Abstract
Clostridium saccharoperbutylacetonicum N1–4 (Csa) is a historically significant anaerobic bacterium which can perform saccharolytic fermentations to produce acetone, butanol, and ethanol (ABE). Recent genomic analyses have highlighted this organism’s potential to produce polyketide and nonribosomal peptide secondary metabolites, but little is known regarding the identity and function of these metabolites. This study provides a detailed bioinformatic analysis of seven biosynthetic gene clusters (BGCs) present in the Csa genome that are predicted to produce polyketides/nonribosomal peptides. An RNA-seq based untargeted transcriptomic approach revealed that five of seven BGCs were expressed during ABE fermentation. Additional characterization of a highly expressed nonribosomal peptide synthetase gene led to the discovery of its associated metabolite and its biosynthetic pathway. Transcriptomic analysis suggested an association of this nonribosomal peptide synthetase gene with butanol tolerance, which was supported by butanol challenge assays.
Keywords: Anaerobe, ABE fermentation, polyketide synthase, nonribosomal peptide synthetase, transcriptomics
Introduction
Clostridium saccharoperbutylacetonicum N1–4 (Csa) is a Gram-positive, spore-forming obligate anaerobe. After its isolation from soil in 1959, it was patented by the Sanraku Distillers Company in 1960 [32] for use in saccharolytic fermentations to produce organic solvents, including acetone, butanol, and ethanol (ABE). In subsequent decades, ABE fermentation largely fell out of favor due to competition from the petrochemical industry [21]. Today, interest in renewably produced butanol as a drop-in biofuel [15] has revived investigations into ABE fermentation as a sustainable source of chemical energy [12, 31, 44]. Despite the advantages of historical precedent [54], substrate flexibility [2, 5, 6, 8, 23, 24, 33, 35, 36, 42, 48, 51, 52, 58, 59], and butanol hyper-production for Csa, challenges remain to improve ABE productivity, titer, and yield of this organism. A deeper understanding of the biology of Csa could provide insights necessary to further improve its industrial fermentation traits [37].
Microbial secondary metabolites are known to possess diverse functions relating to the metabolism, physiology, differentiation, interspecies competition, etc [41]. For example, the plant pathogen, C. puniceum, produced an aromatic polyketide, clostrubin, which enabled C. puniceum to both survive in an oxygen-rich environment and inhibit other plant pathogenic bacteria [39, 46]. An ABE model organism, C. acetobutylicum, produced a glycosylated polyketide, clostrienose, which promoted sporulation and granulose accumulation [18]. A mutant of C. acetobutylicum lacking the capacity to produce clostrienose downregulated these differentiation processes, resulting in improved butanol titer and productivity. The secondary metabolism of clostridia could thus be manipulated to improve traits relevant for industrial applications. Among all secondary metabolites, polyketides (PKs) and non-ribosomal peptides (NRPs) are two major families noted for their chemical diversity, range of potent bioactivities, and well-studied mechanism of biosynthesis [20, 55]. PKs and NRPs are biosynthesized by polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs), the thio-templated assembly-line enzymes, together with diverse tailoring enzymes. The biosynthetic genes for a particular metabolite often co-localize on the genome in a biosynthetic gene cluster (BGC), facilitating discovery in silico [29]. Notably, while Csa has a relatively large genome (6.6 Mb) [9, 40] among clostridia and putative PKS and NRPS encoding genes are widespread in its genome [26], little is known regarding the regulation and function of these genes and no PK and NRP metabolites have been reported.
We here study the PK/NRP-related secondary metabolism in Csa for the first time. First, an in-depth bioinformatic analysis is performed to profile BGCs encoding putative PKSs and NRPSs. Then, an untargeted transcriptomics approach is utilized to probe BGC expression in ABE fermentation culture conditions. Finally, one of the identified highly expressed BGCs is investigated to reveal its associated metabolite and possible role in ABE fermentation.
Materials and methods
Bacterial strains and growth conditions
All strains used in this study are listed in Supplementary Table S1. E. coli strains were cultured in lysogeny broth (LB) containing the appropriate antibiotics at 37°C. Cloning was performed in an E. coli XL1-blue background. Protein work and E. coli in vivo assays were performed in E. coli BAP1 [38]. Csa was cultured at 37°C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) containing an atmosphere of 97% nitrogen and 3% hydrogen. For routine culture and genetics work, Csa was incubated in 2x YTG (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, 10 g/liter glucose, 15 g/liter agar for solid media) supplemented with 40 μg/ml erythromycin when necessary and adjusted to pH 6.5 with 1 N HCl.
Transcriptomic analysis
Fermentation samples were obtained 12 h after inoculation. 1 ml triplicate samples were pelleted and suspended in 1 ml TRIzol reagent (Ambion). These were stored at −80°C before processing based on a procedure modified from a previous method [56]. Samples were thawed and processed according to the manufacturer’s instructions; samples were spun down and supernatants were extracted in chloroform. The aqueous phase of the three-phase mixture was pipetted into a new RNase-free centrifuge tube and mixed with 1 volume 70% ethanol, then applied to a RNeasy Mini Kit (Qiagen North America, Germantown, MD) for cleanup. The mixture was filtered by spinning through another Qiagen mini kit column, followed by the standard RW1 and RPE washes, and RNA was obtained in 60 μl DEPC-treated water. Residual genomic DNA was depleted using RQ1 DNase (Promega, Madison, WI). The samples were adjusted to 100 μl with DEPC-treated water, mixed with 350 μl RLT buffer and 250 μl ethanol, and returned to Qiagen spin columns for desalting according to the manufacturer’s cleanup procedure. This yielded 60 μl samples containing 0.3–2.4 μg total RNA. RNA quality control, library construction, and library sequencing were performed by the University of California-Berkeley QB3 Functional Genomics Laboratory and Vincent J. Coates Genomic Sequencing Laboratory. RNA quality and concentration were assessed using a nanochip on an Agilent 2100 Bioanalyzer. Bacterial 16S and 23S rRNA was depleted using a RiboZero Kit (Illumina, San Diego, CA). The remaining RNA was converted to an RNA-seq library using an Illumina mRNA-Seq library construction kit. RNA library sequencing was performed on an Illumina HiSeq4000 instrument using 100 bp paired-end reads. After adapter trimming, reads were quality controlled using FastQC [4]. Base calls with a Phred score > 30 were kept for downstream analysis. Quality-controlled reads were mapped to the Csa chromosome (Refseq: NC_020291.1) and megaplasmid (Refseq: NC_020292.1) using Bowtie2 [25]. Read counts were extracted using HTSeq [3] and normalized to gene length. Differential expression analysis between wild-type and Δnrps3 strains was carried out using DESeq2 [28].
Plasmid construction
Oligonucleotides were provided by IDT (Coralville, IA). Phusion polymerase (NEB, Ipswich, MA) was used for all PCR reactions. Detailed cloning procedures are provided under Supplementary Methods. All synthetic oligonucleotide sequences are described in Supplementary Table S2. After Gibson assembly [10], the reaction mix was transformed into chemically competent E. coli XL1-blue (Agilent Technologies, Santa Clara, CA). Clones were isolated and DNA was extracted using a plasmid Qiagen Miniprep kit. Constructs were validated by restriction digest patterning and Sanger sequencing.
Generation of Csa nrps3 knockout mutant
Wild-type Csa was transformed with pNK52 using a previously reported electroporation method [18], with some adjustments. Briefly, competent cell stocks of Csa were prepared from overnight cultures (37°C, PL7 media) from glycerol stocks stored at −80°C. After reaching an OD600 of 0.6, overnight cultures were subcultured in 60 ml liquid 2x YTG (10% inoculum) and incubated for 3 to 5 h until reaching an OD600 of 0.6. The subcultures were centrifuged (room temperature, 3,500×g, 15 min), decanted, and the pellet was resuspended in 6 ml room-temperature EPB (270 mM sucrose, 5 mM NaH2PO4 pH 7.4). We found that competent cell stock could be stored at −80°C in 20% glycerol, albeit with some loss of transformation efficiency. 500 μl electrocompetent cells were aliquoted into 4 mm Bio-Rad (Hercules, CA) cuvettes pre-chilled to 4°C, and 2 μg plasmid DNA was added. Electric pulses were delivered by a Bio-Rad Gene Pulser Xcell with parameters as follows: mode, exponential pulse; voltage, 2.0 kV; resistance, 200 Ω; capacitance, 25 μF. Following electroporation (yielding time constants of ~4 ms), cells were immediately resuspended in 10 ml 2x YTG and allowed to recover for 16 h at 37°C. Recovery cultures were centrifuged again and concentrated in 1 ml fresh media. 100 μl cells were plated on 2x YTG plates containing 40 μg/ml erythromycin. Colonies were picked and transferred into 10 ml liquid 2x YTG with antibiotic for 24–48 hours to allow Cas9-nickase mediated homologous recombination to delete 90% of the gene, using the recombination template present on pNK52 (Supplementary Fig. S1a). Dilutions were then plated on nonselective 2x YTG plates to allow curing of pNK52. After two days’ incubation, colonies were replica plated on both nonselective and selective 2x YTG to detect successful loss of the plasmid. Colony PCR was used to screen for the genotype of the Csa Δnrps3 CRISPR mutants (Supplementary Fig. S1b).
Flask fermentations of Csa
Overnight cultures of Csa were prepared in a derivative of clostridial growth medium (CGM) [13], PL7 (30 g/liter glucose, 5 g/liter yeast extract, 2.67 g/liter ammonium sulfate, 1 g/liter NaCl, 0.75 g/liter monobasic sodium phosphate, 0.75 g/liter dibasic sodium phosphate, 0.5 g/liter cysteine-HCl monohydrate, 0.7 g/liter magnesium sulfate heptahydrate, 20 mg/liter manganese sulfate monohydrate, and 20 mg/liter iron sulfate heptahydrate, with the initial pH adjusted to 6.3 using 1 N HCl) from a single colony from a 2x YTG plate. At exponential phase, with OD600 of 0.4 to 0.6, a 4% inoculum was added to 25 ml of PL7G (PL7 with glucose increased to 35 g/liter, and 6 g/liter CaCO3 for pH control) in loosely capped 50 ml centrifuge tubes to avoid pressurization. All fermentations were performed as biological triplicates in static batch culture. 1 ml samples were drawn at intervals of 12, 24, 48, and 72 h after inoculation. An additional 1 ml sample was drawn for metabolome analysis at the same time intervals.
Fermentation analytical procedures
Concentrations of acetone, butanol, ethanol, acetate, butyrate, lactate, and residual glucose were quantified using calibration curves generated on a Shimadzu Prominence UFLC system with refractive index and diode array detection (Shimadzu America, Inc., Columbia, MD). Prior to analysis, samples of culture supernatant were filtered using 0.22 μm polyvinylidene difluoride syringe (PVDF) filters (Restek, Bellefonte, PA). The resulting filtrate was resolved using a Bio-Rad Aminex HPX-87H column (300 mm by 7.8 mm) and detected by refractive index (for glucose, butanol, ethanol, acetate, and lactate), or by UV absorbance (for acetone, 265 nm; for butyrate, 208 nm). The method used a column temperature of 35°C, a 35 min run duration, and the manufacturer-recommended mobile phase (0.01 N H2SO4) at a flow rate of 0.7 ml/min.
Metabolomic analysis
For untargeted metabolomic analyses, samples were analyzed, in biological quadruplicate, using liquid chromatography-high resolution mass spectrometry (LC-HRMS). Samples containing 1 ml culture were extracted in 1 volume ethyl acetate. Mixtures were vortexed and spun down (6000×g, 1 min). The upper phase solvent layer was pipetted into a new centrifuge tube and dried under N2, then resuspended in 100 μl methanol. 10 μl injections were analyzed on an Agilent Technologies 6545 Accurate-Mass QTOF LC-MS instrument fitted with an Agilent Eclipse Plus C18 column (4.6×100 mm). The run method used a linear gradient of 2–98% CH3CN (v/v) over 57 min in H2O with 0.1% formic acid (v/v) at a flow rate of 0.5 ml/min. Metabolomics data were analyzed using XCMS [11] to identify metabolites unique to either strain. The following parameters were used: p-value < 0.0005, fold change > 10, peak intensity > 10,000.
In vitro reconstitution of the CSPA_RS10760 NRPS
For in vitro assays, N-terminally his-tagged NRPS was overexpressed and purified from E. coli BAP1 pXZ247. Protein was purified as follows: A single colony was inoculated into 10 ml LB + 100 μg/ml carbenicillin for overnight growth at 37°C. About 5 ml was used to inoculate 700 ml LB + 100 μg/ml carbenicillin, and the culture was shaken at 240 rpm and 37°C until the OD600 reached 0.4. The culture was iced for ten minutes and isopropyl thio-β-D-1-galactopyranoside (IPTG) was added to a final concentration of 120 μM to induce protein expression. The culture was incubated at 16 °C for 16 h. Cells were harvested by centrifugation (5500×g, 4°C, 20 min), resuspended in 25 ml lysis buffer (50 mM HEPES, pH 8.0, 0.5 M NaCl, 5 mM imidazole), and lysed by homogenization over ice. Cell debris was removed by centrifugation (17,700×g, 4 °C, 60 min), and Ni-NTA agarose resin (Thermo Fisher Scientific, Waltham, MA) was added to the supernatant (2 ml/liter culture). The mixture was nutated at 4 °C for 1 h, loaded onto a gravity flow column, and the NRPS protein was eluted with increasing concentrations of imidazole in Buffer A (50 mM HEPES, pH 8.0, 1 mM DTT). Purified NRPS protein was concentrated and buffer exchanged into Buffer A + 10% glycerol using an Amicon Ultra-15 spin filter (MilliporeSigma, Burlington, MA) with nominal molecular weight cutoff of 100 kDa. Aliquots of purified NRPS protein were aliquoted and flash frozen in liquid nitrogen. The full in vitro reaction mixture contained 50 mM HEPES pH 8, 2 mM MgCl2, 1 mM TCEP, 5 mM ATP, 4 mM NADPH, 5 mM l-valine, 5 mM L-leucine, 10 mM butyric acid, and 5 mM CoA, 0.01 mM Orf35 (a promiscuous CoA ligase) [60] and 0.01 mM NRPS in a 50 μl reaction volume. After 30 min, reactions were quenched with 2 volumes methanol and spun down (14000×g, 1 min) and the supernatant was collected for LC-MS analysis.
Substrate feeding in a heterologous host
E. coli BAP1 pXZ247 was cultured in 30 ml volumes of LB supplemented with 100 μg/ml carbenicillin at 37°C until the OD600 reached between 0.4 and 0.6. Cultures were cooled to 16°C and supplemented with 120 μM IPTG, 1 mM l-valine, 1 mM L-leucine, and 1 mM of various short-chain fatty acids (acetic acid, propionic acid, butyric acid, hexanoic acid, or octanoic acid). After 48 h incubation, cultures were spun down (4000×g, 5 min) and 10 ml of supernatant was extracted twice using 5 ml ethyl acetate. The resulting products were detected by LC-HRMS under the conditions described above.
Butanol challenge assays
Butanol challenge assays were modified from a method used in C. acetobutylicum [53]. Csa wild-type and Δnrps3 cultures (in biological quadruplicate) were incubated overnight at 37°C in 50 ml centrifuge tubes containing 30 ml PL7G. At the onset of exponential growth at 8 h, cultures were challenged with the addition of water or butanol to a final concentration of 0, 2.5, or 5 g/liter butanol. Sampling was performed at 2–3 h time intervals by taking 200 μl volumes. OD600 was monitored in 96-well plates (Corning, NY) using a SpectraMax M2 instrument (Molecular Devices, San Jose, CA).
Results and discussion
Bioinformatic analysis of PK/NRP BGCs in Csa
As an initial step in exploring the secondary metabolism of Csa, we performed in silico analysis of the published genome of the type strain (NCBI accessions NC_020291.1 and NC_020292.1). The Csa genome consists of a 6.53 Mb circular chromosome and a 136 kb circular megaplasmid, giving this organism the largest reported genome of sequenced clostridia to date [9, 40]. The bioinformatic pipelines AntiSMASH [7] and PRISM [49] were used to identify putative BGC loci, and MultiGeneBLAST [30] and BiG-SCAPE [34] were used to query for homologous BGCs in other organisms. We identified seven BGCs which contain genes characteristic of PK or NRP biosynthesis. These can be categorized into four predicted NRPS gene clusters and three hybrid PKS-NRPS gene clusters (Fig. 1). We trimmed the BGCs conservatively to define putative boundaries by removing genes that were not in operon with a biosynthetic gene or that had predicted functions other than biosynthetic/transporter/regulatory/hypothetical. In the case of hyb2 and hyb3, one locus was defined as two adjacent BGCs based on the fractured nature of biosynthetic genes in hyb2, the cis- vs trans-acyltransferase PKS modules, and the presence of distinct termination domains. More in-depth cluster-specific analyses are presented in Supplementary Table S3. The relative abundance of these secondary metabolism genes in Csa compared to other Clostridium species may suggest an important role of secondary metabolites in the evolutionary biology of Csa. Several of the BGCs appear to be conserved in other Firmicutes (Supplementary Fig. S2–S4). Notably, the megasynthetase gene of nrps3 has homologs in two other ABE-producing Clostridia, although their respective neighborhoods are not conserved. This suggests the true boundaries of the BGC encompass the singular gene.
Fig. 1.
BGCs of Csa which contain PK or NRP biosynthetic genes. Gene color indicates putative function: light blue, core biosynthetic genes; dark blue, additional biosynthetic gene; black, transcriptional regulator; light green, transporter; white, other. Domain organization is indicated for PKS/NRPS genes. Abbreviations: C, condensation; A, adenylation; T, thiolation/peptide carrier; E, epimerization; TE, thioesterase; R, reductive release; KR, ketoreductase; CAL, Acyl-CoA ligase; KS, ketosynthase; cMT, C-methyltransferase; ER, enoyl-reductase; DOC, trans-AT docking; ACP, acyl-carrier protein; GNAT, Gcn5-related N-acetyltransferase; DH, dehydratase; PPT, 4-phosphopantetheinyl transferase
Transcriptional profiling of PK/NRP BGCs during ABE fermentation
A transcriptomic approach was used to study gene expression levels of the BGCs in an ABE fermentation context. High quality RNA (RIN ~8) was prepared from Csa batch cultures grown to log phase with four biological replicates. The resulting RNA-seq dataset (SRA: PRJNA551507) was quality controlled using FastQC. An average of 91.4% or 86.3 million reads per biological replicate were used as input for downstream analysis, well above the 5–10 million reads suggested to be sufficient for bacterial transcriptome analysis [16]. Next, input reads were mapped to either the Csa chromosome or the megaplasmid. Reads mapping to multiple loci were counted once for each locus. FPKM values were calculated for each locus and compiled (Table S4) to assess relative gene expression.
Most of the genome is expressed, with 99.8% of genes having at least one mapped read and 97.8% of genes having at least 10 mapped reads. For each BGC, the core PKS/NRPS gene with the minimum average expression level was used to represent overall BGC expression. The gyrB (DNA gyrase subunit B) housekeeping gene was used as an expression benchmark [43]. The resulting profile of expression is shown in Fig. 2. Five of seven BGCs demonstrated some baseline expression (FPKM > 1), with nrps2 and hyb2 falling below the expression cutoff. Two BGCs, nrps3 and nrps4, were expressed at a level comparable to that of gyrB. A detailed compilation of BGC gene expression levels is presented in Supplementary Table S4.
Fig. 2.
Expression level of the BGCs of Csa. Values are normalized to that of gyrB, a housekeeping benchmark.
Characterization of nrps3-associated secondary metabolite
As nrps3 is both highly expressed and conserved in other solvent-producing Clostridium species, we selected this BGC for metabolite interrogation. In order to discover the secondary metabolite associated with nrps3, CSPA_RS10760 was disrupted using a CRISPR/Cas9-nickase targeted homologous recombination strategy [27, 57]. The resulting strain, Δnrps3, was genotyped by PCR to confirm successful editing at the target locus (Supplementary Fig. S1). Next, chemical extracts of the wild-type and Δnrps3 cultures were analyzed using LC-HRMS. Untargeted metabolomic comparison of the strains was performed using XCMS, enabling identification of a new compound, 1, with molecular formula C15H30N2O3 (calculated for C15H31N2O3+: 287.2329; found: 287.2327) (Supplementary Fig. S5). A majority of 1 was found in spent culture medium rather than pelleted cell mass, suggesting that it is secreted upon biosynthesis.
To obtain enough material for structural characterization, three liters of Csa batch culture was extracted with ethyl acetate and purified using a size-exclusion column packed with Sephadex LH-20, followed by several rounds of high-performance liquid chromatography (HPLC) purification. A detailed method is available under Supplementary Methods. This yielded 2 mg of pure compound that was subjected to 1D and 2D NMR analysis for structural elucidation, including 1H, 13C, HSQC, COSY, and HMBC (Supplementary Fig. S6). Compound 1 was determined to be an N-acylated dipeptidyl alcohol derived from butyric acid, valine and leucine monomers. A chemical standard was synthesized (Supplementary Fig. S7) and the proposed structure was verified by HPLC retention time and tandem MS spectrum.
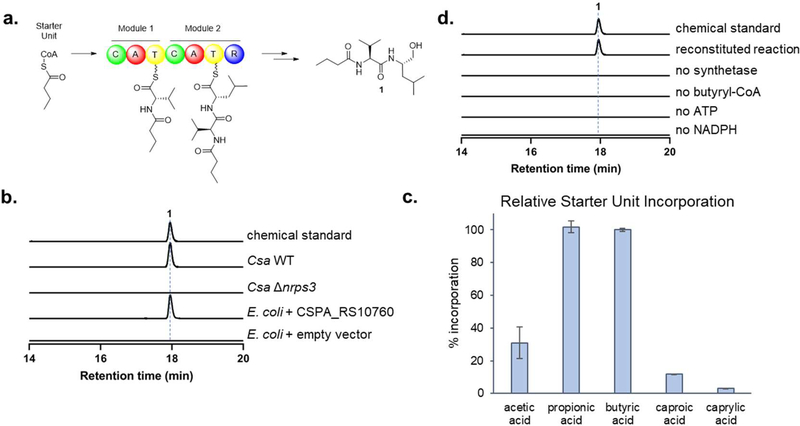
The domain organization of the di-modular NRPS offers insights into the biosynthesis of 1. The proposed mechanism (Fig. 3a) channels substrates through successive modules to form two peptide bonds between the butyryl starting unit and subsequent l-valine and L-leucine monomers. Then, the reductase domain releases the alcohol product from the assembly line through a four-electron reduction. To confirm that the NRPS encoded by CSPA_RS10760 is sufficient to produce the identified compound, we overexpressed the gene in E. coli to probe for possible heterologous metabolite production (Supplementary Fig. S8). The recombinant E. coli strain produced the expected product 1, consistent with the predicted function of the di-modular NRPS (Fig. 3b). We further probed the starter unit substrate promiscuity of the assembly line by feeding fatty acids of varying chain length (Fig. 3c). New products corresponding to the two-, three-, six-, and eight-carbon head groups were identified upon feeding of the corresponding fatty acids, demonstrating that the C domain of the NRPS has a relaxed substrate specificity toward short- to medium-chain fatty acyl groups, albeit with C3–4 substrates preferred. Finally, to unequivocally confirm the function of the NRPS, the intact megasynthetase was purified from E. coli for in vitro activity reconstitution (Fig. 3d). The enzymatic reaction mixture containing butyryl-CoA, l-valine, L-leucine, ATP and NADPH successfully produced compound 1 as a product, confirming that the NRPS is sufficient to make this product.
Fig. 3.
Characterization of nrps3-associated secondary metabolite, 1. a Proposed biosynthesis of the secondary metabolite 1, an N-acylated dipeptidyl alcohol. b HRMS extracted ion chromatograms demonstrating requirement of CSPA_RS10760 in Csa for compound biosynthesis, and heterologous production of 1 in a heterologous E. coli host expressing CSPA_RS10760. c Relative starter unit promiscuity of the NRPS demonstrated by substrate feeding of varying-length short-chain fatty acids in E. coli + CSPA_RS10760. d HRMS extracted ion chromatograms demonstrating biosynthesis of 1 in vitro. The calculated mass for 1 (287.2329) with 10 ppm mass error tolerance was used for each trace in b and d.
The product of CSPA_RS10760 resembles a class of reported dipeptidyl aldehyde compounds associated with various human gut commensal microorganisms [14, 45]. This class of compounds represents potent inhibitors of a variety of human and bacterial proteases. Both these human-gut derived metabolites and 1 are biosynthesized by small NRPS genes terminating in R domains. However, several key differences can be observed between these secondary metabolites. The majority of NRPSs associated with protease inhibition lack the N-terminal C domain, with the resulting lack of N-acylation in the product leading to the formation of pyrazinone shunt metabolites. Moreover, two NRPSs with an identical domain organization to the NRPS encoded by CSPA_RS10760 have been expressed in heterologous E. coli hosts, and N-acylated dipeptidyl aldehydes, instead of alcohols, have been reported as products. In contrast, the major products of CSPA_RS10760 seemed to be alcohols from both anaerobic culture of Csa and aerobic heterologous expression in E. coli, and compound 1 did not have obvious protease inhibition activity toward cathepsin B. Thus, the R domain encoded by CSPA_RS10760 is a more efficient reductase which catalyzes the iterative reduction of the peptidyl carboxyl to a terminal alcohol, distinct from the homologous NRPSs from human gut microbes.
We then examined the possible role of nrps3 in solvent production. Initial batch fermentations of the wild-type Csa and Δnrps3 showed that nrps3 does not have a direct impact on batch ABE titers (Fig. 4a–b) or glucose consumption (Supplementary Fig. S9) after a 48 hour fermentation. In addition, the wild-type Csa and Δnrps3 strains exhibited similar colony morphologies and were indistinguishable in assays assessing for swimming-motility and granulose accumulation (data not shown). We next turned to an untargeted method to identify a possible impact of nrps3 disruption; differential gene expression analysis was carried out using RNA-seq data representing cultures of Csa wild-type and Δnrps3. Filtering the data for two-fold differential gene expression and p-value < 0.001 yielded CSPA_RS10760 (the NRPS) and six protein-coding genes which were revealed by STRING analysis [50] to comprise a putative glycerol metabolism operon (Fig. 4c). While Csa cannot use glycerol as a sole carbon source [22], glycerol metabolism genes have been reported as part of the solvent tolerance response in a wide variety of fermentation hosts, including E. coli (glpBCFQ in response to hexanes stress [17], glpC in response to xylene and cyclohexane [47], and C. acetobutylicum (up-regulation of glpA and glpF in response to 3.7 g/liter butanol) [1]. To probe whether nrps3 mediates a solvent stress response in Csa, we collected growth data comparing wild-type and Δnrps3 after butanol challenge [1]. We found that Δnrps3 exhibited a growth defect during the exponential phase relative to the wild-type (Fig. 4d), albeit the optical densities of Δnrps3 cultures converged with Csa wild-type cultures during the stationary phase. These results support an association between nrps3 and glp-mediated solvent tolerance revealed through transcriptomic analysis, although further work will be required to determine the underlying molecular mechanism.
Fig. 4.
Comparison of wild-type Csa and Δnrps3 in batch cultures under ABE fermentation conditions. a Production of organic acids. b Production of ABE. c STRING network analysis of RNA-seq data comparing wild-type and Δnrps3 fermentations identified a differentially regulated genetic pathway in Δnrps3. The nodes represent the six down-regulated genes, CSPA_RS03935- CSPA_RS03960, summarized in the accompanying table. This glp operon has been associated with solvent stress response in another ABE producer, C. acetobutylicum. The edge colors reflect different lines of evidence for gene functional relationships: light green, gene neighborhood; red, gene fusions; blue, gene co-occurrence; olive green, textmining; black, co-expression. d Butanol challenge assay demonstrating a growth defect in Δnrps3 relative to wild-type Csa. Each assay was repeated at least three times independently.
Conclusions
In summary, this study provides first insights into the secondary metabolism of Csa, an anaerobic bacterium historically valued for industrial solvent production. An in-depth bioinformatic analysis of Csa revealed seven uncharacterized PK/NRP BGCs in its genome, of which five were expressed under solvent-production conditions, as shown by transcriptomic analysis. Further investigation of one of the highly expressed BGCs, nrps3, led to the identification of its associated metabolite, an N-acylated dipeptidyl alcohol, and its biosynthetic mechanism. While phenotypic comparisons between the Csa wild-type and Δnrps3 showed no difference in the batch culture solvent production titers, a comparative transcriptomic analysis followed by butanol challenge assays suggested a possible role of nrps3 in glp-mediated butanol tolerance. This work thus demonstrated another example of a small-molecule secondary metabolite affecting traits relevant for microbial industrial applications.
Supplementary Material
Acknowledgements
We thank J. Pelton (UC Berkeley) for helping with NMR spectroscopic analysis, the University of California-Berkeley QB3 Functional Genomics Laboratory and Vincent J. Coates Genomic Sequencing Laboratory for performing the RNA-Seq library preparation and sequencing. This work was financially supported by the Energy Biosciences Institute, Alfred P. Sloan Foundation, and the National Institutes of Health (DP2AT009148).
References
- 1.Alsaker KV, Paredes C, Papoutsakis ET (2010) Metabolite stress and tolerance in the production of biofuels and chemicals: Gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105:1131–1147. 10.1002/bit.22628 [DOI] [PubMed] [Google Scholar]
- 2.Al-Shorgani NKN, Kalil MS, Yusoff WMW (2012) Biobutanol production from rice bran and de-oiled rice bran by Clostridium saccharoperbutylacetonicum N1–4. Bioprocess Biosyst Eng 35:817–826. 10.1007/s00449-011-0664-2 [DOI] [PubMed] [Google Scholar]
- 3.Anders S, Pyl PT, Huber W (2015) HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 5.Avci A, Kiliç NK, Dönmez G, Dönmez S (2014) Evaluation of hydrogen production by Clostridium strains on beet molasses. Environ Technol (United Kingdom) 35:278–285. 10.1080/09593330.2013.826251 [DOI] [PubMed] [Google Scholar]
- 6.Baba S, Tashiro Y, Shinto H, Sonomoto K (2012) Development of high-speed and highly efficient butanol production systems from butyric acid with high density of living cells of Clostridium saccharoperbutylacetonicum. J Biotechnol 157:605–612. 10.1016/j.jbiotec.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 7.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dada O, Yusoff WMW, Kalil MS (2013) Biohydrogen production from ricebran using Clostridium saccharoperbutylacetonicum N1–4. Int J Hydrogen Energy 38:15063–15073. 10.1016/j.ijhydene.2013.07.048 [DOI] [Google Scholar]
- 9.del Cerro C, Felpeto-Santero C, Rojas A, Tortajada M, Ramón D, García JL (2013) Genome sequence of the butanol hyperproducer Clostridium saccharoperbutylacetonicum N1–4. Genome Announc 1:e0007013 10.1128/genomeA.00071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 11.Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B, Westenskow PD, Wang J, Arkin AP, Deutschbauer AM, Patti GJ, Siuzdak G (2014) Interactive XCMS online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem 86:6931–6939. 10.1021/ac500734c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green EM (2011) Fermentative production of butanol-the industrial perspective. Curr Opin Biotechnol 22:337–343. 10.1016/j.copbio.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 13.Green EM, Boynton ZL, Harris LM, Rudolph FB, Papoutsakis ET, Bennett GN (1996) Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142:2079–2086. 10.1099/13500872-142-8-2079 [DOI] [PubMed] [Google Scholar]
- 14.Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, Craik CS, Cravatt BF, Clardy J, Voigt CA, Fischbach MA (2017) Discovery of Reactive Microbiota-Derived Metabolites that Inhibit Host Proteases. Cell 168:517–526.e18. 10.1016/j.cell.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo M, Song W, Buhain J (2015) Bioenergy and biofuels: History, status, and perspective. Renew Sustain Energy Rev 42:712–725. 10.1016/j.rser.2014.10.013 [DOI] [Google Scholar]
- 16.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J (2012) How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13:734 10.1186/1471-2164-13-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi S, Aono R, Hanai T, Mori H, Kobayashi T, Honda H (2003) Analysis of organic solvent tolerance in Escherichia coli using gene expression profiles from DNA microarrays. J Biosci Bioeng 95:379–383. 10.1263/jbb.95.379 [DOI] [PubMed] [Google Scholar]
- 18.Herman NA, Kim SJ, Li JS, Cai W, Koshino H, Zhang W (2017) The industrial anaerobe Clostridium acetobutylicum uses polyketides to regulate cellular differentiation. Nat Commun 8:1514 10.1038/s41467-017-01809-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman NA, Li J, Bedi R, Turchi B, Liu X, Miller MJ, Zhang W (2017) Development of a high-efficiency transformation method and implementation of rational metabolic engineering for the industrial butanol hyperproducer clostridium saccharoperbutylacetonicum strain N1–4. Appl Environ Microbiol 83:e02942–16. 10.1128/AEM.02942-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertweck C (2009) The biosynthetic logic of polyketide diversity. Angew Chemie - Int Ed 48:4688–4716. 10.1002/anie.200806121 [DOI] [PubMed] [Google Scholar]
- 21.Jones DT, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50:484–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keis S, Shaheen R, Jones DT (2001) Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int J Syst Evol Microbiol 51:2095–2103. 10.1099/00207713-51-6-2095 [DOI] [PubMed] [Google Scholar]
- 23.Kiyoshi K, Furukawa M, Seyama T, Kadokura T, Nakazato A, Nakayama S (2015) Butanol production from alkali-pretreated rice straw by co-culture of Clostridium thermocellum and Clostridium saccharoperbutylacetonicum. Bioresour Technol 186:325–328. 10.1016/j.biortech.2015.03.061 [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi G, Eto K, Tashiro Y, Okubo K, Sonomoto K, Ishizaki A (2005) Utilization of excess sludge by acetone-butanol-ethanol fermentation employing Clostridium saccharoperbutylacetonicum N1–4 (ATCC 13564). J Biosci Bioeng 99:517–519. 10.1263/jbb.99.517 [DOI] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JS, Barber CC, Zhang W (2019) Natural products from anaerobes. J Ind Microbiol Biotechnol 46:375–383. 10.1007/s10295-018-2086-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Chen J, Minton NP, Zhang Y, Wen Z, Liu J, Yang H, Zeng Z, Ren X, Yang J, Gu Y, Jiang W, Jiang Y, Yang S (2016) CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol J 11:961–972. 10.1002/biot.201600053 [DOI] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medema MH, Fischbach MA (2015) Computational approaches to natural product discovery. Nat Chem Biol 11:639–648. 10.1038/nchembio.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medema MH, Takano E, Breitling R (2013) Detecting sequence homology at the gene cluster level with multigeneblast. Mol Biol Evol 30:1218–1223. 10.1093/molbev/mst025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon HG, Jang YS, Cho C, Lee J, Binkley R, Lee SY (2016) One hundred years of clostridial butanol fermentation. FEMS Microbiol Lett 363:fnw001 10.1093/femsle/fnw001 [DOI] [PubMed] [Google Scholar]
- 32.Motoyoshi H (1960) Process for producing butanol by fermentation. US patent 2,945,786 A.
- 33.Nakayama S, Kiyoshi K, Kadokura T, Nakazato A (2011) Butanol production from crystalline cellulose by Cocultured Clostridium thermocellum and Clostridium saccharoperbutylacetonicum N1–4. Appl Environ Microbiol 77:6470–6475. 10.1128/AEM.00706-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Muñoz J, Selem-Mojica N, Mullowney M, Kautsar S, Tryon J, Parkinson E, De Los Santos E, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernàndez-Guerra A, Cappelini LTD, Thomson R, Metcalf W, Kelleher N, Barona-Gomez F, Medema MH (2019) A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. Nat Chem Biol 16:60–68. 10.1038/s41589-019-0400-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noguchi T, Tashiro Y, Yoshida T, Zheng J, Sakai K, Sonomoto K (2013) Efficient butanol production without carbon catabolite repression from mixed sugars with Clostridium saccharoperbutylacetonicum N1–4. J Biosci Bioeng 116:716–721. 10.1016/j.jbiosc.2013.05.030 [DOI] [PubMed] [Google Scholar]
- 36.Oshiro M, Hanada K, Tashiro Y, Sonomoto K (2010) Efficient conversion of lactic acid to butanol with pH-stat continuous lactic acid and glucose feeding method by Clostridium saccharoperbutylacetonicum. Appl Microbiol Biotechnol 87:1177–1185. 10.1007/s00253-010-2673-5 [DOI] [PubMed] [Google Scholar]
- 37.Patakova P, Linhova M, Rychtera M, Paulova L, Melzoch K (2013) Novel and neglected issues of acetone-butanol-ethanol (ABE) fermentation by clostridia: Clostridium metabolic diversity, tools for process mapping and continuous fermentation systems. Biotechnol Adv 31:58–67. 10.1016/j.biotechadv.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C (2001) Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science (80- ) 291:1790–1792. 10.1126/science.1058092 [DOI] [PubMed] [Google Scholar]
- 39.Pidot S, Ishida K, Cyrulies M, Hertweck C (2014) Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew Chemie - Int Ed 53:7856–7859. 10.1002/anie.201402632 [DOI] [PubMed] [Google Scholar]
- 40.Poehlein A, Krabben P, Dürre P, Daniel R (2014) Complete genome sequence of the solvent producer Clostridium saccharoperbutylacetonicum strain DSM 14923. Genome Announc 2:e01056–14. 10.1128/genomeA.01056-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raaijmakers JM, Mazzola M (2012) Diversity and Natural Functions of Antibiotics Produced by Beneficial and Plant Pathogenic Bacteria. Annu Rev Phytopathol 50:403–424. 10.1146/annurev-phyto-081211-172908 [DOI] [PubMed] [Google Scholar]
- 42.Richter H, Qureshi N, Heger S, Dien B, Cotta MA, Angenent LT (2012) Prolonged conversion of n-butyrate to n-butanol with Clostridium saccharoperbutylacetonicum in a two-stage continuous culture with in-situ product removal. Biotechnol Bioeng 109:913–921. 10.1002/bit.24380 [DOI] [PubMed] [Google Scholar]
- 43.Rocha DJP, Santos CS, Pacheco LGC (2015) Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol 108:685–693. 10.1007/s10482-015-0524-1 [DOI] [PubMed] [Google Scholar]
- 44.Sauer M (2016) Industrial production of acetone and butanol by fermentation-100 years later. FEMS Microbiol Lett 363:fnw134 10.1093/femsle/fnw134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider BA, Balskus EP (2018) Discovery of small molecule protease inhibitors by investigating a widespread human gut bacterial biosynthetic pathway. Tetrahedron 74:3215–3230. 10.1016/j.tet.2018.03.043 [DOI] [Google Scholar]
- 46.Shabuer G, Ishida K, Pidot SJ, Roth M, Dahse HM, Hertweck C (2015) Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory. Science (80- ) 350:670–674. 10.1126/science.aac9990 [DOI] [PubMed] [Google Scholar]
- 47.Shimizu K, Hayashi S, Kako T, Suzuki M, Tsukagoshi N, Doukyu N, Kobayashi T, Honda H (2005) Discovery of glpC, an organic solvent tolerance-related gene in Escherichia coli, using gene expression profiles from DNA microarrays. Appl Environ Microbiol 71:1093–1096. 10.1128/AEM.71.2.1093-1096.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukor H, Al-Shorgani NKN, Abdeshahian P, Hamid AA, Anuar N, Rahman NA, Kalil MS (2014) Production of butanol by Clostridium saccharoperbutylacetonicum N1–4 from palm kernel cake in acetone-butanol-ethanol fermentation using an empirical model. Bioresour Technol 170:565–573. 10.1016/j.biortech.2014.07.055 [DOI] [PubMed] [Google Scholar]
- 49.Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA (2017) PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 45:W49–W54. 10.1093/nar/gkx320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Von Mering C (2015) STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tashiro Y, Takeda K, Kobayashi G, Sonomoto K, Ishizaki A, Yoshino S (2004) High butanol production by Clostridium saccharoperbutylacetonicum N1–4 in fed-batch culture with pH-stat continuous butyric acid and glucose feeding method. J Biosci Bioeng 98:263–268. 10.1263/jbb.98.263 [DOI] [PubMed] [Google Scholar]
- 52.Thang VH, Kanda K, Kobayashi G (2010) Production of Acetone-Butanol-Ethanol (ABE) in direct fermentation of cassava by Clostridium saccharoperbutylacetonicum N1–4. Appl Biochem Biotechnol 161:157–170. 10.1007/s12010-009-8770-1 [DOI] [PubMed] [Google Scholar]
- 53.Tomas CA, Beamish J, Papoutsakis ET (2004) Transcriptional Analysis of Butanol Stress and Tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018. 10.1128/JB.186.7.2006-2018.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET (2012) Clostridia: The importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol. 23:364–381 [DOI] [PubMed] [Google Scholar]
- 55.Walsh CT (2016) Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat Prod Rep 33:127–135. 10.1039/c5np00035a [DOI] [PubMed] [Google Scholar]
- 56.Wei H, Fu Y, Magnusson L, Baker JO, Maness PC, Xu Q, Yang S, Bowersox A, Bogorad I, Wang W, Tucker MP, Himmel ME, Ding SY (2014) Comparison of transcriptional profiles of Clostridium thermocellum grown on cellobiose and pretreated yellow poplar using RNA-seq. Front Microbiol 5:142 10.3389/fmicb.2014.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J (2015) Efficient genome editing in clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microbiol 81:4423–4431. 10.1128/AEM.00873-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao D, Dong S, Wang P, Chen T, Wang J, Yue ZB, Wang Y (2017) Robustness of Clostridium saccharoperbutylacetonicum for acetone-butanol-ethanol production: Effects of lignocellulosic sugars and inhibitors. Fuel 208:549–557. 10.1016/j.fuel.2017.07.004 [DOI] [Google Scholar]
- 59.Zhang J, Wang P, Wang X, Feng J, Sandhu HS, Wang Y (2018) Enhancement of sucrose metabolism in Clostridium saccharoperbutylacetonicum N1–4 through metabolic engineering for improved acetone–butanol–ethanol (ABE) fermentation. Bioresour Technol 270:430–438. 10.1016/j.biortech.2018.09.059 [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Bolla ML, Kahne D, Walsh CT (2010) A three enzyme pathway for 2-amino-3-hydroxycyclopent-2-enone formation and incorporation in natural product biosynthesis. J Am Chem Soc 132:6402–6411. 10.1021/ja1002845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.