Abstract
To better understand the genetic diversity, host associations and evolution of coronaviruses (CoVs) in China we analyzed a total of 696 rodents encompassing 16 different species sampled from Zhejiang and Yunnan provinces. Based on reverse transcriptase PCR-based CoV screening of fecal samples and subsequent sequence analysis of the RNA-dependent RNA polymerase gene, we identified CoVs in diverse rodent species, comprising Apodemus agrarius, Apodemus chevrieri, Apodemus latronum, Bandicota indica, Eothenomys cachinus, Eothenomys miletus, Rattus andamanensis, Rattus norvegicus, and Rattus tanezumi. CoVs were particularly commonplace in A. chevrieri, with a detection rate of 12.44 per cent (24/193). Genetic and phylogenetic analysis revealed the presence of three groups of CoVs carried by a range of rodents that were closely related to the Lucheng Rn rat CoV (LRNV), China Rattus CoV HKU24 (ChRCoV_HKU24), and Longquan Rl rat CoV (LRLV) identified previously. One newly identified A. chevrieri-associated virus closely related to LRNV lacked an NS2 gene. This virus had a similar genetic organization to AcCoV-JC34, recently discovered in the same rodent species in Yunnan, suggesting that it represents a new viral subtype. Notably, additional variants of LRNV were identified that contained putative non-structural (NS)2b genes located downstream of the NS2 gene that were likely derived from the host genome. Recombination events were also identified in the open reading frame (ORF) 1a gene of Lijiang-71. In sum, these data reveal the substantial genetic diversity and genomic complexity of rodent-borne CoVs, and extend our knowledge of these major wildlife virus reservoirs.
Keywords: coronavirus, rodents, host-range, heterologous recombination, genetic diversity
1. Introduction
Coronaviruses (CoVs) (family Coronaviridae, order Nidovirales) are important etiological agents of respiratory, enteric, hepatic, and neurological diseases of varying severity that impact a variety of animal species including humans. The first CoV was isolated in chicken embryos in the 1930s (Hudson and Beaudette 1932). Notably, those human CoVs described before 2002 were associated with mild influenza-like symptoms. However, following the emergence of SARS (Severe Acute Respiratory Syndrome) in 2002/03, MERS (Middle East Respiratory Syndrome) in 2012, and COVID-19 (Coronavirus Disease 2019) in 2019, their potential as human pathogens has gained increasing attention. Importantly, all these diseases are associated with zoonotic CoVs, with a variety of bat species recognized as important wildlife reservoirs. In addition to bats, rodents are also a major zoonotic source of emerging viral diseases, including a number of important infectious diseases of humans (Meerburg, Singleton, and Kijlstra 2009; Zhang et al. 2010). Indeed, rodents are highly diverse and some, including mice and rats, often live in close proximity to humans or domestic animals. This increases the risk of disease emergence following direct or indirect exposure to rodent carcasses, faces, urine, and parasites.
CoVs are currently classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (de Groot 2011; https://talk.ictvonline. org/taxonomy/). SARS-CoV, MERS-CoV, and the agent of COVID-19, SARS-CoV-2, are all members of genus Betacoronavirus, itself divided into the subgenera Embecovirus, Hibecovirus, Merbecovirus, Nobecovirus, and Sarbecovirus. Although rodents are important reservoirs for a range of zoonotic pathogens, prior to 2015 the only known rodent CoV was mouse hepatitis virus (MHV) isolated from mice in 1949 (Cheever et al. 1949). Following the discovery of four other distinct rodent CoVs—the alphacoronavirus (alpha-CoV) Lucheng Rn rat CoV (LRNV) and the betacoronaviruses (beta-CoVs; subgenus Embecovirus) ChRCoV_HKU24, Myodes CoV 2JL14 (MrufCoV_2JL14), and Longquan Rl rat CoV (LRLV)— an increasing number of rodent-associated CoVs have been identified in different countries and in a range of rodent species (Lau et al. 2015; Wang et al. 2015; Tsoleridis et al. 2016; Ge et al. 2017; Wu et al. 2018). Hence, rodents are important reservoirs for members of the subgenus Embecovirus (Betacoronavirus) and have likely played a key role in CoV evolution and emergence.
Zhejiang province is located in the southern part of the Yangtze River Delta on the southeast coast of China, from where rodent CoVs have previously been reported (Wang et al. 2015; Lin et al. 2017). Yunnan province is located in southern China, bordering the countries of Myanmar, Laos, and Vietnam, and is often caused the ‘the Kingdom of Wildlife’. A previous study from Yunnan identified a novel SARS-like CoV, Rs-beta-CoV/Yunnan2013, whose ORF8 was nearly identical to ORF8 of SARS-CoVs (98% nt sequence identity; Wu et al. 2016). Recently, two CoVs closely related to SARS-CoV-2 were identified in Rhinolophus sp. (i.e. horseshoe) bats sampled from Yunnan province: RaTG13 (Zhou et al. 2020b) and RmYN02 (Zhou et al. 2020a). However, few rodent CoVs has been documented in Yunnan to date. To explore the diversity and characterization of CoVs in rodents, we performed a molecular evolutionary investigation of CoVs in Zhejiang and Yunnan provinces, China. Our results revealed a remarkable diversity of CoVs in rodents.
2. Materials and methods
2.1 Sample collection
This study was reviewed and approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention of the Chinese CDC. All animals were kept alive after capture and treated in strictly according to the guidelines for the Laboratory Animal Use and Care from the Chinese CDC and the Rules for the Implementation of Laboratory Animal Medicine (1998) from the Ministry of Health, China, under the protocols approved by the National Institute for Communicable Disease Control and Prevention. All dissection was performed under ether anesthesia, and every effort was made to minimize suffering.
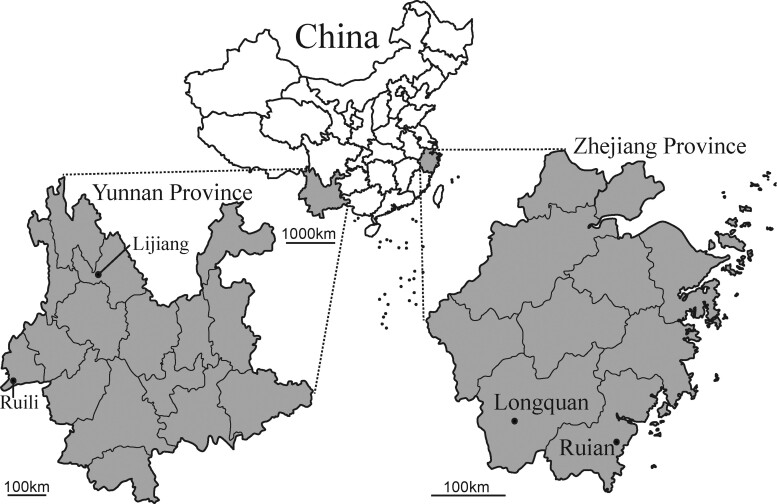
All rodents were collected in 2014 and 2015 from Lijiang and Ruili cities in Yunnan province, and Longquan and Wenzhou cities in Zhejiang province, China. Sampling occurred in cages using fried food as bait set in the evening and checked the following morning. Animals were initially identified by trained field biologists, and further confirmed by sequence analysis of the mitochondrial (mt)-cyt b gene (Guo et al. 2013). Lung samples were collected from animals for the classification of rodent species and alimentary tract samples were collected from animals for the detection of CoVs, respectively.
2.2 DNA and RNA extraction
Total DNA was extracted by using the Cell & Tissue Genomic DNA Extraction Kit (Bioteke Corporation, Beijing, China) from lung samples of rodents according to the manufacturer’s protocol. Total RNA was extracted from fecal samples using the fecal total RNA extraction kit (Bioteke Corporation, Beijing, China) according to the manufacturer’s protocol. The RNA was eluted in 50 μl RNase-free water and was used as a template for further detection.
2.3 CoV detection and complete genome sequencing
The mt-cyt b gene (1,140 bp) was amplified by polymerase chain reaction (PCR) using universal primers for rodents as described previously (Guo et al. 2013). CoV screening was performed using a previously published primer set by a pan-CoV-nested PCR targeted to a conserved region of the RNA-dependent RNA polymerase (RdRp) gene (Wang et al. 2015). First-round reverse transcription PCR (RT-PCR) was conducted by using PrimeScript One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China). A 10 μl reaction mixture contained 5 μl of 2 × 1 Step Buffer, 0.4 μl PrimeScript 1 Step Enzyme Mix, 0.3 μl (10 μmol/l) forward primer, 0.3 μl (10 μmol/l) reverse primer, 3.5 μl RNase Free dH2O, and 0.5 μl of sample RNA. The PCR cycler conditions for the amplification were 50°C for 30 min (RT) then 95°C for 3 min, 35 cycle of 94°C for 45 s (denaturation), 44°C for 45 s (annealing), 72°C for 45 s (extension), then 72°C for 10 min (final extension). The PCR product was then put through a second round PCR which amplify a final PCR product of ∼450 bp.
To recover complete viral genomes, RNA was amplified using several sets of degenerate primers designed by multiple sequence alignments of published CoV genomes. Additional primers were designed according to results of the first and subsequent rounds of sequencing. The 5′ and 3′ end of the viral genome was amplified by rapid amplification of cDNA ends by using the 5′ and 3′ Smarter RACE kit (TaKaRa, Dalian, China).
RT-PCR products of expected size were subject to Sanger sequencing performed by the Sangon corporation (Beijing, China). Amplicons of more than 700 bp were sequenced in both directions. Sequences were assembled by SeqMan and manually edited to produce the final sequences of the viral genomes. Nucleotide (nt) sequence similarities and deduced amino acid (aa) similarities to NCBI/GenBank database sequences were determined using BLASTn and BLASTp.
2.4 Phylogenetic analysis
CoV reference sequences sets representing the RdRp, S and N genes were downloaded from GenBank. Both partial RdRp gene sequences and complete aa sequences of the RdRp, S, and N genes were used to infer phylogenetic trees. All viral sequences were aligned using the L-INS-i algorithm within the MAFFT program (Katoh and Standley 2013). After alignment, gaps and ambiguously aligned regions were removed using Gblocks (v0.91b) with a minimum block length of 10 and no gap positions (Talavera and Castresana 2007). The best-fit model of nt substitution was determined using jModelTest version 0.1 (Posada 2008). Phylogenetic trees were generated using the maximum likelihood (ML) method implemented in PhyML v3.0 (Guindon et al. 2010).
2.5 Genome recombination analysis
Potential recombination events in the history of the LRNV, LRLV, and ChRCoV_HKU24 were assessed using both the RDP4 (Martin et al. 2010) and Simplot (v.3.5.1) programs. The RDP4 analysis was conducted based on an analysis of complete genome sequences, using the RDP, GENECONV, BootScan, maximum chi square, Chimera, SISCAN, and 3SEQ methods. Putative recombination events were identified with a Bonferroni corrected P-value cut-off of 0.01. Similarity plots were inferred using Simplot to further characterize potential recombination events, including the location of possible breakpoints.
3. Results
3.1 Collection of rodents, and the detection of CoV RNA
During 2014 and 2015 a total of 696 rodents from 16 different species were captured in residential areas, farmland and woodland regions from Lijiang city, Ruili city, Yunnan province, and Longquan city, Wenzhou city, Zhejiang province (Fig. 1 and Table 1). RT-PCR was performed to detect CoVs RNA based on partial RdRp sequences. PCR products of the expected size were amplified from one Apodemus agrarius collected from Longquan and two Rattus norvegicus sampled from Wenzhou; twenty-four Apodemus chevrieri, three Apodemus latronum, three Eothenomys miletus from Lijiang; two Bandicota indica, one Eothenomys cachinus, one Rattus andamanensis, and two Rattus tanezumi from Ruili. Overall, 5.6 per cent of rodents were CoV positive. All these sequences exhibited close sequence similarity to published CoVs. Specifically, two CoVs sampled from B. indica and one from R. andamanensis in Ruili shared 92.9–96.0 per cent nt sequence similarity with LRLV; nineteen CoVs from one A. agrarius in Longquan, one E. cachinus and two R. tanezumi in Ruili, as well as one E. miletus, two A. latronum, and twelve A. chevrieri in Lijiang had 93.2–98.4 percent nt sequence similarity to Longquan-343 and ChRCoV HKU24; seventeen CoVs from two R. norvegicus in Wenzhou, one A. latronum, two E. miletus, and twelve A. chevrieri in Lijiang had 83.3–98.9 per cent nt sequence similarity to LRNV.
Figure 1.
Geographic map of Yunnan and Zhejiang provinces, China, showing the location of sampling sites from where the rodents were captured.
Table 1.
Prevalence of rodent CoVs in Yunnan and Zhejiang provinces, China.
| Species | Yunnan |
Zhejiang |
Total (%) | ||
|---|---|---|---|---|---|
| Lijiang | Ruili | Longquan | Wenzhou | ||
| A. agrarius | − | − | 1/44 | 0/5 | 1/49 |
| A. chevrieri | 24/193 | − | − | − | 24/193 |
| A. latronum | 3/6 | − | − | − | 3/6 |
| B. indica | − | 2/5 | − | − | 2/5 |
| E. cachinus | − | 1/1 | − | − | 1/1 |
| E. miletus | 3/119 | 0/12 | − | − | 3/131 |
| Microtus fortis | − | − | 0/10 | − | 0/10 |
| Mus musculus | − | − | − | 0/1 | 0/1 |
| Niviventer eha | − | 0/2 | − | − | 0/2 |
| N. niviventer | − | 0/1 | 0/1 | − | 0/2 |
| R. nitidus | − | 0/2 | − | − | 0/2 |
| R. andamanensis | − | 1/1 | − | − | 1/1 |
| R. norvegicus | 0/3 | − | − | 2/98 | 2/101 |
| R. losea | − | − | 0/3 | 0/12 | 0/15 |
| R. rattus sladeni | − | 0/18 | − | − | 0/18 |
| R. tanezumi | 0/2 | 2/106 | 0/25 | 0/26 | 2/159 |
| Total (%) | 30/323 (9.3) | 6/148 (4.1) | 1/83 (1.2) | 2/142 (1.4) | 39/696 (5.6) |
CoV RNA positive specimens/total specimens; ‘–’ no animals were captured.
3.2 Host range of rodent-associated CoVs
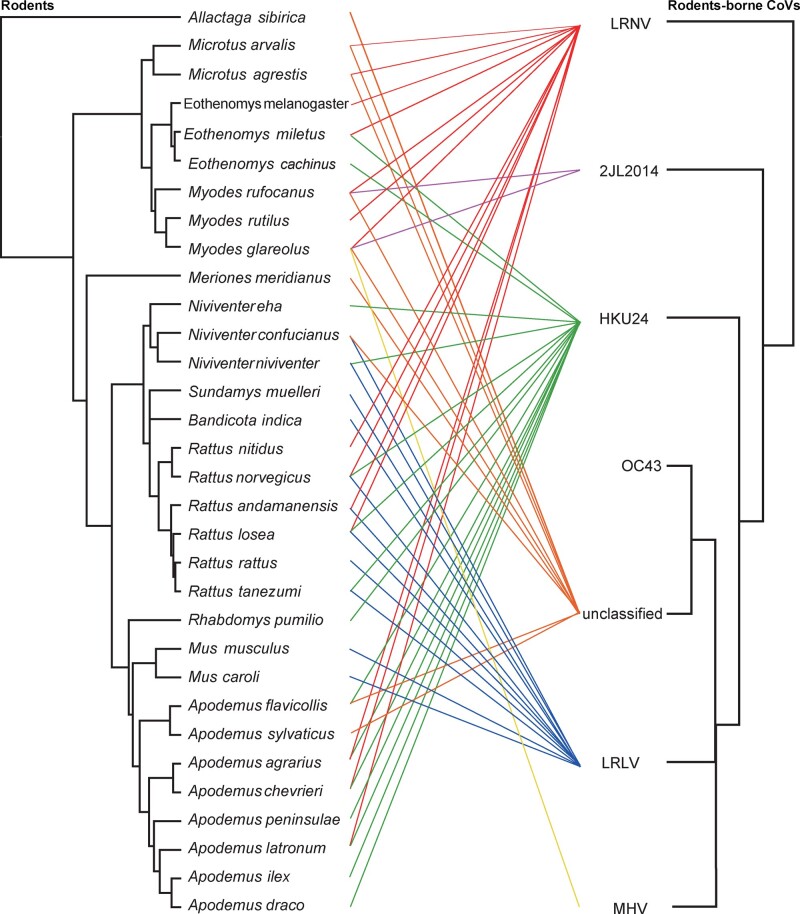
To better understand the relationship between viruses, their hosts and their geographic distributions, we performed a phylogenetic analysis of partial RdRp (381 bp) (Fig. 2A). In total, seventeen virus samples were identified as members of the genus Alphacoronavirus, whereas twenty-two belonged to the genus Betacoronavirus. Our phylogenetic analysis revealed three different clades of rodent-borne CoVs: 1, the first clade fell within the genus Alphacoronavirus and contained a variety of viruses including LRNV; 2, the second clade contained members of the subgenus Embecovirus (genus Betacoronavirus) including LRLV; 3, the third clade also fell withing the subgenus Embecovirus and contained ChRCoV_HKU24. Notably, all three clades contained viruses closely related to CoVs previously identified in rodents from Zhejiang province, China (Wang et al., 2015).
Figure 2.
(A) ML phylogenetic analysis of a 381-nt sequence of the RdRp gene. Different symbols are used to indicate the country from where the viruses were identified. Hosts for each of viruses are marked using abbreviations of the rodent names. The details of viruses and their hosts are provided in Supplementary Table S1. (B) Color block diagram of the diversity of CoVs carried by different rodent species.
The data generated here, along with that published previously, indicate that a total of thirty-seven different species of rodents from nine different countries are currently known to harbor CoVs (Fig. 2). Notably, every virus clade contained different rodent species, sometimes even from different subfamilies, such that there was no rigid host restriction in rodent CoVs (Fig. 3). Indeed, LRNV has been identified in fifteen different species in rodents, comprising A. agrarius, A. chevrieri, A. latronum, Eothenomys fidelis, E. miletus, Eothenomys melanogaster, Microtus agrestis, Microtus arvalis, Microtus glareolus, Microtus rufocanus, Microtus rutilus, R. andamanensis, Rattus lossea, Rattus nitidus, and R. norvegicus (Wang et al. 2015; Tsoleridis et al. 2016; Ge et al. 2017; Wu et al. 2018). In addition, multiple CoVs can be carried by the same rodent species. For example, two Rattus species—R. losea and R. norvegicus—were found to carry three species of rodent-borne CoV (Wang et al. 2015; Wu et al. 2018). This is in contrast to previous studies in which individual CoVs were associated with a single species or genera, including Carollia, Eptesicus, Miniopterus, Scotophilus, and Rhinolipus bats (Anthony et al.2013; Drexler et al.2010; Wacharapluesadee et al. 2015; Fischer et al.2016).
Figure 3.
Tanglegram depicting the evolutionary associations between rodent-associated CoVs and their hosts. The virus tree was estimated using the RdRp gene (right) and the host tree (left) was based on topology implied in the Time tree of life (http://www.timetree.org/).
3.3 Characterization of viral genomes
To better characterize the CoVs found in this study, complete or nearly complete genome sequences data for the three variants of LRNV (Lijiang-71, Lijiang-170, and Wenzhou-83) and four variants of ChRCoV_HKU24 (Lijiang-41, Lijiang-53, Ruili-874, and Longquan-723) as well as the single variant of LRLV (Ruili-66) were obtained by assembly of the sequences of the RT-PCR products from the RNA directly extracted from the individual specimens.
The three genomes of LRNV shared 77.5–92.4 per cent nt sequence similarity with each other. The genome sizes of Wenzhou-83, Lijiang-71, and Lijiang-170 were 28,599, 29,147, and 27,563 nt, respectively, with the G + C contents of 40.29, 39.35, and 40.21 per cent. The genomes of Wenzhou-83, Lijiang-71, Lijiang-170 had 97.7, 92.3, and 77.6 per cent overall nt identity with LRNV Lucheng-19, respectively. The genome organization was similar to that of other LRNVs and had the characteristic gene order 5′-replicase ORF1ab, spike (S), envelope (E), membrane (M), nucleocapsid (N)-3′. Strikingly, however, a major difference among these LRNVs is the additional ORF(s) encoding non-structural (NS) proteins and located between ORF1ab and S gene (Fig. 4). According to the presence and quantity of NS proteins, the virus can be divided into three genomic variants: 1, the first variant comprised Lijiang-170 and AcCoV-JC34 in which no NS protein was observed; 2, the second variant contains Ruian-83, Lucheng-19, and RtRl-CoV/FJ2015 for which there is a putative NS2 gene between their ORF1ab and S gene. The putative NS2 gene lengths for Ruian-83, Lucheng-19, and RtRl-CoV/FJ2015 were 825, 828, and 825 nt, respectively, with 93.5–94.8 per cent sequence identity; 3, the third variant comprises Lijiang-71, RtClan-CoV/GZ2015 and RtMruf-CoV-1/JL2014 and contains two putative NS proteins—NS2 and NS2b—located between the ORF1ab and S gene. The putative NS2 gene of the third variant is 828 nt in length, with 82.9–88.9 per cent sequence identity. Similarly, the putative NS2b has a gene length of 462 nt and exhibits 77.5–96.3 per cent sequence identity among these three viruses. Strikingly, a BLASTp search reveals that the NS2b encodes a putative NS protein of 153 aa residues in length that has no aa sequence similarity to other CoVs; rather, this sequence exhibits ∼43 per cent aa identity to the C-type lectin-like protein within the rodent Microtus ochrogaster genome. Hence, this pattern suggests that the NS2b gene may have originally been acquired from a rodent host genome during evolutionary history. Moreover, the aa sequence identity between Lijiang-170, Lijiang-71, Ruian-83, and LRNV was >90 per cent in RdRp, E, and M genes (as expected from members of the same species), but only 70–88 per cent in ADRP, 3CLpro, ORF1ab, and S (Table 2). Further analysis of the characteristics of Lijiang-170, for which a complete genome sequence is available, shows that it has similar transcription regulatory sequence (TRS) to AcCoV-JC34 (Table 3). Hence, Lijiang-170 and AcCoV-JC34 may represent a novel subtype of LRNV that exhibits marked differences to the prototype strain Lucheng-19.
Figure 4.
Comparison of genome organizations of the different CoVs identified here. All genomes are drawn to scale.
Table 2.
Percent aa sequence identity between Lijiang-170 and known alpha-CoVs.
| ADRP | 3CLpro | RdRp | Hel | ORF1ab | S | E | M | N | |
|---|---|---|---|---|---|---|---|---|---|
| Lijiang-170 | |||||||||
| Lijiang-71 | 74.7 | 88.3 | 94.4 | 95.8 | 85.0 | 69.1 | 93.6 | 92.7 | 80.5 |
| Ruian-83 | 71.3 | 88.3 | 94.6 | 96.5 | 85.0 | 69.5 | 93.6 | 92.3 | 78.5 |
| LRNV Lucheng-19 | 72.4 | 88.0 | 94.7 | 96.3 | 85.1 | 71.0 | 93.6 | 92.3 | 78.1 |
| Rh-BatCoV HKU2 | 44.8 | 59.0 | 74.3 | 72.9 | 53.4 | 41.3 | 36.0 | 56.6 | 30.3 |
| HCoV-229E | 42.5 | 54.7 | 73.0 | 74.2 | 51.6 | 21.0 | 37.3 | 50.0 | 29.2 |
| HCoV-NL63 | 43.7 | 55.7 | 73.3 | 72.7 | 51.7 | 21.4 | 36.0 | 53.8 | 30.3 |
| Mi-BatCoV 1A | 46.0 | 55.0 | 76.2 | 73.5 | 51.3 | 22.1 | 36.5 | 52.3 | 28.5 |
| Ro-BatCoV HKU10 | 43.7 | 55.7 | 74.8 | 73.5 | 52.1 | 23.4 | 32.0 | 57.5 | 31.3 |
| PEDV | 42.5 | 56.0 | 74.1 | 71.7 | 52.6 | 23.0 | 35.1 | 56.9 | 31.0 |
| Sc-BatCoV 512 | 47.1 | 55.3 | 73.0 | 71.7 | 52.3 | 23.4 | 36.5 | 56.6 | 31.0 |
| TGEV | 36.8 | 56.6 | 76.2 | 72.0 | 52.1 | 22.0 | 38.5 | 56.9 | 28.7 |
| MCoV | 34.5 | 59.3 | 77.4 | 72.5 | 52.1 | 22.1 | 42.3 | 52.0 | 28.5 |
| FCoV | 37.9 | 57.7 | 75.1 | 71.5 | 51.5 | 21.5 | 43.6 | 56.1 | 28.7 |
| CCoV | 35.6 | 58.7 | 76.1 | 72.0 | 51.9 | 22.5 | 42.3 | 57.3 | 30.3 |
Table 3.
Locations of predicted ORFs in the genome of Lijiang-170.
| ORF | Location (nt) | Length (nt/aa) | TRS location | TRS sequence(s) (distance to AUG) |
|---|---|---|---|---|
| ORF1ab | 334–20,270 (shift at 12,225) | 19,937/6,646 | 78-83 | AACUAA(250) AUG |
| S | 20,270–23,650 | 3,381/1,127 | 20,200–20,205 | AACUUA(64) AUG |
| NS3 | 23,647–24,291 | 645/215 | 23,607–23,612 | CUAAAC(34) AUG |
| E | 24,291–24,527 | 237/79 | 24,286–24,291 | AACUAA UG |
| M | 24,537–25,283 | 747/249 | 24,530–24,535 | AACUAAAAUG |
| NS6 | 25,295–25,795 | 501/167 | 25,283–25,288 | AACUAA(6) AUG |
| N | 25,797–26,966 | 1,170/390 | 25,790–25,795 | AACUAAAAUG |
| NS8 | 26,968–27,285 | 318/106 | 26,954–26,959 | AACUAA(8) AUG |
Start codons are underlined. The conserved (AACUAA) or variant (AACUUA and CUAAAC) TRS core sequences are highlighted in bold.
In contrast, Lijiang-53, Lijiang-41, Ruili-874, and Longquan-723 were most closely related to ChRCoV_HKU24, exhibiting 94.0–96.1 per cent nt sequence similarity. Strikingly, the length of nsp3 in Lijiang-41 differed from those of ChRCoV_HKU24 as a result of a 75 nt deletion. Ruili-66 was most closed to LRLV and shared 92.7 per cent nt sequence similarity with Longquan-370 and Longquan-189 of LRLV found in Longquan.
3.4 Phylogenetic analysis of viral sequences
To better understand the evolutionary relationships among the CoVs described here and those identified previously, we estimated phylogenetic trees based on the aa sequences of the RdRp, S, and N proteins. The analysis of all three proteins from the LRNV clade again suggests that LRNV can be divided into two phylogenetic Subtypes (I and II); indicated on Fig. 5. Indeed, there was a clear division phylogenetic between the Subtypes I and II LRNV sequences in the RdRp, S, and N aa trees, and while intra-Subtype (I or II) sequences shared high nt sequence identities (92.4–97.7%), inter-subtype sequence identity was only ∼77.5 per cent. Notably, this phylogenetic analysis also suggested that clade LRNV had a recombinant evolutionary history: while the LRNV clade formed a distinct lineage in the RdRp and N gene trees (although with little phylogenetic resolution in the latter), it clustered with Rhinolophus sinicus CoV HKU2, BtRf-alpha-CoV/YN2012, and Sm-CoV X74 in the S gene tree. In contrast, the Clades 2 and 3 rodent CoVs were consistently closely related to LRLV and ChRCoV_HKU24 in the RdRp, S, and N aa trees.
Figure 5.
ML phylogenetic analysis of the RdRp, S, and N proteins of Lijiang-41, Lijiang-53, Lijiang-71, Lijiang-170, Wenzhou-83, Longquan-723, Ruili-66, and Ruili-874. Numbers above individual branches indicate the percentage bootstrap support (1,000 replicates). For clarity, bootstrap support values are shown for key internal nodes only. The scale bar represents the number of aa substitutions per site. The trees were rooted between the alpha- and beta-CoVs.
3.5 Recombination
Multiple methods within the RDP program (Martin et al. 2010) identified statistically significant recombination events in Lijiang-71 (P < 3.05 × 10−23 to P < 7.11 × 10−13; Fig. 6). When Lijiang-71 was used as the query for sliding window analysis with RtClan-CoV/GZ2015 and Lucheng-19 as potential parental sequences, four recombination breakpoints at nt positions 8,188, 8,636, 9,030, and 11,251 in the sequence alignment were identified. This pattern of recombination events is further supported by phylogenetic and similarity plot analyses (Fig. 6). Specifically, in the major parental region (1–8,187, 8,637–9,029, and 11,252–29,349), Lijiang-71was most closely related to RtClan-CoV/GZ2015, whereas in the minor parental region (8,188–8,636 and 9,030–11,251) it was more closely related to Lucheng-19.
Figure 6.
Putative recombination events within the genome of Lijiang-71. (A) The sequence similarity plot reveals four putative recombination breakpoints (black-dashed lines), with their locations indicated at the bottom. The plot shows genome scale similarity comparisons of the Lijiang-71 (query) against RtClan-CoV/GZ2015 (major parent, red) and Lucheng-19 (minor parent, green). The background color of major parental region is white, whereas that of minor parental region is gray. (B) Phylogenies of the major parental region (Positions 1–8,187, 8,637–9,029 and 11,252–29,349) and minor parental region (Positions 8,188–8,636 and 9,030–11,251). Phylogenies were estimated using a ML method and were mid-point rooted for clarity only. Numbers above or below the branches indicate percentage bootstrap values. The scale bar represents the number of substitutions per site.
4. Discussion
We screened CoVs in 696 rodents from 16 different species sampled at four sites in Zhejiang and Yunnan provinces, China. Overall positivity rates were ∼6 per cent, although they ranged from 9.3 per cent in Lijiang city to only 1.2 per cent in Zhejiang province. The latter is lower than the CoV detection rates described in a previous study undertaken in Zhejiang province despite the use of similar methodologies (Wang et al., 2015). We found that A. chevrieri had a relatively high CoV detection rate (24/193, 12.44%) in Lijiang city, Yunnan province, consistent with a previous study showing that A. chevrieri had a high detection rate of CoV (21/98, 21.4%) in Jianchuan county, also in Yunnan province (Ge et al. 2017). As A. chevrieri is a major species in Lijiang city, such a high CoV infection rate highlights the need for ongoing surveillance.
Our analysis of rodent-borne CoVs revealed that all currently recognized viruses fall into six groups—LRNV, LRLV, ChRCoV_HKU24, Myodes CoV 2JL14, HCoV_OC43, MHV, and unclassified members of the genus Embecovirus. Phylogenetic analysis based on the partial RdRp gene reveals that CoVs from different countries (China, UK, Germany, Malaysia, Mexico, The Netherlands, Poland, South Africa, and Thailand) group together with no obvious geographic pattern. Indeed, at least thirty-seven different rodent species are known to carry CoVs, such that they have an extensive host range in these animals, and with frequent cross-species transmission. A growing number of rodent species are found to carry members of the genus Embecovirus, indicating that rodents indeed played an important role for the spread and evolution of embecoviruses, and are the likely reservoir hosts for the human CoV HKU1 (Woo et al. 2005).
Our analysis also provided evidence for multiple variants of rodent-borne CoVs in the genus Alphacoronavirus that differ in the genome organization. For example, although Lijiang-170 had greatest sequence similarity to LRNV variant Lucheng-19, it does not contain a NS2 between ORF1b and S gene as observed in most other alpha-CoVs. More striking was that three viruses (Lijiang-71, RtClan-CoV/GZ2015, and RtMruf-CoV-1/JL2014) had a putative NS protein of 153 aa located downstream of NS2 that likely resulted from a past horizontal gene transfer event involving the rodent host genome. Although the virus-to-host horizontal transfer of genetic material (i.e. endogenization) is commonplace in some eukaryotes, horizontal transfer from host-to-virus is less well documented. However, a number of cases have been documented, with a review on the subject identifying fourteen different gene fragments from seven hosts integrated into ten different viral genomes (Gilbert and Cordaux 2017). In addition, inter-virus recombination events were identified in Lijiang-71. Such mechanisms of genetic transfer may ultimately lead to the creation of novel viruses, perhaps with variable phenotypic properties (Su et al. 2016).
In conclusion, our study revealed a high diversity of CoVs circulating in rodents from Yunnan and Zhejiang provinces, China, including the discovery of a putative novel viral subtype and new rodent host species. Undoubtedly, the larger scale surveillance and analyses of CoV infections in rodents is required to better understand their genetic diversity, cellular receptors, inter-host transmission, and evolutionary history.
Data Availability
The eight complete or near complete CoVs genome sequences, the short RdRp sequences, and the CoV RNA positive rodent mt-cyt b gene sequences (the mt-cyt b of Lijiang-2 was unavailable) generated in this study have been deposited in GenBank under the accession numbers MT820625–MT820632, MW011442–MW011472, and MW023762–MW023799.
Funding
This study was supported by the 12th Five-Year Major National Science and Technology Projects of China (2014ZX10004001-005), the National Science and Technology Major Project of China (2018ZX10305409-003-005), and the National Natural Science Foundation of China (Grants 32041004, 31930001, 81672057, and 81861138003). Edward C. Holmes was supported by the Australian Research Council (grant FL170100022).
Conflict of interest: None declared.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Contributor Information
Wen Wang, Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China.
Xian-Dan Lin, Wenzhou Center for Disease Control and Prevention, Wenzhou, Zhejiang 325001, China.
Hai-Lin Zhang, Yunnan Institute of Endemic Diseases Control and Prevention, Dali 671000, China.
Miao-Ruo Wang, Longquan Center for Disease Control and Prevention, Zhejiang Province, Longquan 323799, China.
Xiao-Qing Guan, Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China.
Edward C Holmes, Shanghai Public Health Clinical Center & School of Life Science, Fudan University, Shanghai 201052, China; Marie Bashir Institute for Infectious Diseases and Biosecurity, School of Life and Environmental Sciences and School of Medical Sciences, The University of Sydney, Sydney, NSW 2006, Australia.
Yong-Zhen Zhang, Shanghai Public Health Clinical Center & School of Life Science, Fudan University, Shanghai 201052, China.
References
- Anthony S. J. et al. (2013) ‘Coronaviruses in Bats from Mexico’, Journal of General Virology, 94: 1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever F. S. et al. (1949) ‘A Murine Virus (JHM) Causing Disseminated Encephalomyelitis with Extensive Destruction of Myelin’, Journal of Experimental Medicine, 90: 181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. J. et al. (2011), ‘Family Coronaviridae’, in King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B. (eds), Ninth Report of the International Committee on Taxonomy of Viruses, pp. 806–828. San Diego, USA: Elsevier. [Google Scholar]
- Drexler J. F. et al. (2010) ‘Genomic Characterization of Severe Acute Respiratory Syndrome-Related Coronavirus in European Bats and Classification of Coronaviruses Based on Partial RNA-Dependent RNA Polymerase Gene Sequences’, Journal of Virology, 84: 11336–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K. et al. (2016) ‘Insectivorous Bats Carry Host Specific Astroviruses and Coronaviruses across Different Regions in Germany’, Infection Genetics and Evolutino, 37: 108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. Y. et al. (2017) ‘Detection of Alpha- and Betacoronaviruses in Rodents from Yunnan, China’, Virology Journal, 14: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Cordaux R. (2017) ‘Viruses as Vectors of Horizontal Transfer of Genetic Material in Eukaryotes’, Current Opinion in Virology, 25: 16–22. [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Guo W. P. et al. (2013) ‘Phylogeny and Origins of Hantaviruses Harbored by Bats, Insectivores, and Rodents’, PLoS Pathogens, 9: e1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C. B., Beaudette F. R. (1932) ‘Infection of the Cloaca with the Virus of Infectious Bronchitis’, Science, 76: 34. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. et al. (2015) ‘Discovery of a Novel Coronavirus, China Rattus Coronavirus HKU24, from Norway Rats Supports the Murine Origin of Betacoronavirus 1 and Has Implications for the Ancestor of Betacoronavirus Lineage A’, Journal of Virology, 89: 3076–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.-D et al. (2017) ‘Extensive Diversity of Coronaviruses in Bats from China’, Virology, 507: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P. et al. (2010) ‘RDP3: A Flexible and Fast Computer Program for Analyzing Recombination’, Bioinformatics, 26: 2462–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg B. G., Singleton G. R., Kijlstra A. (2009) ‘Rodent-Borne Diseases and Their Risks for Public Health’, Critical Reviews in Microbiology, 35: 221–70. [DOI] [PubMed] [Google Scholar]
- Posada D. (2008) ‘jModelTest: Phylogenetic Model Averaging’, Molecular Biology and Evolution, 25: 1253–6. [DOI] [PubMed] [Google Scholar]
- Su S. et al. (2016) ‘Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses’, Trends in Microbiology, 24: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007) ‘Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments’, Systematic Biology, 56: 564–77. [DOI] [PubMed] [Google Scholar]
- Tsoleridis T. et al. (2016) ‘Discovery of Novel Alphacoronaviruses in European Rodents and Shrews’, Viruses, 8: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S. et al. (2015) ‘Diversity of Coronavirus in Bats from Eastern Thailand’, Virology Journal, 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (2015) ‘Discovery, Diversity and Evolution of Novel Coronaviruses Sampled from Rodents in China’, Virology, 474: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. (2005) ‘Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia’, Journal of Virology, 79: 884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. (2016) ‘ORF8-Related Genetic Evidence for Chinese Horseshoe Bats as the Source of Human Severe Acute Respiratory Syndrome Coronavirus’, Journal of Infectious Diseases, 213: 579–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. (2018) ‘Comparative Analysis of Rodent and Small Mammal Viromes to Better Understand the Wildlife Origin of Emerging Infectious Diseases’, Microbiome, 6: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z. et al. (2010) ‘Hantavirus Infections in Humans and Animals, China’, Emerging Infectious Diseases, 16: 1195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. (2020) ‘A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein’, Current Biology, 30: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. et al. (2020) ‘A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin’, Nature, 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The eight complete or near complete CoVs genome sequences, the short RdRp sequences, and the CoV RNA positive rodent mt-cyt b gene sequences (the mt-cyt b of Lijiang-2 was unavailable) generated in this study have been deposited in GenBank under the accession numbers MT820625–MT820632, MW011442–MW011472, and MW023762–MW023799.