Abstract
Objective
A positive result of SARS-CoV-2 nucleic acid detection provides critical laboratory evidence for clinical confirmed diagnosis, pandemic status evaluation, a pandemic prevention plan, treatment of infected people with symptoms, and protection of uninfected people. This study aims to provide a practical reference for SARS-CoV-2 nucleic acid–related research and detection.
Methods
Our laboratory has established policies combining personnel management and quality control practices for SARS-CoV-2 nucleic acid detection during the pandemic.
Results
In this article, we describe cross-department personnel management and key points of personal protection and quality control in the testing process. We also report on the differences in detection and the compatibility between different brand kits.
Conclusion
It is critical to maintain a standard and accurate laboratory operation for nucleic acid testing.
Keywords: SARS-CoV-2, nucleic acid detection, laboratory management, quality control, suspicious cases, external quality assurance
In December 2019, the novel coronavirus outbreak occurred in Wuhan, China. On January 24, 2020, The Lancet reported the epidemiologic and clinical characteristics of the first 41 infected persons.1 Within 3 weeks, the International Committee on Taxonomy of Viruses officially named this new virus the severe acute respiratory syndrome coronavirus 2”, or SARS-CoV-2. The World Health Organization officially named the disease caused by SARS-CoV-2 infection, COVID-19. The novel coronavirus SARS-CoV-2 is a β-coronavirus that spreads mainly through respiratory droplets and by close contact. It may also spread through prolonged exposure to high concentrations of contaminated aerosol in a relatively closed environment. Researchers have learned that COVID-19 may affect people of all ages, but most cases of infection have been found in adults. The highest morbidity occurs in elderly and frail persons.2
A positive result of nucleic acid detection confirms the presence of SARS-CoV-2 in individuals with suspected infection. Since early February 2020, the Hubei (China) Provincial Health and Health Commission has rapidly expanded testing capability by certifying a number of hospitals and third-party laboratories to perform nucleic acid tests.3 Among the first to be approved, our polymerase chain reaction (PCR) laboratory in the department of pathology has carried out large numbers of SARS-CoV-2 tests. At the time of this writing, more than 25,000 specimens have been tested, and we have discovered issues that may affect the accuracy and stability of the test. As such, it has been necessary to establish a system of documentation related to the strict conduct of SARS-CoV-2 nucleic acid testing and management of the laboratory (eg, quality manuals, procedure files, operation instructions, and record forms).
Laboratory Personnel Management
Personnel Access Qualification
An interdepartmental team composed of members of the pathology and laboratory departments was established because of the urgent need for testing preparation. To quickly build a trained and efficient team, we used 3 criteria for staff selection: (i) those who passed the PCR training program organized by the Hubei Center for Clinical Laboratory, (ii) people with previous experience conducting PCR experiments, and (iii) those who showed willingness, responsibility, and a sense of cooperation were permitted to assist in the laboratory.
Training Content
The primary training content dealt with biological safety protection. Specifically, it focused on second- and third-level protection, PCR experimental procedures, laboratory instrument operation, waste disposal, safety management, and other procedures.4-6 A series of assessments and procedures were set up after onsite training and examination. The trained staff were then authorized to conduct testing.
Staff Organization
Staff conducting PCR experiments had to be certified, and each staff member was teamed with 1 or 2 assistants. A full-time staff member was responsible for the management of materials, information, and routine data reporting.
Personal Protection, Mental Health, and Risk Management
Personal protective equipment was in short supply during the early stages of the COVID-19 outbreak. But shortly thereafter, positive-pressure respirators were obtained along with high-quality 3-level standard protective equipment. The availability of these materials improved the sense of security of the staff.
Before working in the lab, all the staff had to test negative for SARS-CoV-2 by nucleic acid testing of oropharyngeal swab specimens. The body temperature of each member was measured every morning and evening of their work cycle. The staff worked every other day to ensure that there was sufficient rest and good working conditions. In addition, expression of feelings or concerns among colleagues was encouraged and psychological counseling was offered.
Key Points of Protection and Quality Control for Nucleic Acid Detection
Protective Measures Implemented during Nucleic Acid Extraction and PCR Amplification
Effective negative pressure and proper airflow were maintained in the PCR laboratory environment. Swabs were processed in a constant 56°C oven for 45 minutes to inactivate the virus and then were transferred to the laboratory. Nucleic acid extraction was carried out in a biological safety cabinet by the operator with strict 3-level biological safety protection.
Disposable sharps containers with a small amount of 0.55% sodium hypochlorite (ie, bleach) were used to keep spent pipette tips that were potentially contaminated. To avoid aerosol contamination, instructions specified that pipette tips containing specimen collection solution were to be placed 2 mm to 3 mm below the lysate liquid surface and slowly and gently pressed. In addition, approximately 5–10 minutes were allowed to elapse after centrifugation, after which the inner and outer chambers were sprayed with 75% alcohol before removing the specimens.
Loading tubes with specimen RNA were transferred to the PCR amplification room through a transfer chamber. Tube surfaces were wiped with 75% alcohol before tests were run. After the specimen plates were removed, the transfer chamber was sterilized for 30 minutes or longer using ultraviolet light.
The disposal of laboratory specimens and medical waste strictly followed the requirements for the handling of live viruses. Items were packed in double-layer medical waste bags and were sealed using a gooseneck knot. After sealing, 75% alcohol was sprayed and the bags were transferred out of the extraction room to be sterilized using an autoclave.
Removal of protective clothing and protective masks was conducted in a buffer room adjacent to the extraction room. If a buffer room was not available, then soiled and clean areas had to be clearly separated. Areas near entryways were kept relatively clean, and efforts were made to avoid polluting the area during testing.
Biological safety cabinets were disinfected with 75% alcohol and sterilized by ultraviolet radiation after each experiment. The experimental tabletop was wiped with a disinfectant solution made of 0.55% sodium hypochlorite or 75% alcohol. A mobile ultraviolet lamp was used to irradiate the operating tabletop and equipment for at least 30 minutes or longer. A medical air filtration machine was used to purify the air in the laboratory.
Negative and Positive Quality Control Settings during Testing
The establishment of strict negative and positive quality control was important to assure the success of a test. Thus, a cell preservation solution of pharyngeal swabs and nucleic acid quality control specimens was used to monitor whether the nucleic acid extraction and PCR reaction system were normal.
Quality Control of Extraction Process: Cell Preservation Solution Swab Specimen
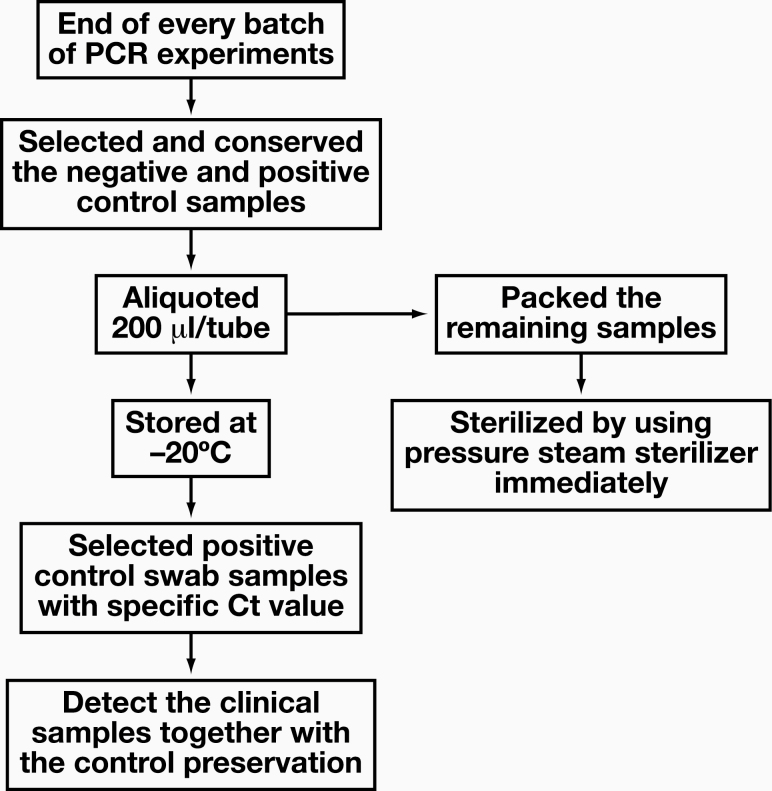
The SARS-CoV-2 viral nucleic acid extraction kit used in our laboratory did not have a positive cell solution control specimen, so the success of the extraction process and comparisons between batches could not be determined. To do so, a positive control of the oropharyngeal/nasopharyngeal swab cell preservation solution was developed: Figure 1 shows the swab quality control specimen retention process.
Figure 1.
The retention process of the control swab cell preservation solution for quality control.
Quality Control of Amplification Process: RNA Specimens
Typically, negative and positive viral RNA control specimens help monitor the stability of each batch of the PCR system. Our testing kit provided plasmids as negative and positive quality controls; however, we observed batch-to-batch and box-to-box variations in the quality control products and reaction systems, which led to unreliable results. These variations were related to the speed at which the kits were developed, produced, and shipped for laboratory use. The performance and stability of the various kits could not be verified and optimized against a large population standard; therefore, we added preprepared negative and positive RNA controls to improve test reliability.
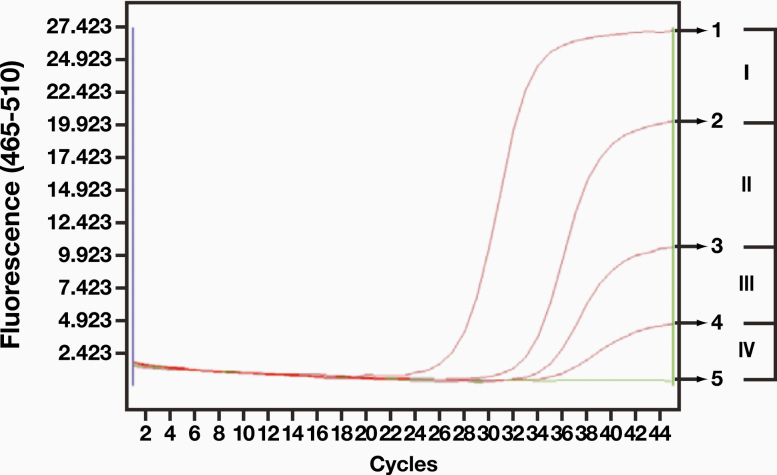
We also found that RNA control specimens were valuable as controls for amplification in weak positive results. The amplification curve and the cycle threshold (Ct) value of clinical RNA amplification in the quality control specimens varied, but they were different from the value of the positive quality control plasmids from the PCR amplification kit. Except for those specimens with the highest virus copy number, few specimens could reach the Ct value and had a typical amplification curve similar to that of a positive plasmid control. Therefore, if only the positive plasmid curve was used as a control, then some weak positive results would likely be missed.
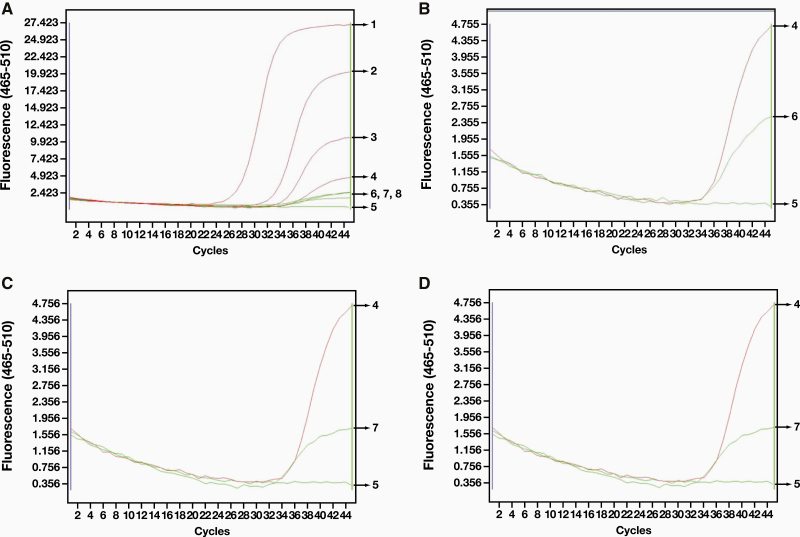
The Ct value of a single channel (FAM) was used as an example of a weak positive result. Several positive swab specimens with different Ct values of RNA were selected. A positive result was defined by kit instructions as a Ct value of ≤ 40 for the amplification curve. We divided the negative and positive quality control curves into 4 zones (Figure 2). If the RNA amplification curve was in zones I to III, then the specimen was judged to be positive; zone IV was negative. We found that in the same batch of 55 swab specimens, the curves of 3 specimens were in zone IV (curves 6, 7, 8 in Figure 3, Image A), but none of them had Ct values. When we compared these curves with the known weak positive curve 4, we found that the curve was amplified but that the characteristics of logarithmic growth were atypical (Figure 3, Image B, Image C, and Image D). The curves remained unchanged after reamplification, and the amplification curve of another channel (VIX) was similar.
Figure 2.
Positive swab collection solution that was used to extract RNA. The positive zone was divided (the Ct value of the FAM channel was used as an example).
Figure 3.
Comparison of specimens with low virus copy infections and weak positive control specimens.
In addition, we were informed by the clinic that the first specimen (swab B) came from someone who was in close contact with the infected person. The other 2 swab specimens (C and D) were from infected patients who had received treatment for 2 weeks. If the weak positive quality control (swab A, curve 4) had not been set, then all 3 specimens might be judged as negative. For prevention and control, we recommended that these 3 specimens be judged as suspicious at minimum and that they should be retested after 24 hours. More important, given that the development of these tests was taking place during a pandemic, there was an expected increase in the number of specimens coming from infected patients who were convalescing, and inaccurate test results for patients could have a negative effect on long-term prevention and treatment.
Stability of Control Specimens under Cold Storage
After aliquoting, all swab specimens and RNA specimens were temporarily stored in a dedicated refrigerator at –20°C. To assess the degradation of RNA, we analyzed the relationship between storage time and the Ct value of 6 positive swab specimens and 3 positive RNA control specimens (Table 1). All the specimens remained positive for up to 3 days, but the Ct value tended to increase on the third day, indicating the degradation of RNA.
Table 1.
Relationship Between Cycle Threshold (Ct) Values and Storage Time of Swab Collection Solution and RNA Positive Quality Control Specimens open reading frame 1ab (ORF1ab)
| Species | First Day | Second Day | Third Day | P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ct Values | Curve | Ct Values | Curve | Ct Values | Curve | |||||
| ORF1ab | N Gene | ORF1ab | N Gene | ORF1ab | N Gene | |||||
| Collection solution | 34.43 ± 1.21 | 32.37 ± 2.41 | Typical | 34.6 ± 1 | 32.15 ± 3.25 | Typical | 35.6 ± 1 | 33.06 ± 1.89 | Typical | >.05 |
| RNA | 34.06 ± 3.73 | 30.42 ± 3.24 | Typical | 31.92 ± 1.68 | 30.41 ± 2.22 | Typical | 35.39 ± 1.7 | 32.97 ± 1.91 | Typical | >.05 |
Quality Control Management with Suspicious Ct Value and Amplification Curve
If a test result did not meet the negative/positive evaluation criteria of the kit or if the linearity of the amplification curve did not match the Ct value, then the result was deemed suspicious. Some kits specify the conditions of a suspicious result; however, they do not fully cover the situations encountered in our laboratory. We reviewed 469 recent specimens and found that 12 were suspicious (2.6%). In the New Coronavirus Pneumonia Diagnosis and Treatment Program (Trial Version 7) issued by the National Health Commission of the People’s Republic of China,2 there was no guidance on how to deal with suspicious nucleic acid test results. The lack of guidance puts pressure on the managers of clinical test classification. To minimize suspicious results, we set up an expert interpretation team. If the original result was ambiguous, then the specimen was submitted to the team for a final positive or negative determination or further verification. Sampled RNA was reamplified immediately. The 2 combined amplification results were then used for a final interpretation, and if still inconclusive, then the kit was replaced and the test was redone. If necessary, it was recommended that the patient be resampled after 24 hours at the same or different anatomical site for retesting and confirmation.
Varying Efficiency of Nucleic Acid Extraction
Differences in Detection Capabilities of Nucleic Acid Extraction Kits
Because of the rapid and sometimes hasty development of tests during the pandemic, the SARS-CoV-2 virus nucleic acid extraction kits have had a widely varying performance. The extraction performance and stability of the certified reagents had not been confirmed when we began to follow these protocols. Early in the pandemic, we needed to use nucleic acid extraction reagents from 3 companies because none were able to fulfill all the demand. We compared and verified old and new kits before replacing the reagents. Even when the same amplification kit was used, positive detection rates of nucleic acid extraction varied (see Group 1 in Table 2).
Table 2.
Impact of Different Kit Detection Capabilities and Matching on Positive Rate of Specimens
| Group | Combinations | Infections | Positive | Positive Rate (%) | P Value |
|---|---|---|---|---|---|
| 1 | S1 + J2 | 8 | 2 | 25 | .19 |
| T1 + J2 | 1 | 12.5 | |||
| 2 | S1 + J2 | 51 | 2 | 3.9 | <.01 |
| S1 + S2 | 12 | 23.5 | |||
| 3 | S1 + D2 | 16 | 10 | 62.5 | <.01 |
| S1 + S2 | 7 | 43.8 | |||
| 4 | D1 + D2 | 10 | 7 | 70 | <.01 |
| S1 + S2 | 6 | 60 |
The capital letters in “Combinations” represent the brands of the kits. The number “1” in “Combinations” represents the extraction kit and “2” represents the amplification kit.
Different Amplification Efficiency and Compatibility of Amplification Kits
The use of reagents produced by different companies was common because there was a limited supply. We tested 3 brands of amplification kit and one brand of nucleic acid extraction kit in different matching modes (Table 2). We found that different amplification kits had different efficiency regarding the amplification of RNA extracted by the same extraction kit (see Groups 2 and 3 in Table 2, where the positive specimens detected by the D2 kit were clinically confirmed patients with infection). There was an even higher detection rate for kits from different brands than for matched kits from the same brand (see Group 3 in Table 2 for details; the positive specimens detected by the D2 kits were clinically confirmed patients with infection).
Based on our findings, we recommended the selection of a high-quality kit with a high correspondence to data from the local large epidemiological surveys and a clear amplification curve before formal testing. In addition, a series of strict performance comparison tests and quality verification experiments were needed for formal testing before replacing reagents of different brands or before purchasing new batches of reagents of the same brand.
External Quality Assurance
In general, it is critical to maintain a standard and accurate laboratory operation for nucleic acid testing. A laboratory should actively participate in the external quality assurance program for COVID-19; doing so should improve the quality of a standardized testing service.
Outlook
We found that SARS-CoV-2 nucleic acid detection requires extreme detection accuracy. The problems we encountered in the testing process were effectively solved through strict ongoing laboratory management and optimized quality control. The system described here provides a useful reference for the rapid development of reliable testing services.
Acknowledgments
We thank TopEdit (http://www.topeditsci.com) for the English-language editing of this manuscript. All authors have reviewed and agreed with the contents of the manuscript, and there is no financial interest to report. We certify that the submission is original work and is not under review by any other publication.
Glossary
Abbreviation
- PCR
polymerase chain reaction
- Ct
cycle threshold
- ORF1ab
open reading frame 1ab
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Diagnosis and Treatment Protocol for COVID-19 (Trial Version 7). National Health Commission of the People’s Republic of China. http://en.nhc.gov.cn/2020-03/29/c_78469.htm. Updated March 29, 2020. Accessed August 18, 2020. [Google Scholar]
- 3. Public announcement on the new coronavirus nucleic acid testing agency in the province. Hubei Provincial Health Commission. February 5, 2020. http://wjw.hubei.gov.cn/fbjd/dtyw/202002/t20200205_2019497.shtml. [Google Scholar]
- 4. Notice of the General Office of the National Health Commission on Printing and Distributing the Guidelines for Biosafety of New Coronavirus Laboratories (Second Edition). National Health Commission of the People’s Republic of China. January 23, 2020. http://www.nhc.gov.cn/xcs/zhengcwj/202001/0909555408d842a58828611dde2e6a26.shtml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laboratory biosafety guidance related to the novel coronavirus (2019-nCoV). Interim guidance. World Health Organization. https://www.who.int/docs/default-source/coronaviruse/laboratory-biosafety-novel-coronavirus-version-1-1.pdf?sfvrsn=912a9847_2. Updated February 12, 2020. Accessed August 18, 2020. [Google Scholar]
- 6. World Health Organization. Laboratory Biosafety Manual. 3rd ed. Geneva, Switzerland: World Health Organization; 2004. [Google Scholar]