 and E values were >0.99 and ≥97%, respectively for all standard curves. These results suggest that the linearity of the method is consistent, corroborating the robustness of the method.
Figure 1.
LC480II plot amplifications and standard curves of the singleplex and multiplex assays. Each assay was tested using related standard ranging from 107 to 10 copies/μL. Polymerase chain reaction curve data are represented by relative fluorescence plotted against cycle threshold (Ct) values. Standard curves were generated from the Ct values obtained against known concentrations. Slope of the regression curve, the coefficient of determination (R2), and efficiency (E) are indicated for each assay.
Specificity and Endpoint Detection of Multiplex Assays
The specificity of primers/probe sets was determined against a panel of pathogens as previously described [7–11]. However, to assess the specificity of each mix reaction and to exclude artificial nonspecific reactivity among combined primer-pairs/probe, oligonucleotides were tested using mixed controls (influenza type A/FriuliVeneziaGiulia/228/2019, influenza type B/Parma/4/2019, and synthetic ssRNA EURM-019), RNA from negative samples, and run mixtures. Each specific set reacted only with its corresponding controls with the expected Ct value, indicating that there was no interference among primers and probes in the multiplex assay (data not shown). The LOQ endpoint of each component of the multiplex assay compared to the single assays showed that multiplexing had no detrimental effect on the endpoint detection limit of each component (Figure 1). On the contrary, the performance of both mix reactions in the multiplex assays highlights an LOQ improvement of Ct value (range, 0.4–0.7 ± SD 0.21), in comparison to the established single assay. The quantification limit of the multiplex ranged overall between 5 and 10 copies/reaction taking an estimating Ct cutoff value between 37.1 and 37.9 (Table 2). All real-time instruments evaluated in this study gave equivalent results with an overall Ct range between 0.4 and 0.9 ± SD 0.57 (P < .05).
Table 2.
Limit of Quantification of the Single and Multiplex Assays for Each Gene Targeta
| Mean Ct ± SD/Mean ± SD Copies for Reaction | |||
|---|---|---|---|
| Mix Reaction | Gene Target/Fluorescent Dye | Singleplex | Multiplex |
| 1 | InfluA/FAM | 37.5 ± 0.98/10.31 ± 1.18 | 37.1 ± 1.08/4.88 ± 1.08 |
| 1 | InfluB/HEX | 37.9 ± 1.06/7.13 ± 1.39 | 37.2 ± 1.11/6.43 ± 1.11 |
| 1 | E-CY5 | 37.6 ± 0.94/9.35 ± 1.15 | 37.3 ± 1.06/5.04 ± 1.15 |
| 2b | N2/FAM | 37.5 ± 1.09/8.24 ± 1.04 | 37.2 ± 1.03/6.98 ± 1.07 |
| 2b | E-HEX | 37.7 ± 1.03/6.36 ± 1.22 | 37.2 ± 1.12/5.55 ± 1.03 |
Abbreviations: Ct, cycle threshold; SD, standard deviation.
aAll data were obtained by the LC480 II Roche real-time polymerase chain reaction platform.
bData of the RP assay, included in the mix 2, were not part of this analysis.
Inter- and Intra-assay Variability
The intra- and interassay precision was evaluated by testing 3 control RNA samples at different concentrations. The precision was determined as a percentage of the coefficient of variation (CV%) of the acquired Ct values. The results are summarized in Table 3, with the mean Ct and CV values. The intraplate variability was in line with predetermined Ct samples tested with a CV% range from 0.30% to 0.66%. As expected, the interrun variability and reproducibility observed with each sample showed a CV% slightly higher (CV% range, 0.66%–0.97%). In the latter case, different factors may play small differences between each assay, but the CV% value remains still lower than 1%. Similar findings were found with other real-time platforms. The results obtained indicate that the developed multiplex assay is highly repeatable, reproducible, and robust even at higher Ct values.
Table 3.
Multiplex Intra- and Interassay Evaluations Based on the Coefficient of Variability Percentage Obtained With 3 Samples of Different Concentrationsa
| Intra-assay Mean Ct ± SD (CV%) | Interassay Mean Ct ± SD (CV%) | ||||||
|---|---|---|---|---|---|---|---|
| Mix | Gene Target | 105 | 104 | 103 | 105 | 104 | 103 |
| 1 | InfluA/FAM | 25.29 ± 0.16 (0.66%) | 28.55 ± 0.12 (0.42%) | 31.70 ± 0.14 (0.44%) | 25.89 ± 0.21 (0.81%) | 29.08 ± 0.23 (0.79%) | 32.22 ± 0.27 (0.84%) |
| 1 | InfluB/HEX | 25.58 ± 0.11 (0.30%) | 28.71 ± 0.13 (0.45%) | 32.04 ± 0.12 (0.47%) | 25.18 ± 0.18 (0.71%) | 28.33 ± 0.24 (0.85%) | 31.68 ± 0.21 (0.66%) |
| 1 | E-CY5 | 25.67 ± 0.15 (0.58%) | 29.05 ± 0.11 (0.38%) | 31.88 ± 0.16 (0.50%) | 26.01 ± 0.19 (0.73%) | 28.73 ± 0.21 (0.73%) | 32.23 ± 0.23 (0.71%) |
| 2b | N2/FAM | 25.17 ± 0.13 (0.51%) | 28.53 ± 0.16 (0.56%) | 32.31 ± 0.12 (0.37%) | 25.78 ± 0.25 (0.97%) | 29.12 ± 0.24 (0.82%) | 31.73 ± 0.28 (0.88%) |
| 2b | E-HEX | 26.07 ± 0.14 (0.54%) | 28.81 ± 0.16 (0.55%) | 31.88 ± 0.14 (0.44%) | 25.98 ± 0.23 (0.88%) | 28.13 ± 0.25 (0.89%) | 32.16 ± 0.26 (0.81%) |
Abbreviations: CV%, coefficient of variation percentage; Ct, cycle threshold; SD, standard deviation.
aAll data were obtained by the LC480 II Roche real-time polymerase chain reaction platform.
bData of the RP assay, included in mix 2, were not part of this analysis.
Detection of Artificial Mixed RNA Control Samples
Influenza viral and SARS-CoV-2 synthetic RNA controls were used in a mixed sample to evaluate the ability of multiplex assay to identify specific viruses. Combinations of different concentration mixtures, from 103 to 100 copy numbers, were tested by using mix 1 and mix 2 reaction assays, with Ct values reported in Table 4. Notably, no detection was found for 100 copy for all mixes, while all primers/probe sets reacted only with their corresponding target gene, suggesting that this assay is able to detect specifically influenza A, influenza B, and SARS-CoV-2 infection from mixed samples. All PCR instruments produced comparable results.
Table 4.
Evaluation of Artificial Mixed RNA Samples Using Triplex Assaysa
| Mean Ct ± SD | ||||
|---|---|---|---|---|
| Combined RNA Copy Numbers | Gene Target/Fluorescent Dye | Influenza A | Influenza B | SARS-CoV-2 |
| 103 | Mix 1 | |||
| InfluA/FAM | 32.24 ± 0.12 | b | … | |
| InfluB/HEX | … | 31.51 ± 0.14 | … | |
| E-CY5 | … | … | 32.17 ± 0.11 | |
| Mix 2 | ||||
| N2/FAM | … | … | 31.44 ± 0.12 | |
| E-HEX | … | … | 31.98 ± 0.16 | |
| RP-CY5c | … | … | … | |
| 102 | Mix 1 | |||
| InfluA/FAM | 35.01 ± 0.23 | … | … | |
| InfluB/HEX | … | 34.43 ± 0.21 | … | |
| E-CY5 | … | … | 35.03 ± 0.19 | |
| Mix 2 | ||||
| N2/FAM | … | … | 34.42 ± 0.22 | |
| E-HEX | … | … | 34.94 ± 0.18 | |
| RP-CY5c | … | … | … | |
| 101 | Mix 1 | |||
| InfluA/FAM | 37.28 ± 1.18 | … | … | |
| InfluB/HEX | … | 37.12 ± 1.12 | … | |
| E-CY5 | … | … | 37.27 ± 1.16 | |
| Mix 2 | ||||
| N2/FAM | … | … | 37.11 ± 1.13 | |
| E-HEX | … | … | 37.18 ± 1.17 | |
| RP-CY5c | … | … | … | |
| 100 | Mix 1 | |||
| InfluA/FAM | … | … | … | |
| InfluB/HEX | … | … | … | |
| E-CY5 | … | … | … | |
| Mix 2 | ||||
| N2/FAM | … | … | … | |
| E-HEX | … | … | … | |
| RP-CY5c | … | … | … | |
Abbreviations: Ct, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation.
aAll data were obtained by the LC480 II Roche real-time polymerase chain reaction platform.
bNo detection.
cData of the RP assay, included in the mix 2, were not part of this analysis.
As alternative for the mix 2, we set up another mix reaction specific for SARS-CoV-2 using N1, N2, and RP primers and probe [12]. Details and results are reported in Supplementary Tables 2–4.
Evaluation of Multiplex Assay With Clinical Samples and Cutoff Value Determination
A total of 800 RNA positive (600 for SARS-CoV-2 and 200 for influenza A/B viruses) and 200 RNA-negative swab samples, previously determined by singleplex gold standard, were tested using the developed multiplex assay. All specimens were confirmed for each specific virus target, and only 2 coinfections (SARS-CoV-2/influenza A and SARS-CoV-2/influenza B) were found.
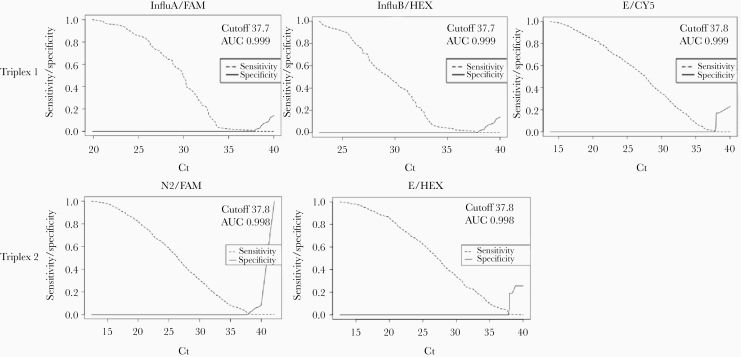
First, the Ct value was empirically fixed at 40, but this arbitrary cutoff may be not representative because it may be either too low (false-negative results) or too high (false-positive results). Consequently, on the basis of the sample status (positive or negative), the optimal cutoff point for each target gene was defined by the ROC analysis.
As shown in Figure 2, the optimal cutoff points of the multiplex assay were estimated to be 37.7 for both InfluA and InfluB, and 37.8 for E in the mix 1 reaction, while 37.8 was estimated for N2 and E in the mix 2 reaction. For each target assay, the area under the ROC curve ranged between 0.998 and 0.999, reflecting an excellent evaluation of the performance of the aggregate quality indicator for the diagnostic assay. Moreover, despite only a few positive SARS CoV-2 specimens at Ct >37.8 that were recognized negative (overall 24/800 [3%]) in multiplex vs singleplex gold standard assays, the test was proficient to discriminate between the 3 virus types. Thus, after the designed cutoff Ct values by ROC analysis, parameters for the sensitivity, specificity, PPV, and NPV revealed a high capacity of the multiplex assay to differentiate a positive from a negative sample, even when low viral loads were present (Table 5).
Figure 2.
Receiver operating characteristic (ROC) curve graphs for each gene target in multiplex assay. The optimal cycle threshold (Ct) cutoff values and the area under the ROC curve (AUC) are indicated.
Table 5.
Performance With Optimal Cutoff Values of Multiplex Versus Singleplex “Gold Standard” Assays by World Health Organization/Centers for Disease Control and Prevention Methods
| Clinical Samplesa, +/– | |||||||
|---|---|---|---|---|---|---|---|
| Assay (Cutoff) | SARS-CoV-2 (n = 600) | Influenza A (n = 120) | Influenza B (n = 80) | Negative (n = 200) | Sensitivity, % | Specificity, % | PPV (%)/NPV (%) |
| Mix 1 | |||||||
| InfluA/FAM (Ct < 37.7) | 0/600 | 120/0 | 0/80 | 0/200 | 100 | 100 | 100/100 |
| InfluB/HEX (Ct < 37.7) | 0/600 | 0/120 | 80/0 | 0/200 | 100 | 100 | 100/100 |
| E/CY5 (Ct < 37.8) | 593/7 | 0/120 | 0/80 | 0/200 | 98.8 | 100 | 100/96.6 |
| Mix 2b | |||||||
| N2/FAM (Ct < 37.8) | 590/10 | 0/120 | 0/80 | 0/200 | 98.3 | 100 | 100/95.2 |
| E/HEX (Ct < 37.8) | 593/7 | 0/120 | 0/80 | 0/200 | 98.8 | 100 | 100/96.6 |
Abbreviations: Ct, cycle threshold; NPV, negative predictive value; PPV, positive predictive value; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation.
aCoinfections are not reported.
bAll specimens were positive for the RP assay (Ct ≤35) included in the mix 2.
DISCUSSION
Several aspects have favored the pandemic spread of SARS-CoV-2: high transmissibility, presence of mild/asymptomatic cases able to spread the disease, the slow development of severe disease, lack of specificity of initial symptoms, and global susceptibility to the infection [17]. After the first wave, which has been interrupted by interventions implemented in many countries in different geographic areas, limited immunity with undefined levels of protection will exist. Thus, until an effective vaccine is developed, the epidemic may reemerge also in areas that have been previously affected. Although lockdown measures have proved to be effective, it would be difficult to again impose such restrictions in a delicate balance between health, the economy, and social life [18].
Meteorological factors do not influence SARS-CoV-2 transmission and the epidemic is supposed to persist after the hot season with a possible increase in virus circulation in autumn and winter time, when seasonal influenza virus is usually more active [19]. This may cause overburdening of hospital services, especially if increasing transmission of both viruses occurs at the same time. Considering that clinical symptoms can be very similar, it is a priority to promote a fast and appropriate differential diagnosis to support clinicians and public health professionals in their public decision-making processes [5, 17, 20].
While the SARS-CoV-2 serology may be used in epidemiological studies, molecular tests remain the gold standard laboratory diagnostic tool to diagnoses the infection. Up to now, no multiplex real-time PCR assay has been available for the simultaneous detection of seasonal influenza A, influenza B, and pandemic SARS-CoV-2.
In our study, we assessed 2 different mix reactions using previously described primes and probes [21, 22]. In particular, the mix 1 assay was designed to amplify highly conserved regions for all viruses, while mix 2 allowed the specific detection of SARS-CoV-2 infection. Targeting >1 region in the viral genome for SARS-CoV-2 detection is important to mitigate the risk of loss of sensitivity due to the mutation rate (currently not quantified but presumed to be of concern as this is an RNA virus) during the SARS-CoV-2 outbreak [23]. The 2 mix reactions (N2/E/RNase P and N1/N2/RNase P) do not have a precise recommendation, but are suggested for laboratories as alternative reaction in the multiplex panel.
The findings presented in this study denote that multiplex assay was highly specific and reproducible, showing an elevated sensitivity. The LOQ of each channel (FAM, HEX, or CY5) ranged between 5 and 10 copies/reaction, while the optimal cutoff values determined by ROC analysis were fixed between 37.7 and 37.8. Moreover, it should be highlighted that the molecular protocol was attained by different real-time instruments with comparable results, obtaining similar performance in terms of specificity and sensitivity on standards and clinical samples.
Compared with the singleplex assay, which identifies only 1 target/reaction, the multiplex test has the advantages of being easy to manage, rapid, and cost-effective. In addition, the human RP gene, as an internal control, allows monitoring of the RNA extraction procedure, checking the errors of real-time PCR handling. The molecular assay is also appropriate to detect coinfections for diagnostic purposes, or to conduct studies to assess whether SARS-CoV-2 and influenza coinfection may affect disease evolution and clinical outcomes.
Samples tested in this study showed only 2 coinfections, consistent with the international literature that shows only a few studies reporting infection with both viruses [24–29].
Despite the evidence of coinfections with other pathogens in COVID-19 patients, little is known about the viral kinetics and dynamics of SARS-CoV-2 coinfections. However, we can hypothesize that mixed infections may be linked to mechanisms of viral interference and to the immunological status of each individual [27].
The main limit of our study was the relatively low sample size. Although we tested about 1000 clinical specimens, the number of samples positive for influenza specimens was very low. In this regard, it should be considered that the influenza active surveillance activities globally decreased because of the COVID-19 pandemic and a low number of samples was collected during the influenza season (2019–2020) in Italy. Moreover, it should be mentioned that also during the SARS pandemic in 2003, only 5% of mixed infection with influenza viruses was reported, though it tended to increase over time [30].
Recently the CDC presented the Flu SC2 diagnostic kit, a quadruplex real-time RT-PCR assay for influenza and SARS-CoV-2 viruses, that detects and differentiates RNA from SARS-CoV-2, influenza A virus, and influenza B in upper or lower respiratory specimens [31]. The multiplex protocol gives full instructions for its use and primer/probe sequences are also listed. Moreover, the Flu SC2 kit is addressed to reference or public health laboratories certified under the Clinical Laboratory Improvement Amendments.
Several companies have announced the development of real-time–based commercial kits, which are expected to detect SARS-CoV-2, influenza A/B, and other respiratory viruses. Generally, these molecular tests are run on specific platforms with dedicated cartridges containing all the necessary reagents. Although this packaging makes them easy to perform in the hospital setting, it also may be a limiting factor in a shortage situation, in which, as previously reported for other aspects, an in-house method could be a useful alternative [32].
In conclusion, the real-time PCR format here described could be a suitable tool for molecular testing with potential for routine surveillance SARS-CoV-2 and influenza viruses. However, it will be necessary to investigate whether the method might be affected by the co-presence of other respiratory pathogens in the clinical samples.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. F. M., F. B., and M. S. developed the real-time polymerase chain reaction (PCR) assays, and contributed to study design and data interpretation. S. F., A. d M., and G. M. performed real-time PCR. S. P., L. C., M. F., G. D. M., and C. F. collected clinical samples and performed nucleic acid extractions. A. B., F. R., P. P., and P. S. coordinated the epidemiological and microbiological COVID-19 integrated surveillance and contributed to the data analysis. G. R. and F. R. provided expert advice, critically reviewed the manuscript, including for aspects related to the English language, and contributed to its content. A. C. had the idea for the study and was responsible for the overall design and writing. All authors reviewed and approved the final version of the manuscript. Members of the Istituto Superiore di Sanità COVID-19 team were involved in the confirmation of cases and provided support to the surveillance activities at national level.
Acknowledgments. The authors thank Alessia Caratelli, Alessia Possenti, Gianluca Marucci, Simona Cherchi, Alessandra Ludovisi, Stefania Orsini, Trentina Di Muccio, Marina Sbattella, and Ambrogio Carlei for giving their time and technical support in sample collection.
Potential conflicts of interest. The authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Contributor Information
Istituto Superiore di Sanità (ISS) COVID-19 Team:
Laura Villa, Daniela Fortini, Angelo Iacobino, Stefano Fiore, Eleonora Benedetti, Antonella Marchi, Giulietta Venturi, Claudia Fortuna, Antonello Amendola, Luciano Toma, Marco Di Luca, and Francesco Severini
References
- 1. Lapostolle F, Goix L, Vianu I, et al. COVID-19 epidemic in the Seine-Saint-Denis department of Greater Paris: one month and three waves for a Tsunami. Eur J Emerg Med 2020; 27:274–8. [DOI] [PubMed] [Google Scholar]
- 2. Riccardo F, Ajelli M, Andrianou X, et al. Epidemiological characteristics of COVID-19 cases in Italy and estimates of the reproductive numbers one month into the epidemic. medRxiv [Preprint]. Posted online 11 April 2020. doi: 10.1101/2020.04.08.20056861. [DOI] [Google Scholar]
- 3. European Centre for Disease Prevention and Control. Coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK—ninth update 23 April 2020. https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-rapid-risk-assessment-coronavirus-disease-2019-ninth-update-23-april-2020.pdf. Accessed 23 April 2020.
- 4. Acter T, Uddin N, Das J, Akhter A, Choudhury TR, Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci Total Environ 2020; 730:138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singer BD. COVID-19 and the next influenza season. Sci Adv 2020; 6:eabd0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization. Influenza update 2020. https://www.who.int/influenza/surveillance_monitoring/updates/latest_update_GIP_surveillance/en/. Accessed 03 August 2020.
- 7. Centers for Disease Control and Prevention. Weekly U.S. influenza surveillance report. https://www.cdc.gov/flu/weekly/index.htm. Accessed 25 July 2020.
- 8. Wu D, Lu J, Ma X, et al. Coinfection of influenza virus and severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). Pediatr Infect Dis J 2020; 39:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization. WHO information for molecular diagnosis of influenza virus in humans—update. Geneva, Switzerland: WHO, 2017. https://www.who.int/influenza/gisrs_laboratory/WHO_information_for_the_molecular_detection_of_influenza_viruses_20171023_Final.pdf?ua=1. Accessed July 2017. [Google Scholar]
- 10. Centers for Disease Control and Prevention. Influenza (flu). https://www.cdc.gov/flu/CLSIS/. Accessed November 2020.
- 11. Wangchuk S, Thapa B, Zangmo S, Jarman RG, Bhoomiboonchoo P, Gibbons RV. Influenza surveillance from November 2008 to 2011; including pandemic influenza A(H1N1)pdm09 in Bhutan. Influenza Other Respir Viruses 2013; 7:426–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention. Novel coronavirus (2019-nCoV) realtime rRT-PCR panel primers and probes. 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf. Accessed 06 June 2020.
- 13. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lalkhen AG, McCluskey A. Clinical tests: sensitivity and specificity. Contin Educ Anaesth Crit Care Pain 2008; 8:221–23. [Google Scholar]
- 15. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611–22. [DOI] [PubMed] [Google Scholar]
- 16. Nutz S, Döll K, Karlovsky P. Determination of the LOQ in real-time PCR by receiver operating characteristic curve analysis: application to qPCR assays for Fusarium verticillioides and F. proliferatum. Anal Bioanal Chem 2011; 401:717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan X, Ojcius DM, Gao T, Li Z, Pan C, Pan C. Lessons learned from the 2019-nCoV epidemic on prevention of future infectious diseases. Microbes Infect 2020; 22:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan A, Liu L, Wang C, et al. Association of public health interventions with the epidemiology of the COVID-19 outbreak in Wuhan, China. JAMA 2020; 232:2186–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao Y, Pan J, Liu Z, et al. No Association of COVID-19 transmission with temperature or UV radiation in Chinese cities. Eur Respir J 2020; 55:2000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flannery B, Meece JK, Williams JV, et al. Systematic testing for influenza and COVID-19 among patients with respiratory illness [manuscript published online ahead of print 20 July 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ ciaa1023.32687197 [Google Scholar]
- 21. Nalla AK, Casto AM, Huang MLW, et al. Comparative performance of SARS CoV-2 detection assays using seven different primer/probe sets and one assay kit. J Clin Microbiol 2020; 58:e00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waggoner JJ, Stittleburg V, Pond R, et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020; 26:1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med 2020; 58:1070–76. [DOI] [PubMed] [Google Scholar]
- 24. Wu X, Cai Y, Huang X, et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg Infect Dis 2020; 26:1324–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu P, Lu W, He L, et al. COVID-19 patients with recent influenza A/B infection: a retrospective study [manuscript published online ahead of print 28 May 2020]. J Infect 2020. 10.1016/j.jinf.2020.05.050. [DOI] [Google Scholar]
- 26. Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients co-infected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol 2020; 96:683–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nowak MD, Sordillo EM, Gitman MR, Paniz Mondolfi AE. Co-infection in SARS-CoV-2 infected patients: where are influenza virus and rhinovirus/enterovirus? [manuscript published online ahead of print 30 April 2020]. J Med Virol 2020. 10.1002/jmv.25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cuadrado-Payán E, Montagud-Marrahi E, Torres-Elorza M, et al. SARS-CoV-2 and influenza virus coinfection. Lancet 2020; 395:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hashemi SA, Safamanesh S, Ghafouri M, et al. Co-infection with COVID-19 and influenza A virus in two died patients with acute respiratory syndrome, Bojnurd, Iran [manuscript published online ahead of print 15 May 2020]. J Med Virol 2020. 10.1002/jmv.26014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang W, Lu E, Zhou X, et al. Influenza virologic and epidemiologic surveillance in Guangzhou, 2003. South China. J Prev Med 2005; 31:10–13. [Google Scholar]
- 31. Centers for Disease Control and Prevention. CDC influenza SARS-CoV-2 (Flu SC2) multiplex assay. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html. Accessed 14 July 2020.
- 32. Mancini F, Barbanti F, Scaturro M, et al. ; ISS COVID-19 Study Group . Laboratory management for SARS-CoV-2 detection: a user-friendly combination of the heat treatment approach and rt-real-time PCR testing. Emerg Microbes Infect 2020; 9:1393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.