Abstract
Background
Tumors of the metastatic colorectal cancer (mCRC) patients that are wildtype (WT) for KRAS or NRAS mutations respond more favorably to anti-epidermal growth factor receptor (EGFR) treatments. Treatment guidelines now recommend that all mCRC patients have WT KRAS and NRAS tumor status confirmed prior to initiating anti-EGFR therapy. Evidence also suggests that BRAF mutations may predict lack of response to anti-EGFR therapy. As such, there is now a need for comprehensive data on the prevalence of KRAS, NRAS, and BRAF mutations among patients with mCRC.
Methods
A systematic literature review was conducted among studies that described the prevalence of KRAS, NRAS, and BRAF gene mutations in mCRC patients. Observational cohort studies and standard of care arm of randomized clinical trials were included. Random effects meta-analysis models were used to create summary prevalence estimates for each of the mutation types. Subgroup analyses were also conducted to identify potential sources of heterogeneity. Exploratory analyses of overall and progression-free survival by mutation status were also conducted.
Results
This systematic review and meta-analysis included 275 studies comprising 77,104 mCRC patients. The summary prevalence estimate was 35.9% for KRAS mutations, 7.1% for BRAF mutations, and 4.1% for NRAS mutations. Female patients had significantly more KRAS and BRAF mutations than males, and significant variation by study location was observed for both KRAS and BRAF mutation prevalence. Overall survival was significantly decreased for patients with KRAS, BRAF, and NRAS mutations compared to those with WT tumors. Progression-free survival was also significantly decreased among patients with KRAS and BRAF mutations.
Conclusions
KRAS, NRAS, and BRAF mutation statuses in patients with mCRC are important predictors of treatment success and may also have prognostic value. In this paper we present the first systematic and comprehensive literature review and meta-analysis of the prevalence of KRAS, BRAF, and NRAS mutations and demonstrate the prognostic impact of mutation status on survival.
Keywords: Metastatic colorectal cancer, KRAS, BRAF, NRAS, Meta-analysis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide in men and the second most in women [1]. Approximately 20-25% of new CRC cases are metastatic (mCRC) at diagnosis and up to 50% of all patients eventually develop metastatic disease [2-5]. During the last decade, improvements in the treatment of patients with mCRC have increased median survival time from 12 to 21 months [6]. This improvement is partly due to the development of therapies that target the epidermal growth factor receptor (EGFR), primarily panitumumab and cetuximab [7]. Numerous studies now show that mutations in codon 12 or 13 of exon 2 of the KRAS gene render tumors essentially insensitive to treatment with EGFR inhibitors [8-17], while anti-EGFR therapies have demonstrated a clear clinical benefit in patients with RAS wildtype (WT) mCRC [8, 18, 19].
KRAS, a GTPase signaling protein, is the human homolog of the Kirsten rat sarcoma-2 virus oncogene and regulates cell proliferation, differentiation, and survival [20]. Approximately 30-50% of CRC tumors have mutations in exon 2, which result in constitutive signaling by decreasing GTPase activity [21-23]. More recent evidence shows that NRAS mutations and KRAS mutations outside of exon 2 also predict treatment failure with either cetuximab or panitumumab, which led the National Comprehensive Cancer Network and other major oncology societies to recommend genotyping of tumor tissue in all patients with mCRC for RAS (KRAS exon 2 and non-exon 2; NRAS) and BRAF at diagnosis of stage IV disease and prior to initiation of anti-EGFR treatment [8, 24-26]. In addition, the Food and Drug Administration (FDA) labels for cetuximab and panitumumab specifically state that these agents are not recommended for the treatment of mCRC characterized by KRAS exon 2 mutations. Although it is known that KRAS/NRAS mutations predict a lack of response to anti-EGFR treatments, many tumors that are WT with respect to both KRAS and NRAS still do not respond to these therapies causing researchers to look downstream of KRAS/NRAS for additional predictive biomarkers [8].
Mutations in the BRAF gene, specifically Val600Glu, are believed to render the protein product constitutively active thereby bypassing EGFR inhibition by anti-EGFR agents; however, the role of BRAF mutations as a predictive marker of anti-EGFR treatment failure is still uncertain [8, 27-29]. Other BRAF mutations, such as Asp594Gly, have not been well-characterized, and their significance in CRC is not well understood [30, 31]. Nevertheless, the presence of BRAF mutations is a strong prognostic factor as mCRC patients with a BRAF mutation have significantly shorter overall survival (OS) than those with WT tumors [32-40].
Results regarding the prognostic value of KRAS and NRAS mutations are mixed [8]. A recent meta-analysis identified that mutations in KRAS exons 3 and 4 and BRAF were associated with significantly worse odds of survival with anti-EGFR treatment compared to WT tumors (odds ratio (OR): 0.26 and 0.29, respectively) [41]. As such, there is now a need for more information on the prevalence of KRAS, NRAS, and BRAF mutations and their impact on survival among patients with mCRC.
The objectives of this systematic review and meta-analysis were to: 1) provide a comprehensive assessment of KRAS, NRAS, and BRAF mutation prevalence among mCRC patients as reported in the scientific literature; 2) identify potential sources of heterogeneity in mutation prevalence by conducting pre-specified subgroup analyses; and 3) conduct an exploratory analysis to describe OS and progression-free survival (PFS) for mCRC patients by KRAS, NRAS, and BRAF mutation status.
Materials and Methods
This review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [42]. Methods and article inclusion criteria were specified in advance and documented in a protocol. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Eligibility criteria
We included peer-reviewed articles published between January 1, 2006 and May 10, 2016 (the most recent 10-year period at time of search) that described the prevalence of KRAS, NRAS, or BRAF gene mutations in mCRC patients in observational studies (either treated or untreated patients) and in the standard of care arm of clinical trials. Though not truly population-based, data on prevalence of the specific genetic mutations from the standard of care arm were considered for the review in order to understand and characterize the prevalence of these mutations in patients who may choose to participate in a clinical trial. Only clinical trials in which the patients were not selected based on mutation status were included.
Eligible analytical study designs were randomized trials, prospective or retrospective observational cohort studies, cross-sectional, and case-control studies. We excluded opinion pieces, editorials, articles without original data (except systematic reviews), case reports and/or case series with less than 20 cases, and experimental animal studies. We excluded studies that did not specifically report prevalence statistics on gene mutations for mCRC patients, studies conducted in pediatric populations, and studies published in languages other than English.
Literature searches and study selection
Electronic literature searches were conducted in the PubMed, Embase, Web of Science, Cochrane Library, and ClinicalTrials.gov databases to identify relevant articles. The bibliography sections from previously published systematic reviews were hand searched to identify additional individual articles of interest not captured by the electronic search engines. A PRISMA flow diagram for study inclusion is shown here (Supplementary Material 1, www.gastrores.org).
A variety of search string parameters were used in an effort to identify all eligible articles, including the following: colon, colorectal, neoplasms, cancer, secondary, metastatic, EGFR, proto-oncogene proteins p21 (RAS), proto-oncogene, proteins, p21 (ras), KRAS, NRAS, BRAF, epidemiology, prevalence, and frequency.
The titles and abstracts of all publications identified via the literature search databases were independently screened by two reviewers based on the inclusion/exclusion criteria outlined above. A standardized abstraction form, which included fields for data elements relevant to the key topics of interest was created in order to maximize reliability. All data elements were entered and then reviewed for quality control by a second researcher. Any disagreements were resolved by a senior epidemiologist. If more than one article from the same study population was published (based on author names, sample sizes, intervention comparisons or outcomes), data from the publication(s) with the longest follow-up or most relevant population and/or outcomes were extracted. Multiple articles from the same study population were included if unique outcomes from the same population (i.e., prevalence of different mutations) were included.
The following qualitative information and quantitative data were abstracted from each of the studies: 1) characteristics of the study (including study design, study dates, name of cohort, NCT number, location (country), study population description, individual study inclusion/exclusion criteria, sample size, number of subjects with mutation results, reason why mutation results were unavailable for some subjects, and the number of subjects with inconclusive results; 2) characteristics of study participants (including sex, age, race/ethnicity, comorbidities, ECOG-PS, location of primary tumor, stage at first diagnosis, number of metastases, and treatment status); 3) characteristics of study outcomes (including length of follow-up, type of mutation, specific mutation location, mutation assessment method, exon/codon numbers, mutation prevalence, whether mutation data were from primary or metastatic tumor, whether mutation data were obtained from tissue or liquid biopsy using plasma, median overall and PFS, and survival hazard ratios (HRs)).
Statistical analysis
Prevalence data were extracted from each study for each mutation and for each of the subgroups of interest where available. Random effects models were used to generate summary prevalence estimates of KRAS, BRAF, and NRAS gene mutations among mCRC patients and within subgroups of interest (specific mutation location, sex, race, median age, study time period, length of follow-up time, treatment status, study location, study design, source of tumor tissue, mutation assessment method, and the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) score) to identify potential sources of heterogeneity. Of note, the analysis for heterogeneity by mutation assessment method evaluated whether the prevalence differed by different assessment method across studies. No direct comparison of testing methods was performed and the results of these analyses should not be interpreted as sensitivity or specificity analyses of mutation assessment method testing. For median age, and length of follow-up time, the median value was calculated across studies and subgroup analyses were conducted for studies above and below the median value. Studies were weighted according to the inverse of their variance using the method proposed by DerSimonian and Laird [43]. Heterogeneity between studies and subgroups in each analysis was evaluated using the Cochran’s Q and I2 statistical tests. Meta-regression analyses were conducted to explore the incremental effect of median age and length of follow-up time on prevalence. All prevalence analyses were performed using Comprehensive Meta-Analysis Software (version 3.0; Biostat, Englewood, New Jersey, USA).
Exploratory survival analysis
In addition to prevalence of KRAS, NRAS, and BRAF gene mutations in mCRC patient population, we also conducted an exploratory analysis of the median OS and PFS in the mutant versus WT populations. Median OS, median PFS, HRs for survival, and corresponding 95% confidence intervals (CIs) were abstracted from the literature if available. This analysis was exploratory in that a specific search was not performed for survival in these populations nor was it necessary for inclusion in the study. Random effects models were used to meta-analyze the HRs of OS and PFS for mutant versus WT populations using Comprehensive Meta-Analysis Software. Random effects models of the log-transformed rates were used for the meta-analyses of median survival, which were performed using the “metan” command in Stata 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC).
Assessment of study quality and reporting bias
Individual study quality was assessed using STROBE guidelines for reporting in observational studies [44]. The STROBE statement is a checklist of 22 items considered as essential for good reporting of observational studies. In our assessment of study quality, individual articles were given 1 point for each of the 22 checklist items addressed in the article.
Publication bias was assessed visually with funnel plots and statistically using Egger’s regression method. For analyses with statistically significant publication bias, the Duval and Tweedie trim and fill method was used to calculate the adjusted estimate after adding in hypothetical “missing” studies due to lack of publication [45].
Results
Study characteristics
The PRISMA flow diagram for study selection is shown in here (Supplementary Material 1, www.gastrores.org). From a total of 786 de-duplicated articles, 275 studies comprising 77,104 mCRC patients were included in the meta-analyses. Individual study characteristics are described in here (Supplementary Material 2, www.gastrores.org). The study designs consisted of randomized and non-randomized clinical trials (n = 64), cross-sectional studies (n = 11), and prospective (n = 66) and retrospective (n = 134) observational studies. Study populations were drawn from all continents apart from Antarctica and Africa. Most studies were conducted in Europe (n = 132), followed by Asia (n = 63), and the USA (n = 41). The studies included in our meta-analysis ranged in STROBE score from 5 to 22 with a mean of 15.2 and median of 16. All studies that received scores of less than 10, except Smith 2014 [46], were studies in which only the abstract was available. Therapies and combination regimens varied substantially across studies, with many studies not reporting specific treatments used by included patients. Thus, treatment status was classified into three groups: partial population treated, complete population treated, and unknown/not treated. Only two studies specified that patients were not treated [47, 48] and thus this category was collapsed with “unknown.”
Synthesis of results
Mutations were categorized inconsistently in the included articles. Some were described only by exon number, only by codon number, or a combination of exon and codon numbers, while other papers provided specific mutation locations. Additionally, the majority of papers placed codons 12 and 13 within exon 2 for KRAS, BRAF, and NRAS, while some papers placed these codons within exon 1 or did not specify an exon number. When papers provided only an exon number, it was not always clear whether the entire exon was evaluated for the presence of mutations or if only the presence of certain hot spot mutations was evaluated. To make the categories as consistent as possible, we only analyzed the prevalence of mutations exactly how they were described in the paper and did not collapse categories. For example, in the table of KRAS mutation prevalence (Table 1), the exon 2 category only includes the papers that specifically described the location of KRAS mutations as occurring within exon 2 and is the most global estimate of exon 2 provided within a paper. There are two categories each for codons 12 and 13. The first category for each includes only prevalence data where the codon was specified as specifically existing within exon 2; and the second category for each of codons 12 and 13 includes the papers that described this codon as existing within exon 1 or 2 or did not specify a particular exon. To identify potential patterns of heterogeneity with regard to mutation prevalence, we performed subgroup analyses on relevant study characteristics (i.e., sex, study location, study design, source, mutation assessment, and study quality). Subgroup analyses were only performed on the overall KRAS, BRAF, or NRAS mutation prevalence estimate for each study rather than by specific mutation location.
Table 1. Overall Prevalence of KRAS Mutations in mCRC and Subgroups.
| N studiesf | Summary prevalence (95% CI) | P-Het; I2 | P value for heterogeneity within subgroups | |
|---|---|---|---|---|
| Overall | 288 | 35.9% (34.6% - 37.3%) | < 0.001; 92.6% | |
| Exon 1a | 14 | 35.5% (30.3% - 41.1%) | < 0.001; 75.4% | |
| Exon 2a | 76 | 39.1% (37.1% - 41.3%) | < 0.001; 85.4% | |
| Codon 12b | 24 | 29.1% (26.3% - 32.1%) | < 0.001; 86.2% | |
| Codon 12c | 50 | 29.0% (26.9% - 31.2%) | < 0.001; 83.9% | |
| Gly12Ala | 59 | 3.3% (2.7% - 3.9%) | < 0.001; 77.8% | < 0.001 |
| Gly12Arg | 36 | 0.9% (0.6% - 1.4%) | < 0.001; 82.9% | |
| Gly12Asp | 85 | 14.6% (13.3% - 16.1%) | < 0.001; 90.8% | |
| Gly12Cys | 61 | 3.5% (3.0% - 4.1%) | < 0.001; 76.1% | |
| Gly12Phe | 5 | 0.2% (0.1% - 0.6%) | 0.035; 61.3% | |
| Gly12Ser | 62 | 2.8% (2.2% - 3.6%) | < 0.001; 88.8% | |
| Gly12Val | 87 | 10.2% (9.1% - 11.4%) | < 0.001; 89.6% | |
| Other codon 12 | 15 | 0.9% (0.3% - 2.9%) | < 0.001; 96.9% | |
| Codon 13b | 19 | 9.0% (7.9% - 10.2%) | < 0.001; 62.7% | |
| Codon 13c | 39 | 8.6% (7.7% - 9.5%) | < 0.001; 55.2% | |
| Gly13Arg | 8 | 0.2% (0.1% - 0.7%) | 0.001; 72.0% | < 0.001 |
| Gly13Asp | 82 | 8.6% (7.6% - 9.6%) | < 0.001; 85.0% | |
| Gly13Cys | 15 | 0.7% (0.4% - 1.2%) | < 0.001; 82.7% | |
| Gly13Ser | 4 | 0.3% (0.0% - 4.8%) | < 0.001; 90.9% | |
| Gly13Val | 8 | 0.4% (0.1% - 1.8%) | < 0.001; 80.2% | |
| Exon 3a | 9 | 2.9% (1.9% - 4.4%) | 0.009; 60.9% | |
| Codon 61 | 14 | 2.7% (2.1% - 3.5%) | 0.001; 62.0% | |
| Gln61Arg | 4 | 0.4% (0.2% - 0.6%) | 0.810; 0.0% | 0.016 |
| Gln61His | 11 | 1.6% (0.7% - 3.5%) | < 0.001; 92.3% | |
| Gln61Leu | 5 | 0.7% (0.2 %- 2.2%) | 0.004; 73.9% | |
| Other codon 61 | 3 | 0.2% (0.0% - 1.8%) | < 0.001; 89.3% | |
| Exon 4a | 8 | 5.0% (4.1% - 6.2%) | 0.228; 25.2% | |
| Codon 146 | 9 | 2.5% (1.9% - 3.3%) | 0.134; 35.5% | |
| P value between exonsd | < 0.001 | |||
| Sex | ||||
| Male | 55 | 37.3% (35.1% - 39.7%) | < 0.001; 76.9% | 0.011 |
| Female | 57 | 42.2% (39.3% - 45.2%) | < 0.001; 81.7% | |
| Median age of study population | ||||
| < 62 | 77 | 37.0% (34.3% - 39.8%) | < 0.001; 92.6% | 0.544 |
| ≥ 62 | 102 | 36.0% (33.8% - 38.1%) | < 0.001; 90.6% | |
| Meta-regression on median age | 179 | 0.004 (-0.012 - 0.020) | N/A | 0.612 |
| Race | ||||
| < 88% White/Caucasian | 21 | 37.8% (32.4% - 43.4%) | < 0.001; 94.1% | 0.401 |
| ≥ 88% White/Caucasian | 17 | 34.1% (28.1% - 40.7%) | < 0.001; 91.1% | |
| Study location | ||||
| Asia | 64 | 31.8% (28.3% - 35.5%) | < 0.001; 93.2% | 0.025 |
| Australia | 7 | 27.3% (19.1% - 37.5%) | < 0.001; 97.9% | |
| Europe | 146 | 37.3% (35.2% - 39.3%) | < 0.001; 92.0% | |
| Multi-country | 21 | 39.4% (36.1% - 42.9%) | < 0.001; 88.2% | |
| North America | 47 | 36.4% (33.4% - 39.6%) | < 0.001; 88.9% | |
| South America | 3 | 37.2% (28.7% - 49.9%) | < 0.001; 70.8% | |
| Study design | ||||
| Observational | 226 | 35.0% (33.5% - 36.6%) | < 0.001; 92.9% | 0.004 |
| Clinical trial | 62 | 39.5% (36.9% - 42.1%) | < 0.001; 89.0% | |
| Treatment status | ||||
| Partial population treated | 10 | 27.5% (18.0% - 39.6%) | < 0.001; 95.0% | 0.115 |
| Complete population treated | 158 | 37.2% (35.6% - 38.9%) | < 0.001; 89.6% | |
| Unknown/not treated | 120 | 35.1% (32.9% - 37.4%) | < 0.001; 94.2% | |
| Source | ||||
| Primary tumor | 60 | 18.9% (15.6% - 22.7%) | < 0.001; 94.6% | 0.012 |
| Metastasis | 37 | 21.2% (16.4% - 27.0%) | < 0.001; 94.3% | |
| Both primary tumors and metastases | 42 | 26.5% (23.0% - 30.4%) | < 0.001; 96.4% | |
| Mutation assessment methode | ||||
| Gel electrophoresis methods | 13 | 41.1% (35.7% - 46.6%) | 0.001; 65.5% | |
| High-resolution melting | 18 | 28.8% (23.7% - 34.6%) | < 0.001; 97.3% | |
| Mass spectrometry | 17 | 36.0% (32.2% - 40.0%) | < 0.001; 92.2% | |
| Multiplex mutation assays | 20 | 36.4% (33.1% - 39.9%) | < 0.001; 87.2% | |
| Mutant allele specific PCR | 77 | 37.4% (35.1% - 39.8%) | < 0.001; 92.2% | |
| Next-generation sequencing | 11 | 29.7% (20.1% - 41.7%) | < 0.001; 97.1% | |
| Pyrosequencing | 38 | 37.2% (33.9% - 40.6%) | < 0.001; 88.9% | |
| Sanger/direct sequencing (PCR) | 144 | 35.5% (34.0% - 37.0%) | < 0.001; 86.3% | |
| Strip assay | 7 | 42.1% (38.9% - 45.3%) | 0.045; 53.4% | |
| Other | 16 | 44.0% (37.4% - 50.7%) | < 0.001; 96.3% | |
| Not reported | 14 | 40.3% (33.1% - 47.9%) | < 0.001; 96.1% | |
| Study quality score | ||||
| ≤ 16 | 172 | 35.0% (33.1% - 37.0%) | < 0.001; 94.5% | 0.073 |
| > 16 | 116 | 37.5% (35.7% - 39.3%) | < 0.001; 85.4% | |
| Study time period | ||||
| Pre-2007 | 22 | 35.5% (32.3% - 38.9%) | < 0.001; 63.9% | 0.105 |
| Includes 2007 | 115 | 36.0% (33.7% - 38.4%) | < 0.001; 90.0% | |
| Post-2007 | 61 | 39.3% (36.7% - 42.0%) | < 0.001; 91.5% | |
| Median length of follow-up time | ||||
| < 25 months | 29 | 34.6% (31.1% - 38.3%) | < 0.001; 80.9% | 0.608 |
| ≥ 25 months | 34 | 33.0% (28.1% - 38.3%) | < 0.001; 91.7% | |
| Meta-regression on follow-up time | 63 | 0.002 (-0.004 - 0.008) | N/A | 0.583 |
aGlobal exon estimate (most global estimate for a given exon contained within a manuscript); bCodon specified in papers as existing within exon 2; cCodon specified in papers as existing within exon 1, exon 2, or unspecified; dP values assess differences between global exon estimates only; eCategories not mutually exclusive as some studies used multiple forms of mutation assessment methods, P value not calculated; fIndividual studies may contribute multiple prevalence estimates derived from independent study arms; so N refers to the number of independent prevalence estimates included in summary measure and may exceed the actual number of studies. mCRC: metastatic colorectal cancer; CI: confidence interval.
Prevalence of KRAS mutations
The overall summary prevalence estimate (Psum) of KRAS mutations was 35.9% (95% CI: 34.6% - 37.3%) based on 288 individual prevalence estimates. KRAS mutations most frequently occurred in exon 2 (Psum = 39.1%, 95% CI: 37.1% - 41.3%) and exon 1 (Psum = 35.5%, 95% CI: 30.3% - 41.1%), with considerably lower prevalence estimates in exon 4 (Psum = 5.0%, 95% CI: 4.1% - 06.2%) and exon 3 (Psum = 2.9%, 95% CI: 1.9% - 4.4%). Within exon 2, mutations in codon 12 were more common than in codon 13 (Table 1). Of the specific codon 12 mutations that were examined, Gly12ASP and Gly12Val were the most prevalent. Of the specific codon 13 mutations, Gly13Asp was the most prevalent. Codon 61 and codon 146 mutations were relatively infrequent. KRAS mutation prevalence was slightly higher in females compared with males (P = 0.011), and in clinical trials compared with observational studies (P = 0.004). KRAS mutation prevalence varied significantly by study location (P = 0.025). KRAS mutations were more prevalent when strip assays or gel electrophoresis methods were used as the assessment method and least prevalent when assessed by next-generation sequencing or high-resolution melting. As studies used multiple assessment methods to identify mutations, a P value could not be calculated for the mutation assessment method subgroup as the groups were not mutually exclusive. The prevalence did not differ significantly by STROBE score, median age of study population, race, treatment status, study time period, or length of follow-up.
Prevalence of BRAF mutations
The overall Psum of BRAF mutations was 7.1% (95% CI: 6.5% - 07.8%). Val600Glu mutations were statistically significantly more common than Asp594Gly mutations (P = 0.002). BRAF mutations were more common in females than in males (P = 0.018) and varied by study location (P = 0.002). BRAF mutation prevalence did not differ significantly by median age, race, study design, treatment status, source of tumor tissue (primary, metastases, both), STROBE score, study time period, or median length of follow-up (Table 2).
Table 2. Overall Prevalence of BRAF Mutations in mCRC and Subgroups.
| N studiesc | Summary prevalence (95% CI) | P-Het; I2 | P value for heterogeneity within subgroups | |
|---|---|---|---|---|
| Overall | 142 | 7.1% (6.5% - 7.8%) | < 0.001; 66.3% | |
| Exon 15a | 44 | 7.0% (6.1% - 8.1%) | 0.001; 45.8% | |
| Val600Glu (V600E) | 75 | 6.8% (5.8% - 7.9%) | < 0.001; 76.9% | 0.002 |
| Asp594Gly | 4 | 0.6% (0.1% - 2.8%) | 0.064; 58.8% | |
| Sex | ||||
| Male | 12 | 7.9% (6.5% - 9.7%) | 0.076; 39.7% | 0.018 |
| Female | 12 | 11.0% (9.2% - 13.1%) | 0.260; 18.7% | |
| Median age of study population | ||||
| < 62 | 35 | 7.8% (6.6% - 9.1%) | < 0.001; 55.5% | 0.970 |
| ≥ 62 | 57 | 7.8% (6.7% - 9.1%) | < 0.001; 68.6% | |
| Meta-regression on median age | 92 | 0.012 (-0.014 - 0.036) | N/A | 0.390 |
| Race | ||||
| < 88% White/Caucasian | 7 | 7.1% (5.5% - 9.2%) | 0.915; 0.0% | 0.855 |
| ≥ 88% White/Caucasian | 5 | 7.6% (4.1% - 13.7%) | 0.007; 71.7% | |
| Study location | ||||
| Asia | 29 | 6.0% (5.1% - 7.0%) | 0.175; 19.6% | 0.002 |
| Australia | 7 | 11.1% (8.6% - 14.2%) | 0.062; 49.9% | |
| Europe | 79 | 7.2% (6.3% - 8.1%) | < 0.001; 68.2% | |
| Multi-country | 8 | 8.3% (5.4% - 12.5%) | < 0.001; 83.3% | |
| North America | 19 | 6.6% (5.1% - 8.4%) | 0.001; 56.5% | |
| Study design | ||||
| Observational | 114 | 7.2% (6.5% - 7.9%) | < 0.001; 65.4% | 0.897 |
| Clinical trial | 28 | 7.1% (5.7% - 8.7%) | < 0.001; 70.5% | |
| Treatment status | ||||
| Partial population treated | 8 | 5.9% (3.9% - 8.9%) | 0.168; 32.6% | 0.659 |
| Complete population treated | 76 | 7.2% (6.3% - 8.3%) | < 0.001; 72.1% | |
| Unknown/not treated | 58 | 7.1% (6.3% - 8.0%) | < 0.001; 57.3% | |
| Source | ||||
| Primary tumor | 34 | 7.5% (6.3% - 8.7%) | 0.152; 20.1% | 0.832 |
| Metastasis | 16 | 6.4% (3.8% - 10.6%) | < 0.001; 68.3% | |
| Both primary tumors and metastases | 20 | 7.1% (5.8% - 8.5%) | 0.039; 39.0% | |
| Mutation assessment methodb | ||||
| Gel electrophoresis methods | 6 | 4.5% (2.5% - 7.9%) | 0.754; 0.0% | |
| High-resolution melting | 13 | 8.0% (5.9% - 10.9%) | < 0.001; 79.0% | |
| Mass spectrometry | 13 | 5.4% (4.4% - 6.6%) | 0.044; 44.2% | |
| Multiplex mutation assays | 10 | 7.5% (5.8% - 9.6%) | 0.170; 29.9% | |
| Mutant allele specific PCR | 35 | 8.1% (7.0% - 9.2%) | < 0.001; 57.0% | |
| Next-generation sequencing | 6 | 7.4% (5.7% - 9.5%) | 0.974; 0.0% | |
| Not reported | 7 | 7.1% (4.6% - 10.9%) | 0.010; 64.2% | |
| Other | 6 | 3.9% (2.3% - 6.4%) | 0.004; 71.2% | |
| Pyrosequencing | 19 | 9.1% (7.2% - 11.4%) | < 0.001; 74.3% | |
| Sanger/direct sequencing (PCR) | 69 | 6.8% (6.1% - 7.6%) | 0.004; 33.6% | |
| Study quality score | ||||
| ≤ 16 | 70 | 7.2% (6.4% - 8.0%) | < 0.001; 58.5% | 0.803 |
| > 16 | 72 | 7.0% (6.1% - 8.1%) | < 0.001; 70.7% | |
| Study time period | ||||
| Pre-2007 | 10 | 6.3% (4.2% - 9.5%) | 0.038; 49.4% | 0.520 |
| Includes 2007 | 59 | 7.4% (6.2% - 8.7%) | < 0.001; 73.1% | |
| Post-2007 | 28 | 6.3% (4.2% - 9.5%) | 0.098; 26.7% | |
| Median length of follow-up time | ||||
| < 25 months | 12 | 9.7% (6.1% - 15.0%) | < 0.001; 71.9% | 0.271 |
| ≥ 25 months | 24 | 7.4% (6.1% - 8.8%) | 0.002; 51.6% | |
| Meta-regression on follow-up time | 36 | -0.005 (-0.015; 0.005) | N/A | 0.314 |
aGlobal exon estimate (most global estimate for a given exon contained within a manuscript); bCategories not mutually exclusive as some studies used multiple forms of mutation assessment methods, P value not calculated; cIndividual studies may contribute multiple prevalence estimates derived from independent study arms, so N refers to the number of independent prevalence estimates included in summary measure and may exceed the actual number of studies. mCRC: metastatic colorectal cancer; CI: confidence interval.
Prevalence of NRAS mutations
The overall prevalence of NRAS mutations was 4.1% (95% CI: 3.5% - 4.8%). Mutation prevalence did not differ significantly by exon number (P = 0.368). Mutations were more prevalent in codons 12 and 61 compared with codon 13. NRAS mutation prevalence by sex was only assessed in one study which determined a similar prevalence in both males and females. NRAS mutation prevalence also did not differ significantly by median age, study location, study design, treatment status, source of tumor tissue (primary, metastases, both), STROBE score, study time period, or median length of follow-up ≥ 25 months (Table 3).
Table 3. Overall Prevalence of NRAS Mutations in mCRC and Subgroups.
| N Studiese | Summary prevalence (95% CI) | P-Het; I2 | P value for heterogeneity within subgroups | |
|---|---|---|---|---|
| Overall | 49 | 4.1% (3.5% - 4.8%) | < 0.001; 56.0% | |
| Exon 2a | 6 | 2.3% (1.4% - 3.7%) | 0.127; 41.8% | |
| Codon 12b | 7 | 2.2% (1.6% - 3.0%) | 0.209; 28.7% | < 0.001f |
| Codon 13b | 4 | 0.7% (0.5% - 1.0%) | 0.927; 0.0% | |
| Exon 3a | 5 | 2.4% (1.1% - 5.0%) | 0.003; 75.5% | |
| Codon 61b | 10 | 3.6% (2.6% - 5.0%) | 0.065; 44.1% | |
| Exon 4a | 3 | 0.6% (0.1% - 3.9%) | 0.991; 0.0% | |
| P value between exonsc | 0.368 | |||
| Sex | ||||
| Male | * | * | * | * |
| Female | * | * | * | |
| Median age of study population | ||||
| < 62 | 12 | 3.7% (3.0% - 4.7%) | 0.863; 0.0% | 0.479 |
| ≥ 62 | 12 | 4.3% (3.2% - 5.6%) | 0.036; 47.1% | |
| Meta-regression on median age | 24 | 0.006 (-0.048 - 0.060) | N/A | 0.822 |
| Race | ||||
| < 88% White/Caucasian | * | * | * | * |
| ≥ 88% White/Caucasian | 3 | 5.8% (3.4% - 9.8%) | 0.954; 0.0% | |
| Study location | ||||
| Asia | 13 | 3.0% (2.0% - 4.5%) | 0.009; 54.5% | 0.108 |
| Australia | * | * | * | |
| Europe | 22 | 4.6% (3.7% - 5.8%) | < 0.001; 64.6% | |
| Multi-country | 3 | 5.8% (3.4% - 9.8%) | 0.954; 0.0% | |
| North America | 10 | 3.6% (2.9% - 4.5%) | 0.980; 0.0% | |
| Study design | ||||
| Observational | 39 | 3.9% (3.3% - 4.7%) | < 0.001; 61.9% | 0.105 |
| Clinical trial | 10 | 5.3% (3.9% - 7.1%) | 0.544; 0.0% | |
| Treatment status | ||||
| Partial population treated | 5 | 3.5% (2.1% - 6.0%) | 0.940; 0.0% | 0.840 |
| Complete population treated | 15 | 4.2% (3.3% - 5.3%) | 0.336; 10.5% | |
| Unknown/not treated | 29 | 4.1% (3.3% - 5.0%) | < 0.001; 69.5% | |
| Source | ||||
| Primary tumor | 8 | 3.4% (2.4% - 4.8%) | 0.026; 55.9% | 0.318 |
| Metastasis | 6 | 4.9% (3.4% - 7.0%) | 0.936; 0.0% | |
| Both primary tumors and metastases | 9 | 3.5% (2.5% - 4.9%) | 0.019; 56.3% | |
| Mutation assessment methodd | ||||
| High-resolution melting | 6 | 3.0% (1.7% - 5.1%) | < 0.001; 84.6% | |
| Mass spectrometry | 11 | 4.0% (3.2% - 5.0%) | 0.023; 51.8% | |
| Multiplex mutation assays | 4 | 4.2% (3.3% - 5.4%) | 0.233; 29.8% | |
| Mutant allele specific PCR | 7 | 4.3% (3.4% - 5.3%) | 0.253; 23.1% | |
| Next-generation sequencing | 8 | 4.9% (3.9% - 6.2%) | 0.309; 15.4% | |
| Other | 3 | 4.5% (2.6% - 7.6%) | 0.064; 63.6% | |
| Pyrosequencing | 9 | 5.1% (3.6% - 7.3%) | < 0.001; 72.4% | |
| Sanger/direct sequencing (PCR) | 20 | 4.5% (4.0% - 5.0%) | 0.483; 0.0% | |
| Strip assay | * | * | * | |
| Study quality score | ||||
| ≤ 16 | 29 | 3.8% (3.0% - 4.8%) | < 0.001; 69.0% | 0.151 |
| > 16 | 20 | 4.7% (4.0% - 5.5%) | 0.535; 0.0% | |
| Study time period | ||||
| Pre-2007 | * | * | * | |
| Includes 2007 | 9 | 3.7% (2.9% - 4.7%) | 0.447; 0.0% | 0.247 |
| Post-2007 | 26 | 4.5% (3.6% - 5.6%) | < 0.001; 63.2% | |
| Median length of follow-up time | ||||
| < 25 months | * | * | * | * |
| ≥ 25 months | 7 | 4.1% (3.0% - 5.5%) | 0.998; 0.0% | |
| Meta-regression on follow-up time | 0.000 (-0.012 - 0.012) | N/A | 0.957 |
aGlobal exon estimate (most global estimate for a given exon contained within a manuscript); bCodon specified in papers as existing within exon 1, exon 2, exon 3 or unspecified; cP values assess differences between global exon estimates only; dCategories not mutually exclusive as some studies used multiple forms of mutation assessment methods, P value not calculated; eIndividual studies may contribute multiple prevalence estimates derived from independent study arms, so N refers to the number of independent prevalence estimates included in summary measure and may exceed the actual number of studies; fP value assesses differences between codon estimates. *N < 3, insufficient studies to run a meta-analysis. mCRC: metastatic colorectal cancer; CI: confidence interval.
OS and PFS by mutation status
In our exploratory analysis of survival based on the studies included in our assessment, a significantly increased risk of mortality was observed among patients with KRAS, BRAF, and NRAS mutations compared to patients with the respective WT tumors (Table 4). The highest risk was observed among patients with BRAF mutations compared to BRAF WT patients (HR = 2.83, 95% CI: 2.23 - 3.58). The risk of disease progression was also significantly higher among patients with BRAF mutations compared to WT (HR = 2.90; 95% CI: 1.84 - 4.56) as well as among patients with KRAS mutations versus WT (HR = 1.62, 95%: 1.33 - 1.97). No significant differences in survival by study population treatment status (complete population treated, partial population treated, unknown/not treated) was observed for OS in the KRAS or BRAF populations. An insufficient number of studies were available to evaluate OS by study population treatment status in the NRAS population or for PFS for any mutation (data not shown).
Table 4. Hazard Ratios and Median Overall and Progression-Free Survival by Mutation Status.
| Overall survival |
Progression-free survival |
|||||
|---|---|---|---|---|---|---|
| N studies | HR (95% CI) | P-Het; I2 | N studies | HR (95% CI) | P-Het; I2 | |
| Survival analysis (mutant vs. wild-type) | ||||||
| KRAS | 47 | 1.49 (1.37 - 1.64) | < 0.001; 50.2% | 25 | 1.62 (1.33 - 1.97) | < 0.001; 83.1% |
| BRAF | 32 | 2.83 (2.23 - 3.58) | < 0.001; 84.1% | 12 | 2.90 (1.84 - 4.56) | < 0.001; 91.3% |
| NRAS | 4 | 1.88 (1.35 - 2.62) | 0.795; 0.0% | * | * | * |
| Median survival | Median months (95% CI) | Median months (95% CI) | ||||
| KRAS mutant | 41 | 13.41 (11.35 - 15.83) | < 0.001; 96.8% | 42 | 4.48 (3.58 - 5.62) | < 0.001; 99.2% |
| KRAS wild-type | 42 | 17.00 (14.52 - 19.89) | < 0.001; 96.8% | 49 | 5.98 (4.74 - 7.54) | < 0.001; 99.3% |
| P value for difference | 0.022 | 0.04 | ||||
| BRAF mutant | 12 | 8.71 (6.54 - 11.59) | < 0.001; 90.1% | 11 | 5.05 (3.67 - 6.96) | < 0.001; 88.6% |
| BRAF wild-type | 13 | 17.19 (13.00 - 22.74) | < 0.001; 96.6% | 12 | 8.26 (6.78 - 10.07) | < 0.001; 93.5% |
| P value for difference | < 0.001 | 0.005 | ||||
*N < 3 (studies), insufficient for meta-analysis. mCRC: metastatic colorectal cancer; HR: hazard ratio; CI: confidence interval.
The median length of survival associated WT status was significantly longer than that associated with mutant status for both KRAS mutations (WT: 17.00 vs. mutant: 13.41 months, P = 0.022) and BRAF mutations (WT: 17.19 vs. mutant: 8.71 months, P < 0.001). Similar trends were observed for PFS for KRAS mutations (WT: 5.98 vs. mutant: 4.48 months, P = 0.04) and BRAF mutations (WT: 8.26 vs. mutant: 5.05 months, P = 0.005).
Risk of bias across studies
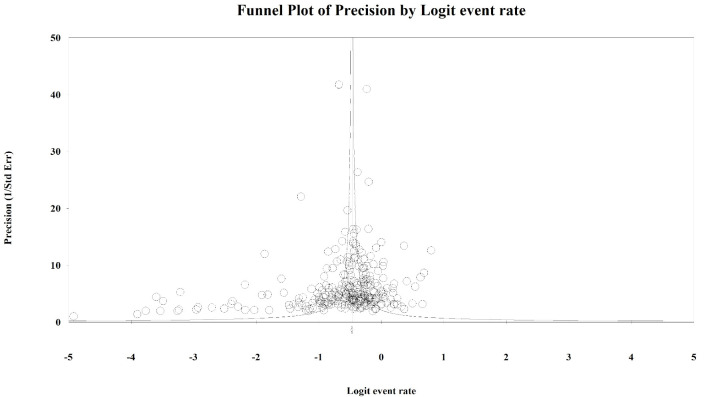
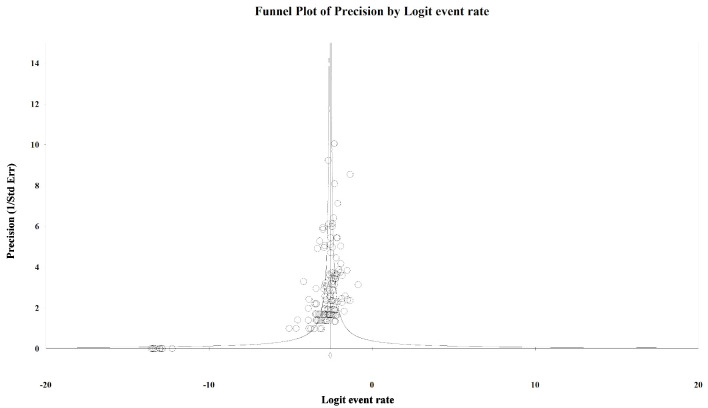
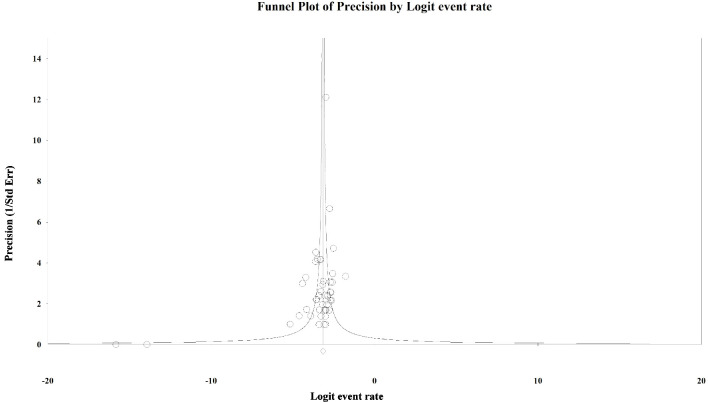
To evaluate the presence of publication bias among the studies, we generated funnel plots for the logit of the event rate by the standard error separately for each of the mutations types. Upon visual inspection of the association of the logit of the prevalence of KRAS mutations by precision for each of the studies, studies with higher precision (typically representing larger study populations) appeared to have a mutation prevalence closer to the mean although the plot was asymmetrical (Egger’s regression test: P value = 0.014, Fig. 1). The Duval and Tweedie method added 64 “missing” (hypothetically unpublished) studies to the left of the mean, resulting in an adjusted prevalence of 31.7% (95% CI: 30.3% - 33.2%), which is slightly less than the unadjusted prevalence of 35.9% (95% CI: 34.6% - 37.3%). This implies that the smaller, less precise studies may have been driving the prevalence up while smaller studies with lower prevalence estimates were not published. However, these are merely theoretical estimates of unknown potential sources of data. Visual inspection of the funnel plot of BRAF mutation prevalence by the precision for each of the studies was also asymmetrical around the summary effect measure (Egger’s regression test: P value = 0.001, Fig. 2). The Duval and Tweedie method added 45 “missing” studies to the right of the mean resulting in an adjusted prevalence estimate of 8.6% (95% CI: 7.8% - 9.5%), which is slightly higher than the unadjusted prevalence of 7.1% (95% CI: 6.5% - 7.8%). This may be due to the smaller studies with higher prevalence estimates being more likely to be published than smaller studies with lower prevalence estimates. Publication bias associated with the NRAS prevalence is unlikely due to the symmetry of the funnel plot (Egger’s regression test: P value = 0.112, Fig. 3).
Figure 1.
Publication bias: KRAS. Egger’s regression test: two-tailed P value = 0.014. Duval and Tweedie method added 64 “missing” studies to the left of the mean, resulting in an adjusted prevalence estimate of 31.7% (30.3% - 33.2%).
Figure 2.
Publication bias: BRAF. Egger’s regression test: two-tailed P value = 0.001. Duval and Tweedie method added 45 “missing” studies to the right of the mean, resulting in an adjusted prevalence estimate of 8.6% (7.8% - 9.5%).
Figure 3.
Publication bias: NRAS. Egger’s regression test: two-tailed P value = 0.112.
Discussion
To the authors’ knowledge, this is first global systemic literature review and meta-analysis of KRAS, BRAF, and NRAS mutation prevalence in mCRC patients. This extensive and systematic review included in 275 studies comprising 77,104 mCRC patients. Our study provided a pooled overall prevalence estimate of 35.9% for KRAS mutations, 7.1% for BRAF mutations, and 4.1% for NRAS mutations. Additionally, our exploratory survival analysis showed increased risk of death associated with KRAS, BRAF, and NRAS mutations when compared with the respective WT tumors. Tumors with KRAS and BRAF mutations were found to have significantly increased risk of disease progression compared to WT tumors.
The subgroup analyses performed in this study identified several potential sources of heterogeneity in mutation prevalence. Female patients had significantly more KRAS and BRAF mutations than males, and significant variation by study location was observed for both KRAS and BRAF mutation prevalence. Additionally, the prevalence of KRAS mutations was higher among patients enrolled in the standard of care arm of clinical trials compared with those in observational studies. The evaluation of potential variation in mutation prevalence by sex, geography, and study design is recommended for future studies. No significant sources of heterogeneity were identified in NRAS prevalence meta-analyses.
This systematic literature review and meta-analysis has many strengths. It includes systematic study selection resulting in a large number of studies and is highly representative of the global prevalence of these mutations. The large number of individual studies included in our analysis provided the opportunity to conduct a number of subgroup analyses to identify potential sources of heterogeneity. We also assessed the risk of bias in individual studies using the STROBE guidelines for reporting observational studies and were then able to use these scores in a subgroup analysis to determine that our prevalence estimates were not significantly impacted by individual study bias.
We were limited in analyzing the data to how they were presented in the individual papers, despite any inconsistencies in nomenclature or mutation assessment methods. This could have resulted in lower overall mutation prevalence estimates as not all studies may have examined the presence of every possible mutation within a given location. There also is some evidence that publication bias may have affected our summary prevalence estimates for KRAS and BRAF mutations.
Our analysis of pooled survival by KRAS, NRAS, and BRAF mutation status can only be considered exploratory in that an exhaustive search of studies that included survival data by mutation status was not conducted. If the studies we identified as part of our comprehensive review happened to also include survival data, they were included in our analysis. The meta-analyses of HRs identified a significant risk of mortality among patients with KRAS, BRAF, and NRAS mutations compared to patients with the respective WT tumors. Patients with BRAF and KRAS mutations were also found to be at an increased risk of disease progression compared to WT tumors. Although subgroup analyses by population treatment status did not show a significant impact on survival, these analyses were limited by the variety of therapeutic regimens and combination therapies, as well as the lack of detail provided on treatment status in many studies, particularly as certain treatments such as anti-EGFR therapies are only effective in WT patients. However, the results of these exploratory analyses may inform future hypotheses on the prognostic impact of these mutations.
In this comprehensive literature review and meta-analysis, we provide the first global, comprehensive, summary prevalence estimates of KRAS, BRAF, and NRAS mutations in mCRC patients. These results provide important insight into the mCRC population for clinicians and researchers, particularly in light of the reduced treatment options for KRAS and NRAS mutant patients. Current clinical guidelines state that mutational testing for BRAF Val600Glu mutations “should be performed in colorectal cancer tissue in patients with colorectal carcinoma for prognostic stratification” [49]. The results of our exploratory survival analyses corroborate the significance of BRAF impact on survival, strengthening the recommendation that BRAF mutation testing should be performed to better inform patient prognosis. Our survival analyses also suggest that KRAS and NRAS mutations may also predict a poorer outcome when compared with their respective WT presentations. This meta-analysis provides representative estimates of mutation prevalence within different subgroups, which is important for making decisions about the implementation of current treatment strategies and for developing new treatments that will be effective for the various patient subgroups.
Supplementary Material
PRISMA flow diagram.
Descriptive Characteristics of Included Studies.
Acknowledgments
The authors would like to thank Human Identity for graphics support. Support for this assistance was funded by Amgen, Inc.
Financial Disclosure
This research project was funded by Amgen, Inc. Amgen was involved in the study design, data collection and analysis, decision to publish, and preparation of the manuscript. All authors had full access to all of the data in this study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
Conflict of Interest
The article processing charges were funded by Amgen, Inc. Individual conflict of interest disclosures are given below: ELS is employed by Epidstat Institute, work contracted through Amgen, Inc. LCB is employed by Epidstat Institute, work contracted through Amgen, Inc. KL is an employee of Amgen, Inc. and owns stock in Amgen, Inc. LS is employed by SimulStat, work contracted through Amgen, Inc. JPF is employed by Epidstat Institute, work contracted through Amgen, Inc. DDA is employed by Epidstat Institute, work contracted through Amgen, Inc.
Informed Consent
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Author Contributions
Elizabeth Levin-Sparenberg: methodology, data curation, investigation, formal analysis, visualization, writing original draft, review and editing. Lauren C. Bylsma: methodology, data curation, investigation, formal analysis, visualization, writing original draft, review and editing. Kimberly Lowe: conceptualization, supervision, visualization, writing review and editing. Laura Sangare: visualization, writing review and editing. Jon P. Fryzek: methodology, visualization, writing-original draft, writing review and editing. Dominik D. Alexander: conceptualization, supervision, methodology, supervision, investigation, writing review and editing.
Data Availability
Any inquiries regarding supporting data availability of this study should be directed to Ms. Lauren C. Bylsma at lbylsma@epidstrategies.com.
References
- 1.Colorectal Cancer: Estimated Incidence, Mortality and PRevalence Worldwide in 2012. Lyon, France: World Health Organization; 2012. International Agency for Research on Cancer. [Google Scholar]
- 2.Win AK, Macinnis RJ, Hopper JL, Jenkins MA. Risk prediction models for colorectal cancer: a review. Cancer Epidemiol Biomarkers Prev. 2012;21(3):398–410. doi: 10.1158/1055-9965.EPI-11-0771. [DOI] [PubMed] [Google Scholar]
- 3.Wong A, Ma BB. Personalizing therapy for colorectal cancer. Clin Gastroenterol Hepatol. 2014;12(1):139–144. doi: 10.1016/j.cgh.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 4.Kindler HL, Shulman KL. Metastatic colorectal cancer. Curr Treat Options Oncol. 2001;2(6):459–471. doi: 10.1007/s11864-001-0068-7. [DOI] [PubMed] [Google Scholar]
- 5.Kurkjian C, Kummar S. Advances in the treatment of metastatic colorectal cancer. Am J Ther. 2009;16(5):412–420. doi: 10.1097/MJT.0b013e3181907ed9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troiani T, Martinelli E, Morgillo F, Capasso A, Nappi A, Sforza V, Ciardiello F. Targeted approach to metastatic colorectal cancer: what comes beyond epidermal growth factor receptor antibodies and bevacizumab? Ther Adv Med Oncol. 2013;5(1):51–72. doi: 10.1177/1758834012462462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moorcraft SY, Smyth EC, Cunningham D. The role of personalized medicine in metastatic colorectal cancer: an evolving landscape. Therap Adv Gastroenterol. 2013;6(5):381–395. doi: 10.1177/1756283X13491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson AB 3rd, Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, Deming D. et al. Colon cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15(3):370–398. doi: 10.6004/jnccn.2017.0036. [DOI] [PubMed] [Google Scholar]
- 9.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G. et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 11.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M. et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 12.Baselga J, Rosen N. Determinants of RASistance to anti-epidermal growth factor receptor agents. J Clin Oncol. 2008;26(10):1582–1584. doi: 10.1200/JCO.2007.15.3700. [DOI] [PubMed] [Google Scholar]
- 13.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S. et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27(5):663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 14.De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B. et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 15.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 16.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW. et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 17.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30(29):3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 18.Pietrantonio F, Cremolini C, Petrelli F, Di Bartolomeo M, Loupakis F, Maggi C, Antoniotti C. et al. First-line anti-EGFR monoclonal antibodies in panRAS wild-type metastatic colorectal cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2015;96(1):156–166. doi: 10.1016/j.critrevonc.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26(1):13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 20.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, Bastit L. et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96(8):1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J. et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF. et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Rubio E, Gomez-Espana A, Massuti B, Sastre J, Reboredo M, Manzano JL, Rivera F. et al. Role of Kras status in patients with metastatic colorectal cancer receiving first-line chemotherapy plus bevacizumab: a TTD group cooperative study. PLoS One. 2012;7(10):e47345. doi: 10.1371/journal.pone.0047345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malapelle U, Bellevicine C, Salatiello M, de Luca C, Rispo E, Riccio P, Sparano L. et al. Sanger sequencing in routine KRAS testing: a review of 1720 cases from a pathologist's perspective. J Clin Pathol. 2012;65(10):940–944. doi: 10.1136/jclinpath-2012-200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y. et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28(31):4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 27.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J. et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 28.Ikenoue T, Hikiba Y, Kanai F, Tanaka Y, Imamura J, Imamura T, Ohta M. et al. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003;63(23):8132–8137. [PubMed] [Google Scholar]
- 29.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM. et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 30.Shinozaki E, Yoshino T, Yamazaki K, Muro K, Yamaguchi K, Nishina T, Yuki S. et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study. Br J Cancer. 2017;117(10):1450–1458. doi: 10.1038/bjc.2017.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaki-Takao M, Yamaguchi T, Natsume S, Iijima T, Wakaume R, Takahashi K, Matsumoto H. et al. Colorectal cancer with BRAF D594G mutation is not associated with microsatellite instability or poor prognosis. Oncology. 2016;91(3):162–170. doi: 10.1159/000447402. [DOI] [PubMed] [Google Scholar]
- 32.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D. et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28(3):466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 33.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I. et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 34.Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S. et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377(9783):2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR, Wang YX. et al. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9(3):e90607. doi: 10.1371/journal.pone.0090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price TJ, Hardingham JE, Lee CK, Weickhardt A, Townsend AR, Wrin JW, Chua A. et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29(19):2675–2682. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 37.Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. PLoS One. 2012;7(10):e47054. doi: 10.1371/journal.pone.0047054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK. et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 39.Saridaki Z, Papadatos-Pastos D, Tzardi M, Mavroudis D, Bairaktari E, Arvanity H, Stathopoulos E. et al. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients' outcome. Br J Cancer. 2010;102(12):1762–1768. doi: 10.1038/sj.bjc.6605694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Q, Xu AT, Zhu MM, Tong JL, Xu XT, Ran ZH. Predictive and prognostic roles of BRAF mutation in patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: a meta-analysis. J Dig Dis. 2013;14(8):409–416. doi: 10.1111/1751-2980.12063. [DOI] [PubMed] [Google Scholar]
- 41.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53(7):852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 42.Moher D, Liberati A, Tetzlaff J, Altman DG. The PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 43.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 44.von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP, Initiative S. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: guidelines for reporting observational studies. Int J Surg. 2014;12(12):1495–1499. doi: 10.1016/j.ijsu.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith BL, Breitfeld P, Cubino J, Weigman V, Richards DP, Chung KY. Abstract 5187: Feasibility study of genomic biomarker profiling for patients with metastatic colorectal cancer. Cancer Research. 2014;74:5187–5187. doi: 10.1158/1538-7445.AM2014-5187. [DOI] [Google Scholar]
- 47.Murata A, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M. et al. Methylation levels of LINE-1 in primary lesion and matched metastatic lesions of colorectal cancer. Br J Cancer. 2013;109(2):408–415. doi: 10.1038/bjc.2013.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenthal MH, Kim KW, Fuchs CS, Meyerhardt JA, Ramaiya NH. CT predictors of overall survival at initial diagnosis in patients with stage IV colorectal cancer. Abdom Imaging. 2015;40(5):1170–1176. doi: 10.1007/s00261-014-0272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE. et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35(13):1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA flow diagram.
Descriptive Characteristics of Included Studies.
Data Availability Statement
Any inquiries regarding supporting data availability of this study should be directed to Ms. Lauren C. Bylsma at lbylsma@epidstrategies.com.