Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load on admission was associated with a significantly increased 30-day mortality (odds ratio [OR], 4.20; 95% CI, 1.62–10.86), and anti-SARS-CoV-2 nucleocapisid IgG seropositivity on admission trended toward a reduced 30-day mortality (OR, 0.43; 95% CI, 0.15–1.26). Reporting of quantitative SARS-CoV-2 viral load and serologic assays may offer prognostic clinical information.
Keywords: quantitative, SARS-CoV-2, serology, viral load
A study of 181 individuals from a single community hospital found SARS-CoV-2 viral load on admission was associated with a significantly increased 30-day mortality (OR 4.20 [95% CI: 1.62-10.86]), and anti-SARS-CoV-2 IgG seropositivity on admission trended toward a reduced 30-day mortality (OR 0.43 [95% CI: 0.15-1.26]).
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus associated with high morbidity and mortality that has rapidly spread across the world. Despite increasing evidence that viral load is associated with clinical outcomes [1–6] and that high viral loads are associated with fewer tissue culture infective viral particles [7, 8], no Food and Drug Administration (FDA) Emergency Use Authorization (EUA) has been issued for a quantitative assay. Similarly, after initial concerns surrounding test characteristics, accurate serological testing for SARS-CoV-2 is increasingly becoming available in the United States. For instance, we have previously shown the Abbott Architect anti-SARS-CoV-2 nucleocapsid IgG assay to be both highly sensitive and specific for detecting prior SARS-CoV-2 infection [9]. Multiple commercial anti-SARS-CoV-2 assays are available across different antibody isotypes and with the ability to detect different antigens. The antibody responses to the different antigens tend to track together and are correlated with neutralization [10–13]. Data are beginning to emerge that the active development of neutralizing antibodies is protective against SARS-CoV-2 infection [14]. Yet, most serology assays are authorized only for the reporting of a qualitative result, even though they return semiquantitative or quantitative results. Data correlating viral load and antibody results to meaningful virologic and clinical outcomes are continuing to emerge.
Here, we examined clinical and virologic features associated with seropositivity and seroconversion to SARS-CoV-2 in a cohort of hospitalized patients in Seattle, Washington. We specifically sought to assess whether detection of anti-SARS-CoV-2 nucleocapsid IgG was associated with a better prognosis, including lower viral load and reduced 30-day all-cause mortality.
METHODS
Patient Consent Statement
The study was approved under a consent waiver by the University of Washington Institutional Review Board.
Study Population and Clinical Laboratory Testing
Patients with positive SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR) results from nasopharyngeal swabs were identified at UW Medicine hospitals, and excess serum and plasma samples were retrieved for SARS-CoV-2 antibody testing. Samples were enriched for patients who had RT-PCR results available on the same calendar date as a remnant serum or plasma sample. A total of 245 patients were identified with at least 1 residual serum/plasma sample and at least 1 clinical note available for chart review to determine days from symptom onset (Table 1). The age by decade was: 10–20 years: 0.4%; 20–29 years: 4.9%; 30–39 years: 6.9%; 40–49 years: 11.0%; 50–59 years: 17.1%; 60–69 years: 18.8%; 70–79 years: 21.2% years; 80–89 years: 12.2%; 90–99 years: 7.3%. The primary clinical encounter was inpatient for 194 patients, emergency department for 39, and outpatient for 12. Eight patients were asymptomatic at the time of initial PCR result; for these patients, day of symptom onset was therefore set at the date of first positive PCR. Thirty-day mortality was determined by manual chart review for all patients who had either an RT-PCR result or an IgG result from the day of admission and calculated from the day of first positive PCR. The number of subjects included in the different analyses is described in the “Results,” as data were not available for all subjects at all time points.
Table 1.
Characteristics of Patients Included in this Study
| Demographic | Percentage (No.) of Applicable Study Population |
|---|---|
| Female | 40 (98/245) |
| Age >65 y | 49.4 (121/245) |
| Inpatient | 79.2 (194/245) |
| Death within 30 d of 1st PCR | 19.0 (45/237) |
| Ever had a positive IgG | 50.6 (124/245) |
| Seropositive by hospital admission | 33.3 (38/114) |
| Ever had a Ct <22 | 34.1 (75/220) |
| Ct <22 on hospital admission | 32.1 (35/109) |
Abbreviations: Ct, cycle threshold; IgG, immunoglobulin G; PCR, polymerase chain reaction.
Anti-SARS-CoV-2 nucleocapsid IgG was determined by the Abbott Architect as previously described [9]. The manufacturer’s suggested cutoff of 1.40 was used for seropositivity. SARS-CoV-2 quantitative RT-PCR (qRT-PCR) was performed using Hologic Panther Fusion, DiaSorin Simplexa, Roche Cobas 6800 platforms, or a Centers for Disease Control and Prevention–based laboratory-developed test (LDT) [15]. Cycle threshold (Ct) values were available from Hologic Panther Fusion, and LDT assays and were treated interchangeably given their close correlation [15]. A Ct of 22 is equivalent to ~2 500 000-copies/mL viral transport media in these assays [16].
Data Analysis and Visualization
The association between SARS-CoV-2 IgG index value and Ct was assessed using a linear mixed-effects model, with significance determined by restricted maximum likelihood ratio using the R packages lme4 and lmerTest [17, 18]. To account for singularity, the model incorporated scaling and a weak Bayesian prior via the R package blme [19]. Multivariate logistic regression to determine the association of Ct value and mortality was performed using the base R function. Visualization was performed using ggplot2 [20].
RESULTS
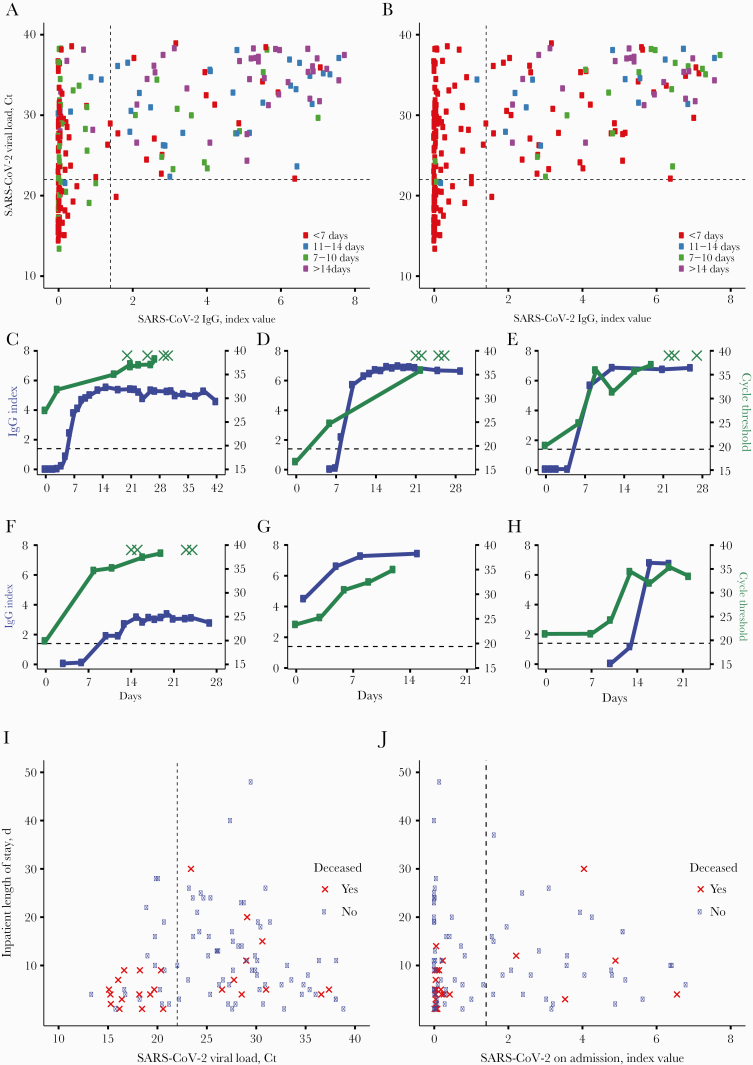
A total of 181 patients had both an Abbott Architect anti-SARS-CoV-2 nucleocapsid IgG index value and a SARS-CoV-2 PCR Ct value available from the same calendar day. Several patients had quantitative PCR and serology data available from multiple days, resulting in a total of 224 total unique patient-days. Comparison of qRT-PCR and serology data revealed only 1 SARS-CoV-2 seropositive individual with a simultaneous SARS-CoV-2 Ct <22 (Figure 1A and B). IgG levels as measured by index values were found to be inversely correlated with SARS-CoV-2 viral load (P < .001). To substantiate this association at the individual patient level, we identified patients with >3 measures of SARS-CoV-2 IgG and viral load. Although the kinetics of SARS-CoV-2 IgG and viral load varied between individual patients, these parameters consistently trended together in individual patients (Figure 1C–H). Lymphocyte counts increased and inflammatory markers decreased over time in these patients (Supplementary Figure 1), concomitant with a decreasing viral load (Figure 1C–H).
Figure 1.
A and B, Unique patient-days with an antinucleocapsid IgG index value and viral load (qRT-PCR Ct value) on the same calendar day. The dashed vertical line indicates the manufacturer’s seropositivity index value cutoff of 1.40. The dashed horizontal line indicates a Ct value of 22. Colors indicate days from symptom onset (A) or days since first positive PCR (B). C–H, Six representative patients with >3 IgG (blue) and Ct (green) results available over the course of their hospital stay. X-axis indicates days from first positive PCR. Dashed horizontal line indicates IgG cutoff of 1.40. Green Xs indicate RT-PCR results with no nucleic acid detected. I, Patients with a Ct value available on the day of admission. Red X data points indicate patients who expired within 30 days of their first positive PCR result. The dashed vertical line indicates the manufacturer’s seropositivity threshold of 1.40. J, Patients with an IgG result available on the day of admission. Red X data points indicate patients who expired within 30 days of first positive PCR result. The dashed vertical line indicates a Ct value of 22. Abbreviations: Ct, cycle threshold; IgG, immunoglobulin G; PCR, polymerase chain reaction; qRT-PCR, quantitative reverse transcription polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction.
To test whether seroconversion or high viral loads were associated with mortality, we examined SARS-CoV-2 viral load (n = 109) and anti-SARS-CoV-2 IgG index values (n = 114) on admission and 30-day all-cause mortality from day of PCR positivity (Figure 1I–J). The viral load on admission was found to be independently associated with mortality, after adjusting for SARS-CoV-2 serostatus, age, and sex (P = .01). Patients with a high viral load on admission (Ct < 22) had a significantly greater odds of mortality (odds ratio [OR], 4.20; 95% CI, 1.62–10.86) compared with patients with lower viral loads (Ct > 22). Thirty-three percent of patients (38/114) seroconverted before admission. Seroconversion on admission trended toward lower mortality, although this relationship was not statistically significant (OR, 0.43; 95% CI, 0.15–1.26) (Figure 1J).
DISCUSSION
Our data provide further support for quantitative viral load assessment, especially on hospital admission. Our results agree with other work that has shown viral load to be associated with disease severity [1, 3, 5, 6, 21]. Currently, the FDA has only authorized reporting of qualitative results from SARS-CoV-2 qRT-PCR tests, despite nearly all tests returning some estimate of viral load. Indeed, our clinical laboratory has reported semiquantitative results for respiratory viruses for more than a decade, and our clinicians are well versed in interpreting semiquantitative Ct values for medical management. Quantitative reporting of SARS-CoV-2 molecular or serologic assays would require significant modifications of existing emergency use authorizations. Our data further suggest that a cycle threshold of 22 may serve as a useful discrete cutoff for significant viral replication that is associated with mortality. We note, however, that sample and swab variability across patient populations may limit the widespread use of a discrete cutoff for quantitative RT-PCR results even if using standard curves to compute copy number/genome equivalents and improve the correlation between assays.
Given our data that indicated a viral load at admission is a significant independent predictor of 30-day mortality, we sought to assess the antinucleocapsid response as a potential biomarker. We demonstrated that detection of anti-SARS-CoV-2 nucleocapsid IgG is associated with lower viral loads in coronavirus disease 2019 (COVID-19) patients. This antibody response also tracked closely with the amount of viral nucleic acid in individual patients over time. Due to the close relationships of both IgG and viral load with days since symptom onset, we could not conclude that the viral load dependence on IgG response was independent from the passage of time, but days since symptom onset was accounted for in our mixed effects model. Individuals who were SARS-CoV-2 antibody positive on admission were less than half as likely to die within 30 days, though this relationship was not statistically significant. The inability to reach statistical significance is likely due to the limited study population size (power = 0.56 for an OR of 0.5). Higher antibody levels have been associated with more significant clinical disease and hospitalization [22, 23]. Nonetheless, our data suggest that admission antibody titers, coupled with molecular testing, may be particularly helpful to assess the disease course for patients who cannot provide a clinical history. This association might only be present at the time of admission, which is when we assessed its prognostic role. While our data do not directly assess the potential for ongoing immunity against future infections of SARS-CoV-2, they indicate that high viral loads almost never coexist with SARS-CoV-2 seropositivity and suggest that persons with anti-SARS-CoV-2 antibodies on admission have a reduced 30-day all-cause mortality.
The main limitation of our study was the retrospective nature in a population enriched for hospitalized patients with acute disease [9, 24]. The retrospective nature precluded analyses of viral clearance and length of stay due to significant confounding factors associated with RT-PCR testing frequency during admission and patient discharge placement. Although we had insufficient sample size to perform separate analyses with patients who only presented to the emergency department or outpatient clinic, results appeared similar to the full data set (Supplementary Figure 2). Our serological test detects IgG antibodies against the nucleocapsid protein of SARS-CoV-2. Whether these antibodies can provide protection themselves or in association with protective responses is clearly unknown. Variability in neutralizing responses between patients not elucidated by our assay may explain some of the variation in our data set. However, neutralizing antibody assays are in vitro methods that may or may not be associated with clinically meaningful outcomes unless performed in concert with challenge studies. In addition, non-neutralizing antibodies may also confer protection against infection in some viral infections [25].
Our work illustrates the importance of quantitative virologic and serological testing for SARS-CoV-2 infection. The association of the presence of anti-SARS-CoV-2 nucleocapsid IgG with lower viral load indicates that antibodies may serve as a biomarker for COVID-19 disease course and infectious risk of the individual to the community.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors would like to thank Nathan Breit of the University of Washington Department of Laboratory Medicine for assistance in obtaining data and Thomas E. Grys of the Mayo Clinic in Arizona for critical review of the manuscript. We also thank the University of Washington Medical Center (UWMC) Northwest Campus clinical laboratory staff and the UWMC Clinical Immunology staff for reserving remnant serum and plasma samples from COVID-19 PCR-positive patients.
Financial support. This work was supported by the Department of Laboratory Medicine at the University of Washington Medical Center.
Potential conflicts of interest. A.L.G. reports personal fees from Abbott Molecular, outside of the submitted work. A.B., S.L.F., M.A.G., G.P., A.C., M.H.W., C.M., K.R.J., and P.C.M. report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 2020; 369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahar B, Jacquot C, Mo YD, DeBiasi RL, Campos J, Delaney M. Kinetics of viral clearance and antibody production across age groups in SARS-CoV-2 infected children. J Pediatr 2020; 227:31–e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Magleby R, Westblade LF, Trzebucki A, et al. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019 [published online ahead of print June 30, 2020]. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rao SN, Manissero D, Steele VR, Pareja J. A narrative systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther 2020; 9:573–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pujadas E, Chaudhry F, McBride R, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med 2020; 8:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prebensen C, Hre PLM, Jonassen C, et al. SARS-CoV-2 RNA in plasma is associated with ICU admission and mortality in patients hospitalized with COVID-19 [published online ahead of print September 05, 2020]. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1338. [DOI] [Google Scholar]
- 7. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 2020; 25:2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullard J, Dust K, Funk D, et al. Predicting infectious SARS-CoV-2 from diagnostic samples [published online ahead of print May 22, 2020]. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 2020; 58:e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng D, Goldgof G, Shy B, et al. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood from the San Francisco Bay Area. medRxiv 2020.05.19.20107482 [Preprint]. 27 May 2020. Available at: 10.1101/2020.05.19.20107482. Accessed 14 September 2020. [DOI] [Google Scholar]
- 11. Patel E, Bloch EM, Clarke W, et al. Comparative performance of five commercially available serologic assays to detect antibodies to SARS-CoV-2 and identify individuals with high neutralizing titers. MedRxiv 2020.08.31.20184788 [Preprint]. 2 September 2020. Available at: 10.1101/2020.08.31.20184788. Accessed 14 September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis 2020; 26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Addetia A, Crawford KHD, Dingens A, et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J Clin Microbiol 2020; 58:e02107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lieberman JA, Pepper G, Naccache SN, Huang M-L, Jerome KR, Greninger AL. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol 2020;58: e00821–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perchetti GA, Nalla AK, Huang ML, et al. Validation of SARS-CoV-2 detection across multiple specimen types. J Clin Virol 2020; 128:104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bates D, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67:1–48. [Google Scholar]
- 18. Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest package: tests in linear mixed effects models. J Stat Softw 2017; 82:1–26. [Google Scholar]
- 19. Chung Y, Rabe-Hesketh S, Dorie V, et al. A nondegenerate penalized likelihood estimator for variance parameters in multilevel models. Psychometrika 2013; 78:685–709. [DOI] [PubMed] [Google Scholar]
- 20. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 21. Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 2020; 130:5235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang P, Liu L, Nair MS, et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect 2020; 9:2091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 24. Corcorran MA, Olin S, Rani G, et al. Prolonged persistence of PCR-detectable virus during an outbreak of SARS-CoV-2 in an inpatient geriatric psychiatry unit in King County, Washington. Am J Infect Control. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ilinykh PA, Huang K, Santos RI, et al. Non-neutralizing antibodies from a Marburg infection survivor mediate protection by Fc-effector functions and by enhancing efficacy of other antibodies. Cell Host Microbe 2020; 27:976–91.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.