Abstract
Nuclear factor erythroid 2-related factor 2 (NRF2) functions as a transcription factor and regulates a wide array of antioxidant and stress-responsive genes. NRF2 has been widely implicated in different types of cancers, but only limited studies concerning the relationship between NRF2 expression and tumour invasion or prognosis in lung cancer. Therefore, we conducted a meta-analysis to determine the prognostic value of NRF2 in patients with non-small cell lung cancer (NSCLC). The relationship between NRF2 expression in NSCLC patients and clinicopathological features was also investigated. Overall survival (OS) and treatment response rate were evaluated using STATA software. Twenty eligible articles with 2530 lung cancer patients were included in this meta-analysis. The results revealed that high expression level of NRF2 was associated with pathologic distant metastasis (odds ratio (OR) = 2.64, 95% confidence interval (CI) 1.62–4.31; P < 0.001), lymph node metastasis (OR = 2.14, 95% CI: 1.53–3.00; P < 0.001), and tumour node metastasis (TNM) stage (OR = 1.95, 95% CI: 1.52–2.49, P < 0.001). High NRF2 expression was associated with low treatment response rate in platinum-based chemotherapy (HR = 0.11, 95% CI 0.02–0.51; P = 0.005). High expression level of NRF2 is predictive for poor overall survival rate (HR = 1.86, 95% CI 1.44–2.41, P < 0.001) and poor progression-free survival (PFS) (HR = 2.27, 95% CI 1.26–4.09, P = 0.006). Compared to patients with a low level of NRF2 expression, patients with high NRF2 expression levels were associated with worse OS and PFS when given the chemotherapy or EGFR-TKI. Together, our meta-analysis results suggest that NRF2 can act as a potential indicator of NSCLC tumour aggressiveness and help the prognosis and design of a better treatment strategy for NSCLC patients.
Introduction
Nuclear factor erythroid 2 like 2 (NRF2), also known as nuclear factor erythroid 2-related factor 2 (NFE2L2), is a transcription factor encoded by the NRF2 gene in humans [1]. NRF2 regulates the transcription of a wide array of genes, including those coding for antioxidant proteins involved in the detoxification of xenobiotics and resistance to oxidative stress [2]. For example, both heme oxygenase-1 (HO-1) and NAD(P)H quinone oxidoreductase 1 (NQO1) are regulated by NRF2. The cytoplasmic NRF2 protein is maintained at a very low level through its selective negative regulator, Kelch-like ECH-associated protein 1 (KEAP1). KEAP1 can sequester NRF2 in the cytoplasm and lead to ubiquitination of CUL3 E3 ligase and subsequent degradation by the proteasome [3,4]. Under oxidative stress or in the presence of NRF2-activated compounds, E3 activity is downregulated and NRF2 is stabilized, thereby increasing the amount of NRF2 protein relative to KEAP1 [5,6]. The free NRF2 translocates to the nucleus, then activates the expression of its downstream antioxidant genes [7,8].
NRF2 signalling is crucial for the initiation and progression of lung cancer, as shown by gene knockout mouse model and clinical studies [9]. An enhanced NRF2 signal activity appears to be correlated with a worse treatment outcome according to the clinical observations [10]. In cancer cells, NRF2 signalling can be activated by endogenous or exogenous stress, accompanied by activation of various cytoprotective genes [11]. Furthermore, crosstalk has been reported between NRF2 and oncogenic signaling pathways such as phosphatidylinositol 3-kinase (PI3K) [12], Kirsten retrovirus-associated DNA sequence (K-RAS) [9], and Notch [13].
Many studies have evaluated whether the positive expression of NRF2 may be a prognostic factor for survival rate among patients with lung cancer. However, the clinical evidence for the relationship between NRF2 expression and tumour invasiveness, prognosis and treatment response rate in NSCLC is not well understood. In this study, a meta-analysis of published data was performed to systematically investigate whether NRF2 expression can be an applicable marker to assist with the prognosis of patients with NSCLC.
Materials and methods
Literature search strategy
This meta-analysis was conducted in accordance with the PRISMA guidelines [14]. The Chinese databases of China National Knowledge Infrastructure (CNKI) as well as English databases of Pubmed, Embase, EBSCO and the web of science were retrieved from inception to May 25, 2020, using combinations of the following keywords: (“NRF2” OR “NFE2L2” OR “nuclear factor erythroid-2-related factor 2”) AND (“Non-Small Cell Lung Carcinoma” OR “lung cancer” OR “lung squamous cell carcinomas” OR “Lung Adenocarcinomas”). Pubmed search terms are shown in S1 Table.
Inclusion criteria
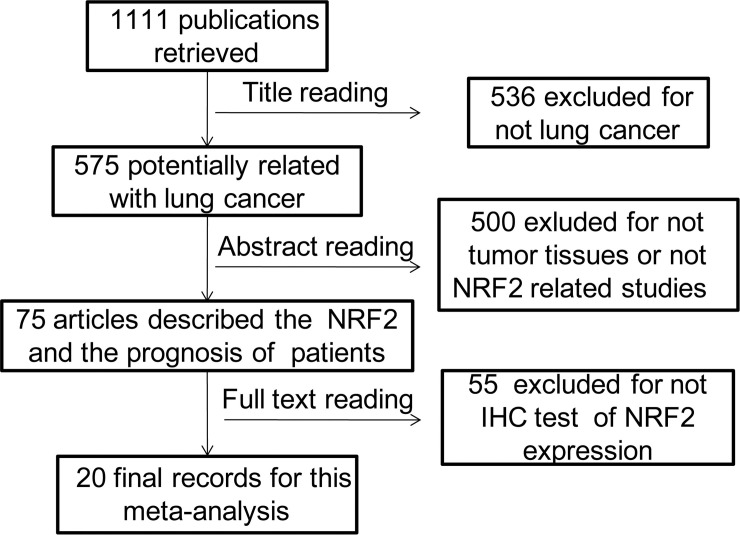
Studies eligible for inclusion in this meta-analysis met the following criteria: (1) measure NRF2 expression in the primary lung cancer with IHC (immunohistochemistry); (2) provide enough clinicopathological parameters or hazard ratio (HR) and 95% confidence interval (CI) between NRF2 expression and OS; (3) the minimum sample size was 30; (4) data specifically focus on NSCLC was extracted. Following the search, 20 articles were selected for our analysis (Fig 1). All the articles are retrospective study.
Fig 1. Flow diagram of study selection process.
Data extraction
Data extraction and information on study design, outcomes were performed by two independent observers (Qingsong Wang and Liang Xu) and disagreements were settled by discussion and consensus with a third author (Bing Yang).
As for each study, the following information was extracted: the name of the first author, year of publication, country, and number of cases, gender, expression location, cut-off value of NRF2, detection method, positive percentage, treatment, clinicopathological features, and the related survival data. Calculation method introduced by Tierney et al [15] and Parmar et al [16] was applied to extract HR with 95% CI where HR was not reported. Quality evaluation was based on the Newcastle-Ottawa quality assessment scale (NOS). The studies with NOS scores ranging from 6 to 9 were deemed as high quality. The summary of included studies can be found in Table 1.
Table 1. The basic information and data of included studies.
| No.of Studies | First Author | Year | Country | Sample Size | Gender(M/F) | Location | Cut-off value | Detection method | NRF2 Positive Percentage | Treatment | NOS Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Luisa M. Solis [11] | 2010 | U.S | 304 | 157/147 | Nuclear | score >0 | IHC(Santa Cruz) | 26.0% | C | 9 |
| 2 | Haihong Yang [10] | 2011 | China | 60 | 40/20 | Cytoplasmic | The cells stained ≥50% | IHC(Beijing Biosynthesis) | 56.7% | C | 9 |
| 3 | Daisuke Inoue [25] | 2012 | Japan | 109 | 78/31 | Nuclear | The cells stained >10% | IHC(Santa Cruz) | 33.9% | N.A | 9 |
| 4 | Heta Merikallio [17] | 2012 | Finland | 289 | N.A | Cytoplasmic | The cells stained≥50% | IHC(Santa Cruz) | N.A | N.A | 8 |
| 5 | Ming-Hsien Chien [25] | 2015 | Taiwan | 167 | 64/103 | Nuclear | The cells stained >20% | IHC(Cell Signaling) | 52.1% | N.A | 9 |
| 6 | Xiang Zhu [19] a | 2014 | China | 31 | 10/21 | Nuclear | score >0 | IHC(Abcam) | 77.4% | T | 9 |
| 6` | Xiang Zhu [19] b | 2014 | China | 31 | 10/21 | Cytoplasmic | score >2 | IHC(Abcam) | 38.7% | T | 9 |
| 7 | Joo-Heon Kim [20] | 2007 | U.S | 89 | 44/45 | Cytoplasmic | The cells stained≥25% | IHC(Abcam) | 61.8% | N.A | 8 |
| 8 | Tinghua Hu [26] | 2014 | China | 66 | 50/16 | Nuclear | IRS ≥ 4 | IHC(Abcam) | 63.6% | N.A | 7 |
| 9 | Baoshan Cao [27] | 2012 | China | 50 | 29/21 | Nuclear | score >0 | IHC(Beijing Biosynthesis) | 34.0% | C | 9 |
| 10 | Jing Wang [28] | 2017 | China | 80 | 42/38 | Nuclear | IRS ≥ 4 | IHC(Beijing Biosynthesis) | 66.2% | N.A | 9 |
| 11 | Shou Yu [29] | 2018 | China | 116 | 60/56 | Nuclear | IRS ≥ 4 | IHC(Abcam) | 62.1% | N.A | 8 |
| 12 | Qingkay Li [30] | 2011 | US | 55 | N.A | Cytolasmic | N.A | IHC(Santa Cruz) | 85.5% | N.A | 7 |
| 13 | Ying-Hui Tong [31] | 2017 | China | 215 | 170/45 | nuclear | The cells stained≥10% | IHC(Santa Cruz) | 68.4% | C | 8 |
| 14 | Jueshi Liu [32] | 2018 | China | 72 | 46/26 | nuclear | ≥2 score | IHC(Beijing Biosynthesis) | 41.7% | N.A | 7 |
| 15 | Yu Xiao [33] | 2018 | China | 104 | 47/57 | nuclear | score >0 | IHC(Abcam) | 71.2% | T | 7 |
| 16 | Xueying Zhu [34] | 2018 | China | 92 | nuclear | score >0 | IHC(Abcam) | 73.9% | N.A | 7 | |
| 17 | Manqing Liu [35] | 2018 | China | 130 | 89/41 | cytoplasmic | The cells stained≥10% | - | 64.6% | C | 9 |
| 18 | Ying E [36] | 2019 | China | 72 | 41/31 | cytoplasmic | score ≥ 4 | IHC(Abcam) | 62.5% | C | 9 |
| 19 | Hongyan Wang [37] | 2019 | China | 95 | 43/52 | nuclear | IRS ≥ 5 | IHC(Santa Cruz) | 60.0% | N.A | 8 |
| 20 | Ming-Jen Chen [24]a | 2020 | Taiwan | 167 | 113/54 | cytoplasmic-nuclear | N.A | IHC(GeneTex) | 19.0% | C | 8 |
| 20` | Ming-Jen Chen [24] b | 2020 | Taiwan | 167 | 113/54 | cytoplasmic | N.A | IHC(GeneTex) | 53.0% | C | 8 |
Abbreviations: C: chemotherapy; T: EGFR-TKI (Epidermal growth factor receptor tyrosine kinase inhibitor)., N.A: not available; IRS = SI (staining intensity) ×PP (percentage of positive cells).
Statistical analysis
All the statistical data were analyzed using STATA software (Version 12.0; Stata Corporation). Pooled odds ratios (ORs) with 95% CIs were calculated to evaluate the association between positive NRF2 expression and clinicopathological features (gender (male vs. female), smoking (current and former vs. never), histopathology (SCC vs. AC), differentiation type (poor/undifferentiated vs. well/moderate), TNM stage (TNM, III~IV vs. I~II), TNM stage (IV vs. III), lymph node metastasis (Yes vs. No), treatment response rate (CR/PR vs. SD/PD). HRs with a 95% CI in OS and PFS were calculated to evaluate the relationships between positive NRF2 expression and the prognosis of lung cancer patients. The heterogeneity among the enrolled studies was evaluated by I2 and Q statistic. The P value> 0.10 and I2 <40% were taken as a lack of heterogeneity. A logistic random-effect model was utilized for the studies with a significant heterogeneity (P≤0.10, I2≥40%). The sensitivity analysis was carried out owing to the relatively significant heterogeneity among the studies. Moreover, subgroup analyses were used to investigate potential sources of heterogeneity. Publication bias was evaluated by Begg test. A value of P<0.05 was considered statistically significant.
Results
Literature search
After the initial search algorithm, there were 507 publications from Web of science, 95 more articles were added from the Pubmed, seven publications were added from EBSCO and 40 were added from EMBASE with duplicates removed. Among them, there were 28 meeting abstracts, so 621 from English database. There were 698 publications from CNKIs. The total of 1111 potentially relevant studies were selected from the English database and CNKI databases using criteria as defined in the methods. Five hundred thirty-six articles were excluded as non-original studies (review) and non-lung-cancer studies. The remaining 575 articles were further assessed by screening the abstracts, 500 of which were excluded because they were concerned with non-human tumour tissue specimens. Seventy-five studies were included for full-text assessment. A further 55 studies were excluded for lacking the immunological histological chemistry (IHC) test of NRF2 expression. Finally, a total of 20 eligible articles with 2530 NSCLC patients were included in this meta-analysis [10,11,17–23]. The schematic process of literature selection is shown in Fig 1.
The primary characteristics of studies
Twenty eligible studies published between 2007 and 2020 used IHC methodology to evaluate the expression level of NRF2 in human NSCLC tissues. The studies were conducted in five countries or regions. Fourteen studies reported the prognostic value of NRF2 status for survival in patients with NSCLC. The location of NRF2 expression within the cell was described differently in the various studies. In the present study, these articles are described as either ‘nuclear location’ or ‘cytosolic location’ depending on the results presented in the source articles. NO.6 Xiang Zhu’s study contains two sets of data including both nuclear and cytosolic locations of NRF2 expression from the same group of patients, so we treated it as two independent studies [19]. NO.20 Ming-Jen Chen’s study contains two sets of data including C+/N+ (NRF2 cytoplasmic/ nuclear both positive immunostaining) and C+/N- (Nrf2 cytoplasmic positive/ nuclear negative immunostaining) two different cytosolic locations of NRF2 expression from the same group of patients, so we also treated it as two independent studies [24]. The sample size ranged from 31 to 304, and the percentage of positive NRF2 expression ranged from 19% to 77.4%. The main characteristics of the twenty included studies were summarized in Table 1. The clinical and pathological parameters of all included studies were showed in Table 2.
Table 2. The summarized data of clinical and pathological parameters from all included studies in the meta-analysis.
| Gender | Smoking history | TNM Stage | Metastasis | Lymph node metastasis | Cancer type(nuclear positive) | Histological differentiation | Treatment | Response rate | OS | PFS | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No.of Studies | First Author | Year | NRF2 expression | male | Female | Never | Current/former | I + II | III+ IV | M0 | M1 | Yes | No | Squamous cell carcinomas | Adenocarcinomas | Well and moderately | Poorly/undifferentiated | Yes | No | CR and PR | SD and PD | HR estimate | 95% CI | HR estimate | 95% CI |
| 1 | Luisa M. Solis | 2010 | High | 157 | 147 | 50 | 253 | - | - | - | - | - | - | 43 | 34 | - | - | 34 | 20 | - | - | 1.747 | 1.12–2.726 | 2.31 | 1.53–3.47 |
| Low | - | - | - | - | - | - | - | - | - | - | 79 | 154 | - | - | 55 | 39 | - | - | - | - | - | - | |||
| 2 | Haihong Yang | 2011 | High | 26 | 8 | 12 | 22 | - | - | - | - | - | - | 5 | 29 | 8 | 17 | 24 | 10 | 4 | 13 | 0.3 | 0.138–0.652 | 0.174 | 0.062–0.448 |
| Low | 14 | 12 | 13 | 13 | - | - | - | - | - | - | 6 | 20 | 8 | 9 | 19 | 7 | 18 | 8 | - | - | - | - | |||
| 3 | Daisuke Inoue | 2012 | High | 31 | 6 | - | - | - | - | - | - | 14 | 23 | 11 | 25 | 27 | 10 | - | - | - | - | 5 | 2.4–10.6 | - | - |
| Low | 47 | 25 | - | - | - | - | - | - | 21 | 51 | 20 | 47 | 49 | 23 | - | - | - | - | - | - | - | - | |||
| 4 | Heta Merikallio | 2012 | High | - | - | - | - | - | - | - | - | - | - | 71 | 57 | - | - | - | - | - | - | 1.49* | 1.23–1.79 | - | - |
| Low | - | - | - | - | - | - | - | - | - | - | 36 | 29 | - | - | - | - | - | - | - | - | - | - | |||
| 5 | Ming-Hsien Chien | 2015 | High | 32 | 55 | - | - | 33 | 54 | 62 | 25 | 55 | 32 | - | - | - | - | - | - | - | - | - | - | - | - |
| Low | 32 | 48 | - | - | 47 | 33 | 64 | 16 | 37 | 43 | - | - | - | - | - | - | - | - | - | - | - | - | |||
| 6 | Xiang Zhu a | 2014 | High | 6 | 18 | 16 | 8 | - | - | - | - | - | - | - | - | 19 | 5 | 24 | 0 | 13 | 11 | 1.352 | 0.487–3.752 | - | - |
| Low | 4 | 3 | 4 | 3 | - | - | - | - | - | - | - | - | 7 | 0 | 7 | 0 | 6 | 1 | - | - | - | - | |||
| 6` | Xiang Zhu b | 2014 | High | 3 | 9 | 8 | 4 | - | - | - | - | - | - | - | - | 9 | 3 | 12 | 0 | 0 | 12 | 5.449 | 1.065–27.873 | 5.944 | 1.912–18.483 |
| Low | 7 | 12 | 12 | 7 | - | - | - | - | - | - | - | - | 17 | 2 | 19 | 0 | 19 | 0 | - | - | - | - | |||
| 7 | Joo-Heon Kim | 2007 | High | 23 | 32 | 14 | 41 | - | - | - | - | - | - | 16 | 33 | 30 | 24 | - | - | - | - | - | - | - | - |
| Low | 21 | 13 | 3 | 31 | - | - | - | - | - | - | 15 | 15 | 12 | 20 | - | - | - | - | - | - | - | - | |||
| 8 | Tinghua Hu | 2014 | High | 35 | 7 | 21 | 21 | 18 | 24 | 28 | 14 | 33 | 9 | 22 | 20 | 15 | 27 | - | - | - | - | - | - | - | - |
| Low | 15 | 9 | 13 | 11 | 17 | 7 | 22 | 2 | 13 | 11 | 14 | 10 | 11 | 13 | - | - | - | - | - | - | - | - | |||
| 9 | Baoshan Cao | 2012 | High | 9 | 8 | 6 | 11 | - | - | 3 | 14 | - | - | 8 | 9 | 17 | 0 | 17 | 0 | 3 | 14 | 1.791 | 0.933–3.438 | 2.067 | 0.649–6.583 |
| Low | 20 | 13 | 13 | 20 | - | - | 17 | 16 | - | - | 15 | 18 | 31 | 2 | 33 | 0 | 14 | 19 | - | - | - | - | |||
| 10 | Jing Wang | 2017 | High | 27 | 27 | - | - | 19 | 35 | - | - | 36 | 18 | 26 | 28 | - | - | - | - | - | - | 2.078 | 1.186–3.641 | 2.623 | 1.229–5.599 |
| Low | 15 | 11 | - | - | 16 | 10 | - | - | 14 | 12 | 10 | 16 | - | - | - | - | - | - | - | - | - | - | |||
| 11 | Shou Yu | 2018 | High | 33 | 39 | 42 | 30 | 30 | 42 | 45 | 27 | - | - | 48 | 24 | 32 | 40 | - | - | - | - | 3.734 | 1.466–9.508 | 0.8 | 0.63–1.01 |
| Low | 27 | 17 | 27 | 17 | 29 | 15 | 37 | 7 | - | - | 34 | 10 | 12 | 32 | - | - | - | - | - | - | - | - | |||
| 12 | Qingkay Li | 2011 | High | - | - | - | 46 | 16 | 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Low | - | - | - | 3 | 27 | 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||
| 13 | Ying-Hui Tong | 2017 | High | 116 | 31 | 34 | 98 | 84 | 63 | - | - | 96 | 51 | 77 | 65 | 77 | 60 | - | - | - | - | 1.55 | 1.2–2.01 | - | - |
| Low | 54 | 14 | 13 | 44 | 44 | 24 | - | - | 28 | 40 | 35 | 32 | 24 | 37 | - | - | - | - | - | - | - | - | |||
| 14 | Jueshi Liu | 2018 | High | 19 | 11 | - | - | 24 | 6 | - | - | 9 | 21 | 17 | 13 | 20 | 10 | - | - | - | - | - | - | - | - |
| Low | 27 | 25 | - | - | 20 | 22 | - | - | 26 | 16 | 23 | 19 | 8 | 34 | - | - | - | - | - | - | - | - | |||
| 15 | Yu Xiao | 2018 | High | 38 | 36 | 45 | 29 | 23 | 51 | - | - | - | - | - | - | 60 | 12 | - | - | - | - | 7.505 | 1.656–34.007 | 8.487 | 2.234–32.239 |
| Low | 9 | 21 | 15 | 15 | 7 | 23 | - | - | - | - | - | - | 22 | 6 | - | - | - | - | - | - | - | - | |||
| 16 | Yingxue Zhu | 2018 | High | - | - | - | - | 31 | 37 | - | - | - | - | 39 | 29 | - | - | - | - | - | - | - | - | - | - |
| Low | - | - | - | - | 18 | 6 | - | - | - | - | 14 | 10 | - | - | - | - | - | - | - | - | - | - | |||
| 17 | Manqing Liu | 2018 | High | 58 | 26 | 47 | 37 | - | 92 | - | - | - | - | 29 | 39 | - | - | - | - | - | - | 6.296 | 1.992–19.899 | - | - |
| Low | 31 | 15 | 32 | 14 | - | 38 | - | - | - | - | 19 | 17 | - | - | - | - | - | - | - | - | - | - | |||
| 18 | Ying E | 2019 | High | 26 | 19 | 17 | 28 | 26 | 19 | - | - | - | - | 26 | 19 | 15 | 30 | - | - | - | - | 2.16 | 0.65–7.18 | - | - |
| Low | 15 | 12 | 9 | 18 | 24 | 3 | - | - | - | - | 17 | 10 | 8 | 19 | - | - | - | - | - | - | - | - | |||
| 19 | Hongyan Wang | 2019 | High | 22 | 35 | - | - | - | - | - | - | - | - | - | - | 17 | 40 | - | - | - | - | - | - | - | - |
| Low | 21 | 17 | - | - | - | - | - | - | - | - | - | - | 12 | 26 | - | - | - | - | - | - | - | - | |||
| 20 | Ming-Jen Chen a | 2020 | High | 15 | 17 | 23 | 9 | 20 | 12 | - | - | - | - | 14 | 18 | - | - | - | - | - | - | 1.638 | 1.059–2.535 | 1.676 | 1.074–2.614 |
| Low | 98 | 37 | 68 | 69 | 102 | 33 | - | - | - | - | 54 | 81 | - | - | - | - | - | - | - | - | - | - | |||
| 20' | Ming-Jen Chen b | 2020 | High | 54 | 34 | 55 | 33 | 64 | 24 | - | - | - | - | 35 | 53 | - | - | - | - | - | - | 1.568 | 1.046–2.349 | 1.609 | 0.874–1.533 |
| Low | 59 | 20 | 36 | 45 | 58 | 21 | - | - | - | - | 33 | 46 | - | - | - | - | - | - | - | - | - | - | |||
Abbreviations: SCC: squamous cell carcinomas, AC: adenocarcinomas, OS: overall survival, PFS: progression-free survival, HR: hazard ratio, OR: odds ratio, RR: relative risk, CI: confidence interval, EGFR-TKI: Epidermal growth factor receptor tyrosine kinase inhibitor, CR: complete response, PR: partial response, PD: progression of disease, SD: stable disease.
Association between NRF2 and clinicopathological features in NSCLC
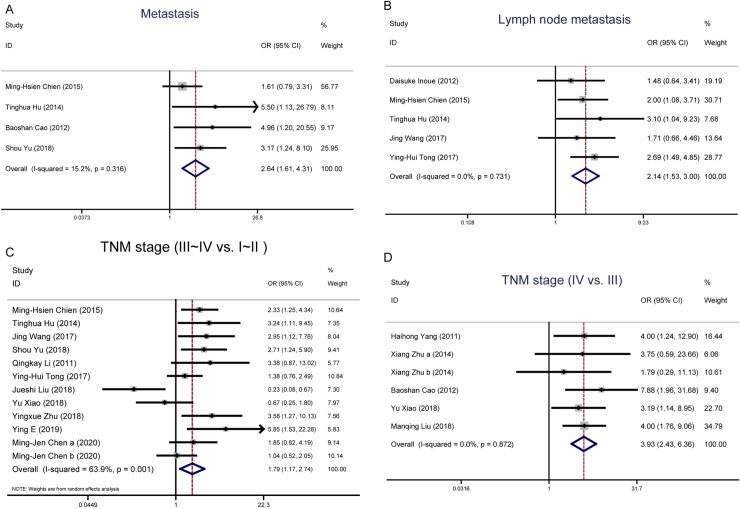
To examine the clinical value of NRF2, we investigated the associations between NRF2 and a number of clinicopathological parameters [18,21,23]. The logistic fixed-effects model was employed because of restricted heterogeneity among the studies (I2 = 15.2%, P = 0.316). As seen in Table 3 and Fig 2A pooled odds ratio (OR) values from the four eligible studies showed that upregulated NRF2 was associated with distant metastasis (OR = 2.64, 95% CI 1.62–4.31; P<0.001).
Table 3. Main results and publication bias for meta-analysis between NRF2 and clinicopathological features, overall survival (OS) and PFS (progression-free survival).
| Correlation between NRF2 and clinicopathological features / OS/PFS | No. of studies | Overall OR/HR (95%CI) | z,POR/HR | Heterogeneity test (I2, Pbias) | Publication bias (Egger’s test) |
|---|---|---|---|---|---|
| (t, Ppublication bias) | |||||
| Metastasis (M1/M0) | 5, 8, 9, 11 | 2.64 (1.62,4.31) | 3.87, < 0.001 | 15.2%, 0.316 | 4.04, 0.056 |
| Lymph node metastasis (Yes vs. No) | 3, 5, 8, 10, 13 | 2.14 (1.53, 3.00) | 4.46 < 0.001 | 0.0%, 0.731 | -0.33, 0.764 |
| TNM stage (III~IV vs. I~II) | 5, 8, 10, 11, 12, 13, 14, 15, 16, 18, 20 20' | 1.79 (1.17, 2.74) | 2.68, 0.007 | 63.9%, 0.001 | 0.53, 0.607 |
| TNM stage (IV vs. III) | 2, 6, 6', 9, 15, 17 | 3.93 (2.43, 6.36) | 5.58, < 0.001 | 0.0%, 0.872 | -0.29, 0.787 |
| Treatment response rate (CR/PR vs. SD/PD) | 2, 6, 6', 9 | 0.11 (0.02, 0.51) | 2.84, 0.005 | 58.0%, 0.067 | -2.06, 0.175 |
| OS | 1, 2, 3, 4, 5, 6, 6', 9, 10, 11, 13, 15, 17, 18, 20, 20' | 1.86 (1.44, 2.41) | 4.73, < 0.001 | 67.9%, < 0.001 | 1.65, 0.122 |
| PFS | 1, 2, 6', 9, 10, 11, 15, 20 | 2.27 (1.26, 4.09) | 2.74, 0.006 | 86.2%, < 0.001 | 3.53, 0.017 |
| Gender (male vs. female) | 2, 4, 5, 6, 6', 7, 8, 9, 10, 11, 13, 14, 15, 17, 18, 19, 20, 20' | 0.90 (0.66, 1.23) | 0.65, 0.515 | 53.6%, 0.004 | 0.78, 0.448 |
| Smoking (current and former vs. never) | 2, 6, 6', 7, 8, 9, 11, 13, 15, 17, 18, 20, 20' | 1.23 (0.96, 1.58) | 1.68, 0.094 | 26.4%, 0.178 | -1.52, 0.157 |
| Histopathology (SCC vs. AC) | 1, 2, 3, 4, 7, 8, 9, 10, 11, 13, 14, 16, 17, 18, 20, 20' | 1.05(0.86, 1.27) | 0.44, 0.657 | 16.6%, 0.264 | -2.46, 0.028 |
| Differentiation type (poor/undifferentiated vs. well/moderate) | 2, 3, 6, 6', 7, 8, 9, 11, 13, 14,15, 18, 19 | 1.48 (0.95, 2.30) | 1.73, 0.083 | 47.3%, 0.035 | -1.23, 0.247 |
Abbreviations: SCC: squamous cell carcinomas, AC: adenocarcinomas, OS: overall survival, PFS: progression-free survival, HR: hazard ratio, OR: odds ratio, CI: confidence interval, EGFR-TKI: Epidermal growth factor receptor tyrosine kinase inhibitor, CR: complete response, PR: partial response, PD: progression of disease, SD: stable disease.
Fig 2. Forest plot of the NRF2 expression level and clinicopathological features.
A. Forest plot of studies evaluating the relationship between NRF2 expression and distant pathological metastasis. B. Forest plot of studies evaluating the relationship between NRF2 expression and lymph node metastasis. C. Forest plot of studies evaluating the relationship between NRF2 expression and pathological tumour-node-metastasis (TNM, III~IV vs. I~II). D. Forest plot of studies evaluating the relationship between NRF2 expression and pathological tumour-node-metastasis (TNM, IV vs. III).
The patients with lymph node metastasis based on different levels of NRF2 expression was reported in 6 studies. The logistic random-effects model was applied because of significant heterogeneity in the studies (I2 = 71.0%, P = 0.004). The analysis showed a pooled OR = 1.55 (95%CI: 0.84–2.87, P = 0.164). Owing to the relatively severe heterogeneity among the studies on lymph node metastasis, the sensitivity analysis and published bias were carried out, in which the Jueshi Liu’s study in 2018 was the cause of statistical heterogeneity (S2 Fig). When this study was removed, the heterogeneity disappeared in the remaining studies (P = 0.731, I2 = 0%). The analysis of these left studies indicated a statistically obvious association between the high NRF2 expression and the lymph node metastasis. The combined OR estimates were 2.14 (95% CI: 1.53–3.00; P < 0.001). This analysis suggests that heterogeneity among different studies should be treated with extra caution when interpreting it (Fig 2B).
The patients with TNM stage (TNM, III~IV vs. I~II) based on different levels of NRF2 expression was reported in 11 studies. The logistic random-effects model was applied because of significant heterogeneity in the studies (I2 = 63.9%, P = 0.001). The analysis showed a pooled OR = 1.79 (95%CI: 1.17–2.74, P = 0.007), as shown in Fig 2C. The sensitivity analysis was carried out owing to the relatively large heterogeneity among the studies on the TNM stage. The pooled OR estimates were consistent without distinct fluctuation. The results demonstrated that high expression level of NRF2 was related to the advanced TNM stage. Then, we further analyzed the NRF2 expression in advanced TNM stage (TNM, IV vs. III). There are five studies with TNM IV and III patients. The logistic fixed-effects model was employed because of restricted heterogeneity among the studies (I2 = 0.0%, P = 0.872). The meta-analysis demonstrated a combined OR = 3.93 (95%CI: 2.43–6.36, P<0.001), as shown in Fig 2D. The results demonstrate that the advanced TNM stage is distinctly related to the high NRF2 expression level.
On the other hand, the relationships between NRF2 expression level and gender (OR = 0.90, 95%CI 0.66–1.23; P = 0.515), smoking (OR = 1.23, 95%CI 0.96–1.58; P = 0.094), histopathology (OR = 1.05, 95%CI 0.86–1.27; P = 0.657), and differentiation type (OR = 1.48, 95%CI 0.95–2.30; P = 0.083) were not significant (Table 3, S1 Fig).
Association between NRF2 expression level and treatment response rate in NSCLC
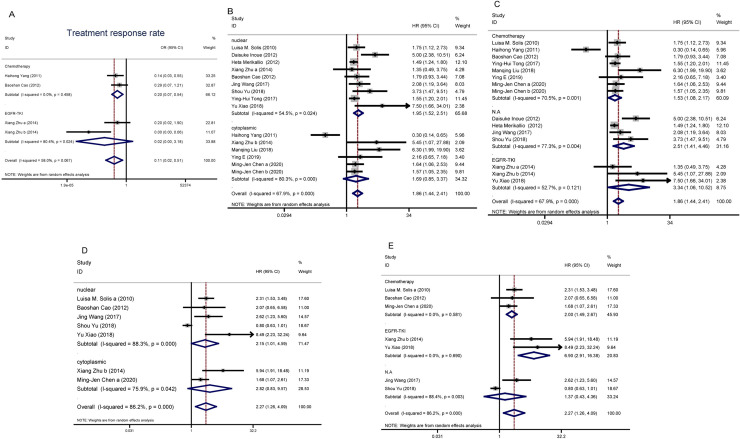
Four studies included data relating to the response to treatment. These studies were assessed for the association between NRF2 and treatment response rate in NSCLC. Two studies elucidated NRF2 expression and outcome in patients treated with platinum-based chemotherapy. The other two studies determined that NRF2 was a good biomarker for predicting response to EGFR-TKI (Epidermal growth factor receptor tyrosine kinase inhibitor) in patients with EGFR gene mutations. All the patients included are in stage III or stage IV NSCLC, and 17 patients had undergone surgery in one of the chemotherapy studies. Our results indicate that the high NRF2 expression level was associated with treatment response rate (OR = 0.11, 95%CI 0.02–0.51; P = 0.005). However, heterogeneity was found to be relatively large (I2 = 58.0%, pbias = 0.067) (Table 3).
Subgroup analysis was performed on treatment method to explore the potential sources of heterogeneity. As seen in Table 4 and Fig 3A, the results showed that there was no relationship between NRF2 expression and treatment response rate (OR = 0.02, 95%CI 0.01–3.18; P = 0.13) in the EGFR-TKI treated group. In the chemotherapy-treated group, upregulated NRF2 was associated with a low treatment response rate (OR = 0.20, 95%CI 0.07–0.54; P < 0.01). Heterogeneity was not a major factor in the chemotherapy treated group (I2 = 0.0%, Pbias = 0.458). Taken together, the heterogeneity of treatment response rate was mainly caused by the different treatment methods. The high NRF2 expression level was associated with a low treatment response rate in platinum-based chemotherapy.
Table 4. Subgroup analysis of treatment response rate, overall survival and progression free survival.
| Subgroups | Studies | OR/HR(95%CI) | z | POR/HR | I2 | Pbias |
|---|---|---|---|---|---|---|
| Treatment response rate | ||||||
| Treatment | ||||||
| Chemotherapy | 2 | 0.20 (0.07, 0.54) | 3.18 | < 0.01 | 0.0% | 0.458 |
| EGFR-TKI | 2 | 0.02 (0.01, 3.18) | 1.52 | 0.13 | 80.4% | 0.024 |
| Overall survival | ||||||
| Location | ||||||
| Nuclear | 9 | 1.95 (1.52, 2.51) | 5.22 | < 0.01 | 54.5% | 0.024 |
| Cytoplasmic | 6 | 1.69 (0.85, 3.37) | 1.50 | 0.13 | 80.3% | < 0.001 |
| Treatment | ||||||
| Chemotherapy | 8 | 1.53 (1.08–2.17) | 2.39 | < 0.01 | 70.50% | 0.001 |
| EGFR-TKI | 3 | 3.34 (1.06–10.52) | 2.06 | 0.04 | 52.70% | 0.121 |
| N.A | 4 | 2.51 (1.41–4.46) | 3.13 | < 0.01 | 77.30% | 0.004 |
| PFS | ||||||
| Location | ||||||
| Nuclear | 5 | 2.15 (1.01,4.59) | 4.77 | 0.048 | 88.3% | < 0.001 |
| Cytoplasmic | 2 | 2.82 (0.83, 9.57) | 0.21 | 0.10 | 75.9% | 0.042 |
| Treatment | ||||||
| Chemotherapy | 3 | 2.00 (1.49–2.67) | 4.65 | < 0.01 | 0.00% | 0.581 |
| EGFR-TKI | 2 | 6.90 (2.91–16.38) | 4.38 | < 0.01 | 0.00% | 0.690 |
| N.A | 2 | 1.37 (0.43–4.36) | 0.53 | 0.60 | 88.40% | 0.003 |
Abbreviations: OR: odds ratio, HR: hazard ratio CI: confidence interval, EGFR-TKI: Epidermal growth factor receptor tyrosine kinase inhibitor. PFS: progression-free survival, N.A: the therapeutic protocol was not clearly defined
Fig 3. The association between NRF2 high expression and treatment response rate, overall survival and progression-free survival in NSCLC.
A. Forest plot for subgroup study about treatment response rate. B. C. Forest plot for the subgroup study about relationship between NRF2 expression and overall survival (OS). D.E. Forest plot of subgroup study about evaluating the relationship between NRF2 expression and progression free survival (PFS).
Association between NRF2 and overall survival, progression-free survival in NSCLC
Fifteen studies were assessed for the association between NRF2 and overall survival (OS). The logistic random-effects model was applied because of significant heterogeneity in the studies (I2 = 67.9%, P < 0.001). The sensitivity analysis was carried out owing to the relatively large heterogeneity among the studies on OS. The pooled HR estimates were consistent without distinct fluctuation. The results indicated that a high NRF2 expression level was associated with inferior OS (HR = 1.86, 95% CI 1.44–2.41, P < 0.001) (Table 3).
Subgroup analysis based on NRF2 signal localization was also used to explore whether these potential sources of heterogeneity had an effect on overall survival. As seen in Table 4 and Fig 3B, NRF2 signal localization impacts the correlation of higher NRF2 expression with worse OS (nuclear: HR = 1.95, 95%CI 1.52–2.51, P < 0.01; cytoplasmic: HR = 1.69, 95%CI 0.85–3.37, P = 0.13). Although there was high heterogeneity within the nuclear subgroup (I2 = 54.5%) and cytosolic subgroup (I2 = 80.3%). These results indicate that the patients with a high level of nuclear NRF2 expression had a lower survival rate.
Meanwhile, we separated the studies into three subgroups according to the different treatments, including chemotherapy, EGFR-TKI, and N.A group without clearly defined treatment. The treatments impact the association of NRF2 expression with OS (chemotherapy: HR = 1.53, 95%CI 1.08–2.17, P < 0.01; EGFR-TKI: HR = 3.34, 95%CI 1.06–10.52, P = 0.04; N.A: HR = 2.51, 95%CI 1.41–4.46, P < 0.01), as shown in Table 4 and Fig 3C. Because various chemotherapies were employed in different studies and the therapeutic protocol was not clearly defined in the N.A group, the OS showed high heterogeneity among different studies. Nevertheless, the higher NRF2 expression level within these three groups of patients were inversely correlated with their OS.
Eight studies were included in the meta-analysis of progression-free survival (PFS). The random-effects model was applied because of significant heterogeneity in the studies (I2 = 88.1%, P < 0.001). The influence analysis was carried out showed in S2 Fig, the Haihong Yang’s study in 2011 was removed data. The heterogeneity still was high (I2 = 86.2%) so the logistic random-effects model was applied. The results indicated that positive NRF2 expression was associated with poor PFS (HR = 2.27, 95% CI 1.26–4.09, P = 0.006) (Table 3).
Subgroup analysis based on NRF2 signal localization and treatment was also used to explore whether these potential sources of heterogeneity had an effect on PFS. As seen in Table 4 and Fig 3D, NRF2 signal localization impacts the association of NRF2 expression with PFS (nuclear: HR = 2.15, 95% CI 1.01–4.59, P = 0.048; cytoplasmic: HR = 2.82, 95% CI 0.83–9.57, P = 0.10). The high NRF2 level in nucleus was associated with poor PFS. As shown in Fig 3E and Table 4, the treatments impact the association of NRF2 expression with PFS (chemotherapy: HR = 2.00, 95%CI 1.49–2.67, P < 0.01; EGFR-TKI: HR = 6.90, 95%CI 2.91–16.38, P < 0.01; N.A: HR = 1.37, 95%CI 0.43–4.36, P = 0.60). In the N.A group, because the therapeutic protocol was not clearly defined, the heterogeneity was high. That also caused the high heterogeneity of PFS. NRF2 higher expression was not associated with poor PFS in N.A group.
Publication bias
Funnel plots, as well as Begg's test, was performed to evaluate potential publication bias in this meta-analysis. Most of the plots were symmetric, indicating that publication bias was low (S3 Fig). There was no evidence of significant publication bias by inspection of the formal statistical tests (Tables 3 and 4).
Discussion
NRF2 is a transcription factor that acts as the main regulator of various antioxidant genes [38]. Several studies have shown that dysregulation of NRF2 is closely related to human cancer [39,40]. NRF2 is involved in various tumour processes, mainly by interfering with cell proliferation and apoptosis, causing resistance to conventional chemotherapy and radiotherapy. In the case of lung cancer, clinical evidence on the relationship between NRF2 positive expression and tumour invasion or prognosis has not been thoroughly investigated.
NRF2 activators have been used in clinical trials for cancer treatment and the treatment of diseases related to oxidative stress. On the other hand, constitutive activation of NRF2 contributes to the growth of cancer cells in many types of tumours, leading to the resistance to anticancer therapy [1,41]. Considering the inconsistent reports in the literature, we conducted the meta-analysis and found that a high expression level of NRF2 was related to poor survival rate among lung cancer patients. This meta-analysis is the first systematic study to evaluate the association between NRF2 expression and clinicopathological features and overall survival in NSCLC patients. Through combined 20 publications including 2530 patients with NSCLC, our results indicate that positive NRF2 expression is correlated with high pathological metastasis, high TNM stage and increased lymph node metastasis. These findings are consistent with previous report that NRF2 has a significant impact on neoplasm invasiveness-associated features [42]. The higher expression level of NRF2 appears to be an indication of worse OS and RFS, which is consistent with the results of Wang et al. on the solid tumour [43]. The NRF2 sequences of 103 NSCLC patients was studied by Hu et al., and it was found that the NRF2 mutation rate of current and former smokers was significantly higher than that of non-smokers [44]. According to Hu, Sasaki et al., sequenced NRF2 in 262 surgically resected lung tumours confirmed that NRF2 mutations were more common in squamous cell carcinoma and smokers [45]. However, we did not detect any association between high NRF2 expression and smoking history. Since our study was entirely based on IHC data, representing the protein level of NRF2 expression, no information regarding the genetics of NRF2 can be obtained. On the other hand, only a limited number of studies were available for this meta-analysis, and such associations may become apparent with increased sample size.
There are some studies about NRF2 mutation [4,46] as a prediction for lung cancer survival. KEAP1/NRF2 mutant lung cancer is a microenvironmentally distinct, biologically heterogeneous and clinically underestimated disease that increased radioresistance in NSCLC [47–50]. The higher NRF2 expression level was significantly correlated with EGFR gene mutation in NSCLC [32]. Previous studies using microarray data to analyze the NRF2-associated genes [4,51] found that they were biomarkers for poor prognosis in NSCLC cohorts. But, the mRNA level of NRF2 alone was not correlated with the clinicopathology in NSCLC [36,52]. Thus, it is important to consider NRF2 gene mutations, mRNA level and protein level altogether in order to provide more comprehensive understanding of the prognosis among different NSCLC patients.
KEAP1/NRF2 signalling regulates glutaminolysis metabolism by inhibition of glutaminase in KRAS-KEAP1 mutant lung cancer [53] and regulates the sensitivity to EGFR-TKI. It has been reported that mutations in KEAP1/NRF2 [54] and higher expression level of DJ1 [17], NQO1 [31], TP53 [55], CUL3 [56] and PRDX5 [57] are associated with poor survival of patients with NSCLC. The synergy between the KEAP1/NRF2 and PI3K pathways drives NSCLC with an altered immune microenvironment and achieves tumour regression through suppression of immune checkpoint [58]. Patients with NSCLC usually received targeted therapy (EGFR/ALK mutation patient) or chemotherapy with cisplatin [59], with or without combined radiotherapy. The KEAP1/NRF2 mutation may define a molecular subtype that is resistant to chemotherapy [47] and therefore may rapidly develop into NSCLC. Among NSCLC patients with EGFR mutations, if KEAP1/NRF2/CUL3 co-mutation existed, the EGFR-TKI treatment showed a significantly reduced effective time window [56]. Our results also indicated that higher NRF2 expression was associated with the poor OS and PFS in both chemotherapy and EGFR-TKI treatment group. Therefore, in order to increase the sensitivity of chemotherapy and EGFR-TKI, NRF2 can be developed as a therapeutic target to benefit the NSCLC patients.
In the current study, we observed that positive NRF2 expression was associated with low treatment response rate in platinum-based chemotherapy. However, the patients underwent different chemotherapy strategies and 17 of them received surgical interventions. This suggests that chemotherapy alone may not be an effective therapy for NRF2 positive NSCLC patients [15]. The NRF2/KEAP1 pathway controls the localization of NRF2 in the nucleus and cytoplasm. The dual roles of NRF2 in tumorigenesis might therefore be caused by NRF2 shuttling between the nucleus and cytoplasm [24]. Therefore, the NRF2 location should be considered during analysis for the clinicopathological features.
There were some limitations in this meta-analysis. Firstly, in this study, the NRF2 expression was based on IHC staining data. Therefore, the choice of primary antibody and the dilution adopted could give rise to inconsistent NRF2 detection. Secondly, due to the strict selection criteria and the limited number of published studies concerning NRF2 expression and lung cancer prognosis, we were only able to include 20 published articles in this meta-analysis. Finally, the various definitions of the cut-off value of NRF2 expression in the original studies might cause additional heterogeneity and bias.
In conclusion, despite the limitations mentioned above, our meta-analysis is the first study to systematically evaluate the association between NRF2 expression and NSCLC survival. The results support an association between high expression level of NRF2 and aggressive tumor pathology in NSCLC patients. Therefore, NRF2 has the potential to become a molecular signature predicting NSCLC survival.
Supporting information
(DOC)
A. Forest plot of studies evaluating the relationship between NRF2 expression and gender. B. Forest plot of studies evaluating the relationship between NRF2 expression and smoking. C. Forest plot of studies evaluating the relationship between NRF2 expression and histopathology. D. Forest plot of studies evaluating the relationship between NRF2 expression and tumour differentiation type.
(TIF)
A., Lymph node metastasis; B., PFS
(TIF)
A., Metastasis; B., Lymph node metastasis; C., TNM stage (III~IV vs. I~II); D.,TNM stage (IV vs. III); E., Treatment response rate; F., OS; G., PFS; H., Gender; I., Smoking; J. Histopathology; K. Differentiation type.
(TIF)
(DOCX)
Acknowledgments
We thank Xiyue Jing (Tianjin Medical University) for technical assistance during the data analysis. The authors acknowledge Dr. Thomas C. Roberts (Department of Paediatrics, University of Oxford, Oxford, United Kingdom) and Jonathan R. Hart (Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA 92037) for critical reading of the manuscript.
Abbreviations
- NSCLC
non-small cell lung cancer
- NRF2
Nuclear factor erythroid 2-Related Factor 2
- NFE2L2
Nuclear factor, erythroid 2 like 2
- NQO1
NAD(P)H quinone oxidoreductase 1
- HO-1
Heme oxygenase-1
- GST
glutathione S-transferase
- KEAP1
Kelch-like ECH-associated protein 1
- PI3K
phosphatidylinositol 3-kinase
- K-RAS
Kirsten retrovirus-associated DNA sequence
- IHC
immunological histological chemistry
- TNM stage
pathologic tumour-node-metastasis
- SCC
squamous cell carcinomas
- AC
adenocarcinomas
- OS
overall survival
- PFS
progression-free survival
- HR
hazard ratio
- OR
odds ratio
- CI
confidence interval
- EGFR-TKI
Epidermal growth factor receptor tyrosine kinase inhibitor
- CR
complete response
- PR
partial response
- PD
progression of disease
- SD
stable disease
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The National Natural Science Foundation of China (#81472052 to Qingsong Wang, #81501988 to Xiangli Jiang). The Tianjin Municipal Science and Technology Commission ((# 16JCQNJC10300 to Bing Yang and #16JCQNJC11500 to Xuewen Wang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, et al. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420 10.1371/journal.pmed.0030420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, et al. (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47: 1475–1481. 10.1038/ng.3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sporn MB, Liby KT (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564–571. 10.1038/nrc3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian Z, Zhou T, Gurguis CI, Xu X, Wen Q, et al. (2015) Nuclear factor, erythroid 2-like 2-associated molecular signature predicts lung cancer survival. Sci Rep 5: 16889 10.1038/srep16889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaramillo MC, Zhang DD (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27: 2179–2191. 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotblat B, Melino G, Knight RA (2012) NRF2 and p53: Januses in cancer? Oncotarget 3: 1272–1283. 10.18632/oncotarget.754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian Y, Liu Q, He X, Yuan X, Chen Y, et al. (2016) Emerging roles of Nrf2 signal in non-small cell lung cancer. J Hematol Oncol 9: 14 10.1186/s13045-016-0246-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XJ, Li Y, Luo L, Wang H, Chi Z, et al. (2014) Oxaliplatin activates the Keap1/Nrf2 antioxidant system conferring protection against the cytotoxicity of anticancer drugs. Free Radic Biol Med 70: 68–77. 10.1016/j.freeradbiomed.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 9.Satoh H, Moriguchi T, Takai J, Ebina M, Yamamoto M (2013) Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res 73: 4158–4168. 10.1158/0008-5472.CAN-12-4499 [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Wang W, Zhang Y, Zhao J, Lin E, et al. (2011) The role of NF-E2-related factor 2 in predicting chemoresistance and prognosis in advanced non-small-cell lung cancer. Clin Lung Cancer 12: 166–171. 10.1016/j.cllc.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 11.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, et al. (2010) Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 16: 3743–3753. 10.1158/1078-0432.CCR-09-3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abazeed ME, Adams DJ, Hurov KE, Tamayo P, Creighton CJ, et al. (2013) Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res 73: 6289–6298. 10.1158/0008-5472.CAN-13-1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparaneo A, Fabrizio FP, Muscarella LA (2016) Nrf2 and Notch Signaling in Lung Cancer: Near the Crossroad. Oxid Med Cell Longev 2016: 7316492 10.1155/2016/7316492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6: e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8: 16 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parmar MK, Torri V, Stewart L (1998) Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 17.Merikallio H, Pääkkö P, Kinnula VL, Harju T, Soini Y (2012) Nuclear factor erythroid-derived 2-like 2 (Nrf2) and DJ1 are prognostic factors in lung cancer. Hum Pathol 43: 577–584. 10.1016/j.humpath.2011.05.024 [DOI] [PubMed] [Google Scholar]
- 18.Chien MH, Lee WJ, Hsieh FK, Li CF, Cheng TY, et al. (2015) Keap1-Nrf2 Interaction Suppresses Cell Motility in Lung Adenocarcinomas by Targeting the S100P Protein. Clin Cancer Res 21: 4719–4732. 10.1158/1078-0432.CCR-14-2880 [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Liang L, Liu C, Yin W, Chen S, et al. (2014) [Role of the expression level of Nrf2 in predicting response of EGFR-TKIs in lung adenocarcinoma patients with EGFR gene mutations]. Zhongguo Fei Ai Za Zhi 17: 155–162. 10.3779/j.issn.1009-3419.2014.02.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JH, Bogner PN, Ramnath N, Park Y, Yu J, et al. (2007) Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clin Cancer Res 13: 3875–3882. 10.1158/1078-0432.CCR-06-2893 [DOI] [PubMed] [Google Scholar]
- 21.Hu T, Yao Y, Yu S, Guo H, Tian T, et al. (2014) [CXCR4 and Nrf2 expressions in non-small cell lung cancer and their clinical implications]. Nan Fang Yi Ke Da Xue Xue Bao 34: 153–158. [PubMed] [Google Scholar]
- 22.CAO B-s, ZHU X, YIN W-c, CHEN S, WANG M-p, et al. (2012) The role of expression of Nrf2 in predicting chemoresistance and prognosis in advanced non-small cell lung cancer receiving platinum-based i rst-line chemotherapy. Tumor 32: 6. [Google Scholar]
- 23.Inoue D, Suzuki T, Mitsuishi Y, Miki Y, Suzuki S, et al. (2012) Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci 103: 760–766. 10.1111/j.1349-7006.2012.02216.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen MJ, Lin PL, Wang L, Cheng YM, Chen CY, et al. (2020) Cytoplasmic, but not nuclear Nrf2 expression, is associated with inferior survival and relapse rate and response to platinum-based chemotherapy in non-small cell lung cancer. Thoracic Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ming-Hsien C, Wei-Jiunn L, Feng-Koo H, Chia-Feng L, Tsu-Yao C, et al. (2015) Keap1–Nrf2 Interaction Suppresses Cell Motility in Lung Adenocarcinomas by Targeting the S100P Protein. American Association for Cancer Research 21. [DOI] [PubMed] [Google Scholar]
- 26.Tinghua H, Yu Y, Shuo Y, Hui G, Tao T, et al. (2014) CXCR4 and Nrf2 expressions in non-small cell lung cancer and their clinical implications. J South Med Univ 34: 153–158. [PubMed] [Google Scholar]
- 27.Cao BS, Zhu X, Yin WC, Chen S, Wang MP, et al. (2012) The role of expression of Nrf2 in predicting chemoresistance and prognosis in advanced non-small cell lung cancer receiving platinum-based first-line chemotherapy. Tumor 32: 919–924. [Google Scholar]
- 28.Wang J, Liu Z, Hu T, Han L, Yu S, et al. (2017) Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Taylor & Francis 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu S, Cheng C, Wang J, Wang J, Qu Z, et al. (2018) Loss of Beclin1 Expression and Nrf2 Overexpression are Associated with Poor Survival of Patients with Non-Small Cell Lung Cancer. Anti-Cancer Agents in Medicinal Chemistry 18 10.2174/1871520618666180830110700 [DOI] [PubMed] [Google Scholar]
- 30.Li QK, Singh A, Biswal S, Askin F, Gabrielson E (2011) KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. Nature Publishing Group UK 56 10.1038/jhg.2010.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong YH, Zhang B, Yan YY, Fan Y, Yu JW, et al. (2017) Dual-negative expression of Nrf2 and NQO1 predicts superior outcomes in patients with non-small cell lung cancer. Oncotarget 8: 45750–45758. 10.18632/oncotarget.17403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LIU J, ZHANG Y, LONG L, XIANG H (2018) Expression and Clinical Significance of Nrf-2 and Rsf-1 in Lung Cancer. Anti-tumor Pharmacy 8: 223–226. [Google Scholar]
- 33.XIAO Y, ZHU X, GU Y, CHEN S, LIANG L, et al. (2018) Nrf2 and Keap1 Abnormalities in 104 Lung Adenocarcinoma Cases and Association with Clinicopathologic Features. Chin J Lung Cancer 21: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ZHU X-y, MA D-c, WANG B-m, ZHU F, CAO W, et al. (2018) Expression and clinicopathological correlation of Keap 1-Nrf 2 signal pathway protein in non-small cell lung cancer. Chin J Dis Control Prev 22: 938–942. [Google Scholar]
- 35.LIU M HUANG J (2018) Prognostic value of different Nrf2 and ERCC1 levels in advanced NSCLC patients receiving platinum chemotherapy regimen. Anhui Medical and Pharmaceutical Journal 22: 2369–2372. [Google Scholar]
- 36.Ying E, Gao SD, Sheng Y (2019) Expression and clinical significance of Nrf2 in non—small cell lung cancer. Journal of Clinical and Experimental Medicine 18: 379–382. [Google Scholar]
- 37.Wang H, Liu K, Chi Z, Zhou K, Ren G, et al. (2019) Interplay of MKP-1 and Nrf2 drives tumor growth and drug resistance in non-small cell lung cancer. Aging 11: 11329–11346. 10.18632/aging.102531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loboda A, Was H, Jozkowicz A, Dulak J (2008) Janus face of Nrf2-HO-1 axis in cancer—friend in chemoprevention, foe in anticancer therapy. Lung Cancer 60: 1–3. 10.1016/j.lungcan.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 39.Jaiswal AK (2004) Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med 36: 1199–1207. 10.1016/j.freeradbiomed.2004.02.074 [DOI] [PubMed] [Google Scholar]
- 40.Nguyen T, Nioi P, Pickett CB (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284: 13291–13295. 10.1074/jbc.R900010200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes JD, McMahon M (2009) NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 34: 176–188. 10.1016/j.tibs.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 42.E-Deek HEM, Ahmed AM, Mohammed RAA (2019) Aberration of Nrf2-Bach1 pathway in colorectal carcinoma; role in carcinogenesis and tumor progression. Annals of Diagnostic Pathology 38: 138–144. 10.1016/j.anndiagpath.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 43.Wang LL, Zhang CZ, Qin LT, Xu JY, Li XB, et al. (2018) The prognostic value of NRF2 in solid tumor patients: a meta-analysis. Oncotarget 9: 1257–1265. 10.18632/oncotarget.19838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y, Ju Y, Lin D, Wang Z, Huang Y, et al. (2012) Mutation of the Nrf2 gene in non-small cell lung cancer. Mol Biol Rep 39: 4743–4747. 10.1007/s11033-011-1266-4 [DOI] [PubMed] [Google Scholar]
- 45.Sasaki H, Suzuki A, Shitara M, Hikosaka Y, Okuda K, et al. (2013) Genotype analysis of the NRF2 gene mutation in lung cancer. Int J Mol Med 31: 1135–1138. 10.3892/ijmm.2013.1324 [DOI] [PubMed] [Google Scholar]
- 46.Cescon DW, She D, Sakashita S, Zhu CQ, Pintilie M, et al. (2015) NRF2 Pathway Activation and Adjuvant Chemotherapy Benefit in Lung Squamous Cell Carcinoma. Clin Cancer Res 21: 2499–2505. 10.1158/1078-0432.CCR-14-2206 [DOI] [PubMed] [Google Scholar]
- 47.Goeman F, Nicola FD, Scalera S, Sperati F, Gallo E, et al. (2019) Mutations in the KEAP1-NFE2L2 Pathway Define a Molecular Subset of Rapidly Progressing Lung Adenocarcinoma. Journal of Thoracic Oncology 14 10.1016/j.jtho.2019.07.003 [DOI] [PubMed] [Google Scholar]
- 48.Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, et al. (2018) Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated Non–Small Cell Lung Carcinoma (NSCLC). Clinical Cancer Research 24: 3087–3096. 10.1158/1078-0432.CCR-17-3416 [DOI] [PubMed] [Google Scholar]
- 49.Xu X, Yang Y, Liu X, Cao N, Zhang P, et al. (2020) NFE2L2/KEAP1 Mutations Correlate with Higher Tumor Mutational Burden Value/PD-L1 Expression and Potentiate Improved Clinical Outcome with Immunotherapy. Oncologist. 10.1634/theoncologist.2019-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong Y, Hellyer JA, Stehr H, Hoang NT, Niu X, et al. (2020) Role of KEAp1/NFE2L2 mutations in the chemotherapeutic response of patients with non–small cell lung cancer. Clinical Cancer Research 26: 274–281. 10.1158/1078-0432.CCR-19-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Namani A, Cui QQ, Wu Y, Wang H, Wang XJ, et al. (2017) NRF2-regulated metabolic gene signature as a prognostic biomarker in non-small cell lung cancer. Oncotarget 8: 69847–69862. 10.18632/oncotarget.19349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tung MC, Lin PL, Wang YC, He TY, Lee MC, et al. (2015) Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 6: 41692–41705. 10.18632/oncotarget.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, et al. (2017) Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23: 1362–1368. 10.1038/nm.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai MC, Chen M, Ma P, Wu J, Lu H, et al. (2019) Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2‐mutant lung cancer John Wiley & Sons, Inc 144. [DOI] [PubMed] [Google Scholar]
- 55.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, et al. (2017) Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov 7: 86–101. 10.1158/2159-8290.CD-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellyer JA, Stehr H, Das M, Padda SK, Ramchandran K, et al. (2019) Impact of KEAP1/NFE2L2/CUL3 mutations on duration of response to EGFR tyrosine kinase inhibitors in EGFR mutated non-small cell lung cancer Elsevier BV; 134. [DOI] [PubMed] [Google Scholar]
- 57.Chen X, Cao X, Xiao W, Li B, Xue Q (2020) PRDX5 as a novel binding partner in Nrf2-mediated NSCLC progression under oxidative stress. Aging 12: 122–137. 10.18632/aging.102605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Best SA, De Souza DP, Kersbergen A, Policheni AN, Dayalan S, et al. (2018) Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab 27: 935–943 e934. 10.1016/j.cmet.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 59.Wang N, Song L, Xu Y, Zhang L, Wu Y, et al. (2019) Loss of Scribble confers cisplatin resistance during NSCLC chemotherapy via Nox2/ROS and Nrf2/PD-L1 signaling. EBioMedicine 47: 65–77. 10.1016/j.ebiom.2019.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
A. Forest plot of studies evaluating the relationship between NRF2 expression and gender. B. Forest plot of studies evaluating the relationship between NRF2 expression and smoking. C. Forest plot of studies evaluating the relationship between NRF2 expression and histopathology. D. Forest plot of studies evaluating the relationship between NRF2 expression and tumour differentiation type.
(TIF)
A., Lymph node metastasis; B., PFS
(TIF)
A., Metastasis; B., Lymph node metastasis; C., TNM stage (III~IV vs. I~II); D.,TNM stage (IV vs. III); E., Treatment response rate; F., OS; G., PFS; H., Gender; I., Smoking; J. Histopathology; K. Differentiation type.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.